Abstract
Multiple Imputation, Maximum Likelihood and Fully Bayesian methods are the three most commonly used model-based approaches in missing data problems. Although it is easy to show that when the responses are missing at random (MAR), the complete case analysis is unbiased and efficient, the aforementioned methods are still commonly used in practice for this setting. To examine the performance of and relationships between these three methods in this setting, we derive and investigate small sample and asymptotic expressions of the estimates and standard errors, and fully examine how these estimates are related for the three approaches in the linear regression model when the responses are MAR. We show that when the responses are MAR in the linear model, the estimates of the regression coefficients using these three methods are asymptotically equivalent to the complete case estimates under general conditions. One simulation and a real data set from a liver cancer clinical trial are given to compare the properties of these methods when the responses are MAR.
Keywords and phrases: Missing data, Multiple imputation, Maximum likelihood, Fully Bayesian, Missing response, Missing at random
1. INTRODUCTION
Missing data is very common in various experimental settings, including clinical trials, sample surveys and environmental studies. There are essentially three major likelihood-based approaches for handling missing data in a regression problem. These are i) Maximum Likelihood (ML), ii) Multiple Imputation (MI), and iii) Fully Bayesian (FB). The EM algorithm is a technique often used to obtain ML estimates and is useful when the likelihood function of the observed data has no closed form. The recent developments of missing data approaches also include empirical likelihood method [18], parametric fractional imputation [10], among others. In this paper, we investigate theoretical connections between MI, ML (especially within the EM framework), and FB approaches in the linear regression model when the response variable is missing at random (MAR).
It is well known that when the response variable is MAR and the covariates are fully observed, the likelihood function of the observed data is the same as the complete case likelihood (i.e. the likelihood obtained by omitting all cases with missing values), and therefore the ML estimates are identical to the complete case (CC) estimates. However, this result is not obvious under the MI and FB approaches. Although the CC estimates are unbiased and efficient under MAR responses, the MI and FB methods are still used in practice since many researchers are unaware of this special property of the CC estimates. To study the relationships between the three methods in this context, we consider MAR responses in the linear model and investigate the small and large sample properties of the estimates, and derive analytic and asymptotic expressions of the estimates and standard errors for the MI, ML and FB approaches. Under noninformative priors for the MI and FB methods, we show that the estimates and their standard errors under these three approaches are asymptotically equivalent to the CC estimates.
There is much literature on MI, ML and FB, respectively. [21] provide asymptotic results for MI with MAR responses in linear models. [17], [15], and [20] discuss theoretical properties of proper and improper MI. [22] and [19] propose a consistent variance estimator for MI. For ML, one of the earliest references is [12]. [6], [11], and [8] proposes the “EM by the method of weights” and the Monte Carlo EM algorithm (MCEM) for the ML framework in generalized linear models (GLM). [16] examine the problem of using EM to obtain the asymptotic covariance matrix of the parameter estimates. [7] discuss FB methods for MAR covariates in GLM’s. There are two major differences of our work in this paper from previous literature. First, for MAR responses, we derive both the small and large sample properties of the estimates, while the previous work mainly focuses on large sample properties. Secondly, the main purpose of this paper is to investigate the theoretical relationships between MI, ML and FB, as this was only investigated only through simulations before.
The rest of this paper is organized as follows. In Section 2.1, we derive the small sample and asymptotic expressions of the estimates and standard errors for proper MI. In Section 2.2, we derive the expressions for ML via the EM algorithm. The expressions for FB are derived in Section 2.3. In Section 3, we conduct a simulation to demonstrate our results. A real data analysis of a liver cancer clinical trial is given in Section 4, and we conclude the paper with a brief discussion in Section 5.
