Abstract
Background
Culex quinquefasciatus mosquitoes can be found in almost every major city of Brazil and are vectors of filariasis and several arboviruses. Microsatellite markers have been widely used to uncover the genetic structure of various groups of insect populations. The aim of this study was to glimpse the genetic structure of Cx. quinquefasciatus in Brazil.
Methods
Nine populations were sampled across Brazil (one of them from a laboratory colony - COL) and another one from Argentina and process regarding the variability of six microsatellite loci.
Results
The analyzed loci revealed moderate population genetic structure (mean Fst = 0.12). Dendrograms of genetic distances evidenced two major population clusters, respectively corresponding to the northern and southern populations. The hybrid population Cx. pipiens/quinquefasciatus (from La Plata, Argentina) and the colony population fell outside the major clusters. Those clusters were substructured and there was a significant correlation between genetic and geographic distances and environmental variables (r = 0.51; p > 0.001 and r = 0.46; p > 0.004).
Conclusions
Multilocus cluster Bayesian analysis confirmed that populations are mutually distinct, and the set of results point to genetic differences among populations. The presumable low gene flow among them may be due to the large geographic distances (>1000 km) and to the environmental heterogeneity of the sampled areas. The genetic structure observed in this study may lead to the best understanding of Cx. quinquefasciatus demographical diversity as well as their genetic variations patterns in Brazil so far unknown.
Background
Culex pipiens species complex sharing morphological similarities can be found in urban areas and are responsible for the transmission of several pathogens [1,2]. One member of this complex, Culex quinquefasciatus, is adapted to live in tropical and subtropical areas while Cx. pipiens mosquitoes lives in temperate regions [3]. Cx. quinquefasciatus is well established in Brazil and can be found in almost all major cities [4]. This mosquito transmits lymphatic filariasis caused by Wuchereria bancrofti (Spirurida: Onchocercidae), most cases occur in tropical regions of the planet, 800 million people live in endemic areas and 120 million people are infected [5]. There is active transmission of filariasis in the state of Pernambuco, Brazil [5,6]. This species can also transmit several arboviruses such as West Nile Virus and Saint Louis encephalitis [7-11].
Cx. quinquefasciatus mosquitoes are able to survive in polluted waters where there are no natural predators, which leads to excessive growth of the population [12]. This phenomena may be due to phenotypic plasticity and might have a role in environmental adaptation and insecticide resistance with implications that made chemical interventions no longer effective for the control of mosquito populations [13-22]. Therefore, a better knowledge of the genetic structure of insect populations is required for the development of effective strategies for vector control. Molecular markers have been widely used at the resolution of taxonomic studies and population genetics issues of several insect groups [23-27].
Microsatellites were utilized to seek for population variations in Cx. quinquefasciatus mosquitoes on Hawaii that are associated with landscape altitude variations which lead to population structures caused by spatial interactions among vector, host and parasite. Disease patterns are interconnected with elevation gradient structuration and therefore there are epidemiologically important outcomes [28]. Hybrid Cx. quinquefasciatus/pipiens mosquitoes can be found in Uruguay, northern Argentina and western of Brazil while Cx. pipiens can be found in the southern regions of Latin America [29].
Morais et al. [30] found that Brazilian populations’ of Cx. quinquefasciatus from tropical regions had wing shapes distinct from subtropical populations, sorting out the northern populations (tropical zone) of southern populations (sub-tropical zone), also demonstrated by the use of ace-2 molecular marker. This event can be explained by greater gene flow among populations of the same region than between regions, indicating a barrier, yet to be confirmed [30].
It is still not well known how Cx. quinquefasciatus mosquitoes are demographically distributed, in addition to the paucity of information on their genetic variations. Recent studies indicate that this species varies regionally and thus different control approaches are needed [29,30]. A better understanding of the mosquito population genetic structure might be useful to anticipate vector borne disease distribution patterns and play a decisive role in epidemiological interventions [31]. Herein, we seek to estimate the genetic diversity of Cx. quinquefasciatus populations in Brazil by analyzing ten populations from several regions with distinct climatic and geographical characteristics.
Methods
Collection of specimen - mosquito samples
Adult Cx. quinquefasciatus mosquitoes were captured from ten sampling localities (Table 1, Figure 1) using a battery powered aspiration device near breeding sites during February and March (rain season) in 2008 [32]. Each captured mosquito was identified by taxonomic keys and stored in silica until processed for DNA extraction [2,30], they were then processed with taxon-specific PCR primers, ace-2 locus to distinguish and characterize populations and zones of hybridization [30]
Table 1.
Mosquito populations, collecting sites and geographic coordinates
| Population | Origin | Coordinates |
|---|---|---|
| RBR | Rio Branco - AC | 10°1′17.47″S/67°46′36.43″W |
| BEL | Belém - PA | 1°34′30.83″S/48°27′58.97″W |
| COL | São Paulo - SP | 23°34′0.44″S/46°43′57.61″W |
| LPL | La Plata - Argentina | 34°55′2.86″S/57°56′57.13″W |
| TER | Teresina - PI | 5°6′22.28″S/42°46′28.18″W |
| PLA | Pontes e Lacerda - MT | 15°15′42.29″S/59°17′59.74″W |
| SVI | Santa Vitória - RS | 33°30′33.51″S/53°19′16.71″W |
| CHA | Chapecó - SC | 27°3′23.01″S/52°35′9.32″W |
| PET | São Paulo - SP | 23°29′1.07″S/46°30′14.41″W |
| PIN | São Paulo - SP | 23°38′48.31″S/46°43′37.07″W |
Figure 1.
