Abstract
Individuals infected with HIV-1 non-B subtypes are understudied in the United States. Their characterization may augment prevention and treatment interventions. We examined the regional molecular epidemiology of non-B subtypes using a combined phylogenetic and geospatial approach. HIV-1 pol sequences and clinical data obtained for routine clinical care were aggregated from 2004–2011 at the largest HIV center in Rhode Island. Subtyping was performed by neighbor-joining and maximum-likelihood phylogeny and compared across eight commonly used tools (HIVdb, REGA, RIP, NCBI, Geno2Pheno, EuResist, jpHMM and STAR) using proportional odds ordinal regression. Individuals with non-B subtypes were characterized according to demographics and risk factors for infection, intra-subtype clustering by maximum-likelihood phylogeny, and geospatial hotspot analysis using Getis-Ord Gi* statistics. Of 1,277 unique sequences, phylogenetic subtyping demonstrated 8.3% (N=106, 95% CI 6.8%–10%) non-B subtypes and circulating recombinant forms (CRFs): CRF02_AG=46; A=15; C=15; CRF01_AE=6; CRF06_CPX=5; CRF14_BG=5; G=3; CRF43_02G=3; D=3; CRF24_BG=3; CRF11_CPX=1; F1=1. Compared to phylogeny, Geno2Pheno was the most concordant (86% exact match) followed by REGA (85%), EuResist (85%) and STAR (82%). Of 106 individuals with non-B subtypes, 50% were male, 71% acquired infection through heterosexual transmission; 76%, were born in Africa, 6% Southeast Asia, 5% the United States, 3% Central America, 1% Europe, and 9% unknown. Eighty percent of CRF02_AG, 93% of A and 87% of C sequences were from African-born individuals. Twenty-two percent of non-B subtypes formed transmission clusters, including a significant number of younger individuals with perinatally-acquired infection. Geospatial analyses revealed hotspots of B and non-B subtypes in the state capital with a more concentrated focus among non-B subtypes. Molecular examination of regional HIV diversity revealed a larger than expected non-subtype B infected population, mostly born in Africa, with low ongoing regional transmission. Phylogenetic and geospatial characterization of infection clusters is helpful to identify targets for treatment and prevention interventions.
Keywords: HIV, NONSUBTYPE B, PHYLOGENY, GEOGRAPHY
1. INTRODUCTION
The HIV epidemic continues to affect diverse racial and ethnic populations across the world, including the United States (US) (Johnson et al., 2013). HIV is broadly classified as type 1 or 2, of which type 1 forms several groups (M, N, O, P) (Robertson et al., 2000). Most infections worldwide are caused by HIV type 1 group M which is further divided into discrete clades, or subtypes (A–D, F–H, J, K) and many circulating recombinant forms (CRFs) (Hemelaar et al., 2011). Subtype C is the most common worldwide and other subtypes are found across much of Africa and Asia. Greater than 95% of US infections are due to HIV-1 subtype B (Bennett, 2005; Pyne et al., 2013), which is also common in Europe, Australia, and Central/South America. HIV-1 diversity has important ramifications for clinical practice including diagnostics (Apetrei et al., 1996), viral load and drug resistance monitoring (Chan et al., 2008; Luft et al., 2011), blood donor screening (Delwart et al., 2012), antiretroviral (ARV) treatment (Kantor, 2006), clinical disease course (Pant Pai et al., 2012), and vaccine development (Taylor et al., 2008). Surveillance and investigation of HIV-1 subtype diversity is therefore important, especially since the prevalence of non-subtype B strains in the US has been increasing over the last decade (Brennan et al., 2009; Carr et al., 2010; Delwart et al., 2012; Lin et al., 2006; Wheeler et al., 2010).
Previous studies in the US of individuals with non-B infection have been conducted among military personal (Brodine et al., 1999; Wegner et al., 2000), state public health departments (Sides et al., 2005), small case studies (Womack et al., 2001), urban centers (Achkar et al., 2004; Carr et al., 2010), blood donors (Brennan et al., 2009; Delwart et al., 2012) and other molecular epidemiology based studies (Gonzales et al., 2001). Among US blood donors from 1999–2009, 2.5–4.7% had non-B infections (Brennan et al., 2009; Delwart et al., 2012). Large reference laboratories have found a similar prevalence (Pyne et al., 2013). The Centers for Disease Control and Prevention (CDC) Variant, Atypical, and Resistant HIV Surveillance Group (VARHS), the major surveillance network in the US for HIV genetic diversity, demonstrated non-B subtypes in 3.8% of infections in 2006 (Wheeler et al., 2010). The VAHRS network encompasses a subset of states across the country (Colorado, Illinois, Louisiana, Michigan, Minnesota, Mississippi, North Carolina, Pennsylvania-excluding Philadelphia, Washington, South Carolina, Virginia), but does not include many of the major urban centers in the US Northeast.
The gold-standard comprehensive classification of HIV-1 subtypes is based on phylogenetic analyses of whole genomes (Robertson et al., 2000), despite their rare availability. Conversely, HIV pol sequences, collected as routine clinical care to assess for drug resistance mutations, have enough phylogenetic signal to accurately determine the pol subtype (Hué et al., 2004; Pasquier et al., 2001; Snoeck et al., 2002; Yahi et al., 2001). Other subtyping methods include commonly used algorithms such as the Stanford HIV Database (Liu and Shafer, 2006), the REGA HIV Subtyping Tool (de Oliveira et al., 2005), Los Alamos Recombinant Identification Program (RIP) (Siepel et al., 1995), the National Center for Biotechnology Information (NCBI) tool (Rozanov et al., 2004), Geno2Pheno (Beerenwinkel et al., 2002), EuResist (Rosen-Zvi et al., 2008), Jumping Profile Hidden Markov Model (jpHMM) (Schultz et al., 2006), and Subtype Analyzer (STAR ) (Gale et al., 2004). These approaches use a combination of similarity scores, statistical analyses and phylogenetic inference to determine subtype.
Subtype diversity may have important implications for HIV treatment and prevention in the US. Individuals with non-B infection are more likely to be foreign-born and have different risk factors for HIV acquisition than US-born individuals infected with HIV-1 subtype B (Prosser et al., 2012). Over 60% of new HIV infections in the US are among men who have sex with men (MSM) (Johnson et al., 2013). The major risk factor for HIV acquisition among foreign-born individuals with non-B infection is heterosexual contact (Prosser et al., 2012). This discordance may be attributable to regional, discrete sub-epidemics with differing characteristics. In combination with other cultural and language barriers, especially stigma surrounding sex and HIV/AIDS, such differences might be relevant to the design and implementation of HIV treatment and prevention strategies. Common programs and interventions for education and testing that target the most affected HIV-infected US populations such as MSM and injection drug users (IDU) may need to be modified for other populations. Characterization of non-B infections may identify populations and regions that would benefit from HIV treatment and prevention interventions.