2. MAR RESPONSES IN THE LINEAR MODEL
Consider the linear model
| (1) |
where β is a p × 1 vector of unknown parameters, X is an n×p full rank matrix of explanatory variables including an intercept, and e is an n × 1 vector of random errors with e ~ Nn(0, σ2I), where σ2 is assumed unknown throughout. We assume throughout that X is fully observed and the components of y are MAR. For simplicity, we rearrange the data so that y1 = (y1, …, yn1)′ are fully observed and y2 = (yn1+1, …, yn)′ are MAR, and assume that the corresponding n1 × p and n2 × p matrices of fixed covariates X1 and X2 for y1 and y2 are full-rank, n1+n2 = n and p < n1. Therefore, we write and .
As shown in [13], the maximum likelihood estimates of β and σ2 are the same as the CC estimates in the linear regression model, in which cases with any missing values are simply discarded. In fact, this is true for any regression model with MAR responses satisfying conditional independence between y1 and y2 given X and γ, where γ = (β, σ2), since the likelihood function of the observed data Dobs = (y1, X) is given by
which is the CC likelihood.
The standard results for the linear regression model with MAR responses are
| (2) |
which is independent of
| (3) |
with E(β̂) = β and . The variances of the estimates are
| (4) |
and
| (5) |
Clearly, we can adjust the estimate of σ2 to get an unbiased estimate by letting . It is worth noting that if we apply the EM algorithm in this setting and use Louis’s method [14] to get the variance estimates, we get same variance estimates of β̂ML as in Eq. (4), and the variance estimate of is equal to , which is larger than Eq. (5) in small samples but asymptotically equivalent when n1 → ∞. In the next three subsections, we explore the small and large sample properties of the estimators under MI, ML via the EM algorithm, and FB methods for MAR responses under model (1).
2.1 Multiple imputation (MI)
Multiple imputation has emerged as a very popular technique for inference in missing data problems. In this section, we consider the precision parameter instead of the variance parameter for the development of MI. Therefore, we assume γ* = (β, τ), where τ = 1/σ2. Proper MI is based on creating imputed datasets in which the missing values are sampled from the posterior predictive distribution of the missing data given the observed data, given by
| (6) |
where Dobs = (y1, X1, X2), and Dmis = y2 for the current setting. π(γ*|Dobs) is the posterior distribution of γ* based on the observed data, given by π(γ*|Dobs) ∝ {∫ L(γ*|Dmis, Dobs)dDmis}π(γ*), where L(γ*|Dmis, Dobs) is the likelihood function of the complete-data, ∫ L(γ*|Dmis, Dobs)dDmis is the likelihood based on the observed data, and π(γ*) is the prior distribution of γ*. Assume , l = 1, …, m, are draws of Dmis from the posterior predictive distribution p(Dmis|Dobs) given in Eq. (6). Let and Vl denote the posterior mean and covariance matrix of γ* based on π(γ*|Dobs) calculated for the lth imputed data set (y1, ). Then, the MI estimate of γ* is , and the estimate of the variance of is
| (7) |
where and is the between-imputation variance. There are several imputation methods that have been proposed for the MI method. In this paper, we concentrate on proper MI using the improper prior,
| (8) |
We note that in MI, the imputation model can be different from the analysis model, but in this paper we only consider the case in which the two models are the same.
Theorem 1 gives the small sample behavior of the estimates of β and σ2 for proper MI assuming the improper prior π(γ*) ∝ τ−1. Large sample properties of the estimates are also given under some general conditions. To derive these properties, we need the following lemma.
Lemma 2.1
If the n×1 random vector z has a multivariate t distribution, denoted Sn(v, μ, V), with density proportional to , and A and B are matrices of constants, then
, when v > 2
, when v > 2
, when v > 4.
The proof of Lemma 2.1 is given in the Appendix. For the linear regression model (1) with prior as Eq. (8), the posterior distribution of γ* based on the observed data is
and the posterior predictive distribution is
where and . Since , and are of full-rank, it can be shown that H is positive definite with inverse . Hence, the posterior predictive distribution of [y2|y1] is a multivariate t distribution given by
| (9) |
Theorem 2.1 establishes the small and large sample properties of the estimates based on the MI method.