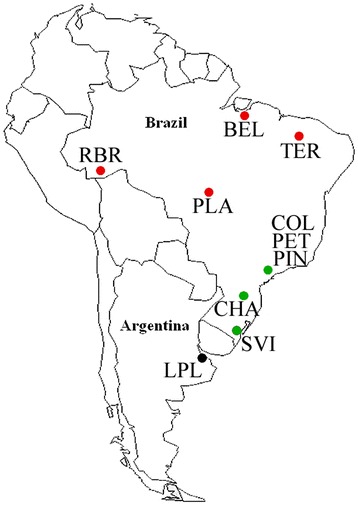
Map of South America showing mosquito population capture sites (n = 10). Coloured dots indicate microsatellite-based genetic clustering of populations.
Populations
All mosquito populations were collected from urban areas with different climate and urbanization characteristics. RBR’ mosquito inhabit an equatorial region with temperatures ranging between 25–40°C, located in the Amazon region. The BEL population is located in the banks of the Amazon River, with only 6.5% of home sewage connected to the collection network resulting in an abundance of breeding sites. COL is the laboratory colony, highly monomorphic as a result of high levels of inbreeding due to isolation in the laboratory since 1980. LPL population comprises of hybrid mosquitoes from a hybridization zone between Culex quinquefasciatus/pipiens in La Plata, Argentina. The TER population resides in a semi humid tropical area, located in the transition zone between the semi-arid northeast of Brazil and the Amazon rainforest, as well as PLA located in a transition zone between Cerrado and the Amazon rainforest. SVI is located in the extreme south of Brazil, and has a temperate climate. CHA is located in the Uruguay River basin with an annual average temperature of 19.60°C. PET mosquitoes were collected in a linear park with 12.5 million m2 located within the city of São Paulo. PIN population was collected in a highly urbanized area, subject to selective pressures caused by humans and is highly anthropophilic.
DNA extraction and microsatellite amplifications
Thirty adult females of each population were used to perform the genomic DNA extraction, according to the DNeasy Blood and Tissue kit (Qiagen, California, USA). Six microsatellite primers previously utilized in Cx. quinquefasciatus were selected (Table 2) [31,33]. These six primers, named: CA-115 - GT-14 - GT-108 - GA-12 - CA-118 - ATG-09, were fluorescent-labeled (FAM - HEX - NED) and tested in our Cx. quinquefasciatus populations.
Table 2.
Selected Culex quinquefasciatus primers
| Primers | Sequence 5′-3′ | Allele Size (bp) | Number of Alleles |
|---|---|---|---|
| CxqA115 | F: GTCGTCAAACTGCCAATAA | 86-296 | 30 |
| R: GCGGAAATAGAACAAACG | |||
| CxqGT14 | F: TGTTAGCCTAGTGGGAAGGTG | 106-200 | 18 |
| R: AATCCACCATGCACGGATAC | |||
| CxqGT108 | F: CGTGTTTTATAGGCTTCTTTC | 106-316 | 27 |
| R: TCTTCCTTAACTTTACCCACTC | |||
| CxqGA12 | F: ACCCGTTCTGGCAACACTG | 110-188 | 17 |
| R: TGGTGCGGATGGACGTT | |||
| CxqA118 | F: ACCCCGAGCCAACCTTAT | 112-254 | 26 |
| R: CCCCCATTTCACACCTGT | |||
| CxqATG9 | F:CCACTCAAACTAAAACACCACA | 108-300 | 26 |
| R: AATGCCATAACCATCGTCAT |
Amplification reactions (PCR) were performed as in Edillo et al. [31] and Smith et al. [33], in an AG-22331 Thermocycler Eppendorf (Hamburg Germany). After the PCR amplification multiplex dilutions were performed using three fluorescent-labeled primer (FAM, HEX and NED), where 3 μL of PCR product of each primer was added to 21 μL ultra-pure water for each sample for a final volume of 30 μL. A second dilution was performed with 2 μL of diluted PCR product resuspended in 7.5 μL of Formamide HI-DI (Applied Biosystems, Warrington, UK), 0.5 μL of molecular size standard GeneScan 500 LIZ (Applied Biosystems, Warrington, UK) was added for a final volume of 10 μL. Samples were processed in the ABI 3730 automatic sequencer (Applied Biosystems, Foster, CA, USA).
Fragment size determination and statistical analyses
Fragment sizes were determined with GeneMarker software package (Softgenetics). A parametric t-test was performed for each population, aiming to quantify genetic differentiation. Hardy-Weinberg equilibrium deviations and genetic variation indices were obtained utilizing GENEPOP V4.0 (http://genepop.curtin.edu.au/) [34,35]. Multilocus genotypes of each individual were then processed by the program Arlequin [36]; Fst values were generated to survey the level of genetic structure between populations. Genetic isolation by geographic distances and pairwise multilocus number of migrants (Nm) between populations per generation were estimated in GENEPOP 4.0 utilizing regression of pairwise Fst/(1-Fst) and the geographical distances were shown for a straight-line in kilometers [35]. The selected environmental variables allowed us to examine how environmental factors might be associated with genetic variations of Cx. quinquefasciatus. The selected parameters incorporate annual trends and environmental characteristics of collection sites (mean annual temperature, annual precipitation, altitude, inhabitants and Human Development Index) [37]. Correlations between genetic, geographic distances and environmental variables were estimated with Statistica v7.0 software.
