Abstract
The adoptive transfer of T cells is a promising approach to treat cancers. Primary human T cells can be modified using viral and non-viral vectors to promote the specific targeting of cancer cells via the introduction of exogenous T-cell receptors (TCRs) or chimeric antigen receptors (CARs). This gene transfer displays the potential to increase the specificity and potency of the anticancer response while decreasing the systemic adverse effects that arise from conventional treatments that target both cancerous and healthy cells. This review highlights the generation of clinical-grade T cells expressing CARs for immunotherapy, the use of these cells to target B-cell malignancies and, particularly, the first clinical trials deploying the Sleeping Beauty gene transfer system, which engineers T cells to target CD19+ leukemia and non-Hodgkin's lymphoma.
Keywords: Chimeric antigen receptors, Sleeping Beauty, gene therapy, T cells, immunotherapy, CD19, receptor tyrosine kinase orphan-like receptor 1 (ROR1)
Allogeneic hematopoietic stem cell transplantation (HSCT) was initially performed to treat hematologic malignancies in order to restore hematopoiesis after myeloablative radiotherapy and/or chemotherapy. However, the attack of residual tumor cells by donor-derived lymphocytes exerts a beneficial effect, defined as the graft-versus-tumor (GVT) effect[1]. The T cells that participate in the GVT effect also cause toxicity due to the inadvertent recognition of major and minor histocompatibility antigens, causing graft-versus-host disease (GVHD). A central role of engrafted T cells in both the beneficial and damaging effects is supported by the finding that recipients exhibiting GVHD were less likely to relapse than patients who did not exhibit GVHD[2],[3]. Even in the absence of GVHD, the targeting of leukemia by the T-cell-mediated alloresponse exerts a beneficial effect following HSCT. Patients with leukemia who received T-cell-depleted grafts were more likely to relapse than patients who did not exhibit GVHD after allogeneic HSCT[4]–[6]. Furthermore, patients who did not exhibit GVHD after receiving human leukocyte antigen (HLA)-matched bone marrow from a sibling donor were less likely to relapse than patients who received bone marrow from their genetically identical twins[5].
Donor leukocyte infusions (DLI) have been administered after transplantation to promote the regression of relapsed disease by exploiting these T-cell-mediated GVT effects. This strategy has been successfully used to treat chronic myelogenous leukemia (CML), resulting in prolonged and stable remission[7]. DLI has also been employed to treat other hematologic malignancies and solid tumors, with mixed results[8]–[10]. The major limitation of DLI can be ascribed to the lack of specificity for tumor-associated antigens (TAAs); thus, recipients are exposed to the potential of inducing or exacerbating GVHD, which outweighs the beneficial effects of DLI for most tumor types.
The engrafted T cells cultured from donor-derived hematopoietic stem cells (HSCs) or infused T cells from DLI are stimulated by antigen-presenting cells (APCs). In the microenvironment in which T cells engage APCs, the αβ T-cell receptor (TCR) recognizes an antigen presented in the context of the major histocompatibility complex (MHC). Mismatched HLA and the related minor histocompatibility antigens between allogeneic individuals stimulate T cells in an antigen-independent manner. The results from animal models suggest that recipient-derived APCs[11], rather than epithelial cells[12], are crucial for inducing GVHD. Similarly, patient-derived APCs also induce a GVT effect[13]–[15]. Thus, interactions between donor-derived T cells and recipient-derived APCs lead to divergent outcomes: a beneficial graft-versus-leukemia (GVL) effect and detrimental GVHD[16]. In addition to GVHD, other potential limitations of allogeneic HSCT include the high cost of the transplantation procedure and subsequent supportive care as well as the risk of infection during immunosuppressive treatment.
The task of generating an effective antitumor response is inherently difficult, as the genetic instability of tumors promotes adaptations to escape immune surveillance. This genetic instability is apparent, as patients with ostensibly healthy immune systems develop cancers. Tumors can generate an immunosuppressive microenvironment by increasing the expression of immunomodulatory molecules, such as programmed cell death ligand-1 (PD-L1)[17], or secreting immunosuppressive cytokines, such as transforming growth factor-β (TGF-β)[18]–[20]. In addition, tumors can render themselves functionally invisible to T cells by down-regulating the expression of MHC molecules and the antigen-processing machinery, which are necessary for antigen recognition by an αβ TCR[21],[22]. Indeed, a study of primary breast cancer samples detected total loss of MHC class I molecules in greater than 50% of the samples[23]. Further increasing the difficulty of tumor recognition by T cells is that malignant cells present TAAs which are perceived by the immune system as “self.” During the process of thymic education, the T cells that display an αβ TCR which is strongly reactive to a self-antigen are deleted from the repertoire to avoid the subsequent generation of an autoimmune response. Thus, the destruction of cancer is dependent on T cells overcoming the immunomodulatory adaptations of tumors and the propensity to ignore self-antigens while maintaining the ability to recognize and respond to TAAs.