Two approaches to characterize HIV molecular epidemiology in a community include phylogenetic and geospatial cluster analyses, which attempt to identify groups of HIV-infected individuals that share certain features. Phylogenetic characterization of HIV pol sequences can identify clusters of sequences with common ancestors that may be indicative of ongoing local HIV transmission in a community (Brenner et al., 2011; Chan et al., 2012, 2011; Lewis et al., 2008). Lack of, or limited, clustering in a well-represented epidemic suggests unlinked infections and multiple, independent viral introductions. Geospatial analyses can identify “hotspots” of infection by a combination of geographic information system (GIS) and spatial statistical methods, which imply concentrated regions of HIV-infected individuals living within a defined geographic area (Hixson et al., 2011; Shepard et al., 2011). Given that foreign-born populations tend to segregate by ethnicity and/or race (Fischer et al., 2004), determination of geospatial clusters may assist in targeting HIV testing, prevention, linkage to care and treatment interventions in these populations.
In this study, we characterize populations with diverse HIV-1 subtypes at the largest HIV center in Rhode Island, a state of 1.1 million people with approximately 4,000 individuals infected with HIV (Rhode Island Department of Health Surveillance Report). We use a combined phylogenetic and geospatial approach to identify targets for future treatment and prevention interventions. We determine subtype diversity and associated features in our regional HIV epidemic and evaluate transmission networks in individuals infected with non-B subtypes by assessing intra-subtype phylogenetic clustering and performing geospatial analyses to determine geographic concentrations of HIV infection.
2. MATERIAL AND METHODS
2. 1 Study Population
HIV-1 pol sequences (protease amino acids 4–99 and reverse transcriptase amino acids 38–248) were collected from January 2004 to December 2011 at the Lifespan Hospital System, a Brown University teaching affiliate, encompassing several major academic hospitals and outpatient clinics in Rhode Island, which provide care for approximately 75% of the state’s HIV population (Gillani et al., 2009). Sequences were obtained as part of routine clinical care to evaluate for ARV drug resistance from individuals who were both ARV treatment-naïve and treatment-experienced. Demographic and clinical data were reviewed from available records of all individuals with available sequences attending the clinic. These data included age, gender, biologic sex, race, ethnicity, address (street, city, state, and zip code of residence during 2011), risk factor(s) for HIV acquisition, date of HIV diagnosis, ARV treatment history, and country of origin. Risk factors for HIV acquisition included MSM, males who have sex with females (MSF), females who have sex with males (FSM), IDU, mother-to-child-transmission (MTCT), and other/unknown. Men who identified as MSM and MSF were categorized as MSM. IDUs who reported MSF or FSM were categorized as IDU as the likely route of HIV acquisition.
The study was approved by the Lifespan institutional review board.
2.2 HIV-1 Subtyping
Sequence quality control was performed using the sequence quality analysis tool (SQUAT) (Delong et al., 2011). Sequences were aligned by multiple sequence comparison by log-expectation (MUSCLE) (Edgar, 2004), manually edited to remove gaps and trimmed to identical sequences lengths in BioEdit (Hall, 1999). To accommodate the large sequence number, phylogenetic trees were initially constructed using a Kimura 2-parameter model of evolution and neighbor-joining algorithm in MEGA v5.2 (Tamura et al., 2011). Drug-resistance mutations were removed to minimize convergent evolution.
HIV-1 subtype was determined by phylogenetic analyses of pol sequences together with a comprehensive set of HIV-1 subtype reference sequences (Los Alamos HIV Reference Sequence Database, 2010) (Table S1). In cases where one individual had multiple sequences, only the earliest sequence was used. Branch support was evaluated with bootstrapping for 1000 replicates. Subtype was assigned as the clustering reference sequence with bootstrap support ≥70%, or as the reference sequence with the lowest genetic distance if bootstrap support was <70%. A separate tree was constructed for sequences with closest distance to a subtype A containing reference sequence and <70% bootstrap support, in an attempt to clearly differentiate between them, given their close similarity in pol (Los Alamos HIV Reference Sequence Database, 2010). Mutations associated with transmitted drug resistance (TDR) based on the World Health Organization (WHO) surveillance drug resistance mutation list (Bennett et al., 2009) were determined in sequences from treatment-naïve individuals using Stanford HIV Sequence Database tools (hivdb.stanford.edu).
To evaluate concordance among available tools, subtyping of sequences determined as non-B by phylogeny was performed by eight commonly available algorithms: Stanford HIV Database (Liu and Shafer, 2006), REGA v3.0 (de Oliveira et al., 2005), Los Alamos RIP v3.0 (Siepel et al., 1995), NCBI (Rozanov et al., 2004), Geno2Pheno (Beerenwinkel et al., 2002), EuResist (Rosen-Zvi et al., 2008), jpHMM (Schultz et al., 2006), and STAR (Gale et al., 2004). Variations between phylogeny and each tool’s subtype determination were assessed by a similarity score that was determined for each phylogeny-tool pair. An exact match of subtype determination between phylogeny and each tool was given 3 points, a minor variation 2 points (one subtype determination encompassed by the other, e.g. CRF01_AE by phylogeny, subtype A by tool), a moderate variation 1 point (incomplete overlap of subtype determination, e.g. CRF01_AE and CRF02_AG), and a major variation 0 points (no subtype determination overlap, e.g. subtypes A and G). Scores for all non-B sequences were summarized by tool and the proportion of sequences with an exact match versus inexact match were compared among tools using methods for repeated measures of binomial data, fit with generalized estimating equations and assuming an unstructured correlation structure.
2.3 Phylogenetic Clustering
To characterize intra-subtype clusters among non-B sequences, JModeltest 2.1.4 (Darriba et al., 2012) was first used to determine the best model of evolution based on the Akaike Information Criterion (AIC). Phylogenetic trees were constructed using maximum likelihood and the GTR+I+G model of evolution with four discrete gamma categories plus proportion of invariable sites and subtree pruning resection (SPR) heuristics. Branch support was evaluated using bootstrapping with 1000 replicates. To overcome sequence clustering due only to subtype similarity, strict criteria were used to define intra-subtype clusters as sequences with branch support ≥95% and short genetic distances (≤1.5%). Maximum likelihood trees were constructed with all non-B sequences together, as well as for each subtype separately, with reference sequences and other sequences from regions in the US and across the world (Los Alamos HIV Sequence Database). Simian immunodeficiency viruses (SIV; Genbank accession numbers U42720, DQ373066, AF103818) were used as outgroup given their known divergence from HIV-1 group M sequences.
Statistical analyses were performed using SPSS (IBM SPSS Statistics for Windows, Version 20.0, 2011) and the statistical package R (R foundation for statistical computing, 2013). Bivariate analyses included the use of the chi-square test for categorical variables to detect significant differences between different subtypes. Significance was defined as p-values less than 0.05.
2.4 Geospatial Clustering
ArcMap 10.0 (ESRI, 2010) was used for spatial data input, preparation, analyses and mapping. The last known street address of individuals with available sequences was mapped within Rhode Island (geocoded). The spatial units of analysis used were the 2010 US Census Tract block groups (www.census.gov), small subdivisions of cities/towns with a population between 600–3,000 individuals. The total number of individuals with available HIV sequences living within each block group was calculated and Getis-Ord Gi* statistics (Getis et al., 1992) within ArcMap were used to identify concentrated areas where individuals with either subtype B or non-B infection live, i.e. geographical hotspots. The number of infections in a given block group was evaluated within the context of neighboring block groups. A statistically significant hotspot was defined as a block group with a high number of infections surrounded by other block groups with high values as well. The local sum of infections in a given block group and surrounding ones was compared proportionally to the sum of all infections in the city (subtype B or non-B). Statistical significance was determined based on whether the actual local sum differed from the expected local sum and was too large to be the result of random chance. The null hypothesis of random geographic distribution of infections was rejected when z-scores were high (>1.96), corresponding to low p-values (<0.05), which indicated significant spatial clustering. In order to protect confidentiality, only information on statistical outcomes is shown and not actual data.