Theorem 2.1
For the linear regression model (1) with prior (8), let , l = 1, …, m, be the samples of y2 from [y2|y1] in Eq. (9). Then
-
the multiple imputation (MI) estimate of β is
(10) with mean E(β̂MI) = β and variance(11) -
The MI estimate of σ2 is
(12) with meanand variance(13) where .
From Theorem 2.1, it can be shown that the MI estimate of β and σ2 as well as their variances are asymptotically equivalent to the CC estimates. Furthermore, after some algebra, we can show that when n > n1 > p+ 2
and
where is given in Eq. (3). We note here that throughout this paper, we do not consider the situation in which the number of regression coefficients p increases as n increases, so p is either fixed or increases at a slower rate than n.
Remark 2.1
is independent from m while is a function of m, therefore, increasing the number of imputations, m, does not reduce the bias of , but it reduces the variance of .
Remark 2.2
(β̂MI|y1)/β̂CC → 1, as m → ∞, where and are unbiased estimates of σ2. However, this is not true for a fixed m.
We also note that Var(β̂MI) > Var(β̂CC) and , which imply that the MI estimates are less efficient than the CC estimates. This is because the ML estimates for MAR responses in the linear model are the same as the CC estimates, and the ML estimates are most efficient if the model is correct. The extra variability of the MI estimate is induced by the sampling involved in finding the estimator. Even though we are able to improve the MI estimates under the setting of MAR responses in linear regression with small samples, this is not the main aim of this paper. The goal of this paper is to investigate the relationships between MI, ML, and FB approaches. The small sample properties of MI have been studied under more general settings in [1] and [9].
2.2 Maximum likelihood (ML)
As shown in equations (2) and (3), there are closed form estimates of β and σ2 using the ML method when the response variable is MAR in the linear model, and those estimates are precisely the CC estimates. However, the ML method is more generally carried out using the EM algorithm, which can be either directly solved when the E-step has a closed form, or it may be obtained using Monte Carlo methods when it does not have a closed form. This latter version of the EM algorithm is referred to as the Monte Carlo EM (MCEM) algorithm and is a more general method of carrying out ML since for most regression models with missing data, the E-step does not have a closed form. We will study ML via MCEM in this subsection in order to study the connections between ML, MI, and FB, and to shed light on examining the properties of the MCEM method when closed form estimates under ML do not exist. ML via MCEM will be the basis of our development in this subsection. In particular, we will derive expressions for the estimates, and their associated variances for both the small sample and large sample situations using MCEM. Following [6] and [8], the Monte Carlo E-Step at the (t + 1)st EM iteration can be written as
where l(γ | Dobs, Dmis, γ(t)) is the log-likelihood function based on the complete data given the parameter estimates at the tth iteration, Dobs = (y1, X1, X2) is the observed data, Dmis = y2 and the ’s are the missing values replaced by their jth sampled values from the full conditional distribution p(Dmis | Dobs, γ(t)). The M-Step at the (t+1)st EM iteration maximizes Q(γ | γ(t)). Standard errors can be calculated by using Louis’s method and the estimated observed information matrix of γ based on Louis’s method is given by
where γ̂ is the ML estimate at MCEM convergence and Q̈(γ̂|γ̂) is the second derivative matrix of the Q function. The estimate of the asymptotic covariance matrix of γ̂ is therefore [
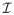 (γ̂)]−1.
(γ̂)]−1.
Note that unlike the MI method, which creates m pseudo complete datasets by replacing the missing values with each of the m sets of imputed values, ML via MCEM calculates the estimates from a single dataset and assigns a weight of 1 for complete observations and a weight of 1/m for each sampled value. In order to explore the connections between MI and ML, we consider the imputation distribution [y2|y1, β̂] of MCEM, given by
| (14) |
where . Theorem 2.2 gives the estimates of β and σ2 along with their small and large sample properties.