Gene Therapy
Allogeneic T cells mediate GVT effects in the specialized environment of allogeneic HSCT. It is unclear how to extrapolate these clinical data to infuse autologous T cells to avoid GVHD and induce an antitumor response. In gene therapy, a promising approach for cancer treatment, cells are genetically modified ex vivo for in vivo administration to patients. Due to their natural effector functions and established role in mediating anti-leukemia responses, T cells are an attractive vehicle to be engineered for targeted cancer immunotherapy. One goal of combining T-cell therapy with gene therapy is to abolish tolerance of the body's immune system to the tumor, thereby leading to recognition and eradication of cancer cells. In addition, such modifications must be compliant with current Good Manufacturing Practices (GMP) to achieve human application of the genetically modified T-cell product. GMP-complaint manufacture and release of T cells can be accomplished using viral and non-viral methods.
Viral vectors have been effectively used to promote the integration of exogenous DNA into T cells. Both recombinant lentivirus and γ-retrovirus stably introduce transgenes into primary human T cells and have been successfully used in clinical trials. However, there are drawbacks to this approach. First, the construction of GMP-compliant viral vectors requires extensive validation and involves considerable cost in terms of expense, specialized reagents and labor. There is also a significant turnaround time for viral production, partially due to a bottleneck in GMP-compliant viral production facilities. In addition, the size of the viral cargo can be restricted due to the necessary inclusion of viral packaging components and limited size of the viral capsid. Moreover, considerable safety concerns remain due to the nature of the viral vector, which may be assuaged by assessing each T-cell product for its replication competency, but this release test is expensive and time consuming.
Studies have also described a potential for mutagenesis associated with the integration of genetic material delivered by a recombinant viral particle. For example, γ-retroviruses based on the murine leukemia virus are prone to integration near transcriptional start sites of actively transcribed genes[24]. Likewise, lentiviral vectors prefer integration into certain genetic loci, with 57%[24] and 69%[25] of integration events occurring within genes, which is higher than what is expected due to random integration. These risks were illustrated by the development of T-cell leukemia in 25% of patients infused with HSCs that were transduced with γ-retrovirus to treat X-linked severe combined immunodeficiency disease[26]. These cases of induced leukemia were traced to viral integration near the LMO2 proto-oncogene[27],[28]. However, it should be emphasized that the cell type transduced impacts the potential for insertional mutagenesis. In contrast to HSCs, T cells appear to be significantly more resistant to oncogenic transformation after infection with retrovirus[29],[30] and have been successfully and safely transduced hundreds of times for use in clinical trials[31].
In contrast to the production of clinical-grade virus, naked DNA plasmids are manufactured in a much faster turnaround time due to a greater number of GMP-approved vendors and the relative simplicity of their production. In addition, the production of plasmids occurs in the absence of eukaryotic cells, reducing the manufacturing burden and post-production validation, all of which contributes to the reduced cost of producing DNA compared to virus for human application. In addition, naked DNA plasmids do not exhibit the same size constraints as plasmids that must be packaged into capsid particles. The major limitation of naked DNA is its low efficiency of stable transfection into primary T cells. This limitation can now be overcome using transposon/transposase systems. Multiple Class II DNA transposons display activity in human cells, including Tol2, piggyBac, and Sleeping Beauty (SB). Class II DNA transposons are advantageous, as they typically insert a transposon using a copy/paste mechanism, minimizing disruption to the surrounding DNA.
Tol2 is a fish-derived transposon containing an autonomous transposase that retains activity in human cells[32]. Tol2 is advantageous due to its ability to catalyze the integration of large DNA sequences (greater than 10 kb) without a substantial loss in transposition efficiency[33],[34]. However, Tol2 displays preference to integrate near transcriptional start sites[35]. In addition, Tol2 does not display the enzymatic activity of piggyBac, a transposable element derived from the cabbage looper moth[36]. Similar to Tol2, piggyBac is capable of catalyzing the transposition of large elements (greater than 14 kb) of DNA without a detrimental loss of efficiency[37]. Integration by piggyBac targets TTAA sites, rarely resulting in mutations to the surrounding sequences[37]–[39]. Moreover, overexpression of the piggyBac transposase does not greatly inhibit its transposase activity[39], reducing the need for the optimization of transposase expression based on the transposase activity. This characteristic contrasts that of other transposases, including SB (discussed below). However, the use of piggyBac is potentially compromised by its propensity to integrate transposons in and around actively transcribed genes[37],[40], which increases the probability of deleterious effects.
