Abstract
Epigenetic processes, defined as heritable changes in gene expression that occur without changes to the DNA sequence, have emerged as a promising area of cardiovascular disease research. Epigenetic information transcends that of the genotype alone, and provides for an integrated etiologic picture of cardiovascular disease pathogenesis because of the interaction of the epigenome with the environment. Epigenetic biomarkers, which include DNA methylation, histone modifications, and RNA-based mechanisms, are both modifiable and cell-type specific, which makes them not only responsive to the environment, but also an attractive target for drug development. However, the enthusiasm surrounding possible applications of cardiovascular epigenetics currently outpaces available evidence. In this review, we synthesize the evidence linking epigenetic changes with cardiovascular disease, emphasizing the gap between the translational potential and the clinical reality of cardiovascular epigenetics.
Despite its initial promise, the clinical utility of genetic markers for prediction and prevention of heart disease has proven to be limited.1 Following this translational disappointment, the attention of the cardiovascular research community has turned to epigenetics as the new frontier for risk stratification, prevention, and treatment. Broadly defined, epigenetics is the study of heritable changes in gene expression that are not coded in the DNA sequence itself.2 Epigenetic variation falls into three interconnected categories: DNA methylation, RNA-based mechanisms including microRNAs and non-coding RNAs, and post-translational histone modifications1.3 Although DNA methylation is the most common epigenetic modification in the mammalian genome,4 a growing body of evidence suggests all three types of epigenetic changes are involved in the pathogenesis of cardiovascular disease (CVD).5
The field of CVD epigenetics is rapidly growing,6 although the current enthusiasm about its clinical applications far exceeds available evidence. In 2013, 353 of the 8026 manuscripts published in the field of epigenetics (4.4%) were related to CVD. However, of those, only 125 reported findings in humans, and of those 125, 66 (52.8%) were reviews rather than original research, and only 4 (0.6%) were clinical trials or population-level studies. Therefore, the need for more translational studies that would evaluate specific epigenetic modifications as both prognostic markers and therapeutic targets remains pressing. The clinical potential for epigenetic markers in CVD is underscored by several factors, chief among them 1) animal and in vitro model evidence linking epigenetic changes to CVD pathways such as atherosclerosis; 2) increased availability of epigenetic analysis technologies at a decreasing cost; and 3) translational success of epigenetics in other chronic disease settings, most notably cancer. The study of epigenetics is also conceptually appealing because processes such as DNA methylation and histone modifications link the genotype and the environment, helping elucidate the mechanisms underlying CVD.
However, the very nature of epigenetic processes makes their study and their interpretation difficult for both researchers and clinicians. Unlike genetic variation, epigenetic changes are cell-type specific, reversible, and susceptible to both inherited and environmental influences.7 On one hand, these characteristics describe attractive targets for interventions; on the other hand, the relevance of epigenetic biomarkers as definitive tools for diagnosis or risk stratification is thoroughly confounded by other variables such as age, genotype, and lifestyle, particularly diet and smoking. Unlike studies of genetic markers, epigenetics research must address the problem of reverse causality: just as epigenetic processes may underlie CVD pathogenesis, there is evidence that subclinical or clinical disease influences epigenetic variation.6 The causality discussion is further complicated by evidence suggesting that epigenetic changes may be secondary to other disease processes, e.g. inflammation,8 and thus represent an epiphenomenon rather than a genuine disease mechanism.9 Another challenge to clinical applications is posed by tissue specificity: CVD-relevant epigenetic changes may occur in hard to access tissues, e.g. the myocardium, and not be reflected in blood, which is commonly used as a diagnostic tissue. Furthermore, DNA methylation patterns can change rapidly and are reversible, so the optimal timing of the measurement relative to disease onset remains unclear. Finally, there is the practical issue of storing and interpreting epigenetic data as part of the patient’s medical record, which has become more commonplace in oncology but has yet to translate to the CVD context.
Despite these challenges, in recent years a number of epigenetic tags have emerged as promising biomarkers for CVD. Because most studies of RNA-based epigenetic changes and histone modifications are currently limited to animal and in vitro models, the following review of potential clinical applications of epigenetic data will focus on DNA methylation. DNA methylation is a covalent chemical modification of DNA that typically involves adding a methyl group to cytosine residues at cytosine-phosphate-guanine (CpG) nucleotides, although non-CpG methylation can also occur,10 DNA methylation plays a role in mammalian development, transcription, chromatin structure, cellular homeostasis, genomic imprinting, and disease pathogenesis.11 Commonly a genomic region with methylated DNA becomes inaccessible to transcriptional machinery, and as a result gene expression is suppressed.12 There exists cross-talk between DNA methylation and other epigenetic modifications, notably histone acetylation, which can contribute to aberrant gene regulation and disease.13 Methylation and the resulting expression changes are generally stable and heritable during mitosis, although stochastic events or environmental factors can alter methylation patterns throughout lifetime.14
Global methylation studies
The first studies of epigenetic biomarkers in the context of CVD focused on global DNA methylation, mostly because of the prominent role that homocysteine, an independent vascular disease risk factor, plays in the methylation process. Global DNA methylation refers to a pattern unique to humans and other vertebrates, in whom genomes are heavily methylated in most cell types and developmental stages and genome-wide hypomethylation is often associated with the risk of disease.15,16 Several human and animal studies have linked increased plasma homocysteine with decreased global methylation, likely occurring due to accumulation of S-adenosyl homocysteine, which, in turn, inhibits transmethylation reactions.17 However, the evidence linking global methylation patterns, usually measured in blood cells, with cardiovascular outcomes remains conflicting and comes mostly from cross-sectional studies, limiting causal inference. While some studies showed that increased homocysteine and decreased global DNA methylation was associated with vascular disease,18 others reported associations of coronary heart disease with elevated homocysteine but increased global DNA methylation.19 (See Table 1 for a summary of epigenetic markers noted in this review.) A subsequent study failed to demonstrate a correlation between global DNA methylation and plasma homocysteine or folate, suggesting that non-folate related mechanisms such as systemic inflammation may lead to increased global DNA methylation in the setting of CVD.42 Consistent with this paradigm, an analysis of methylation patterns in dialysis patients demonstrated a relationship between global DNA hypermethylation with systemic inflammation and increased mortality from chronic kidney disease.20
Table 1.