Theorem 2.2
For the linear regression model (1), let , l = 1, …, m, be the Gibbs samples of y2 from [y2|y1, β̂] in Eq. (14). Then
-
the maximum likelihood estimate of β using MCEM is
(15) with variance(16) β̂ML2 is an unbiased estimator of β.
-
The ML estimate of σ2 is
(17) with meanand variance(18) where and .
, as n1 → ∞ and m → ∞.
Again from Theorem 2.2, it can be easily shown that the estimate of β and its variance based on MCEM are asymptotically equivalent to the CC estimates. In particular, after some algebra, it can be shown that
as n1 or m → ∞. The condition that tr(M) → K, 0 ≤ K < ∞, as n → ∞, implies that the information contained in the covariates corresponding to the missing responses is finite compared to the total information in the covariates. The variance of in Eq. (18) can also be written as
and hence as n1 or m go to infinity.
Note that the variance of β̂ML2 in Eq. (16) is smaller than the variance of β̂MI in Eq. (11), however, the derivation of Theorem 2.2 is based on the assumption that the imputation distribution of the missing responses yields the ML estimates, which may not be true in practice. Again, note that although we write the estimates of (β, σ2) and their variance as if there were m data sets in order to compare the MI and ML methods, in practice, ML via MCEM calculates the estimates from only one dataset with different weights assigned to the observed and sampled values. In this sense, MCEM augments the data “vertically” and MI augments the data “horizontally”.
Remark 2.3
Both and are functions of m, the number of Gibbs samples, and therefore, increasing m reduces the bias and variance of .
2.3 Fully Bayesian (FB)
FB methods for the missing data problem are based on specifying priors for all of the parameters and then the missing data are sampled from their full conditional distributions within the Gibbs sampler. Clearly, ML and MI have Bayesian connections, since ML can be viewed as a large sample Bayesian method, and in many cases, the implementation of Bayesian methods using uniform improper priors on all parameters leads to ML estimates. In this subsection, we consider the FB method under conjugate priors, which yield closed form expressions for the posterior mean and variance of the parameters.
Note that observed data likelihood for MAR responses is the CC likelihood and thus the posterior distribution of γ* based on the observed data is p(γ*|y1, X) ∝ p(y1|X; γ*)π(γ*). Theorem 2.3 provides the properties of the fully Bayesian estimates of β and τ.
Theorem 2.3
For the linear regression model (1), assume that the prior for γ* = (β, τ) is π(γ*) = π(β|τ)π(τ), where π(β|τ) = N(μ0, τ−1Σ0) and π(τ) = Gamma(δ0/2, λ0/2). Then
-
the fully Bayesian estimate of β iswhere βm is the m sample from the posterior distribution
with β̃ = Λμ0 + (I − Λ)β̂, , and .
-
The posterior mean and variance of β based on the observed data areand
-
The fully Bayesian estimate of τ = 1/σ2 is
where p(τ |Dobs) ~ Gamma((n1 + δ0)/2, (n1 + δ0)s̃2/2) with s̃2 defined in (i).
- The posterior mean and variance of τ are
The proof of Theorem 2.3 is straightforward and can be found in most Bayesian textbooks. We state it as a Theorem here only to be consistent with other sections.
Remark 2.4
When the prior for γ* is an improper prior, π(γ*) ∝ τ−1, s̃2 reduces to and the posterior mean and variance of (β, τ) are then equal to the CC estimates given in equations (2) and (3).
Therefore, the CC analysis is recommended over the MI, MCEM, and FB methods for MAR responses in the linear regression model, unless additional information is available to specify informative priors for the MI and FB methods, or the imputation model of MI includes covariates not specified in the analysis model. On the other hand, the loss of efficiency of MI, MCEM, and FB methods can be significantly reduced by increasing the number of imputations for MI or the number of Gibbs samples for MCEM and FB.
3. SIMULATION STUDY
In this section, we will compare inferences about β using the four methods, MI, CC, MCEM and FB using the formulas we developed in Section 2 for a small sample size n and various values of m for MI and MCEM.