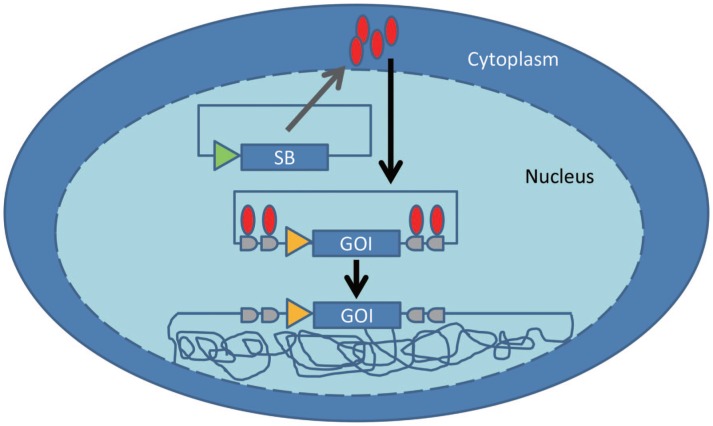
The transposase SB (Figure 1) was reconstructed from an extinct transposase found in salmonid fish[41]. Since the “awakening” of SB, molecular phylogenetics coupled with mutagenesis has been used to increase the activity of the SB system. The activity of the original transposase was increased[42] and rendered 100-fold more potent in the SB100X mutant[43]. Approaches have also been taken to optimize the inverted terminal repeat sequence of the transposon[44]. Compared to genetic engineering using a virus or other transposons, SB is appealing because it can efficiently integrate a gene of interest (flanked by the appropriately inverted and direct repeat sequences) into primary T cells at one (or more) of the 2 × 108 TA dinucleotide sites in the human genome[45]–[47]. Based on analysis of the resulting integration sites, SB-mediated transposition appears to be less likely to alter gene expression than other vectors that preferentially integrate near transcriptional sites. However, there are some disadvantages of the SB system. Chief among these is an apparent decrease in the efficiency of transposition with increasing cargo load, particularly for transposons greater than 6 kB[42],[48],[49]. Decreased transposition of large vectors by SB can be ameliorated by increasing the number of transposase-binding sites in the transposon vector[44]. Further complicating the SB system is that the ratio of SB transposase to SB transposon must be optimized, as excess transposase inhibits the transposition reaction[42].
Figure 1. Sleeping Beauty (SB)-mediated DNA transposition.
The SB transposase (red ovals) expressed by a transiently transfected plasmid binds to the inverted repeat sequences (in gray) flanking the gene of interest (GOI) and catalyzes the cut-and-paste transposition of the GOI into the genome of the target cell (tangled lines) at thymine and adenine dinucleotide base pairs. Transposon integration and loss of the plasmid expressing SB transposase results in stable gene expression. The triangles denote promoter sequences, which often vary between the transposase and the GOI.
Transposon-mediated gene delivery presents a risk of remobilization. In contrast to infection with replication-deficient viral particles, active introduced transposase could theoretically promote the continuous excision and reintegration of transposons. The likelihood that each integration event induces a deleterious effect is the same as that of the original transposition. However, excision of a SB transposon typically results in a 5-bp insertion, although other small insertions and deletions are possible[50]. Insertions and deletions resulting from excision of the SB plasmid vary depending on the cell type and may reflect the preferred DNA repair mechanisms in each cell type[50]. This “footprint” can lead to a frameshift mutation in the unlikely event that SB integrates into and is mobilized from an exon. Nonetheless, excision of a transposon is estimated to be rare (probability less than 1 × 10−4 transpositions), and the probability of an adverse event is even lower[51],[52]. Risks from continual transposition are limited by transient expression of the transposase by a non-integrating vector. This risk is further diminished by expressing the SB transposase using electro-transferred, in vitro transcribed mRNA[53].
Strategies to Target TAAs
The goal of virus- or transposon-mediated gene therapy for cancer is to abolish tolerance of the immune system to cancer cells and promote the specific recognition and destruction of tumor cells. This therapeutic strategy involves targeting T cells to TAAs. Ideally, TAAs are presented on tumor cells but not healthy cells, ensuring that TAA-specific T cells achieve antitumor effects without causing on-target off-tissue toxicity. TAA-specific T cells can be numerically expanded ex vivo by culturing tumor-infiltrating T cells, or those from the peripheral blood, in the presence of autologous tumors and stimulatory cytokines[54],[55] to promote propagation in the absence of the immunoinhibitory environment that may exist in vivo. This approach has demonstrated clinical success, particularly for expanding melanoma-specific T cells[56]. Gene therapy can be used to re-direct T cells to specific TAAs by genetically manipulating the T cells ex vivo via the introduction of an exogenous TCR or a novel chimeric antigen receptor (CAR).
Introduction of an exogenous cloned αβ TCR is advantageous in that it should signal as a conventional TCR and mimic an endogenous immune response. The two chains can be co-expressed by a single vector using an internal ribosome entry site (IRES) or a pircornavirus 2A peptide sequence. Equimolar α/β chain partners are required for efficient expression of the exogenous TCR, rendering the 2A method more desirable[57], as genes expressed downstream of an IRES are typically not expressed as efficiently as the upstream gene[58]. Although vectors have been developed that stably express TCR genes, this field has been limited by obtaining immunoreceptors that are specific for TAAs.