Epigenetic markers and their associated cardiovascular disease or phenotype.
| Epigenetic marker | Associated condition/phenotype | Ref |
|---|---|---|
| Global methylation studies | ||
| increased homocysteine, ↓ GM | ↑ vascular disease, coronary heart disease | 18,19 |
| ↑ GM | ↑ systemic inflammation, mortality from chronic kidney disease |
20 |
| ↑ Alu | environmental pollution impact on inflammatory markers | 21 |
| ↓ LINE-1 | ↑ inflammatory marker, ischemic heart disease, stroke, total mortality, LDL, metabolic syndrome, ↓ HDL |
22-25 |
| Candidate gene studies | ||
| ↑ FTO | ↑ obesity | 26 |
| ↓ CERCAM, ↓ DPYD, ↑ IL12A, ↓ ZNF35, ↓ ZNF362, ↑↓ Others |
↑ obesity | 27 |
| ↑ F2RL3 | ↑ CVD risk factors, total mortality | 28 |
| ↑ INS, GNASAS | ↑ MI risk in females | 29 |
| ↑ PLA2G7 | ↑ CHD in females | 30 |
| ↑ IGF2 | ↑ obesity development, TG/HDL ratio | 31,32 |
| ↑ POMC | ↑ childhood TG, insulin | 33 |
| ↑ TLR2, NOS2 | ↑ blood pressure | 34 |
| ↑ IFNG | ↓ blood pressure | 34 |
| ↑ HSD11B2 promoter | ↑ hypertension risk | 35 |
| Epigenome-wide studies | ||
| ↑ CPT1A | ↑ TG, VLDL | 36 |
| ↑ TNNT1 promoter | ↑ HDL particle size, HDL-C | 37 |
| ↑ at multiple loci | ↑ obesity | 38 |
| ↑ ABCG1 | ↑ fasting insulin | 39 |
| ↑ LY75, ↓ ERBB3, ↓ HOXB13, ↓ ADORA2A, ↑ DUX4 |
↑ cardiomyopathy | 40,41 |
↑, increased methylation at locus or increase in phenotype metric; ↓, decreased methylation at locus or decrease in phenotype metric; CHD, coronary heart disease; CVD, cardiovascular disease; GM, global methylation; HDL-C, high-density lipoprotein cholesterol; HDL, highdensity lipoprotein; LDL, low-density lipoprotein; LINE-1, long interspersed nucleotide element-1; MI, myocardial infarction; Ref, reference; TG, triglycerides; VLDL, very low-density lipoprotein
Global DNA methylation studies often interrogate repetitive sequences found across the genome, e.g. long interspersed nucleotide element-1 (LINE)-1 and Alu elements. These DNA segments are generally hypermethylated and, as a result, not transcribed, yet they are abundant; for example, LINE-1 retrotransposon sequences account for approximately 17% of the human genome.43 Therefore, quantifying DNA methylation in repetitive elements provides a useful picture of global genomic methylation44 and may indicate susceptibility to disease. Data from male participants of the VA Normative Aging Study showed that hypermethylation of the Alu repetitive element exacerbated the effect of environmental pollution on a range of systemic inflammation markers like C-reactive protein (CRP), intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1).21 In the same cohort, LINE-1 element hypomethylation in blood DNA was associated with cardiovascular risk factors such as higher serum vascular cell adhesion molecule22 as well as with prevalent and incident ischemic heart disease, stroke, and total mortality.23 This is concordant with evidence from a cross-sectional study of Samoan Islanders that showed that lower LINE-1 methylation was associated with higher low-density lipoprotein (LDL) and lower high-density lipoprotein (HDL) cholesterol.24 In visceral adipose tissue samples from severely obese individuals, LINE-1 hypomethylation was found to be associated with higher prevalence of metabolic syndrome, and therefore elevated risk for CVD.25 Although data presented by Baccarelli et al.23 suggest that LINE-1 hypomethylation precedes disease diagnosis and could be used for early identification of at-risk individuals, this suggestion has yet to be translated into clinical applications.
Candidate gene methylation studies
Complementing the global methylation studies, several reports have assessed relationships between the methylation status of a priori-selected, biologically relevant genomic regions and cardiovascular risk. Of particular interest among candidate gene studies are investigations of FTO, a prominent obesity-associated gene that has also been linked to CVD risk independently of its effect on body mass index.45 Several studies revealed associations between sequence variation in FTO and levels of DNA methylation, both at the FTO locus26 as well as in KARS, TERF2IP, DEXI, MSI1, STON1 and BCAS3, 27 suggesting that the association between FTO mutations and cardiometabolic outcomes may be underpinned by epigenetics. Interestingly, the effects of FTO variation are mediated by regular physical activity, 46 but a recent epigenome-wide DNA methylation study in adipocytes47 did not identify FTO loci as differentially affected by a six-month exercise intervention, raising the possibility of involvement for other mechanisms. Specifically, recent studies show that the FTO protein can also catalyze modifications of RNA through oxidative demethylation of N6-methyladenosine.48,49 While the evidence from both human cells and animal tissue is compelling, the implications of these RNA modifications for disease risk warrant further study. Most recently, evidence emerged that FTO directly interacts at long range with the promoter of the homeobox gene IRX3 in humans, mice, and zebrafish, enhancing IRX3 expression; in turn, IRX3 has a direct effect on body mass and composition, as demonstrated by knockout mouse models.50 Such rigorous studies are poised to fill in the mechanistic gaps in our current understanding of cardiometabolic genetics and epigenetics.