We generate N = 1,000 replicates with each simulation consisting of n = 250 independent response variables yi from the linear regression model as
where the ei’s are independent and identically distributed (i.i.d.) as N(0, σ2). The values chosen for the parameters are (β0, β1, β2) = (1.0, 1.5, −1.0) and σ2 = 1.0. The covariates (xi1, xi2) are i.i.d. and simulated as
where (α0, α1) = (1.0, 1.0) and .
We assume that yi is MAR for some i’s and xi1 and xi2 are completely observed throughout. In this setting, the model for the missing data mechanism of yi is given by
where (ϕ01, ϕ11, ϕ21) = (−5.5, 1.0, 1.0) and ri1 = 1 if yi is missing, 0 otherwise.
Table 1 gives the results using the four methods, MI, CC, MCEM and FB, and also gives the estimates based on the full data (i.e., no missing values), as these estimates serve as a benchmark for comparison. From the N = 1,000 simulations, the average number of observations with yi missing is 19%. We chose the number of samples m equal to 30 and 3 in both the MI and MCEM methods in order to compare the results. [13] note that for proper MI, m, the number of imputed datasets, can be as small as m = 5. However, m in MCEM, the number of Gibbs samples, is usually large, say m = 100 or more, in order to accurately represent the sampling distribution in the E-step, especially in complex models with large missing data fractions. This is consistent with the simulation results. When m = 3 in the MI method, the two forms of the variance estimates give similar coverage rates because both of them adjust well for small m, and when m = 3 in MCEM, equations (16) and (18) give much better coverage rates than the Louis method. On the other hand, for the MI method, considering that the estimates with m = 30 always have smaller variances than the estimates with m = 3 with better coverage probabilities, larger values of m may need to be considered if the computational burden is not heavy. The simulation results confirm the theorems in Section 2, and show that the three methods (MI, ML via MCEM, FB) produce consistent estimates with valid inferences and all are asymptotically equivalent to the CC estimates when the response variable is MAR.
Table 1.
Simulation with MAR responses in the linear regression model. The 95% CR is the coverage rate of a 95% confidence interval. γ̂F (full data), (Multiple Imputation with covariance matrix as Eq. (7), (Multiple Imputation with covariance matrix as Eq. (11) and Eq. (13), γ̂CC (CC estimates), (MCEM with covariance matrix using Louis’s method, (MCEM with covariance matrix as Eq. (16) and Eq. (18), and γ̂FB (fully Bayesian). The number m is defined in the section discussing the corresponding method
| Method | m | Estimate (var(×10−3))[95% CR] | ||||
|---|---|---|---|---|---|---|
| β0 = 1.00 | β1 = 1.50 | β2 = −1.00 | σ2 = 1.0 | |||
| γ̂F | - | 0.994(12)[94] | 1.499(8)[96] | −0.998(4)[96] | 0.989(8)[94] | |
|
| ||||||
|
|
30 | 0.995(14)[95] | 1.496(11)[95] | −0.998(5)[95] | 1.000(10)[95] | |
|
|
30 | 0.995(14)[95] | 1.496(10)[95] | −0.998(5)[95] | 1.000(10)[95] | |
|
|
3 | 0.993(14)[95] | 1.496(11)[95] | −0.998(6)[94] | 1.002(11)[94] | |
|
|
3 | 0.993(14)[95] | 1.496(11)[96] | −0.998(6)[95] | 1.002(10)[94] | |
|
| ||||||
| γ̂CC | - | 0.995(14)[95] | 1.497(10)[95] | −0.9984(5)[95] | 0.987(10)[94] | |
|
| ||||||
|
|
30 | 0.995(14)[95] | 1.497(10)[95] | −0.998(5)[95] | 0.987(10)[94] | |
|
|
30 | 0.995(14)[95] | 1.497(10)[95] | −0.998(5)[95] | 0.987(10)[94] | |
|
|
3 | 0.993(14)[92] | 1.497(10)[90] | −0.998(5)[89] | 0.987(10)[90] | |
|
|
3 | 0.993(14)[95] | 1.497(10)[95] | −0.998(5)[93] | 0.987(10)[93] | |
|
| ||||||
| γ̂FB | - | 0.995(14)[95] | 1.497(10)[95] | −0.998(5)[95] | 0.992(10)[95] | |
4. LIVER CANCER DATA
To further illustrate the CC, MI, ML and FB methods, we consider a real dataset on n = 174 patients from two Eastern Cooperative Oncology Group clinical trials, EST 2282 [3] and EST 1286 [4]. We are interested in how the number of cancerous liver nodes (CNT) when entering the trials is predicted by six other baseline characteristics: body mass index (BMI, defined as weight in kilograms divided by the square of height in meters); age (in years); associated jaundice (yes, no); and time since diagnosis of the disease (TSD, in weeks). Thirty four out of 174 (19.5%) patients have a missing response variable (CNT). Throughout, we assume that the response variable CNT is MAR. The square root transformation on CNT and TSD was used in the analyses.