The TCRs of T cells which are responsive to tumor cells can be isolated and cloned for transfection/infection into peripheral blood T cells to enhance the tumor-specific immune response. Such techniques have been used to produce T cells specific to melanoma antigens, such as melanoma antigen recognized by T cells (MART-1)[59],[60] and testis antigen NY-ESO-1, which are found in synovial cell sarcoma and metastatic melanoma[61],[62], and to general TAAs, such as p53[63],[64] and survivin[65]. T cells expressing TAA-specific TCRs have successfully promoted clinical responses in patients, including complete responses (CRs) in 2 of 11 patients with metastatic melanoma treated with T cells expressing a NY-ESO-1-recognizing TCR[66]. However, there are drawbacks to this approach. First, the introduced TCR requires a specific TAA-derived peptide to be presented in the context of a particular HLA molecule. In addition, the affinity of the cloned TCR appears to correlate with the therapeutic potential of the genetically modified T cells. There are multiple approaches to increase the association and dissociation rate constants of a TCR[67],[68]. However, ex vivo alterations of TCR affinity may lead to inadvertent recognition of alternative processed antigens, as was the case for patients receiving melanoma antigen family A, 3 (MAGE-A3)-specific T cells who experienced cardiac toxicity due to the recognition of titin by the infused genetically modified T cells[69]. In addition, the exogenous TCR α/β pairs could partner with the endogenous TCR components, reducing the expression of the desired TCR complex. Moreover, these altered TCR pairings could result in the recognition of self-antigen and the development of autoimmune disease[70]. The latter effect could be diminished by silencing the endogenous TCR using short hairpin RNA (shRNA)[71] or by permanently removing the endogenous TCR using artificial nucleases, such as transcription activator-like effector nucleases (TALENs)[72] or clustered regularly interspaced short palindromic repeat (CRISPR)-associated proteins (CRISPR/Cas9)[72]–[75]. However, T cells expressing an exogenous TCR specific for a TAA would continue to be restricted by HLA expression on tumors, which may be down-regulated[21]–[23].
In contrast, CARs recognize TAAs independent of HLA. The typical CAR consists of four separate domains that are assembled from individual units: 1) the single chain variable fragment (scFv) domain, 2) the extracellular scaffold/linker, 3) the transmembrane domain, and 4) the signaling endodomain[76]. The specificity of the CAR is imparted by the scFv domain, which is directed against a TAA. Other molecules can be substituted for the scFv domain, including cytokines[77], pattern-recognition receptors[78], and cell surface molecules[79], to dock the CAR to the respective ligands of these molecules. The scFv domain is attached to the transmembrane region via an extracellular scaffold. Optimal recognition of the TAA is affected by the extracellular length of the CAR. TAAs presented proximal to the plasma membrane may be more efficiently recognized by a CAR containing a longer scaffold, and conversely, TAAs presented further from the cell membrane may be more efficiently recognized by a CAR containing a shorter scaffold[80],[81]. Moreover, the source of the scaffold may greatly affect the effectiveness and in vivo function of the CAR. CARs targeting the B-cell antigen CD19 that have been used for clinical trials contain scaffolds derived from CD28, CD8, IgG1, or IgG4. The IgG motifs found in CARs may bind to endogenous Fc receptors, leading to clearance of CAR+ T cells expressing the IgG motifs and the non-specific activation of NK cells[82]. In vivo binding between CAR+ T cells containing the IgG scaffolds and endogenous cells expressing Fc receptors can apparently be reduced by mutagenesis of the IgG scaffolds[82]. The transmembrane domain links the extracellular scFv domain and scaffold to the signaling endodomain. Oligomerization of the transmembrane domain may favor CAR signaling, but few studies have systematically analyzed different transmembrane domains[83]. Signaling via the CAR molecule occurs via the addition of various endodomains to the cytoplasmic portion of the chimeric molecule. First-generation CARs signaled via immunoreceptor tyrosine-based activation motifs (ITAMs) from the CD3-ζ chain or FcR-γ[84]. A second co-stimulatory signal is included in second-generation CARs: the signaling endodomains of proteins such as CD28, 4-1BB, OX-40, and ICOS[85]. CARs containing two or more co-stimulatory molecules in addition to CD3-ζ are commonly referred to third-generation CARs. A competitive repopulation experiment demonstrated a survival advantage for second-generation CAR+ T cells infused into lymphoma patients compared to first-generation designs[86]. The ability to infuse more than one population of CAR+ T cells into a given recipient is appealing to help understand the immunobiology which favors the sustained persistence and, thus, the enhanced therapeutic activity of a given CAR species. However, the expense associated with generating two infusion products from two recombinant viral vectors diminishes the enthusiasm for this otherwise appealing experimental approach.