3. RESULTS
3.1 Characteristics of Non-Subtype B Infection
A total of 1,277 individuals had at least one HIV-1 pol sequence available from 2004–2011. Based on phylogenetic analyses, 8.3% (106/1,277; 95% CI 6.8% to 10%) were infected with 12 different HIV-1 non-B subtypes (TABLE 1; FIGURE S1). The most common non-B subtypes were CRF02_AG (43% of non-B subtypes), A (14%), and C (14%). Less common subtypes included CRF01_AE (6%), D (3%), F1 (1%), G (3%), CRF14_BG (5%), CRF06_CPX (6%), CRF24_BG (3%), CRF43_02G (3%), and CRF11_CPX (1%).
Table 1.
Characteristics of HIV-1 infected patients according to specific subtypes
| Charact eristic |
No - Subt ype B (Tot al, N=1 06) |
02_ AG (N= 46) |
C (N= 15) |
A (N= 15) |
01_ AE (N= 6) |
06_ CPX (N= 5) |
14_ BG (N= 5) |
Oth er1 (N= 14) |
P val ue |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||||||||||
| Male | 53 | 50% | 20 | 43% | 10 | 67% | 6 | 40% | 4 | 67% | 2 | 40% | 3 | 60% | 8 | 57% | 0.62 |
| Female | 53 | 50% | 26 | 57% | 5 | 33% | 9 | 60% | 2 | 33% | 3 | 60% | 2 | 40% | 6 | 43% | |
| Age at Diagnosis (years) | |||||||||||||||||
| 0–17 | 15 | 14% | 5 | 11% | 6 | 40% | 2 | 13% | 0 | 0% | 1 | 20% | 0 | 0% | 1 | 7% | 0.08 |
| 18–29 | 30 | 28% | 15 | 33% | 3 | 20% | 4 | 27% | 1 | 17% | 2 | 40% | 3 | 60% | 2 | 14% | 0.48 |
| 30–39 | 29 | 27% | 12 | 26% | 3 | 20% | 4 | 27% | 3 | 50% | 2 | 40% | 0 | 0% | 5 | 36% | 0.57 |
| 40–49 | 23 | 22% | 11 | 24% | 2 | 13% | 2 | 13% | 1 | 17% | 0 | 0% | 2 | 40% | 5 | 36% | 0.49 |
| >=50 | 7 | 7% | 2 | 4% | 0 | 0% | 3 | 20% | 1 | 17% | 0 | 0% | 0 | 0% | 1 | 7% | 0.28 |
| Unknown | 2 | 2% | 1 | 2% | 1 | 7% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0.84 |
| Transmission Risk2 | |||||||||||||||||
| MSM | 5 | 5% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 17% | 0 | 0% | 0 | 0% | 4 | 29% | <0.01 |
| MSF | 34 | 32% | 17 | 37% | 2 | 13% | 5 | 33% | 2 | 33% | 1 | 20% | 3 | 60% | 4 | 29% | 0.53 |
| FSM | 41 | 39% | 19 | 41% | 1 | 7% | 8 | 53% | 2 | 33% | 3 | 60% | 2 | 40% | 6 | 43% | 0.17 |
| IDU | 6 | 6% | 4 | 9% | 1 | 7% | 1 | 7% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0.85 |
| Perinatal | 11 | 10% | 3 | 7% | 6 | 40% | 1 | 7% | 0 | 0% | 1 | 20% | 0 | 0% | 0 | 0% | <0.01 |
| Unknown | 8 | 8% | 3 | 7% | 5 | 33% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0.006 |
| Other | 1 | 1% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 17% | 0 | 0% | 0 | 0% | 0 | 0% | 0.01 |
| Place of Birth | |||||||||||||||||
| Africa | 81 | 76% | 37 | 80% | 13 | 87% | 14 | 93% | 0 | 0% | 5 | 100% | 4 | 80% | 8 | 57% | <0.01 |
| Asia | 6 | 6% | 0 | 0% | 0 | 0% | 0 | 0% | 6 | 100% | 0 | 0% | 0 | 0% | 0 | 0% | |
| Europe | 1 | 1% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 7% | |
| Central America | 3 | 3% | 1 | 2% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 2 | 14% | |
| United States | 5 | 5% | 2 | 4% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 20% | 2 | 14% | |
| Unknown | 10 | 9% | 6 | 13% | 2 | 13% | 1 | 7% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 7% | |
| Treatment | |||||||||||||||||
| Naive | 67 | 63% | 29 | 63% | 8 | 53% | 9 | 60% | 3 | 50% | 3 | 60% | 3 | 60% | 12 | 86% | 0.64 |
| Residence | |||||||||||||||||
| Providence | 64 | 60% | 31 | 67% | 12 | 80% | 8 | 53% | 3 | 50% | 2 | 40% | 0 | 0% | 8 | 57% | 0.052 |
| Phylogeny | |||||||||||||||||
| Clusters | 21 | 20% | 4 | 9% | 8 | 53% | 2 | 13% | 0 | 0% | 2 | 40% | 2 | 40% | 3 | 20% | <0.01 |
Other refers to subtypes and recombinants with less than five sequences, including CRF24_BG(3), CRF43_02G(3), D(3), F1(1), G(3), CRF11_CPX(1).
Transmission risk: MSM=men who have sex with men; MSF=males who have sex with females; FSM=females who have sex with males; IDU=Injection drug use. ‘Other’ risk factor includes one transfusion–related infection.
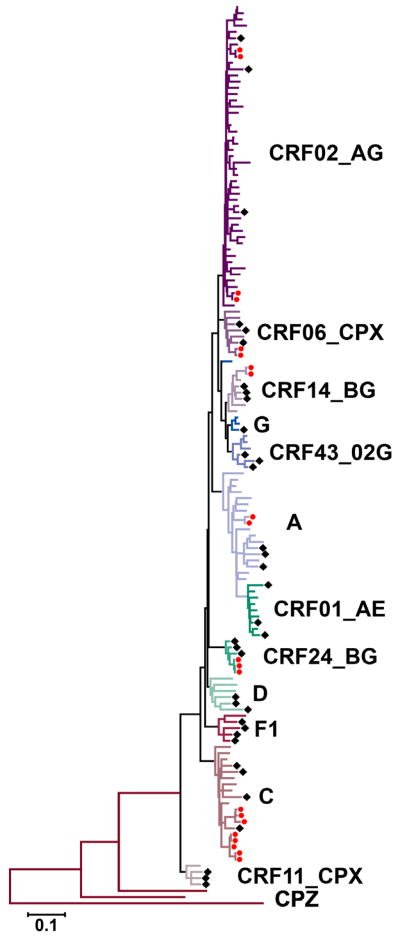
Of 106 non-B subtypes, 31% were defined by ≥70% bootstrap support on the neighbor-joining phylogenetic tree, including subtypes C, F1, G (two of the three sequences), CRF01_AE, CRF11_CPX, CRF06_CPX, and CRF24_BG. Sixty-nine percent of non-B subtypes were defined by closest distance to a reference sequence. These included (i) Subtype A, closest distances to reference A1 (for four sequences, average distance 5.4%) and other A-containing recombinants (11 sequences, average distance 6.2%); (ii) Subtype D, closest distances to reference D (for two sequences, average distance 6.8%) and CRF10_CD (for one sequence, distance 6.3%; assigned as subtype D based on similarity to subtype D in pol) (Koulinska et al., 2001); (iii) subtype G (for one of the three sequences), closest distance to reference G (distance 5.5%); (iv) CRF14_BG, closest distances to reference CRF14_BG (average distance 3.3%); (v) CRF02_AG, closest distance to reference CRF02_AG (for 24 sequences, average distance 4.3%) and CRF36_CPX (for 22 sequences, average distance 4.1%); CRF36_CPX is a CRF01_AE and CRF02_AG recombinant, with CRF02_AG comprising the pol region (Powell et al., 2007); and (vi) CRF43_02G, closest distance to reference CRF43_02G (average distance 4.4%) (FIGURE 1).