The software Structure 2.3.4 [38] was utilized to appoint the amount of genetic clusters enclosed into the data. To define the value of K, from 1–12 twelve runs were made using default settings, the formula ΔK = m ([L”K])/s [L (K)] was applied [39].
Results
Genetic diversity
Hardy-Weinberg Equilibrium Test was performed for all six microsatellite loci. It was observed after Bonferroni correction [40] that heterozygosity was lower than expected in 36 of 60 tests that could be conducted (p < 0.00083) and the mean Fis value was 0.34. Out of the 150 possible tests, significant linkage disequilibrium was revealed only between locus ATG-09 and GT-14 in the RBR sample suggesting the loci were not in linkage (Table 3).
Table 3.
Six microsatellite loci in Culex quinquefasciatus mosquitoes’ genetic diversity
| Locus | Population | No. of alleles | Observed heterozygosity (%) | Expected heterozygosity (%) | Fis | P |
|---|---|---|---|---|---|---|
| GT-108 | ACR | 11 | 50 | 78 | 0.3636 | 0 |
| BEL | 13 | 63.3 | 87 | 0.2764 | 0.0003 | |
| COL | 3 | 36.7 | 39.7 | 0.0754 | 0.0203 | |
| PLA | 11 | 66.7 | 88.7 | 0.2521 | 0 | |
| TER | 12 | 56.7 | 83.7 | 0.327 | 0.0019 | |
| PLA | 11 | 56.7 | 88 | 0.3614 | 0 | |
| SVI | 9 | 63.3 | 80.7 | 0.2173 | 0.0622 | |
| CHA | 10 | 50 | 85.7 | 0.4141 | 0 | |
| PET | 10 | 70 | 82 | 0.1488 | 0.0778 | |
| PIN | 11 | 50 | 83.3 | 0.4049 | 0 | |
| ATG-09 | ACR | 8 | 60 | 69.7 | 0.1414 | 0.0487 |
| BEL | 10 | 36.7 | 70.7 | 0.4855 | 0 | |
| COL | 3 | 0 | 12.7 | 1,000 | 0.0003 | |
| PLA | 14 | 60 | 85.7 | 0.304 | 0 | |
| TER | 7 | 46.7 | 69.7 | 0.335 | 0.006 | |
| PLA | 9 | 76.7 | 73 | −0.0529 | 0.1283 | |
| SVI | 8 | 76.7 | 84 | 0.0907 | 0.1593 | |
| CHA | 14 | 50 | 86.7 | 0.4284 | 0 | |
| PET | 8 | 86.7 | 79 | −0.0983 | 0 | |
| PIN | 12 | 70 | 83 | 0.16 | 0.0296 | |
| CA-118 | ACR | 6 | 36.7 | 53 | 0.311 | 0.0245 |
| BEL | 13 | 53.3 | 88 | 0.399 | 0 | |
| COL | 3 | 0 | 12.7 | 1,000 | 0.0002 | |
| PLA | 7 | 40 | 60.3 | 0.3409 | 0 | |
| TER | 6 | 20 | 64 | 0.692 | 0 | |
| PLA | 11 | 40 | 82.3 | 0.5177 | 0 | |
| SVI | 9 | 46.7 | 82 | 0.4349 | 0 | |
| CHA | 11 | 46.7 | 77.3 | 0.4007 | 0 | |
| PET | 7 | 26.7 | 38.3 | 0.3085 | 0.0027 | |
| PIN | 9 | 56.7 | 75.7 | 0.2536 | 0.0003 | |
| GT-14 | ACR | 10 | 26.7 | 71.3 | 0.6297 | 0 |
| BEL | 13 | 43.3 | 74 | 0.4178 | 0 | |
| COL | / | / | / | / | / | |
| PLA | 5 | 5.9 | 48.8 | 0.8824 | 0 | |
| TER | 5 | 34.5 | 66.2 | 0.4834 | 0.0004 | |
| PLA | 5 | 36.7 | 70.7 | 0.4863 | 0.0003 | |
| SVI | 5 | 17.8 | 62.8 | 0.7202 | 0 | |
| CHA | 5 | 28 | 70.4 | 0.6084 | 0 | |
| PET | 5 | 35.7 | 62.5 | 0.4334 | 0.0002 | |
| PIN | 5 | 34.5 | 71.7 | 0.5246 | 0 | |
| GA-12 | ACR | 6 | 70 | 73.7 | 0.0492 | 0.502 |
| BEL | 10 | 70 | 74.7 | 0.0624 | 0.0247 | |
| COL | 2 | 46.7 | 50.3 | 0.0794 | 0.7243 | |
| PLA | 4 | 75.9 | 61 | −0.2457 | 0.0002 | |
| TER | 5 | 53.3 | 73 | 0.2716 | 0.0001 | |
| PLA | 7 | 56.7 | 75 | 0.2485 | 0 | |
| SVI | 6 | 56.7 | 58.3 | 0.0286 | 0.1255 | |
| CHA | 7 | 43.3 | 67.7 | 0.3653 | 0.0011 | |
| PET | 8 | 66.7 | 71.7 | 0.0698 | 0.2929 | |
| PIN | 5 | 50 | 68.3 | 0.2738 | 0.0085 | |
| A-115 | ACR | 12 | 57.7 | 82.3 | 0.3023 | 0.0009 |
| BEL | 13 | 76.7 | 83.7 | 0.0863 | 0.2186 | |
| COL | 3 | 0 | 61.3 | 1,000 | 0 | |
| PLA | 12 | 53.3 | 82.3 | 0.3564 | 0 | |
| TER | 5 | 28.6 | 66.8 | 0.5765 | 0.0002 | |
| PLA | 5 | 56.7 | 75.3 | 0.2519 | 0.0621 | |
| SVI | 6 | 30 | 64 | 0.5364 | 0 | |
| CHA | 9 | 40 | 64 | 0.3797 | 0.0005 | |
| PET | 5 | 50 | 58.3 | 0.1454 | 0.0017 | |
| PIN | 6 | 36.7 | 57.3 | 0.3652 | 0.0219 |
Inbreeding coefficient (Fis). In bold, significant P value (α = 0.05) after Bonferroni correction rejects the Hardy-Weinberg equilibrium. p <0.00083.