To help reduce the barriers to combining gene therapy with T-cell therapy, our laboratory at MD Anderson Cancer Center in Houston, Texas has employed electroporation using a commercially available Nucleofector device (Lonza Inc, Allendale, NJ, USA) to stably express CARs in primary human T cells for clinical trials[52]. Circulating T cells are electro-transferred with both the CAR-containing transposon and a hyperactive SB transposase, which catalyzes the cut-and-paste insertion of the transposon into the host genome. The apparent transient expression of the SB transposase in manufactured T cells helps to minimize the genotoxicity that arises via the continuous reintegration of the transposon. Transient expression of this fish-derived transposase also reduces the risk of the recipient developing an immune response to the transposase after the cells are infused. This improvement is significant, as an anti-transgene immune response could prompt the clearance and rejection of the genetically modified cells[87]. After gene transfer, the T cells are selectively propagated on activating and propagating cells (AaPCs) in the presence of stimulatory soluble cytokines for 4 weeks. The clinical-grade AaPCs are genetically modified K-562 cells that are available as a master cell bank. The AaPCs support the selective numeric expansion of CAR+ T cells by expressing a TAA and the co-stimulatory molecules CD86, 4-1BB, and membrane-bound interleukin (IL)-15[88].
Currently, to maintain patient safety, the manufacture of clinical-grade SB-modified CAR+ T cells occurs over a period of 3 to 5 weeks to enable the loss of the DNA plasmid containing the SB transposase[88]–[90]. Indeed, polymerase chain reaction (PCR) is used to confirm the absence of the SB transposase prior to infusion. Redirected T-cell specificity mediated by the CAR is evaluated via a chromium release assay. Sterility is verified via visual inspection, a negative result for mycoplasma based on a PCR assay, and a negative result for endotoxin based on the limulus amoebocyte lysate (LAL) test. The absence of genotoxicity is determined by the lack of growth of the genetically modified T-cell population upon removal of the AaPCs and cytokines, as well as by demonstrating a normal karyotype. The electroporated and propagated T cells are evaluated via flow cytometry for 1) viability, 2) the expression of the CAR, 3) the memory phenotype, 4) the absence of AaPCs, 5) exhaustion markers, and 6) telomere length. The presence of a polyclonal TCR repertoire is validated by flow cytometry or using bar-coded probes to confirm the lack of skewing of the numerically expanded T cells[91].
CAR+ T Cells in Clinical Trials
CAR+ T cells typically engineered by introducing DNA via a virus or a SB system have been successfully infused into patients with either solid or hematologic malignancy (Table 1). Transfer of autologous T cells transduced with retrovirus to enforce the expression of a CD19-specific CAR containing the CD28 and CD3-ζ endodomains after a preparative chemotherapy regimen led to the prolonged eradication of CD19+ cells in a follicular lymphoma patient[92]. Later studies conducted using CD19-specific CAR+ T cells in 8 lymphoma and chronic lymphocytic leukemia (CLL) patients found favorable responses in 6 of these patients, including 1 CR lasting more than 15 months and 5 partial responses (PRs) lasting at least 7 months[93]. The expected adverse events associated with targeting CD19 include long-term B-cell aplasia in about half of the patients. In addition to these long-term toxicities, acute adverse events manifested as symptoms of inflammation, such as fever, fatigue, and hypotension. The loss of humoral immunity can be life-threatening, as 1 patient has died of culture-verified influenza 18 days after receiving an infusion of CAR+ T cells[93].
Table 1. Chimeric antigen receptor (CAR) T cells in clinical trials.