We use linear regression to model the response variable, , as , where the ei’s are i.i.d. normally distributed as ei~ N(0, σ2).
Table 2 show the results for the CC analysis, MI with m = 3 and m = 30 using equations (11) and (13), MCEM with m = 30 and m = 300 using equations (16) and (18), and the FB method discussed in Section 3. Moreover, the variance estimates are 0.775, 0.715, 0.715, 0.724, 0.739, and 0.712 for CC, MI with m = 3 and m = 30, ML via MCEM with m = 30 and m = 300, and FB, respectively. As shown in the table, the MI, MCEM and FB methods yield very similar estimates with very little differences from the CC estimates. In particular, the p-values of all the covariates except Age are smaller with larger m. The results show that the age of the patients is significantly associated with the number of cancerous liver nodes controlling for body mass index, associated jaundice, and time since diagnosis.
Table 2.
Estimates for liver cancer data
| Effect | Method | β̂ | Std | P-value | |
|---|---|---|---|---|---|
| Intercept | CC | 2.826 | 0.445 | < .001 | |
| MI m = 3 | 2.801 | 0.446 | < .001 | ||
| MI m = 30 | 2.763 | 0.408 | < .001 | ||
| ML2 m = 30 | 2.782 | 0.440 | < .001 | ||
| ML2 m = 300 | 2.766 | 0.395 | < .001 | ||
| FB | 2.770 | 0.408 | < .001 | ||
| BMI | CC | −0.008 | 0.015 | 0.595 | |
| MI m = 3 | −0.007 | 0.015 | 0.632 | ||
| MI m = 30 | −0.005 | 0.013 | 0.726 | ||
| ML2 m = 30 | −0.005 | 0.014 | 0.740 | ||
| ML2 m = 300 | −0.005 | 0.013 | 0.715 | ||
| FB | −0.005 | 0.013 | 0.720 | ||
| Age | CC | −0.012 | 0.005 | 0.018 | |
| MI m = 3 | −0.012 | 0.005 | 0.016 | ||
| MI m = 30 | −0.012 | 0.005 | 0.007 | ||
| ML2 m = 30 | −0.012 | 0.005 | 0.012 | ||
| ML2 m = 300 | −0.012 | 0.004 | 0.006 | ||
| FB | −0.012 | 0.005 | 0.008 | ||
| Jaundice | CC | 0.190 | 0.152 | 0.212 | |
| MI m = 3 | 0.231 | 0.146 | 0.115 | ||
| MI m = 30 | 0.217 | 0.134 | 0.107 | ||
| ML2 m = 30 | 0.210 | 0.144 | 0.147 | ||
| ML2 m = 300 | 0.204 | 0.129 | 0.116 | ||
| FB | 0.204 | 0.134 | 0.129 | ||
|
|
CC | 0.002 | 0.034 | 0.964 | |
| MI m = 3 | 0.000 | 0.034 | 0.998 | ||
| MI m = 30 | −0.002 | 0.032 | 0.957 | ||
| ML2 m = 30 | −0.003 | 0.034 | 0.933 | ||
| ML2 m = 300 | −0.003 | 0.030 | 0.926 | ||
| FB | −0.002 | 0.032 | 0.943 |
5. DISCUSSION
It is known in the missing data literature that when the responses are MAR, the CC analysis is unbiased and efficient. However, MI, ML via MCEM, and FB are still commonly used in practice in this setting. This may be due to the fact that (a) the unbiasedness and efficiency properties of the CC method in this setting is not known to general researchers; (b) MI, ML, and FB, as well as some other methods including parametric fractional imputation [10] and empirical likelihood [18] outperform CC in a general setting. To overcome these barriers, it is important to inform researchers and practitioners about these important results. Moreover, we also showed in this paper that the loss of efficiency of MI, ML via MCEM, and FB can be significantly reduced by increasing the number of imputations for MI