The COL population was monomorphic, sharing the same allele in most individuals, in the loci CA-118 and ATG-9, 56 alleles from 60 have the same fragment size. Population RBR had thirty alleles with the same size (106 bp) for the locus GT-14 and was not found in any other population. COL showed the same phenomenon with A-115 locus, where 28 alleles have 296 bp and can only be found in this population.
Genetic differentiation
In order to quantify levels of genetic structure among populations the Fst index (genetic heterogeneity) was used. The Fst mean value was 0.12 indicating moderate genetic structuring, pairwise Fst value comparisons between populations ranged from 0.08 to 0.29 and all of them were statistically significant (P < 0.01; Table 4).
Table 4.
Lower diagonal shows pairwise F st estimates
| Population | RBR | BEL | COL | LPL | TER | PLA | SVI | CHA | PET | PIN |
|---|---|---|---|---|---|---|---|---|---|---|
| RBR | - | |||||||||
| BEL | 0.046 | - | ||||||||
| COL | 0.236 | 0.310 | - | |||||||
| LPL | 0.136 | 0.075 | 0.344 | - | ||||||
| TER | 0.052 | 0.058 | 0.281 | 0.146 | - | |||||
| PLA | 0.052 | 0.033 | 0.290 | 0.090 | 0.045 | - | ||||
| SVI | 0.116 | 0.087 | 0.353 | 0.089 | 0.137 | 0.064 | - | |||
| CHA | 0.100 | 0.107 | 0.270 | 0.105 | 0.129 | 0.052 | 0.052 | - | ||
| PET | 0.050 | 0.116 | 0.263 | 0.146 | 0.106 | 0.067 | 0.099 | 0.043 | - | |
| PIN | 0.098 | 0.083 | 0.293 | 0.086 | 0.116 | 0.035 | 0.046 | 0.027 | 0.050 | - |
The statistical significance of Fst estimates was assessed using 10000 permutations. All comparisons were statistically significant (p < 0.01).
Genetic distance
Statistically significant correlation was detected between genetic distance (estimated as Fst/(1 - Fst)) and geographic distance between populations (r = 0.51; r2 = 0.26; p = 0.001) as well as statistically significant correlation between genetic distance and Environmental Variables (r = 0.46; r2 = 0.21; p > 0.004) (Figure 2). The Pairwise multilocus estimated number of migrants (Nm) between populations per generation was 2.60736.
Figure 2.
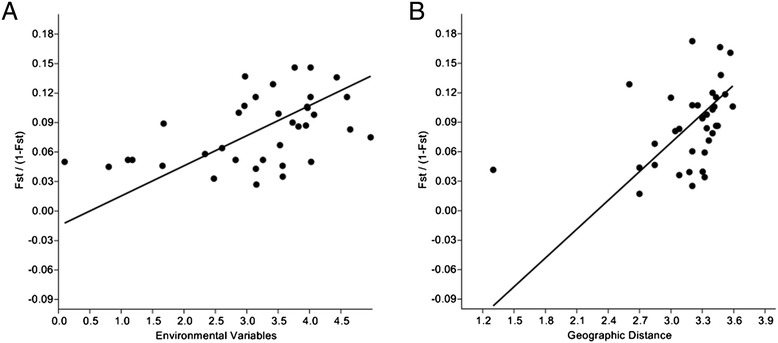
Fst (Fst/(1-Fst) upon environmental variables and linear distance. A-Linearized Fst values (Fst/(1-Fst) upon environmental variables between pairs of populations, showing a significant IBD effect (r = 0.46; p > 0.004) between analyzed populations. B-Linearized Fst values (Fst/(1-Fst) upon linear distance (in km) between pairs of populations, showing a significant IBD effect (r = 0.51; p > 0.001) between analyzed populations. (COL is not shown).