| Reference | Cancer | CAR target | CAR endodomains | Number of patients | Clinical outcomes |
| Kochenderfer et al., 2010[92] | Lymphoma | CD19 | CD28 and CD3-ζ | 1 | PR |
| Kochenderfer et al., 2012[93] | Lymphoma | CD19 | CD28 and CD3-ζ | 4 | 3 PR, 1 died of influenza |
| CLL | CD19 | CD28 and CD3-ζ | 4 | 1 CR, 2 PR, 1 SD | |
| Kochenderfer et al., 2013[101] | CLL | CD19 | CD28 and CD3-ζ | 4 | 1 CR, 1 PR, 2 PD |
| Lymphoma | CD19 | CD28 and CD3-ζ | 6 | 1 PR, 5 SD | |
| Brentjens et al., 2011[94] | CLL | CD19 | CD28 and CD3-ζ | 8 | 1 PR, 2 SD, 3 NR, 1 PD, 1 died of sepsis-like disease (symptoms preceded T-cell transfer) |
| ALL | CD19 | CD28 and CD3-ζ | 1 | CR | |
| Brentjens et al., 2013[96] | ALL | CD19 | CD28 and CD3-ζ | 5 | 5 CR |
| Davila et al., 2014[97] | ALL | CD19 | CD28 and CD3-ζ | 16 | 10 CR, 4 CRi, 2 NR |
| Porter et al., 2011[98] | CLL | CD19 | 4-1BB and CD3-ζ | 1 | CR |
| Kalos et al., 2011[99] | CLL | CD19 | 4-1BB and CD3-ζ | 3 | 2 CR, 1 PR |
| Grupp et al., 2013[100] | ALL | CD19 | 4-1BB and CD3-ζ | 2 | 2 CR |
| Jensen et al., 2010[87] | Lymphoma | CD19 | CD3-ζ | 2 | 2 NR |
| Lymphoma | CD20 | CD3-ζ | 2 | 2 NR | |
| Till et al., 2008[102] | Lymphoma | CD20 | CD3-ζ | 7 | 1 PR, 4 SD, 2 NED maintained |
| Till et al., 2012[103] | Lymphoma | CD20 | CD28,4-1BB,CD3-ζ | 3 | 1 PR, 2 NED maintained |
| Kershaw et al., 2006[104] | Ovarian cancer | αFR | CD3-ζ | 14 | 14 NR |
| Lamers et al., 2012[106] | Renal cell carcinoma | CAIX | CD3-ζ | 12 | 12 NR |
| Park et al., 2007[107] | Neuroblastoma | CD171 | CD3-ζ | 6 | 1 PR, 5 PD |
| Louis et al., 2011[108] | Neuroblastoma | GD2 | CD3-ζ | 11 | 3 CR, 3 PR, 1 SD, 4 PD |
| Morgan et al., 2010[109] | Colorectal cancer | HER2 | CD28,4-1BB,CD3-ζ | 1 | Died of cytokine release syndrome |
CLL, chronic lymphocytic leukemia; ALL, acute lymphocytic leukemia; PR, partial response; CR, complete response; SD, stable disease; PD, progressive disease; NR, no objective response; CRi, complete response with incomplete count recovery; NED, no evidence of disease.
A separate group has also demonstrated success in treating patients with CLL and B-lineage acute lymphocytic leukemia (ALL) with CAR+ T cells that target CD19 and signal via the CD28 and CD3-ζ endodomains. Out of 8 patients with CLL who were infused with CAR+ T cells, 1 was partially responsive to therapy, and 2 maintained stable disease[94]. Early in the study, 1 patient with CLL died 48 h post T-cell infusion; however, serum cytokine abnormalities observed prior to this administration suggested that T-cell therapy was not the primary cause of this patient's death[95]. Treatment of ALL with CAR+ T cells was more successful and has resulted in clinical responses in 20 of 22 treated patients reported in three studies[94],[96],[97]. In the earliest study, the sole ALL patient demonstrated protracted B-cell aplasia following infusion of CD19-specific CAR+ T cells. To consolidate the antitumor effect, this recipient underwent allogeneic HSCT[94]. A subsequent study demonstrated that 4 of 5 patients exhibited CRs following infusion of CAR+ T cells, and these patients also went on to receive allogeneic HSCT[96]. The fifth recipient with ALL in that study initially responded to CAR+ T cells but was ineligible for HSCT and relapsed 13 weeks later before succumbing to the disease[96]. The most recent study found responses in 14 of 16 patients, including clinical CRs in 10 patients[97].
Similarly, patients with CD19+ CLL or ALL treated with autologous CD19-specific CAR+ T cells that signal via the 4-1BB and CD3-ζ endodomains after lymphodepleting chemotherapy experienced prolonged remissions[98]–[100]. Porter et al.[98] administered CAR+ T cells to a patient with CLL, resulting in CR lasting longer than 10 months. A related study transferred CAR+ T cells to 3 CLL patients, leading to 2 CRs and 1 PR lasting at least 7 months[99]. CRs were also observed in 2 patients with ALL; however, CD19− leukemia cells were detected in 1 patient 2 months after treatment[100]. Toxicities associated with this treatment include tumor lysis syndrome, elevated serum cytokine levels, fever, and hypotension. One of the patients with ALL displayed acute vascular leak syndrome, requiring pressor support, and acute respiratory distress syndrome, requiring intubation[100].
CD19-specific CAR+ T cells have also demonstrated success in reducing the tumor burden after allogeneic HSCT in patients with tumors resistant to DLI. A single dose of donor-derived CAR+ T cells mediated tumor regression in 3 of 10 patients, including 1 who exhibited CR[101]. Importantly, none of these recipients exhibited GVHD.
CD20 represents another target for the treatment of B-cell malignancies with infused CAR+ T cells. PRs were observed in patients who received first- or third-generation CAR+ T cells targeting this B-lineage antigen[102],[103]. Only 3 patients were treated with the third-generation CAR signaling via the CD28, 4-1BB, and CD3-ζ endodomains: 2 maintained no evidence of disease for at least 1 year, and 1 exhibited PR prior to relapse 12 months after the infusion. Significantly, PCR revealed that these third-generation CAR+ T cells persisted for at least 9 months in all 3 patients[103]. A lack of persistence of first-generation CAR+ T cells, which contain the signaling endodomain of only one molecule (CD3-ζ), has been attributed to a failure to control the cancer in some clinical studies[86],[87].