Figure 1. Phylogenetic clustering of HIV-1 non-subtype B sequences.
The shown maximum likelihood tree is based on the GTR+I+G4 model of evolution of HIV-1 non-subtype B pol sequences. Subtype assignments are shown by letters to the right of the tree and different branch colors. Subtype and circulating recombinant form (CRF) reference sequences are indicated by black diamonds (see Table S1 for details on specific sequences used). Transmission clusters are shown by red circles and are based on high bootstrap values (≥95%) and short genetic distances (<1.5%). Simian immunodeficiency virus (SIV) CPZ sequences (Genbank accession numbers U42720, DQ373066, AF103818) were used as an outgroup. A distance scale is shown at the bottom of the tree.
Place of birth was known for 91% (96/106) of individuals infected with non-B subtypes. These individuals were much more likely to be born outside of North America compared to those with subtype B (95% versus 5%, p<0.01). Place of birth for individuals with non-B subtypes included Africa (81%), Asia (6%), North America (5%), Central America (3%) and Europe (1%). Sixteen different countries in Africa were represented with Liberia (42%) and Cape Verde (10%) the most common. Compared to individuals infected with subtype B, individuals with non-B subtypes were more likely female (50% versus 27%, p<0.01), diagnosed less than 18 years of age (14% versus 3%, p<0.01), identified heterosexual transmission as the most likely route of HIV acquisition (MSF: 32% versus 11%; FSM: 29% versus 15%; p<0.01), and less likely to have TDR (0% versus 6%, p<0.01).
Comparison of examined characteristics across all non-B subtypes revealed no significant differences in gender or age at diagnosis (TABLE 1). The majority of individuals reported heterosexual transmission (71%) and there was no difference in sexual orientation across non-B subtypes with ≥5 available sequences. However, a subset of less common subtypes/CRFs reported MSM as the main risk factor for HIV acquisition. These four individuals were infected with CRF24_BG (3) and subtype F1 (1), and were born in the US and Central America, respectively. Individuals with subtype C were more likely to have been infected perinatally (40%, 6/15) compared to other subtypes (p<0.01). Overall, the majority of individuals with non-B subtype (60%) lived in Providence, the State capital.
3.2 Subtyping Tool Concordance
Compared to phylogeny, the algorithm that was the most concordant for subtype determination was Geno2Pheno (86% exact matches), followed by REGA (85%), Euresist (85%), STAR (82%), HIVdb (70%), jpHMM (53%), RIP (32%), and NCBI (8%) (TABLE 2). Exact matches were common for many of the pure subtypes including C (82% of subtype C sequences), F (75%), A (60%), G (58%), and D (50%). CRF01_AE and CRF02_AG also displayed high proportions of exact matches, 75% and 71% respectively. Exact matches were less common (24%), and minor (54%), moderate (18%) and major (4%) variations were more common, among the less prevalent non-AE/AG CRFs. Compared to the most phylogeny-concordant subtyping tool (Geno2Pheno), HIVdb, jpHMM, RIP, and NCBI were significantly more likely to assign a different subtype (HIVdb: OR 2.6, 95%CI[1.7–4.1]; jpHMM: OR 5.4, 95%CI[2.8–10.6]; RIP: OR 12.8, 95%CI[6.7–24.5]; NCBI: OR 65.4, 95%CI[26.5–161.5]; all p-values <0.01).
Table 2.
Concordance between phylogeny and eight available HIV subtyping tools
| Exact Match | Minor Variation | Moderate Variation | Major Variation | OR | 95% CI | P value | Reference | |
|---|---|---|---|---|---|---|---|---|
| Geno2Pheno | 91 (86%) | 12 (11%) | 3 (3%) | 0 (0%) | Ref | (Beerenwinkel et al., 2002) | ||
| REGA | 90 (85%) | 14 (13%) | 1 (1%) | 1 (1%) | 1.1 | 0.6, 2.0 | 0.81 | (de Oliveira et al., 2005) |
| EuResist | 90 (85%) | 12 (11%) | 1 (1%) | 3 (3%) | 1.1 | 0.7, 1.8 | 0.76 | (Rosen-Zvi et al., 2008) |
| STAR | 87 (82%) | 14 (13%) | 1 (1%) | 4 (4%) | 1.3 | 0.7, 2.4 | 0.35 | (Gale et al., 2004) |
| Stanford | 74 (70%) | 26 (25%) | 6 (6%) | 0 (0%) | 2.6 | 1.7, 4.1 | <0.001 | (hivdb.stanford.edu) |
| jpHMM | 56 (53%) | 39 (37%) | 11 (10%) | 0 (0%) | 5.4 | 2.8, 10.6 | <0.001 | (Schultz et al., 2006) |
| RIP | 34 (32%) | 65 (61%) | 7 (7%) | 0 (0%) | 12.8 | 6.7, 24.5 | <0.001 | (Siepel et al., 1995) |
| NCBI | 9 (8%) | 97 (92%) | 0 (0%) | 0 (0%) | 65.4 | 26.5, 161.5 | <0.001 | (Rozanov et al., 2004) |
OR=Odds Ratio, odds of making any subtyping mistake compared to the best performing tool; CI=confidence interval.
3.3 Phylogenetic clustering
Twenty-percent (21/106) of non-subtype B sequences formed eight distinct phylogenetic clusters on a maximum-likelihood tree that contained all non-subtype B sequences (FIGURE 1). All clusters were verified in single subtype phylogenetic trees that were created with study sequences as well as sequences from other areas of the US and countries across the world (Figure S2). The average cluster size included 2.6 individuals (range 2–5). Of 11 individuals who acquired HIV through perinatal infection, six (64%) were part of transmission clusters. Fifty-three percent (8/15) of subtype C sequences formed clusters, 40% (2/5) of CRF06_CPX, 40% (2/5) of CRF14_BG, 13% (2/15) of A, 9% (4/46) of CRF02_AG, 0% (0/6) of CRF01_AE, and 20% (3/14) of other less common subtypes/recombinants (p<0.01 across all subtypes) (TABLE 1).