The dendrogram of genetic distances based on the allelic variability of the six microsatellite loci (GT-108, ATG-09, CA-118, GT-14, GA-12, CA-115) for 10 populations of Cx. quinquefasciatus is presented in Figure 3. The dendrogram revealed two clusters, which corresponded geographically to Northern and Southern sampling sites. Population samples LPL (hybrid population) and COL (laboratory colony) were not grouped with any other populations, and the population COL proved to be the most distinct from the other populations, all results were statistically significant.
Figure 3.
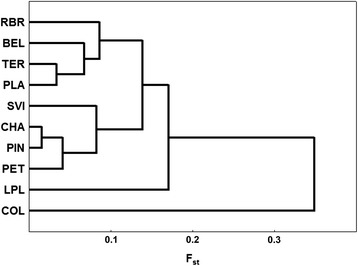
Genetic distance dendrogram of Culex quinquefasciatus populations. Statistically significant for all iterations.
Bayesian cluster analyses
The multilocus cluster Bayesian analysis of all 10 population samples together indicate genetic structuration among Cx. quinquefasciatus’ populations (delta k = 10) (K value = 10) (Figure 4A). A second multilocus cluster Bayesian analysis was made without COL because it was no longer considered as a sylvatic population (colony was originated in 1980) and LPL was removed because it is actually a hybrid population between Cx. pipiens X Cx. quinquefasciatus (delta k = 8) (K value = 2) [30] (Figure 4B).
Figure 4.
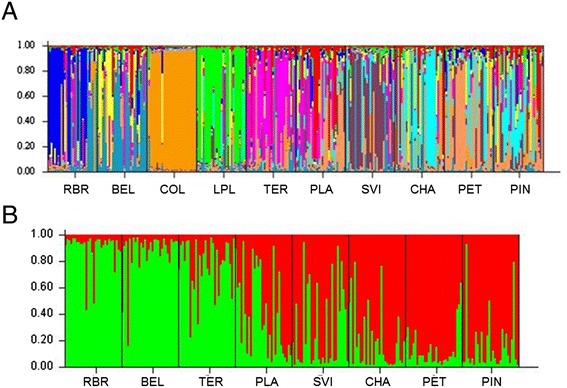
Multilocus cluster Bayesian analysis of microsatellite genotypes. A-Multilocus cluster Bayesian analysis of microsatellite genotypes (K = 10). Each of the 300 individuals included in the analysis is represented by a vertical line (each population is composed of 30 mosquitoes) divided into segments of colors that represent the probability of each individual to belong to any of the genetic clusters. 1- RBR, 2- BEL, 3- COL, 4- LPL, 5- TER, 6- PLA, 7- SVI, 8- CHA, 9- PET and 10- PIN. B-Multilocus cluster Bayesian analysis of microsatellite genotypes (K values = 2). Each individual is represented by a vertical line and each population is composed of 30 mosquitoes. The populations COL and LPL have not been included in this analysis. Each of the 240 individuals from the remaining populations was represented by a vertical line, divided into segments of colours that represent the probability of each individual to belong to any of the genetic clusters. 1- RBR, 2- BEL, 5- TER, 6- PLA, 7- SVI, 8- CHA, 9- PET and 10- PIN.
Discussion
Microsatellite markers were polymorphic and exposed a significant regional differentiation, which is compatible with the population structuration hypothesis for Cx. quinquefasciatus. Population structure comprised a clear North–south dichotomy in clustering. Such interpretation is in accordance to Morais et al. [30], who first noted that this species varies geographically in the Neotropics using morphometric wing characters [29].
The presence of correlation between genetic and geographic distances suggests that genetic isolation by distance might occur but considering that samples came from different biomes, ecological components are also an influential factor on population structure genetic characteristics as seen in Figure 2. However, our interpretations are yet limited since Brazilian territory was not homogeneously covered, and thus, it may represent several distinct scenarios.
Concerning the northern cluster the geographic and genetic distances did not correlate. For example, TER and PLA although being 2100 km apart, they appeared adjoining in the dendrogram. This discrepancy may be an indirect result of environmental constraints; because both are located in similar ecosystems (transition between semi-arid to rain forest). RBR population is located in a remote region of Brazil where urban mobility is mostly by boat or plane, leading to an isolated scenario, which might have increased the genetic differentiation found in that population.
Located in the southern branch, the SVI population presented lower genetic variability probably because it is located in the southernmost border of geographic distribution (temperature fluctuations may be limitrophe for Cx. Quinquefasciatus) [30]. Comparatively, CHA and PIN populations were highly polymorphic, arguably because of an ancestral polymorphism retention, favoured by the fact that environmental constraints are not so restrictive as those of SVI.
Apart from the main dichotomy in the dendrogram, one cannot make deep interpretations regarding the lower clusters up to the present time. Concerning the discrepant samples, COL is a highly inbred and monomorphic laboratory colony and thus appeared to be eccentric; LPL is a hybrid population between Cx. quinquefasciatus/pipiens [29] and was genetically distinct from all Brazilian natural populations.
Bayesian analysis confirmed the results obtained by the dendrogram and Fst correlation showed significant differences among populations and thus low genic flow. The great distances (>1000 of range in km between populations) and distinct biomes might block migratory flows, if there is gene flow it occurs in a smaller geographic scale.