Other candidate gene studies have identified a number of promising epigenetic targets in CVD. Notably, a large-scale prospective cohort study of patients with stable coronary heart disease established that methylation (in whole blood samples) of F2RL3, a known locus linked with tobacco use, is associated with both CVD risk factors (e.g. CRP and body mass index) at baseline as well as with overall mortality but not with CVD mortality at 8 years of follow-up.28 Interestingly, F2RL3 methylation status was a better prognostic factor than self-reported and cotinine-validated smoking behavior.28 In another candidate gene study, higher leukocyte DNA methylation at INS and GNASAS, two loci previously reported to be sensitive to prenatal environment, was prospectively associated with the risk of (MI) infarction in women but not in men.29 Cross-sectionally, hypermethylation of PLA2G7 was associated with coronary heart disease (CHD) among females30 and hypermethylation of a locus on 9p21, a region of robust genetic associations with CVD, was more prevalent among coronary artery disease cases in a Chinese population.51
Other candidate gene studies have investigated intermediate cardiovascular risk phenotypes such as obesity, dyslipidemia, or hypertension. For example, the methylation extent of IGF2, which encodes a key growth factor in fetal development, predicts development of obesity31 and cross-sectionally associates with a higher triglyceride/HDL cholesterol ratio.32 Additionally, hypermethylation of POMC, a gene encoding proopiomelanocortin, in umbilical cord blood was associated with higher blood triglycerides and insulin during childhood.33 In animal models, changes in methylation and expression of FADS2, a gene previously linked to cardiometabolic disease, affected arterial biosynthesis of polyunsaturated fatty acids and induced vascular dysfunction.52,53 Another analysis of the VA Normative Aging Study related small changes in blood pressure with the methylation of genes encoding pro-inflammatory proteins such as toll-like receptor 2, inducible nitric oxide synthase, and interferon-gamma.34 Similarly, increased methylation in the promoter region of the HSD11B2 gene was shown to be associated with the increased activity of the 11-betaHSD2 enzyme and the risk of hypertension.35
Most epigenetic markers discovered through the candidate gene approach have proven to be useful in elucidating the underlying pathophysiological mechanisms, but their clinical potential, especially in comparison with traditional cardiovascular risk factors, remains unclear. Moreover, is likely that the narrow scope of candidate gene studies precludes detection of many novel methylation loci that could be germane to CVD.54
Epigenome-wide studies
With the introduction of the Infinium Human Methylation 450K array (Illumina, San Diego, CA) that measures DNA methylation at approximately 450,000 cytosine-phosphate-guanine (CpG) sites across the genome, the focus of cardiovascular epigenetics has shifted from candidate regions to epigenome-wide studies. A recent study from our group, conducted in CD4+ T-cells from 991 metabolically healthy European American participants of the Genetics of Lipid Lowering Drugs and Diet Network Study, has identified robust associations between the methylation status of two markers in CPT1A, the expression of CPT1A, and the levels of plasma triglycerides and very low-density lipoprotein cholesterol.55 CPT1A encodes the carnitine palmitoyltransferase enzyme, which controls fatty acid flux in the liver via the beta-oxidation and esterification pathways, thus providing strong biological plausibility for the observed association. Furthermore, the direction of the association was replicated in an independent cohort.55 Using a lower-resolution platform (Illumina Infinium Human Methylation 27), another group has identified new loci associated with HDL cholesterol in the setting of familial hypercholesterolemia.37 Epigenome-wide studies of cardiomyopathy found distinct patterns of DNA methylation and histone-3 lysine-36 trimethylation in left ventricle tissue between cases and controls, particularly in LY75, ERBB3, HOXB13, ADORA2A, and DUX4; of those, DUX4, LY75 and ADORA2A also exhibited changes in gene expression and functional relevance.40,41 In animal models of hypertrophy, epigenome-wide studies of cardiomyocytes identified signature histone modifications and transcriptional changes, linking epigenetic processes to gene expression reprogramming implicated in heart failure.56
In addition to CVD phenotypes, epigenome-wide studies have shown differential methylation status of various loci associated with such CVD risk factors as age,57 obesity,38 air pollution,58 measures of fasting insulin and insulin resistance,39 and smoking,59,60 lending support to the view of epigenetic changes as the process by which environmental factors influence heritable genetic variation. Currently several other large-scale cohorts are in process of analyzing their own genome-wide methylation data and establishing consortia for meta-analyses, suggesting that more epigenetic biomarkers of cardiovascular risk will be uncovered in the months to come. The International Human Epigenome Consortium is in process of producing reference maps of 1,000 human epigenomes for a range of cell types, which could significantly accelerate epigenome-wide studies for many common diseases.54 Additionally, as the results from gene expression consortia suggest, these large-scale projects may identify surrogate tissues (and/or proxy epigenetic loci) to facilitate clinical implementation of the methylation markers.
However, the findings of the epigenome-wide association studies must be interpreted in light of several important limitations. First, all of the observed associations to date have come from cross-sectional studies; thus, it is impossible to establish either temporality or causality of the relationship between cardiovascular risk markers and DNA methylation. Second, from the clinical perspective, the epigenetic biomarkers are only as good as the intermediate risk factor they represent (e.g. triglycerides) because, unlike candidate gene studies, no epigenome-wide study to date has investigated the CVD endpoints of myocardial infarction, stroke, or death from related causes. Finally, as epigenome-wide studies represent an agnostic research approach, independent replication of epigenetic findings remains the gold standard for reducing false positive results.