Specific epidemiological linkage information was not available for cluster members. Both subtype C clusters included individuals from different African countries, the smaller one comprised of a male/female/child and the larger of four children and one female. The CRF06_CPX cluster was comprised of an African female/child pair. There was one individual from Africa and one from the US of Hispanic ethnicity in the CRF14_BG cluster. The subtype A cluster was comprised of an African male/female pair from the same country. The two CRF02_AG clusters comprised of a male/female and female/child pair, all from the same country in Africa. The CRF24_BG cluster contained only MSM (two born in the US, one with an unknown birth place). In comparison across all non-subtype B clusters, individuals who formed clusters were more likely to be younger than 18 years of age compared to those who did not form clusters (38% versus 8%, p<0.01) and more likely to have perinatal transmission as the most likely risk factor for acquisition of HIV (33% versus 5%, p<0.01). Individuals with subtype C were more likely to have been infected through perinatal transmission compared to other subtypes. Individuals who formed clusters were less likely to identify MSF as the risk factor for HIV acquisition (10% versus 38%, p=0.02). There was no significant difference between individuals that were part of a transmission cluster and gender or place of birth.
3.4 Geospatial clustering
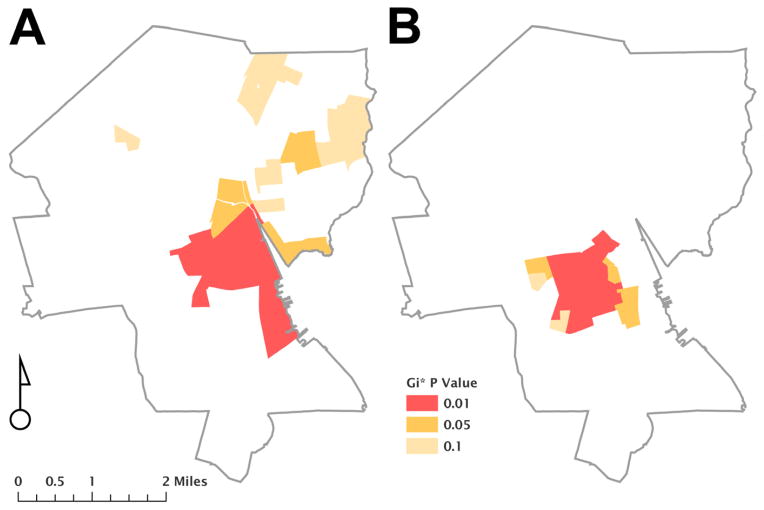
Ninety-nine percent (105/106) of individuals with non-B subtypes had addresses that were successfully geocoded. One individual lived outside Rhode Island and was not included in this analysis. Fifty-eight percent (60/104) of individuals with non-subtype B infection lived in Providence, the capital city. Spatial analyses revealed a cluster of 15 block groups that contained a higher likelihood of having individuals with non-subtype B infection compared to 11 block groups for subtype B (p<0.05; FIGURE 2). Six hotspot block groups overlapped between non-subtype B and subtype B maps. Block groups with non-subtype B infections demonstrated lower p-values compared to subtype B infections, indicating that non-subtype B infections are geographically more tightly clustered than subtype B infection (David O’Sullivan and David Unwin, 2010).
Figure 2. Geospatial representation of HIV-infected individuals living in Providence, Rhode Island.
The figure shows two maps of Providence, Rhode Island based on 2010 US Census tract block groups and representing the geographic location of individuals infected with HIV-1 subtypes B (part A) and non-B (part B). Getis-Ord Gi* analysis of geocoded addresses reveals clusters of infections, which differs among subtypes, as represented by color shading.
4. DISCUSSION
We report high prevalence (8.3%) of phylogenetically-defined, diverse non-subtype B infections at the largest HIV Center in Rhode Island, the most common being CRF02_AG, C, and A. Ninety-five percent of individuals with non-subtype B infections and known places of birth were born outside of North America. The majority of these individuals were from Africa (81%), specifically Liberia (42%) and Cape Verde (10%). The majority of non-B infected individuals (71%) reported heterosexual transmission as the risk factor for HIV acquisition. Twenty-percent of non-subtype B sequences formed nine phylogenetic clusters with a high-degree of clustering among subtype C (53%) and less among CRF02_AG (9%). Geospatial analyses revealed a defined geographical region of individuals with non-subtype B infection that differed from those with subtype B. These results have important implications for the understanding of HIV-1 subtype diversity in our region, as well as for treatment, outreach and prevention strategies.
The prevalence of non-subtype B infections in our community is approximately twice that of large-scale surveillance studies in the US (2.5–4.7%) (Brennan et al., 2009; Delwart et al., 2012; Pyne et al., 2013; Wheeler et al., 2010). However, this estimate varies widely between urban centers (Carr et al., 2010; Karchava et al., 2006; Parker et al., 2007) and is largely dependent on immigration and racial/ethnic communities, given that most HIV-infected individuals with non-B subtypes are foreign-born. For example, investigation of 83 African-born HIV-infected individuals in Minnesota revealed non-subtype B in 95% of individuals (Sides et al., 2005). Other studies have also documented the high prevalence of non-subtype B infection among immigrants in the US (Achkar et al., 2004; Carr et al., 2010; Karchava et al., 2006; Parker et al., 2007). The most common subtypes in these studies vary by population, but have been reported as subtypes A, C, or CRF02_AG (Achkar et al., 2004; Carr et al., 2010; Sides et al., 2005; Wheeler et al., 2010), which is a similar distribution to the common subtypes in our region, and suggests immigration from endemic countries. Other than one study with only two non-subtype B sequences in our state (Pyne et al., 2013), the current study is the first to provide a detailed analysis of subtype diversity in Rhode Island. The majority of individuals with non-subtype B infection (92%) were diagnosed after 2000, and 69% after 2003, consistent with other surveillance studies suggesting recent increases in non-B subtypes in the US, likely due to rises in travel and immigration (Brennan et al., 2009; Carr et al., 2010; Delwart et al., 2012; Wheeler et al., 2010).
Rhode Island is a main immigration destination for refugees and other foreign born individuals from many African countries (Statistical Yearbook of the Immigration and Naturalization Service, www.dhs.gov), specifically West Africa and Liberia (Beckwith et al., 2009; Desjardins et al., 2007; Lacourse et al., 2013; Watts et al., 2012). Individuals from these regions may also be at increased risk for other endemic diseases which may potentiate HIV infection, such as tuberculosis (TB) or viral hepatitis. Given that HIV screening is no longer required for entry into the US, clinicians and public health officials should be aware of at-risk immigrant populations in their region and offer routine HIV testing to individuals from these high-prevalent regions, including testing and treatment for HIV-2 (Chan et al., 2008; Hollenbeck and Beckwith, 2013). Notably, three non-B infected adults in our patient cohort were born in the US (one female and two MSM), and though time of infection could not be determined, this observation suggests that increased awareness of HIV-1 subtype and its impact should not be limited to foreign-born individuals. HIV treatment and prevention strategies should address behavioral risk factors that are more common in non-subtype B infection (e.g. heterosexual transmission), which are different from other common US risk groups (e.g. MSM). Based on the retrospective nature of our study, a limitation of most studies, it is impossible to determine whether HIV transmission occurred in the US or in the individual’s native country. Culturally competent care may be important for effective prevention and treatment strategies.