The deficit of heterozygotes, found in all loci, can be explained by a number of non-mutually excludable factors: Population substructure (Wahlund effect), inbreeding and genetic drift [41]. The possibility that some loci are under selective pressure, although remote, cannot be discarded. Our interpretation of the results is equivalent to those of da Costa-Ribeiro et al. [42].
Taken together, our exploratory findings suggest that Cx. quinquefasciatus has a complex population structure and a broad genetic variability. This thought is compatible with the available epidemiological and biological data of the species. Cx. quinquefasciatus mosquitoes can be found in urban habitats where there is a great thermal range, manmade selective pressures and several kinds of available breeding sites, features that can lead to temporal variations of clustering and population differentiation [42]. Insects of epidemiological importance might be dispersed, expanding its borders when associated with humans. Aedes aegypti mosquitoes can be disseminated by ground transportation as roads and railways are likely to spread dengue virus beyond the normal reach of mosquitoes but are unlikely to spread dengue virus over large areas [42].
Interactions between vector, host and pathogen can change patterns of disease transmissibility resulting in selection, genetic drift and hybridization [43-45]. Parasite and host bond can undergo effects caused by vectors, the most usual effect is the genetic diversity of parasites caused by the variation in the genetic structure of vectors. Parasite strains may be transmitted beyond primary to secondary host populations increasing population numbers of parasites, thus resulting in the decrease of genetic diversity caused by genetic drift or the recurrence of more virulent strains, or the outgrowth of resistance proportioned by the high levels of genetic diversity [46]. Cx. quinquefasciatus vector competence is genetically interconnected to Wuchereria bancrofti causing significant differentiation among populations [43].
Population genetic analyses can be useful to enlighten demographic and dispersal trends in mosquitoes, leading to uncovering disease patterns and epidemiological changes in transmission dynamics [47,48].
Sunil et al. [49] found that environmental and ecological factors are not the main causes for the genetic differentiation between populations of Anopheles culicifacies, an important vector of malaria in Southeast Asia.
Kothera et al. [44] suggest that models of disease transmission can be achieved by population genetics and epidemiological data, therefore, improving the efficacy of mosquito control methods. Venktesan & Rasgon [46] state that population dynamics and spread of Culex tarsalis mosquitoes may have an association in the invasion of North America by WNV. Fonseca et al. [50] showed that mosquitoes from the Culex pipiens complex from North America and Europe have distinct epidemiological patterns that can lead to changes in their vectorial capacity, becoming new efficient vectors if introduced into new areas.
Conclusions
The understanding of population structure of Cx. quinquefasciatus mosquitoes, as well as their territorial variations and genetic characteristics might help to predict the introduction of this mosquito into new areas that can lead to disease outbreaks, it could also help with the development of new control strategies. Further studies should aim at better understanding of population and demographical dynamics for smaller geographic scales where there is the possibility of migration and a more intense gene flow.
Acknowledgements
We thank Sirlei Antunes de Morais for the mosquito samples and Antônio Ralph Medeiros de Souza for the assistance with the analysis. A.B.B.W is a fellow of FAPESP Grant 2008-57468-6. P.O.V. is a fellow of FAPESP Grant 2010/15039-1. L.S. is recipient of the research Grant “CAPES 1275/2011”.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ABBW conceived the experiments. ABBW, POV, LS and MTM designed the experiments. ABBW and POV performed the experiments. ABBW, POV, LS and MTM analyzed the data and wrote the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
André Barretto Bruno Wilke, Email: andrebw.br@gmail.com.
Paloma Oliveira Vidal, Email: lomavidal@gmail.com.
Lincoln Suesdek, Email: lincoln.suesdek@butantan.gov.br.
Mauro Toledo Marrelli, Email: mtmarrelli@gmail.com.