Crosstalk between genetic markers, DNA methylation, and gene expression
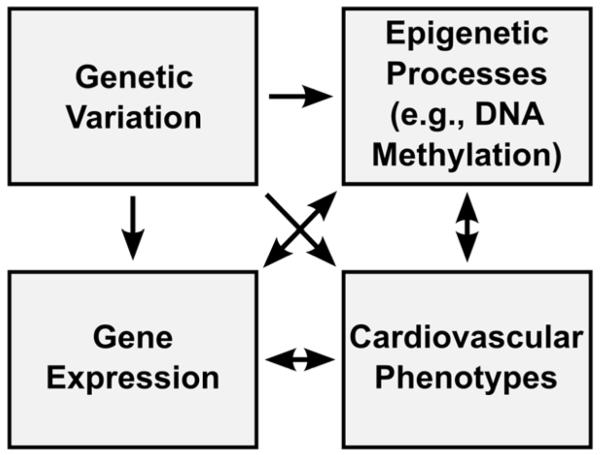
One of the factors hindering clinical implementation of the findings from candidate gene or epigenome-wide methylation studies is the complexity of the relationship between sequence variation, methylation, and gene expression patterns. The correlation between genetic polymorphisms and DNA methylation levels is well-established and predominantly accounted for by variants located at CpG sites.61-63 Similarly, genetic variation has been shown to underlie variation in gene expression.64 The traditional paradigm holds that DNA methylation is also associated with changes in gene expression, specifically gene repression, due to interference with the binding of transcription factors or recruitment of histone deacetylases or other repressors.65,66 In contrast, recent studied showed that DNA methylation can affect expression positively as well as negatively, and other factors (e.g. genetic variation or environmental inputs such as drug treatment) have been postulated to affect both methylome and transcriptome.62,65-67 It is also notable that both methylation and gene expression are dynamic processes, so their effects may be mutual and bidirectional; disentangling the causal web would require repeated measurements and considerable expenses. Because of the complexity of these interactions (Figure 1), any novel methylation markers cannot be interpreted or used outside of their genomic context, data on which are not always available. While genotype information has become commonplace with the advent of genome-wide associations studies, most population or clinical methylation studies of cardiovascular traits, with few exceptions, 37,40 do not have expression data. As a result, the mechanistic insights provided by such studies are limited, especially in light of the evidence supporting the connection between methylation and expression in recent in vitro studies of cardiovascular phenotypes.68,69 However, as large-scale expression profiling techniques such as RNA-Seq become more accessible, studies will be able to better integrate data on epigenetic changes, DNA sequence variation, and gene expression, identifying the most promising targets for screening and intervention.
Figure 1.
An integrated perspective on genomic variation, gene expression, and epigenetic modification in cardiovascular phenotypes. While DNA sequence variation necessarily precedes changes in both gene expression and epigenetic processes, all other relationships are bidirectional.
The aging process, which affects the methylation status of as many as 29% of CpG sites located on the Illumina 450K array in white blood cells,70 introduces further complexity into the epigenetic paradigm. Specifically, over the course of the human lifespan the DNA methylation content decreases and patterns become less correlated between neighboring regions.71 Although associated with normal aging processes, such changes may also underlie the development of some pathological conditions, including CVD. Finally, the consequences of aging may be mediated by trans-generational effects, with methylation at highly heritable regions shown to be more stable over time and functionally relevant.72 Trans-generational effects also occur with other CVD risk factors, most notably smoking and other environmental chemicals,73 via in utero exposure or the paternal germline.74 As a result, the influences of inherited (as in the case of transgenerational effects) or environmental factors on CVD risk are difficult to distinguish from the cardiovascular implications of epigenetic processes themselves.
Epigenetic markers in CVD prevention and treatment
Because DNA methylation is intrinsically reversible, it presents a lucrative target for interventions in cardiovascular disease. These interventions can be either lifestyle-related or pharmacological. In the lifestyle domain, gene-diet interactions are often underpinned by epigenetic mechanisms, giving rise to the idea of nutrition as “epigenetic medicine.”75,76 The nutriepigenetic paradigm was first proposed in the context of atherosclerosis, suggesting that global hypomethylation due to reduced folic acid and vitamin B12 levels increases cardiovascular risk;77 however, subsequent studies have failed to convincingly support this hypothesis.78 An emerging body of data points to the role of other dietary factors (e.g. polyphenols, catechins, bioflavonoids, or selenium) in the methylation process.79-81 For example, a recent study showed that consuming cocoa, a rich source of polyphenols, decreases the global DNA methylation of peripheral leukocytes in humans with cardiovascular risk factors.82 While an intriguing possibility, the epigenetic link between consumption of certain nutrients and cardiovascular risk remains to be directly tested. Most likely, because of their relatively low concentrations in common foods and low bioavailability, the effect of any given compound on CVD risk is negligible.79 Despite the lack of supporting evidence, many functional foods or supplements that market themselves as nutriepigenetic modulators are currently under development or available to consumers.76
Studies of associations between DNA methylation patterns and phenotypic responses to diet and/or dietary changes represent another nutriepigenetic facet of cardiovascular risk. Two trials from the same research group have identified that differential methylation of seven genes (ATP10A, CD44, AQP9, DUSP22, HIPK3, TNNI3, and TNNT1) as prognostic biomarkers of response to a weight loss intervention, and also reported diet-induced changes in methylation of WT1 and ATP10A at the end of the study.83,84 Because these studies were conducted in adolescents, the effect of the same markers on CVD endpoints was not investigated. In a small study of obese adults, hypermethylation of SERPINE1, a gene encoding plasminogen activator inhibitor 1 that had previously been associated with metabolic dysfunction, was prospectively associated with not only weight loss but also more drastic reductions in total cholesterol and triglycerides.85 These findings provide important etiological clues to the pathogenesis of chronic disease, but their clinical implications are not straightforward. First, some of the strongest predictors of both DNA methylation and dietary response are non-modifiable factors like age and sex,86,87 which could confound the observed associations and limit the prognostic value of the biomarkers. Second, none of the above-referenced studies measured gene expression, further limiting insights into the underlying mechanisms. Third, larger sample sizes and longer intervention periods would enhance precision and clinical relevance of the findings.