Phylogenetic investigation of non-subtype B infection provided insight on local HIV transmission. The overall low intra-subtype clustering across most of the non-subtype B sequences from unrelated individuals suggests either missing sequences from patients in care, or multiple, unlinked introductions of non-subtype B HIV-1 into our region. Since our HIV center is the largest in the state, representing approximately 75% of the state’s HIV infected individuals, the latter scenario is more likely. Alternatively, phylogenetic analyses may be underpowered to detect transmission clusters (Stürmer et al., 2004). Ongoing regional transmission would have been expected to result in more observed transmission clusters, but could also be limited by the existence of undiagnosed infections, mandating intensified ongoing prevention efforts. The largest subtype group (CRF02_AG) only formed transmission clusters in 9% of individuals which is low compared to other population-based studies in our community (17–30% in subtype B) (Chan et al., 2012, 2011). Individuals with subtype C formed a high proportion of phylogenetically-linked transmission clusters, likely due to mother-to-child transmission. In Rhode Island, all pregnant females are screened for HIV and are started on ARVs if HIV positive, with resulting few cases of documented perinatal infection. Therefore, most perinatal transmission among foreign-born individuals likely occurs outside of the US. The absence of observed TDR in treatment-naïve individuals with non-B infections also suggests infection transmission outside of the US, since TDR in our community is estimated to be over 10% (Chan et al., 2012, 2011). TDR in many African countries is still less common due to later introduction of ARVs (Chan and Kantor, 2009).
GIS mapping, endorsed by the CDC as an effective tool for developing HIV testing and treatment interventions (www.AIDSVU.org), can help identify communities to target such interventions. In our cohort, geospatial analyses of HIV-1 subtypes identified a statistically significant concentration of individuals with subtype B and non-B infections in Providence, Rhode Island’s capital city. Although there was some overlap, individuals with non-subtype B infection were found to concentrate in 15 block groups, a racially diverse area of Providence with higher poverty rates than other sections of the city (profiles.provplan.org). This geospatial concentration of non-subtype B HIV infections, combined with major risk factor differences for HIV acquisition among B (MSM) and non-B (heterosexual) infections, as well as additional religious and cultural factors, can all help target HIV testing campaigns, improve access to support services, and provide insight on basic epidemiologic factors contributing to the epidemic in these populations (Hixson et al., 2011; Shepard et al., 2011). Targeted HIV testing, treatment and prevention interventions have the potential to reach these geographic regions in our state with a high prevalence and density of infection.
Subtyping methodology is an important factor to consider in the interpretation and comparison of data from diverse HIV variants. Understanding limitations of available subtyping tools is important to avoid erroneous conclusions. Phylogenetic analyses of pol sequences was used in our study as the gold-standard for subtype determination, as is commonly reported (Holguín et al., 2008; Hué et al., 2004; Pineda-Peña et al., 2013). However, the observation that phylogenetic subtype assignment for the majority of sequences in this cohort was not supported by high bootstraps, and the use of the whole sequence for subtype determination, emphasize the limitations of this methodology. Understanding these limitations, phylogeny is used here only as a reference and its subtyping outcomes are only used for comparison across the tools examined.
Most automatic, publicly available subtyping tools are concordant in identifying pure subtypes (Gifford et al., 2006; Holguín et al., 2008; Pineda-Peña et al., 2013; Yebra et al., 2011). Discordance is usually with closely related subtypes (such as B and D) and less well-defined recombinant forms. Two major reasons for discordance among subtyping tools include the reference datasets that are used, some of which do not include representative references from all recombinant forms (e.g. STAR and Stanford), and the method the tool is based on (e.g. phylogeny for REGA; scoring matrices for STAR; similarity-based algorithms for Geno2pheno, EuResist, Stanford, NCBI; and probability for jpHMM). Additional explanation for discordant subtyping includes current definitions of CRFs, atypical breakpoints and absence of breakpoints in pol (Pineda-Peña et al., 2013). In our cohort, despite these differences, exact matches or minor variations were seen for 96% of sequences. The remaining 4% produced varying results with some of the less common recombinant forms (55% of moderate or major variations were among CRF06_CPX or CRF11_CPX). The limitations of each methodology should be understood when performing subtyping across non-B subtypes, especially minor recombinant forms.
The current study was limited due to the retrospective nature of data collection and incomplete available data. Future studies should focus on prospective evaluation of individuals with non-subtype B infection to obtain accurate and detailed epidemiologic data, and on larger (beyond pol only) genomic regions to more accurately represent circulating HIV diversity. Second, timing of infection, important in order to determine if transmission occurs within or outside the US, was not examined here. Lastly, due to the small numbers of individual subtypes available, it is difficult to adequately evaluate characteristics of patients infected with specific subtypes. Our study did include a comprehensive, although not complete, subset of HIV-infected individuals in our state resulting in a large and detailed evaluation of non-subtype B infections.
5. CONCLUSION
In summary, evaluation of HIV diversity at the major HIV center in Rhode Island highlights the important need for continued surveillance, monitoring and characterization of non-subtype B infections given that in our cohort: (1) Individuals with non-subtype B infection have different risk factors for HIV acquisition (heterosexual versus MSM transmission), suggesting the need for targeted or augmented approaches to HIV treatment and prevention; (2) Individuals with non-subtype B infection form less phylogenetic transmission clusters across subtypes hinting at less local transmission and more imported cases, though achieving complete representation of HIV infections is ideal to substantiate this statement; and (3) Non-subtype B infections form geospatial clusters in select urban neighborhoods, suggesting specific areas for outreach, treatment and prevention.
Supplementary Material
Research Highlights.
We evaluated HIV-1 non-B subtype epidemiology using a phylogenetic and geospatial approach.
Individuals with non-B subtypes had different risk factors for infection than those with subtype B.
Non-B subtypes formed fewer phylogenetic transmission clusters suggesting more imported cases.
Non-B subtypes formed geospatial clusters in select urban neighborhoods.
Epidemiology of non-B subtypes suggests specific areas for outreach, treatment, and prevention.
Acknowledgments
Philip A. Chan and Rami Kantor are supported by grants (1K23AI096923-01 and R01AI66922, respectively) from the National Institute of Allergy And Infectious Diseases at the National Institutes of Health. Additional support was provided by the Lifespan/Tufts/Brown Center for AIDS Research funded by the National Institute of Allergy And Infectious Diseases (P30AI042853, PI: Charles Carpenter).