References
- 1.Barr AR. Distribution of Culex p. pipiens and C. p. quinquefasciatus in North America. Am J Trop Med Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- 2.Forattini OP. Culicidologia médica. São Paulo: EDUSP; 2002. [DOI] [PubMed] [Google Scholar]
- 3.Huang S, Molaei G, Andreadis TG. Reexamination of Culex pipiens hybridization zone in the Eastern United States by ribosomal DNA-based single nucleotide polymorphism markers. Am J Trop Med Hyg. 2011;85:434–441. doi: 10.4269/ajtmh.2011.10-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morais SA, Marrelli MT, Natal D. Aspectos da distribuição de Culex (Culex) quinquefasciatus Say (Diptera, Culicidae) na região do rio Pinheiros, na cidade de São Paulo, estado de São Paulo, Brasil. Rev Bras Entomol. 2006;50:413–418. doi: 10.1590/S0085-56262006000300012. [DOI] [Google Scholar]
- 5.Cartaxo MF, Ayres CF, Weetman D. Loss of genetic diversity in Culex quinquefasciatus targeted by a lymphatic filariasis vector control program in Recife, Brazil. Trans R Soc Trop Med Hyg. 2011;105:491–499. doi: 10.1016/j.trstmh.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 6.WHO-Global programme to eliminate lymphatic filariasis (GPELF) Progress Report 2000–2009 and Strategic Plan 2010–2020. 2010. [Google Scholar]
- 7.CDC - Epidemic/Epizootic West Nile Virus in the United States . Guidelines for Surveillance, Prevention, and Control. Fort Collins, Colorado: U.S. Department of Health and Human Services Public Health Service Centers for Disease Control and Prevention National Center for Infectious Diseases Division of Vector Borne Infectious Diseases; 2003. [Google Scholar]
- 8.Diaz LA, Flores FS, Beranek M, Rivarola ME, Almirón WR, Contigiani MS. Transmission of endemic St Louis encephalitis virus strains by local Culex quinquefasciatus populations in Cordoba, Argentina. Trans R Soc Trop Med Hyg. 2013;107:332–334. doi: 10.1093/trstmh/trt023. [DOI] [PubMed] [Google Scholar]
- 9.Fontes G, Brito AC, Calheiros CML, Antunes CMF, Rocha EMM. Situação Atual da Filariose Bancroftiana na Cidade de Maceió, Estado de Alagoas. Brasil Cad Saúde Públ. 1994;10:293–300. doi: 10.1590/S0102-311X1994000800007. [DOI] [PubMed] [Google Scholar]
- 10.Sardelis MR, Turell MJ, Dohm DJ, O’Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7:1018–1022. doi: 10.3201/eid0706.010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO - TDR . Planning meetings on Progress and Prospects for the Use of Genetically Modified Mosquitoes to Prevent Disease Transmission: Meeting 1. 2009. [Google Scholar]
- 12.Dibo MR, Menezes RM, Ghirardelli CP, Mendonça AL, Chiaravalloti Neto F. The presence of Culicidae species in medium-sized cities in the State of São Paulo, Brazil and the risk of West Nile fever and other arbovirus infection. Rev Soc Bras Med Trop. 2011;44:496–503. doi: 10.1590/S0037-86822011000400019. [DOI] [PubMed] [Google Scholar]
- 13.Amorim LB, Helvecio E, Fontes de Oliveira CM, Junqueira Ayres CF. Susceptibility status of Culex quinquefasciatus (Diptera: Culicidae) populations to the chemical insecticide temephos in Pernambuco, Brazil. Pest Manag Sci. 2013;69:1307–1314. doi: 10.1002/ps.3502. [DOI] [PubMed] [Google Scholar]
- 14.Bracco JE, Dalbon M, Marinotti O, Barata JM. Resistance to organophosphorous and carbamates insecticides in a population of Culex quinquefasciatus. Rev Saude Publica. 1997;31:182–183. doi: 10.1590/S0034-89101997000200013. [DOI] [PubMed] [Google Scholar]
- 15.Govindarajan M, Sivakumar R. Larvicidal, ovicidal, and adulticidal efficacy of Erythrina indica (Lam.) (Family: Fabaceae) against Anopheles stephensi, Aedes aegypti, and Culex quinquefasciatus (Diptera: Culicidae) Parasitol Res. 2014;113:777–791. doi: 10.1007/s00436-013-3709-4. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson GM. Fighting the global pest problem: preface to the special toxicon issue on insecticidal toxins and their potential for insect pest control. Toxicon. 2007;49:413–422. doi: 10.1016/j.toxicon.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Pocquet N, Milesi P, Makoundou P, Unal S, Zumbo B, Atyame C, Darriet F, Dehecq JS, Thiria J, Bheecarry A, Iyaloo DP, Weill M, Chandre F, Labbé P. Multiple insecticide resistances in the disease vector Culex p. quinquefasciatus from Western Indian Ocean. PLoS One. 2013;21:e77855. doi: 10.1371/journal.pone.0077855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds A. Why EPA Thinks Resistance Monitoring for Bt Crops has been Important and where do we go from here. San Diego: Entomological Society of America Annual Meeting; 2007. [Google Scholar]
- 19.Shearman DC. The evolution of sex determination systems in dipteran insects other than Drosophila. Genetica. 2002;116:25–43. doi: 10.1023/A:1020955507978. [DOI] [PubMed] [Google Scholar]
- 20.Taipe-Lagos CB, Natal D. Abundância de culicídeos em área metropolitana Preservada e suas Implicações Epidemiológicas. Rev Saude Publica. 2003;37:275–279. doi: 10.1590/S0034-89102003000300002. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Liu SL, Liu YY, Qiao CL, Chen SL, Cui F. Over-transcription of genes in a parathion-resistant strain of mosquito Culex pipiens quinquefasciatus. Insect Sci. 2014;00:1–7. doi: 10.1111/1744-7917.12106. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Liu N. Permethrin resistance variation and susceptible reference line isolation in a field population of the mosquito, Culex quinquefasciatus (Diptera: Culicidae) Insect Sci. 2013;00:1–8. doi: 10.1673/031.013.8501. [DOI] [PubMed] [Google Scholar]