Non-hematopoietic cell surface antigens have also been targeted using CAR+ T cells in humans. In aggregate, these trials targeting solid tumors have not displayed the same level of success as those targeting hematologic malignancies. These results may be partially related to the deployment of first-generation CAR+ T cells in some of these trials. No patients exhibited a clinical response after receiving first-generation CAR+ T cells targeting the α-folate receptor (αFR) (0 of 14)[104] or carbonic anhydrase IX (CAIX) (0 of 12)[105],[106] for the treatment of ovarian or renal cell carcinoma, respectively. Some positive results were obtained for the treatment of neuroblastoma using first-generation CAR+ T cells targeting CD171 or GD2. One of 6 patients treated with CAR+ T cells targeting CD171 exhibited PR[107], and 3 of 11 patients with active disease exhibited CRs following the infusion of CAR+ T cells targeting the ganglioside GD2[108]. Of these 3 responders, 1 remained disease-free for 6 weeks prior to relapse, whereas the responses of the other 2 lasted longer than 21 and 60 months[108].
Thus, CAR+ T cells can expand and remain persistent in recipients in a variety of clinical settings, resulting in encouraging clinical outcomes. Despite the relative safety of CAR+ T-cell therapy in these studies, the risk remains that the hyper-physiologic activation of CAR+ T cells when synchronously activated by a large bio-burden of antigen-expressing cells could lead to toxicities resulting from systemic cytokine release, referred to as cytokine release syndrome[93],[96],[97],[100],[109]. Indeed, rapid and systemic cytokine release following the infusion of third-generation CAR+ T cells targeting HER2 into a colorectal cancer patient directly resulted in the patient's death[109]. Furthermore, not all patients respond to this therapy, and there remains considerable room for improvement in both the efficacy and specificity of infused CAR+ T cells.
Improving the Specificity and Effectiveness of CAR+ T Cells
A major obstacle is that the expression of some targeted TAAs is not restricted to tumor cells. Clinical trials targeting B-cell malignancies use an scFv domain specific for the kappa light chain[110], CD19[76], CD20[102],[103],[111], or CD22[112]. These antigens are expressed on both healthy and malignant B cells. Thus, targeting these TAAs may lead to partial or complete B-cell aplasia and compromised humoral immunity[93]. Although the clinical issues caused by loss of normal B cells can be circumvented by intravenous immunoglobulin infusion, TAAs targeted in association with non-hematopoietic malignancies are also often expressed on healthy tissues[113], potentially leading to severe adverse events, including death[109]. Much effort is being devoted to the discovery of TAAs that are indeed tumor-specific. One promising candidate TAA is the receptor tyrosine kinase orphan-like receptor 1 (ROR1). ROR1 is an appealing TAA because it is apparently absent from the cell surface of most adult tissues[114]. However, ROR1 is expressed on a variety of cancers, including B-cell malignancies, such as CLL and mantle cell lymphoma[115], pancreatic cancer, ovarian cancer, and lung cancer[116]. In contrast to targeting CD19, targeting ROR1 should lead to the specific destruction of cancerous B cells while avoiding damage to healthy B cells and the resulting hypogammaglobulinemia.
The functional specificity of a CAR can be increased by separating the activation and co-stimulatory endodomains into two CARs that are co-expressed on a single T cell. Thus, the activation of CAR+ T cells occurs upon co-recognition of two TAAs by separate scFv domains: one provides the activation signal, and one provides co-stimulation[117]. This technique has demonstrated promise but has yet to be evaluated in a clinical setting. A related technique co-expresses a CAR and an inhibitory CAR (iCAR)[118]. The iCAR targets an antigen expressed on normal tissue and contains an inhibitory, as opposed to an activating, endodomain. Recognition of the target antigen by the iCAR inhibits signaling via the separate activating CAR, should that CAR recognize a TAA on a healthy cell[118].
A separate approach to limit off-target damage caused by CAR+ T cells involves the co-expression of a death switch capable of rapidly shutting down the CAR+ T-cell effector response. One such switch involves inducible caspase-9 (iCasp9). iCasp9 remains dormant until the administration of a biologically inert, cell-permeable molecule that dimerizes and subsequently activates iCasp9[119],[120], inducing the genetically modified T cells to undergo apoptosis[121]. A single dose of the dimerizing drug administered to patients whose donor-derived T cells expressed iCasp9 resulted in the rapid and near complete removal of the infused T cells, causing GVHD[122]. The herpes simplex virus thymidine kinase (TK), which promotes death upon administration of the pro-drug ganciclovir, has also been used in human cells[123]. However, the expression of a viral antigen in CAR+ T cells likely leads to an anti-transgene immune response and the rejection of the infused products[87]. Moreover, killing mediated by ganciclovir in TK+ T cells apparently takes longer than killing via dimerized caspase-9[124], which may be detrimental in cases in which an induced immune response must be rapidly and efficiently halted. Alternatively, non-enzymatic death switches include the surface expression of CD20, which can be targeted using antibodies such as rituximab to induce cell destruction[125],[126]. Similar to iCasp9, the targeting of CD20-expressing cells with an antibody promotes rapid cell death[124]; however, in vivo administration of this antibody would also deplete healthy B cells.