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
All authors declare no conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Getis A, Ord K. The analysis of spatial association by use of distance statistics. Geographical Analysis. 1992;24:189–206. [Google Scholar]
- Achkar JM, Burda ST, Konings FAJ, Urbanski MM, Williams CAU, Seifen D, Kahirimbanyi MN, Vogler M, Parta M, Lupatkin HC, Zolla-Pazner S, Nyambi PN. Infection with HIV type 1 group M non-B subtypes in individuals living in New York City. J Acquir Immune Defic Syndr. 2004;36:835–844. doi: 10.1097/00126334-200407010-00011. [DOI] [PubMed] [Google Scholar]
- Apetrei C, Loussert-Ajaka I, Descamps D, Damond F, Saragosti S, Brun-Vézinet F, Simon F. Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS. 1996;10:F57–60. doi: 10.1097/00002030-199612000-00002. [DOI] [PubMed] [Google Scholar]
- Beckwith CG, DeLong AK, Desjardins SF, Gillani F, Bazerman L, Mitty JA, Ross H, Cu-Uvin S. HIV infection in refugees: a case-control analysis of refugees in Rhode Island. Int J Infect Dis. 2009;13:186–192. doi: 10.1016/j.ijid.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerenwinkel N, Schmidt B, Walter H, Kaiser R, Lengauer T, Hoffmann D, Korn K, Selbig J. Diversity and complexity of HIV-1 drug resistance: a bioinformatics approach to predicting phenotype from genotype. Proc Natl Acad Sci USA. 2002;99:8271–8276. doi: 10.1073/pnas.112177799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D. HIV [corrected] genetic diversity surveillance in the United States. J Infect Dis. 2005;192:4–9. doi: 10.1086/430329. [DOI] [PubMed] [Google Scholar]
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CAB, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Stramer SL, Holzmayer V, Yamaguchi J, Foster GA, Notari EP, IV, Schochetman G, Devare SG. Identification of human immunodeficiency virus type 1 non-B subtypes and antiretroviral drug-resistant strains in United States blood donors. Transfusion. 2009;49:125–133. doi: 10.1111/j.1537-2995.2008.01935.x. [DOI] [PubMed] [Google Scholar]
- Brenner BG, Roger M, Stephens D, Moisi D, Hardy I, Weinberg J, Turgel R, Charest H, Koopman J, Wainberg MA. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. J Infect Dis. 2011;204:1115–1119. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodine SK, Shaffer RA, Starkey MJ, Tasker SA, Gilcrest JL, Louder MK, Barile A, VanCott TC, Vahey MT, McCutchan FE, Birx DL, Richman DD, Mascola JR. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann Intern Med. 1999;131:502–506. doi: 10.7326/0003-4819-131-7-199910050-00004. [DOI] [PubMed] [Google Scholar]
- Carr JK, Osinusi A, Flynn CP, Gilliam BL, Maheshwari V, Zhao RY. Two independent epidemics of HIV in Maryland. J Acquir Immune Defic Syndr. 2010;54:297–303. doi: 10.1097/QAI.0b013e3181e0c3b3. [DOI] [PubMed] [Google Scholar]
- Chan PA, Kantor R. Transmitted drug resistance in nonsubtype B HIV-1 infection. HIV Ther. 2009;3:447–465. doi: 10.2217/hiv.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PA, Kazi S, Rana A, Blazar I, Dejong CC, Mayer KH, Huard TK, Carleton K, Gillani F, Alexander N, Parillo Z, Flanigan TP, Kantor R. New HIV Infections at Southern New England Academic Institutions: Implications for Prevention. AIDS research and human retroviruses. 2012 doi: 10.1089/AID.2012.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PA, Tashima K, Cartwright CP, Gillani FS, Mintz O, Zeller K, Kantor R. Transmitted Drug Resistance and Molecular Epidemiology in Antiretroviral Naive HIV Type 1-Infected Patients in Rhode Island. AIDS Res Hum Retroviruses. 2011;27:275–281. doi: 10.1089/aid.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PA, Wakeman SE, Flanigan T, Cu-Uvin S, Kojic E, Kantor R. HIV-2 diagnosis and quantification in high-risk patients. AIDS Res Ther. 2008;5:18. doi: 10.1186/1742-6405-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan David, Unwin David. Geographic Information Analysis. 2. Wiley; 2010. [Google Scholar]
- De Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AMJ, van de Vijver DA, Boucher CA, Camacho R, Vandamme AM. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- Delong AK, Wu M, Bennett D, Parkin N, Wu Z, Hogan JW, Kantor R. Sequence Quality Analysis Tool for HIV Type 1 Protease and Reverse Transcriptase. AIDS Res Hum Retroviruses. 2011 doi: 10.1089/AID.2011.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E, Slikas E, Stramer SL, Kamel H, Kessler D, Krysztof D, Tobler LH, Carrick DM, Steele W, Todd D, Wright DJ, Kleinman SH, Busch MP NHLBI-REDS-II Study Group. Genetic diversity of recently acquired and prevalent HIV, hepatitis B virus, and hepatitis C virus infections in US blood donors. J Infect Dis. 2012;205:875–885. doi: 10.1093/infdis/jir862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins S, Beckwith CG, Ross H, Bazerman L, Mitty JA. Caring for HIV-infected refugees in Rhode Island. Med Health R I. 2007;90:360–362. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CS, Stockmayer G, Stiles J, Hout M. Distinguishing the geographic levels and social dimensions of U.S. metropolitan segregation, 1960–2000. Demography. 2004;41:37–59. doi: 10.1353/dem.2004.0002. [DOI] [PubMed] [Google Scholar]
- Gale CV, Myers R, Tedder RS, Williams IG, Kellam P. Development of a novel human immunodeficiency virus type 1 subtyping tool, Subtype Analyzer (STAR): analysis of subtype distribution in London. AIDS Res Hum Retroviruses. 2004;20:457–464. doi: 10.1089/088922204323087697. [DOI] [PubMed] [Google Scholar]
- Gifford R, de Oliveira T, Rambaut A, Myers RE, Gale CV, Dunn D, Shafer R, Vandamme AM, Kellam P, Pillay D Collaborative Group on HIV Drug Resistance UK. Assessment of automated genotyping protocols as tools for surveillance of HIV-1 genetic diversity. AIDS. 2006;20:1521–1529. doi: 10.1097/01.aids.0000237368.64488.ae. [DOI] [PubMed] [Google Scholar]
- Gillani FS, Zaller ND, Zeller K, Rich JD, Cu-Uvin S, Flanigan TP, Carpenter CCJ. Changes in demographics and risk factors among persons living with HIV in an academic medical center from 2003–2007. Med Health R I. 2009;92:237–240. [PMC free article] [PubMed] [Google Scholar]
- Gonzales MJ, Machekano RN, Shafer RW. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a northern California clinic-based population. J Infect Dis. 2001;184:998–1006. doi: 10.1086/323601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S WHO-UNAIDS Network for HIV Isolation and Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hixson BA, Omer SB, del Rio C, Frew PM. Spatial clustering of HIV prevalence in Atlanta, Georgia and population characteristics associated with case concentrations. J Urban Health. 2011;88:129–141. doi: 10.1007/s11524-010-9510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguín A, López M, Soriano V. Reliability of rapid subtyping tools compared to that of phylogenetic analysis for characterization of human immunodeficiency virus type 1 non-B subtypes and recombinant forms. J Clin Microbiol. 2008;46:3896–3899. doi: 10.1128/JCM.00515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck BL, Beckwith CG. HIV-2 infection in Providence, Rhode Island from 2002 to 2011. HIV Med. 2013;14:115–119. doi: 10.1111/j.1468-1293.2012.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hué S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- IBM. SPSS Statistics for Windows, Version 200. IBM Corporation; Armonk, NY: 2011. [Google Scholar]
- Johnson AS, Beer L, Sionean C, Hu X, Furlow-Parmley C, Le B, Skarbinski J, Hall HI, Dean HD National Center for HIV/AIDS Viral Hepatitis STD and TB Prevention CDC. HIV Infection - United States, 2008 and 2010. MMWR Surveill Summ. 2013;62(Suppl 3):112–119. [PubMed] [Google Scholar]
- Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19:594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- Karchava M, Pulver W, Smith L, Philpott S, Sullivan TJ, Wethers J, Parker MM. Prevalence of drug-resistance mutations and non-subtype B strains among HIV-infected infants from New York State. J Acquir Immune Defic Syndr. 2006;42:614–619. doi: 10.1097/01.qai.0000225871.87456.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulinska IN, Ndung’u T, Mwakagile D, Msamanga G, Kagoma C, Fawzi W, Essex M, Renjifo B. A new human immunodeficiency virus type 1 circulating recombinant form from Tanzania. AIDS Res Hum Retroviruses. 2001;17:423–431. doi: 10.1089/088922201750102508. [DOI] [PubMed] [Google Scholar]
- Lacourse S, Rybak N, Lewis C, Gartman J, Larkin J, McLaughlin S, Toll ET. Health screening of newly resettled refugees in a primary care setting. R I Med J (2013) 2013;96:28–32. [PMC free article] [PubMed] [Google Scholar]
- Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-H, Gaschen BK, Collie M, El-Fishaway M, Chen Z, Korber BT, Beatrice ST, Zhang L. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr. 2006;41:399–404. doi: 10.1097/01.qai.0000200663.47838.f1. [DOI] [PubMed] [Google Scholar]
- Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los Alamos HIV Reference Sequence Database, 2010.