- 23.Demari-Silva B, Vesgueiro FT, Sallum MA, Marrelli MT. Taxonomic and phylogenetic relationships between species of the genus Culex (Diptera: culicidae) from Brazil inferred from the cytochrome c oxidase I mitochondrial gene. J Med Entomol. 2011;48:272–279. doi: 10.1603/ME09293. [DOI] [PubMed] [Google Scholar]
- 24.Monteiro FA, Wesson DM, Dotson EM, Schofield CJ, Beard CB. Phylogeny and molecular taxonomy of the Rhodniini derived from mitochondrial and nuclear DNA sequences. Am J Trop Med Hyg. 2000;62:460–465. doi: 10.4269/ajtmh.2000.62.460. [DOI] [PubMed] [Google Scholar]
- 25.Vesgueiro FT, Demari-Silva B, Malafronte RS, Sallum MA, Marrelli MT. Intragenomic variation in the second internal transcribed spacer of the ribosomal DNA of species of the genera Culex and Lutzia (Diptera: Culicidae) Mem Inst Oswaldo Cruz. 2011;106:1–8. doi: 10.1590/S0074-02762011000100001. [DOI] [PubMed] [Google Scholar]
- 26.Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Harnessing genomics and genome biology to understand malaria biology. Nat Rev Genet. 2012;13:315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H, Zhu X, Wu Z, Zhao T. Identifying the main mosquito species in china based on DNA barcoding. PLoS One. 2012;7:47051. doi: 10.1371/journal.pone.0047051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyghobadi N, Lapointe D, Fleischer RC, Fonseca DM. Fine-scale population genetic structure of a wildlife disease vector: the southern house mosquito on the island of Hawaii. Mol Ecol. 2006;15:3919–3930. doi: 10.1111/j.1365-294X.2006.03069.x. [DOI] [PubMed] [Google Scholar]
- 29.Morais SA, Almeida FD, Suesdek L, Marrelli MT. Low genetic diversity in Wolbachia-Infected Culex quinquefasciatus (Diptera: Culicidae) from Brazil and Argentina. Rev Inst Med Trop. 2012;54:325–329. doi: 10.1590/S0036-46652012000600007. [DOI] [PubMed] [Google Scholar]
- 30.Morais SA, Moratore C, Suesdek L, Marrelli MT. Genetic-morphometric variation in Culex quinquefasciatus from Brazil and La Plata, Argentina. Mem Inst Oswaldo Cruz. 2010;105:672–676. doi: 10.1590/S0074-02762010000500012. [DOI] [PubMed] [Google Scholar]
- 31.Edillo FE, Tripet F, McAbee RD, Foppa IM, Lanzaro GC, Cornel AJ, Spielman A. A set of broadly applicable microsatellite markers for analyzing the structure of Culex pipiens (Diptera: Culicidae) populations. J Med Entomol. 2007;44:145–149. doi: 10.1603/0022-2585(2007)44[145:ASOBAM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 32.Nasci RS. A light weight battery powered aspirator for collecting resting mosquitoes in the field. Mosq News. 1981;41:808–811. [Google Scholar]
- 33.Smith JL, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Mol Ecol Notes. 2005;5:697–700. doi: 10.1111/j.1471-8286.2005.01034.x. [DOI] [Google Scholar]
- 34.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 35.Rousset F, Genepop’007 A complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 36.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 37.McGaughran A, Morgan K, Sommer RJ. Environmental variables explain genetic structure in a beetle-associated nematode. PLoS One. 2014;9:e87317. doi: 10.1371/journal.pone.0087317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 40.Rice TK, Schork NJ, Rao DC. Methods for handling multiple testing. Adv Genet. 2008;60:293–308. doi: 10.1016/S0065-2660(07)00412-9. [DOI] [PubMed] [Google Scholar]
- 41.Waples RS, Antao T, Luikart G. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics. 2014;114:e164822. doi: 10.1534/genetics.114.164822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Da Costa-Ribeiro MC, Lourenço-de-Oliveira R, Failloux AB. Low gene flow of Aedes aegypti between dengue-endemic and dengue-free areas in southeastern and southern Brazil. Am J Trop Med Hyg. 2007;77:303–309. [PubMed] [Google Scholar]
- 43.Fonseca DM, Smith JL, Wilkerson RC, Fleischer RC. Pathways of expansion and multiple introductions illustrated by large genetic differentiation among worldwide populations of the southern house mosquito. Am J Trop Med Hyg. 2006;74:284–289. [PubMed] [Google Scholar]
- 44.Kothera L, Godsey MS, Jr, Doyle MS, Savage HM. Characterization of Culex pipiens complex (Diptera: Culicidae) populations in Colorado: USA using microsatellites. PLoS One. 2012;7:e47602. doi: 10.1371/journal.pone.0047602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciota AT, Chin PA, Kramer LD. The effect of hybridization of Culex pipiens complex mosquitoes on transmission of West Nile virus. Parasit Vectors. 2013;6:305. doi: 10.1186/1756-3305-6-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol. 2010;19:1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novikov YM, Vaulin OV. Expansion of Anopheles maculipennis s.s. (Diptera: Culicidae) to northeastern Europe and northwestern Asia: Causes and Consequences. Parasit Vectors. 2014;7:389. doi: 10.1186/1756-3305-7-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demari-Silva B, Suesdek L, Sallum MA, Marrelli MT. Wing geometry of Culex coronator (Diptera: Culicidae) from South and Southeast Brazil. Parasit Vectors. 2014;7:174. doi: 10.1186/1756-3305-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunil S, Singh OP, Nanda N, Raghavendra K, Reddy BP, Subbarao SK. Analysis of population genetic structure of Indian Anopheles culicifacies species A using microsatellite markers. Parasit Vectors. 2013;6:166. doi: 10.1186/1756-3305-6-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi FM, Fleischer RC, Wilkerson RC. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]