In contrast to the uncertain nature of nutriepigenomic findings, the cardiovascular implications of pharmacoepigenetics appear to be more direct and far-reaching. A growing body of evidence shows that epigenetic modifications could be reversible through both novel pharmaceutical approaches such as bromodomain and extra-terminal (BET) protein inhibitors as well as commonly used drugs.88,89 Some examples of common drugs with newly discovered epigenetic effects are vasodilators such as hydralazine, which has been shown to inhibit DNA methylation by either interfering with DNA methyltransferase directly or by reducing its gene expression, and procainamide, a sodium channel blocker that inhibits DNA methyltransferase I.90-92 A serious side effect common to both hydralazine and procainamide is an autoimmune disease similar to lupus, believed to be triggered by extensive hypomethylation across the genome.90,93 Both of these therapeutic approaches are now considered a promising avenue for treating cancer,94 but the cardiovascular implications of their epigenetic effects remain unclear.
The evidence in support of epigenetic effects of statins paints a complex picture. One group of studies demonstrated that trichostatin A can inhibit histone deacetylases (HDACs) in cancer cells and mouse models, reducing deacetylation and thus increasing transcription.7,95,96 Initially described as repressors of transcription because of their effects on histones, HDACs have since been shown to act on other cellular substrates and activate transcription as well, e.g. by deacetylating a key member of the mitogen-activated kinase (MAP) family.97-99 Because the cardioprotective potential of such gene upregulation has not been demonstrated and animal studies documented adverse morphological effects instead, drug development to date has focused on downregulating HDACs.98 A number of preclinical trials have highlighted the potential of HDAC inhibitors, particularly in the context of heart failure and myocardial injury.95 In animal and cell culture models, HDAC inhibitors such as trichostatin A have been shown to reduce inflammation and block adverse cardiac remodeling.100 While the underlying mechanism remains largely unknown, recent studies implicated signaling molecules MKK3 and Akt-1 as well as gp-91, a subunit of HADPH-oxidase, in HDAC-inhibition-induced cardioprotection.101,102 The anti-inflammatory effect of HDAC inhibitors may be due to induction of regulatory T-cells shown with statin therapy in patients with acute coronary syndrome.100,103 Furthermore, HDAC inhibitors may be beneficial in treatment of cardiac fibrosis associated with activation and proliferation of fibroblasts.104 However, further mechanistic questions must be answered for effective design of pharmaceutical interventions, e.g. which of the over a dozen HDAC genes and their many mRNA isoforms are particularly salient in the setting of CVD?100,105 As most clinical data on HDAC inhibitors come from cancer trials, the requisite dose for effective treatment of heart failure with minimal side effects remains to be determined as well, although studies of HDAC inhibition in autoimmune disease suggest that it may be lower than in the cancer setting.100,106
In contrast to the findings of increased gene expression as a result of HDAC inhibition, a study of cultured human endothelial cells107 showed that statin-induced histone modification can also reduce gene expression, accounting for some of the anti-inflammatory benefits of statin therapy. However, while promising in the context of cancer treatment,108 the epigenetic effects of statins in the setting of CVD are likely to be pleiotropic, leading to both positive and negative consequences. For example, one such effect was shown to be decreased expression of the atrogin-1 gene, which causes muscle fiber damage, a common side effect of statin therapy.88,109,110 It is evident that any therapeutic approaches that aim to harness the epigenetic potential of statins and other HDAC inhibitors will need to address this problem of unspecific effects, requiring many more studies of safety and efficacy.
At the cutting edge of epigenetic therapies for CVD is RVX-208, a small molecule BET bromodomain inhibitor that targets HDL cholesterol levels by inducing apolipoprotein A-I (apoA-I).111 In addition to its effects on apoA-I, BET inhibition suppresses cardiomyocyte hypertrophy in vitro and pathological cardiac remodeling in vivo by mediating transcriptional pause release in heart failure, suggesting pleiotropic effects on cardiovascular health.112 Two clinical trials have now been completed evaluating the safety and efficacy of RVX-208 in patients with CVD who were simultaneously receiving statin therapy, and the results were conflicting. The Study of Quantitative Serial Trends in Lipids with Apolipoprotein A-I stimulation (SUSTAIN) assessed safety as well as changes in HDL cholesterol and apoA-I levels and found increases in both parameters compared with placebo over 24 weeks.113,114 There was no evidence of additional hepatotoxicity in that study population.114 On the other hand, the results of the ApoA1 Synthesis Stimulation and Intravascular Ultrasound for Coronary Atheroma Regression Evaluation (ASSURE) trial, which used intravascular ultrasonography to assess changes in atheroma volume after 26 weeks of RVX-208 therapy, failed to detect statistically significant differences between the treatment and placebo group.115 Similarly, the investigators of ASSURE did not observe the beneficial changes in apoA-I or HDL-cholesterol that were reported in the SUSTAIN study.115 Most recently, when SUSTAIN and ASSURE data on major adverse cardiovascular events (CVD deaths, nonfatal myocardial infarctions, angina pectoris, or revascularizations for progressive coronary artery disease) were analyzed jointly, the event rate was considerably lower (6% vs. 21%, P=0.007) in the RVX-208 group compared with placebo.116 While these findings are undoubtedly promising, they are far from conclusive and require independent replication. Additionally, it is unclear whether the beneficial effects of RVX-208 would translate to the setting of primary prevention.