- Luft LM, Gill MJ, Church DL. HIV-1 viral diversity and its implications for viral load testing: review of current platforms. Int J Infect Dis. 2011;15:e661–670. doi: 10.1016/j.ijid.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Pant Pai N, Shivkumar S, Cajas JM. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996–2010. J Acquir Immune Defic Syndr. 2012;59:382–388. doi: 10.1097/QAI.0b013e31824a0628. [DOI] [PubMed] [Google Scholar]
- Parker MM, Gordon D, Reilly A, Horowitz HW, Waters M, Bennett R, Hallack R, Smith J, Lamson D, Aydemir A, Dvali N, Agins BD, Drusano GL, Taylor J Resistance Study Group. Prevalence of drug-resistant and nonsubtype B HIV strains in antiretroviral-naïve, HIV-infected individuals in New York State. AIDS Patient Care STDS. 2007;21:644–652. doi: 10.1089/apc.2006.0172. [DOI] [PubMed] [Google Scholar]
- Pasquier C, Millot N, Njouom R, Sandres K, Cazabat M, Puel J, Izopet J. HIV-1 subtyping using phylogenetic analysis of pol gene sequences. J Virol Methods. 2001;94:45–54. doi: 10.1016/s0166-0934(01)00272-5. [DOI] [PubMed] [Google Scholar]
- Pineda-Peña A-C, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, Gómez-López A, Camacho RJ, de Oliveira T, Vandamme A-M. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol. 2013;19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- Powell RLR, Zhao J, Konings FAJ, Tang S, Nanfack A, Burda S, Urbanski MM, Saa DR, Hewlett I, Nyambi PN. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- Prosser AT, Tang T, Hall HI. HIV in persons born outside the United States, 2007–2010. JAMA. 2012;308:601–607. doi: 10.1001/jama.2012.9046. [DOI] [PubMed] [Google Scholar]
- Pyne MT, Hackett J, Jr, Holzmayer V, Hillyard DR. Large-scale analysis of the prevalence and geographic distribution of HIV-1 non-B variants in the United States. J Clin Microbiol. 2013;51:2662–2669. doi: 10.1128/JCM.00880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R foundation for statistical computing. R Development Core Team; 2013. [Google Scholar]
- Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- Rosen-Zvi M, Altmann A, Prosperi M, Aharoni E, Neuvirth H, Sönnerborg A, Schülter E, Struck D, Peres Y, Incardona F, Kaiser R, Zazzi M, Lengauer T. Selecting anti-HIV therapies based on a variety of genomic and clinical factors. Bioinformatics. 2008;24:i399–406. doi: 10.1093/bioinformatics/btn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov M, Plikat U, Chappey C, Kochergin A, Tatusova T. A web-based genotyping resource for viral sequences. Nucleic Acids Res. 2004;32:W654–659. doi: 10.1093/nar/gkh419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz AK, Zhang M, Leitner T, Kuiken C, Korber B, Morgenstern B, Stanke M. A jumping profile Hidden Markov Model and applications to recombination sites in HIV and HCV genomes. BMC Bioinformatics. 2006;7:265. doi: 10.1186/1471-2105-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard CW, Gortakowski HW, Nasrallah H, Cutler BH, Begier EM. Using GIS-based density maps of HIV surveillance data to identify previously unrecognized geographic foci of HIV burden in an urban epidemic. Public Health Rep. 2011;126:741–749. doi: 10.1177/003335491112600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sides TL, Akinsete O, Henry K, Wotton JT, Carr PW, Bartkus J. HIV-1 subtype diversity in Minnesota. J Infect Dis. 2005;192:37–45. doi: 10.1086/430322. [DOI] [PubMed] [Google Scholar]
- Siepel AC, Halpern AL, Macken C, Korber BT. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- Snoeck J, Van Dooren S, Van Laethem K, Derdelinckx I, Van Wijngaerden E, De Clercq E, Vandamme A-M. Prevalence and origin of HIV-1 group M subtypes among patients attending a Belgian hospital in 1999. Virus Res. 2002;85:95–107. doi: 10.1016/s0168-1702(02)00021-7. [DOI] [PubMed] [Google Scholar]
- Stürmer M, Preiser W, Gute P, Nisius G, Doerr HW. Phylogenetic analysis of HIV-1 transmission: pol gene sequences are insufficient to clarify true relationships between patient isolates. AIDS. 2004;18:2109–2113. doi: 10.1097/00002030-200411050-00002. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Friedman JF, Vivier PM, Tompkins CEA, Alario AJ. Health care utilization of refugee children after resettlement. J Immigr Minor Health. 2012;14:583–588. doi: 10.1007/s10903-011-9530-1. [DOI] [PubMed] [Google Scholar]
- Wegner SA, Brodine SK, Mascola JR, Tasker SA, Shaffer RA, Starkey MJ, Barile A, Martin GJ, Aronson N, Emmons WW, Stephan K, Bloor S, Vingerhoets J, Hertogs K, Larder B. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naïve HIV-1 infected US military personnel. AIDS. 2000;14:1009–1015. doi: 10.1097/00002030-200005260-00013. [DOI] [PubMed] [Google Scholar]
- Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, Mahle KC, Heneine W, Johnson JA, Hall HI, Variant Atypical Resistant HIV Surveillance Group. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS. 2010;24:1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- Womack C, Roth W, Newman C, Rissing JP, Lovell R, Haburchak D, Essex M, Bond VC. Identification of non-B human immunodeficiency virus type 1 subtypes in rural Georgia. J Infect Dis. 2001;183:138–142. doi: 10.1086/317649. [DOI] [PubMed] [Google Scholar]
- Yahi N, Fantini J, Tourres C, Tivoli N, Koch N, Tamalet C. Use of drug resistance sequence data for the systematic detection of non-B human immunodeficiency virus type 1 (HIV-1) subtypes: how to create a sentinel site for monitoring the genetic diversity of HIV-1 at a country scale. J Infect Dis. 2001;183:1311–1317. doi: 10.1086/319859. [DOI] [PubMed] [Google Scholar]
- Yebra G, de Mulder M, Martín L, Pérez-Cachafeiro S, Rodríguez C, Labarga P, García F, Tural C, Jaén A, Navarro G, Holguín A Cohort of Spanish AIDS Research Network (CoRIS) Sensitivity of seven HIV subtyping tools differs among subtypes/recombinants in the Spanish cohort of naïve HIV-infected patients (CoRIS) Antiviral Res. 2011;89:19–25. doi: 10.1016/j.antiviral.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.