and the number of Gibbs samples for MCEM and FB. It would be of interest to extend our theoretical results to MAR responses for models other than linear regression. This is a topic of current investigation. It would also be interesting to accommodate missingness in the predictors. Unfortunately, even for linear regression models with normally distributed MAR covariates, no closed form expressions are available for the estimates of the three methods, which makes the comparisons between the methods very hard. A special scenario of it, assuming unit variances for response variable and missing covariates, was investigated in [2] for the maximum likelihood approach.
Acknowledgments
The authors wish to thank the editor, the associate editor and two referees for several suggestions and editorial changes which have greatly improved the paper.
APPENDIX
Proof of Lemma 2.1
If the n × 1 random vector z has a multivariate t distribution as Sn(v, μ, V), then we can write , where x is an n × 1 random vector that has a multivariate normal distribution N(0, V), y is a random variable which has a distribution, x and y are independent. Therefore, (i) is straightforward and, to get (ii), we have
We substitute and calculate the double expectation in the first equality. Because of independence between x and y, we can substitute in expressions for multivariate normal moments in the second equality, and therefore
In the second equality, the two zero components correspond to the first and third moments of x. In the last equality, we use the second and fourth moments of x, which are available in [5] with a modification for nonsymmetric matrices A and B.
Proof of Theorem 2.1
(i) and (ii): It is straightforward to get the estimate of β as in Eq. (10). It is also straightforward to use double expectations to get E(β̂MI) = β. To find the variance of β̂MI, we have
(iii) and (iv): It is straightforward to get the estimate of σ2 as in Eq. (12). In order to find , we write , where , and . Let and . Then after some algebra, we have
by noting that y1 ~ N(X1β, σ2I), A − D = PX1, and .
To find , we write . First we have
Then we obtain
where and s2 is defined as in Theorem 2.1. The last equality holds because is independent of given y1, when j ≠ k. Then we use Lemma 2.1 and get
and therefore, we can get as in (13). Since .
Proof of Theorem 2.2
(i) and (ii): It is straightforward to get the estimate of β as in Eq. (15). It is straightforward to use double expectations to get E(β̂MI) = β. To find the variance of β̂ML2, we have
(iii) and (iv): It is straightforward to get the estimate of σ2 as in Eq. (17). In order to find , we write , where the symbols are same as the proof of Theorem 2.2. Then we have
where . To find , we have
Then we have
Using Chapter 10.9 of [5] and after some algebra, we get
Therefore, after some algebra, we can get as in Eq. (18), and therefore .
Contributor Information
Qingxia Chen, Email: cindy.chen@vanderbilt.edu, Department of Biostatistics, Department of Biomedical Informatics, Vanderbilt University, Nashville, TN, 3723, USA.