In summary, the development of clinical applications of cardiovascular epigenetics is still in its infancy. While candidate gene and epigenome-wide studies of DNA methylation have yielded promising findings (Table 1) that have elucidated underlying biological pathways, these markers have yet to be translated into meaningful prognostic markers or intervention targets, mostly because of their temporal instability and the complexity of the relationships between genetic variation, gene expression, and epigenetic processes. Therapeutic approaches that target epigenetic processes in the cardiovascular system are emerging, but clinical evidence of their safety and effectiveness in humans is extremely limited. Several research directions represent particularly promising avenues of investigation: 1) testing the prognostic ability of epigenetic tags against or as a supplement to traditional CVD risk stratification tools, e.g. the Framingham Risk Score; 2) development of more sensitive and specific pathophysiologic markers of CVD, e.g. microRNAs as diagnostic tools for acute MI,117,118 and 3) development of therapeutic and preventative approaches specifically targeting epigenetic processes in the cardiovascular system. On balance, the clinical potential of cardiovascular epigenetics still has a long road to fulfillment.
Acknowledgements
All authors have read this journal’s policy on the disclosure of potential conflicts of interest: all authors declare that they have no conflicts of interest. All authors have read this journal’s authorship agreement, and all authors have read, reviewed, and approved this manuscript.
Footnotes
Throughout this review, we use “epigenetic” to refer to variation in these three categories and “genetic” to refer to polymorphisms in the DNA sequence.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Di Angelantonio E, Butterworth AS. Clinical utility of genetic variants for cardiovascular risk prediction: a futile exercise or insufficient data? Circ Cardiovasc Genet. 2012;5:387–90. doi: 10.1161/CIRCGENETICS.112.964148. [DOI] [PubMed] [Google Scholar]
- 2.Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–40. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duygu B, Poels EM, da Costa Martins PA. Genetics and epigenetics of arrhythmia and heart failure. Front Genet. 2013;4:219. doi: 10.3389/fgene.2013.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Udali S, Guarini P, Moruzzi S, et al. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol Aspects Med. 2013;34:883–901. doi: 10.1016/j.mam.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–73. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010;7:510–9. doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grabiec AM, Reedquist KA. Histone deacetylases in RA: epigenetics and epiphenomena. Arthritis Res Ther. 2010;12:142. doi: 10.1186/ar3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratan RR. Epigenetics and the nervous system: epiphenomenon or missing piece of the neurotherapeutic puzzle? Lancet Neurol. 2009;8:975–7. doi: 10.1016/S1474-4422(09)70276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647:30–8. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 13.Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–8. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Heijmans BT, Kremer D, Tobi EW, et al. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–54. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 15.Huh I, Zeng J, Park T, et al. DNA methylation and transcriptional noise. Epigenetics Chromatin. 2013;6:9. doi: 10.1186/1756-8935-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 17.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–56. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castro R, Rivera I, Struys EA, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–6. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 19.Sharma P, Kumar J, Garg G, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–65. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 20.Stenvinkel P, Karimi M, Johansson S, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–99. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 21.Bind MA, Baccarelli A, Zanobetti A, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–40. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baccarelli A, Tarantini L, Wright RO, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5 doi: 10.4161/epi.5.3.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–28. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cash HL, McGarvey ST, Houseman EA, et al. Cardiovascular disease risk factors and DNA methylation at the LINE-1 repeat region in peripheral blood from Samoan Islanders. Epigenetics. 2011;6:1257–64. doi: 10.4161/epi.6.10.17728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turcot V, Tchernof A, Deshaies Y, et al. LINE-1 methylation in visceral adipose tissue of severely obese individuals is associated with metabolic syndrome status and related phenotypes. Clin Epigenetics. 2012;4:10. doi: 10.1186/1868-7083-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell CG, Finer S, Lindgren CM, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almen MS, Jacobsson JA, Moschonis G, et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–7. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Breitling LP, Salzmann K, Rothenbacher D, et al. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J. 2012;33:2841–8. doi: 10.1093/eurheartj/ehs091. [DOI] [PubMed] [Google Scholar]
- 29.Talens RP, Jukema JW, Trompet S, et al. Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int J Epidemiol. 2012;41:106–15. doi: 10.1093/ije/dyr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang D, Zheng D, Wang L, et al. Elevated PLA2G7 gene promoter methylation as a gender-specific marker of aging increases the risk of coronary heart disease in females. PLoS One. 2013;8:e59752. doi: 10.1371/journal.pone.0059752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins E, Murphy SK, Murtha AP, et al. Insulin-like growth factor 2/H19 methylation at birth and risk of overweight and obesity in children. J Pediatr. 2012;161:31–9. doi: 10.1016/j.jpeds.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deodati A, Inzaghi E, Liguori A, et al. IGF2 methylation is associated with lipid profile in obese children. Horm Res Paediatr. 2013;79:361–7. doi: 10.1159/000351707. [DOI] [PubMed] [Google Scholar]
- 33.Yoo JY, Lee S, Lee HA, et al. Can proopiomelanocortin methylation be used as an early predictor of metabolic syndrome? Diabetes Care. 2014;37:734–9. doi: 10.2337/dc13-1012. [DOI] [PubMed] [Google Scholar]
- 34.Alexeeff SE, Baccarelli AA, Halonen J, et al. Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes. Int J Epidemiol. 2013;42:270–80. doi: 10.1093/ije/dys220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friso S, Pizzolo F, Choi SW, et al. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199:323–7. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 36.Irvin MR, Zhi D, Aslibekyan S, et al. Genomics of post-prandial lipidomic phenotypes in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) PloS ONE. doi: 10.1371/journal.pone.0099509. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guay SP, Voisin G, Brisson D, et al. Epigenome-wide analysis in familial hypercholesterolemia identified new loci associated with high-density lipoprotein cholesterol concentration. Epigenomics. 2012;4:623–39. doi: 10.2217/epi.12.62. [DOI] [PubMed] [Google Scholar]
- 38.Xu X, Su S, Barnes VA, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–33. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hidalgo B, Irvin MR, Sha J, et al. Epigenome-Wide Association Study of Fasting Measures of Glucose, Insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network Study. Diabetes. 2014;63:801–7. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas J, Frese KS, Park YJ, et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med. 2013;5:413–29. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Movassagh M, Choy MK, Knowles DA, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M, Long TI, Arakawa K, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–39. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Mou S, Pan C. The FTO gene rs9939609 polymorphism predicts risk of cardiovascular disease: a systematic review and meta-analysis. PLoS One. 2013;8:e71901. doi: 10.1371/journal.pone.0071901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vimaleswaran KS, Li S, Zhao JH, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90:425–8. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- 47.Ronn T, Volkov P, Davegardh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Y, Jia G, Pang X, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smemo S, Tena JJ, Kim KH, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhuang J, Peng W, Li H, et al. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS One. 2012;7:e47193. doi: 10.1371/journal.pone.0047193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelsall CJ, Hoile SP, Irvine NA, et al. Vascular dysfunction induced in offspring by maternal dietary fat involves altered arterial polyunsaturated fatty acid biosynthesis. PLoS One. 2012;7:e34492. doi: 10.1371/journal.pone.0034492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoile SP, Irvine NA, Kelsall CJ, et al. Maternal fat intake in rats alters 20:4n-6 and 22:6n-3 status and the epigenetic regulation of Fads2 in offspring liver. J Nutr Biochem. 2013;24:1213–20. doi: 10.1016/j.jnutbio.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rakyan VK, Down TA, Balding DJ, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irvin MR, Zhi D, Joehanes R, et al. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid Lowering Drugs and Diet Network Study. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.009158. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papait R, Cattaneo P, Kunderfranco P, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110:20164–9. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Florath I, Butterbach K, Muller H, et al. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, et al. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120:296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–51. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 60.Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhi D, Aslibekyan S, Irvin MR, et al. SNPs located at CpG sites modulate genomeepigenome interaction. Epigenetics. 2013;8 doi: 10.4161/epi.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics. 2010;5:578–82. doi: 10.4161/epi.5.7.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pickrell JK, Marioni JC, Pai AA, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 66.van Eijk KR, de Jong S, Boks MP, et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics. 2012;13:636. doi: 10.1186/1471-2164-13-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D, Cheng L, Badner JA, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu CE, Cudaback E, Foraker J, et al. Epigenetic signature and enhancer activity of the human APOE gene. Hum Mol Genet. 2013;22:5036–47. doi: 10.1093/hmg/ddt354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joo JE, Hiden U, Lassance L, et al. Variable promoter methylation contributes to differential expression of key genes in human placenta-derived venous and arterial endothelial cells. BMC Genomics. 2013;14:475. doi: 10.1186/1471-2164-14-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson A, Enroth S, Gyllensten U. Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLoS One. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaminsky ZA, Tang T, Wang SC, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 73.Marczylo EL, Amoako AA, Konje JC, et al. Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics. 2012;7:432–9. doi: 10.4161/epi.19794. [DOI] [PubMed] [Google Scholar]
- 74.Laubenthal J, Zlobinskaya O, Poterlowicz K, et al. Cigarette smoke-induced transgenerational alterations in genome stability in cord blood of human F1 offspring. FASEB J. 2012;26:3946–56. doi: 10.1096/fj.11-201194. [DOI] [PubMed] [Google Scholar]
- 75.Zaina S, Lund G. Epigenetics: a tool to understand diet-related cardiovascular risk? J Nutrigenet Nutrigenomics. 2011;4:261–74. doi: 10.1159/000334584. [DOI] [PubMed] [Google Scholar]
- 76.Szarc vel Szic K, Ndlovu MN, Haegeman G, et al. Nature or nurture: let food be your epigenetic medicine in chronic inflammatory disorders. Biochem Pharmacol. 2010;80:1816–32. doi: 10.1016/j.bcp.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 77.Newman PE. Can reduced folic acid and vitamin B12 levels cause deficient DNA methylation producing mutations which initiate atherosclerosis? Med Hypotheses. 1999;53:421–4. doi: 10.1054/mehy.1998.0794. [DOI] [PubMed] [Google Scholar]
- 78.Esteller M. Epigenetics in biology and medicine. CRC Press; Boca Raton, FL: 2008. p. 212. [Google Scholar]