Joseph G. Ibrahim, Email: ibrahim@bios.unc.edu, Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
References
- 1.Barnard J, Rubin D. Small-sample degrees of freedom with multiple imputation. Biometrika. 1999;86:948–955. [Google Scholar]
- 2.Chen Q, Ibrahim JG, Chen MH, Senchaudhuri P. Theory and inference for regression models with missing response and covariates. Journal of Multivariate Analysis. 2008;99:1302–1331. doi: 10.1016/j.jmva.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkson G, Cnaan A, Simson IW. A randomized phase II study of acivicin and d́eoxydoxorubicin in patients with hepatocellular carcinoma in an eastern cooperative oncology group study. American Journal of Clinical Oncology. 1990;13:510– 515. doi: 10.1097/00000421-199012000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Falkson G, Lipsitz S, Borden E, Simson IW, Haller D. A ECOG randomized phase i1 study of beta interferon and menogoril. American Journal of Clinical Oncology. 1995;18:287–292. [PubMed] [Google Scholar]
- 5.Graybill FA. Matrices with Applications in Statistics. 2. Duxbury Press; 1983. p. MR0682581. Wadsworth Statistics/Probability Series. [Google Scholar]
- 6.Ibrahim JG. Incomplete data in generalized linear models. Journal of the American Statistical Association. 1990;85:765–769. [Google Scholar]
- 7.Ibrahim JG, Chen MH, Lipsitz SR. Bayesian methods for generalized linear models with covariates missing at random. Canadian Journal of Statistics. 2002;30:55–78. [Google Scholar]
- 8.Ibrahim JG, Lipsitz SR, Chen MH. Missing covariates in generalized linearmodels when the missing data mechanism is nonignorable. Journal of the Royal Statistical Society: Series B. 1999;61:173–190. [Google Scholar]
- 9.Kim JK. Finite sample properties of multiple imputation estimators. The Annals of Statistics. 2004;32:766–783. [Google Scholar]
- 10.Kim JK. Parametric fractional imputation for missing data analysis. Biometrika. 2011;98:119–132. [Google Scholar]
- 11.Lipsitz SR, Ibrahim JG. A conditional model for incomplete covariates in parametric regression models. Biometrika. 1996;83:916–922. [Google Scholar]
- 12.Little RJA. Inference about means from incomplete multivariate data. Biometrika. 1976;63:593–604. [Google Scholar]
- 13.Little RJA, Rubin DB. Statistical Analysis With Missing Data. 2. John Wiley; 2002. p. MR1925014. [Google Scholar]
- 14.Louis T. Finding the observed information matrix when using the EM algorithm. Journal of the Royal Statistical Society: Series B. 1982;44:226–233. [Google Scholar]
- 15.Meng XL, Romero M. Discussion to S. F. Nielsen: Efficiency and self-efficiency with multiple imputation inference. International Statistical Review. 2003;71:607–618. [Google Scholar]
- 16.Meng XL, Rubin DB. Using EM to obtain asymptotic variance-covariance matrices: The SEM algorithm. Journal of the American Statistical Association. 1991;86:899–909. [Google Scholar]
- 17.Nielsen SF. Proper and improper multiple imputation. International Statistical Review. 2003;71:593–607. [Google Scholar]
- 18.Qin J, Zhang B, Leung DHY. Empirical likelihood in missing data problems. Journal of the American Statistical Association. 1999;104:1492–1503. [Google Scholar]
- 19.Robins JM, Wang N. Inference for imputation estimators. Biometrika. 2000;87:113–124. [Google Scholar]
- 20.Rubin D. Discussion to S. F. Nielsen: Discussion on multiple imputation. International Statistical Review. 2003;71:619–625. [Google Scholar]
- 21.Schenker N, Welsh AH. Asymptotic results for multiple imputation. The Annals of Statistics. 1988;16:1550–1566. [Google Scholar]
- 22.Wang N, Robins JM. Large-sample theory for parametric imputation procedures. Biometrika. 1998;85:935–948. [Google Scholar]