- 79.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223s–8s. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 80.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–30. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 81.Pilsner JR, Hall MN, Liu X, et al. Associations of plasma selenium with arsenic and genomic methylation of leukocyte DNA in Bangladesh. Environ Health Perspect. 2011;119:113–8. doi: 10.1289/ehp.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crescenti A, Sola R, Valls RM, et al. Cocoa Consumption Alters the Global DNA Methylation of Peripheral Leukocytes in Humans with Cardiovascular Disease Risk Factors: A Randomized Controlled Trial. PLoS One. 2013;8:e65744. doi: 10.1371/journal.pone.0065744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Milagro FI, Campion J, Cordero P, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25:1378–89. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 84.Moleres A, Campion J, Milagro FI, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J. 2013;27:2504–12. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 85.Lopez-Legarrea P, Mansego ML, Zulet MA, et al. SERPINE1, PAI-1 protein coding gene, methylation levels and epigenetic relationships with adiposity changes in obese subjects with metabolic syndrome features under dietary restriction. J Clin Biochem Nutr. 2013;53:139–44. doi: 10.3164/jcbn.13-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73:770–80. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 89.Napoli C, Infante T, Casamassimi A. Maternal-foetal epigenetic interactions in the beginning of cardiovascular damage. Cardiovasc Res. 2011;92:367–74. doi: 10.1093/cvr/cvr201. [DOI] [PubMed] [Google Scholar]
- 90.Arce C, Segura-Pacheco B, Perez-Cardenas E, et al. Hydralazine target: from blood vessels to the epigenome. J Transl Med. 2006;4:10. doi: 10.1186/1479-5876-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee BH, Yegnasubramanian S, Lin X, et al. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–56. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cornacchia E, Golbus J, Maybaum J, et al. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–200. [PubMed] [Google Scholar]
- 93.Zhou Y, Lu Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev. 2008;7:376–83. doi: 10.1016/j.autrev.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 94.Singh V, Sharma P, Capalash N. DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr Cancer Drug Targets. 2013;13:379–99. doi: 10.2174/15680096113139990077. [DOI] [PubMed] [Google Scholar]
- 95.Lin YC, Lin JH, Chou CW, et al. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008;68:2375–83. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- 96.Mitro N, Godio C, De Fabiani E, et al. Insights in the regulation of cholesterol 7alpha-hydroxylase gene reveal a target for modulating bile acid synthesis. Hepatology. 2007;46:885–97. doi: 10.1002/hep.21819. [DOI] [PubMed] [Google Scholar]
- 97.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lehmann LH, Worst BC, Stanmore DA, et al. Histone deacetylase signaling in cardioprotection. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi MC, Cohen TJ, Barrientos T, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012;47:122–32. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McKinsey TA. Therapeutic potential for HDAC inhibitors in the heart. Annu Rev Pharmacol Toxicol. 2012;52:303–19. doi: 10.1146/annurev-pharmtox-010611-134712. [DOI] [PubMed] [Google Scholar]
- 101.Zhao TC, Du J, Zhuang S, et al. HDAC inhibition elicits myocardial protective effect through modulation of MKK3/Akt-1. PLoS One. 2013;8:e65474. doi: 10.1371/journal.pone.0065474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao TC, Zhang LX, Cheng G, et al. gp-91 mediates histone deacetylase inhibition-induced cardioprotection. Biochim Biophys Acta. 2010;1803:872–80. doi: 10.1016/j.bbamcr.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meng X, Zhang K, Li J, et al. Statins induce the accumulation of regulatory T cells in atherosclerotic plaque. Mol Med. 2012;18:598–605. doi: 10.2119/molmed.2011.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tao H, Shi KH, Yang JJ, et al. Histone deacetylases in cardiac fibrosis: Current perspectives for therapy. Cell Signal. 2014;26:521–7. doi: 10.1016/j.cellsig.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 105.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–93. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 106.Vojinovic J, Damjanov N. HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol Med. 2011;17:397–403. doi: 10.2119/molmed.2011.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dje N’Guessan P, Riediger F, Vardarova K, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380–6. doi: 10.1161/ATVBAHA.108.178319. [DOI] [PubMed] [Google Scholar]
- 108.Chen JB, Chern TR, Wei TT, et al. Design and synthesis of dual-action inhibitors targeting histone deacetylases and 3-hydroxy-3-methylglutaryl coenzyme A reductase for cancer treatment. J Med Chem. 2013;56:3645–55. doi: 10.1021/jm400179b. [DOI] [PubMed] [Google Scholar]
- 109.Hanai J, Cao P, Tanksale P, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–51. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buettner C, Lecker SH. Molecular basis for statin-induced muscle toxicity: implications and possibilities. Pharmacogenomics. 2008;9:1133–42. doi: 10.2217/14622416.9.8.1133. [DOI] [PubMed] [Google Scholar]
- 111.Bailey D, Jahagirdar R, Gordon A, et al. RVX-208: a small molecule that increases apolipoprotein A-I and high-density lipoprotein cholesterol in vitro and in vivo. J Am Coll Cardiol. 2010;55:2580–9. doi: 10.1016/j.jacc.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 112.Anand P, Brown JD, Lin CY, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–82. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nicholls SJ, Gordon A, Johannson J, et al. ApoA-I induction as a potential cardioprotective strategy: rationale for the SUSTAIN and ASSURE studies. Cardiovasc Drugs Ther. 2012;26:181–7. doi: 10.1007/s10557-012-6373-5. [DOI] [PubMed] [Google Scholar]
- 114.Corp R . Resverlogix’s BET Protein Inhibitor RVX-208 Meets Primary Endpoint in SUSTAIN Clinical Trial in Patients With High Risk Cardiovascular Disease. Resverlogix Corp; 2012. Web Site. [Google Scholar]
- 115.Nicholls SJ, Chapman J. ASSURE: effect of an oral agent inducing Apo A-I synthesis on progression of coronary atherosclerosis: Results of the ASSURE Study. European Society of Cardiology Congress; Amsterdam. 2013. [Google Scholar]
- 116.Corp R . Resverlogix Announces Combined RVX-208 findings from SUSTAIN & ASSURE Phase 2b Trials. Resverlogix Corp; 2014. Web Site. [Google Scholar]
- 117.Jaguszewski M, Osipova J, Ghadri JR, et al. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur Heart J. 2013 doi: 10.1093/eurheartj/eht392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raizman JE, Diamandis EP, Rayner K, et al. Novel Biomarkers for Acute Myocardial Infarction: Is MicroRNA the New Kid on the Block? Clin Chem. 2013 doi: 10.1373/clinchem.2013.215491. [DOI] [PubMed] [Google Scholar]