Perhaps the greatest opportunity to increase the therapeutic effectiveness of CAR+ T cells is the modification of the CAR endodomain. Most current clinical trials targeting CD19 employ CAR designs that contain the CD3-ζ with either CD28 or CD137 signaling domains. There is apparent equipoise in the literature regarding the potential of these two second-generation CARs to eliminate malignant B cells. The endodomain used in a CAR may impact the nature of the induced immune response. Signaling via CD28 has been shown to favor the production of IFN-γ and inhibit the production of IL-17, whereas signaling via inducible costimulator (ICOS) favors the secretion of IL-17[127]. As both IFN-γ and IL-17 can alter the tumor microenvironment[128], modifying the CAR endodomain to favor production of different cytokines may change the in vivo antitumor activity of a CAR+ T cell. It is likely that different combinations of endo- domains (e.g., CD3-ζ with CD28, CD137, ICOS, 4-1BB, or OX-40) diffe- rentially affect the CAR+ T-cell response to the corresponding TAA.
Even in the case of efficient targeting and killing by CAR+ T cells, tumors can escape clearance by the immune system via the down-regulation of the targeted TAA. Indeed, a recent clinical trial observed the emergence of CD19− ALL cells after CD19-specific CAR+ T cells induced tumor regression[100]. Similar observations have been made in CD20-expressing tumors targeted using rituxamab[129],[130]. To circumvent tumor escape via the down-regulation of the targeted TAA, multiple CARs targeting separate TAAs can be expressed in a single cell in two populations of T cells. In one study, a tandem CAR specific for CD19 and HER2 remained responsive to each individual antigen and displayed increased activity against targets expressing both TAAs[131].
Future Directions
Thus far, the CARs used for immunotherapy in clinical trials have been expressed in T cells expressing αβ TCR, which is partially due to the relative abundance of αβ T cells in the peripheral blood compared to other lymphocyte populations, such as γδ T cells. However, γδ T cells may have distinct advantages over αβ T cells for adoptive immunotherapy. For example, γδ T-cell populations exhibit endogenous antitumor activity mediated by the γδ TCR. The tumor specificity of some αβ TCRs is becoming apparent based on antigens that have already been described, such as the cellular stress indicator MHC Class I chain-related A (MICA), which is targeted by Vγ1Vδ1 TCR[132]. Both MICA and MHC Class I chain-related B (MICB) expressed on tumor cells activate γδ T cells[133]. γδ T cells are also reactive to other stress indicators, such as heat shock proteins[134] and members of the UL16-binding protein (ULBP) family[135], and expression of these markers by tumor cells triggers lysis via γδ T cells[135]–[138]. In addition, γδ T cells express invariant innate immune receptors with antitumor activity, such as NKG2D[139]. Another apparent advantage of γδ T cells over αβ T cells is that antigen recognition by γδ T cells is not restricted by HLA[140],[141]. This advantage has implications for the development of “off-the-shelf” CAR+ T cells for immunotherapy. Current T-cell immunotherapy relies on harvesting T cells either from the patient, who may produce few or defective T cells due to the malignancy itself and prior treatment of the malignancy, or from an HLA-matched allogeneic donor, which includes the risk of GVHD. Due to their inherent antitumor activity and lack of HLA restriction, γδ T cells are an intriguing candidate therapy that can be pre-prepared from unrelated third-party donors and infused on demand.
The practical difficulties in using γδ T cells for CAR therapy include their relative low frequency in the peripheral blood[142] and the lack of a clinically appealing approach to propagate functional polyclonal γδ T cells. Until recently, γδ T cells were expanded for human use via the exogenous addition of phosphoantigens, such as zoledronic acid, which is only recognized by the Vγ9Vδ2 TCR[143],[144]. As a result, γδ T cells responsive to phosphoantigens have been infused in clinical settings[145]–[151]. We have successfully expanded polyclonal CAR+[152] and CAR−[153] γδ T cells on modified K-562-derived AaPCs. These CAR+ γδ T cells retain the expression of receptors displaying inherent antitumor activity, such as the γδ TCR and natural killer (NK) receptors, potentially enabling the tumor to trigger γδ T-cell activation via both engineered and innate mechanisms. Future studies will compare the effectiveness of γδ CAR+ T cells to that of αβ CAR+ T cells.
Artificial nucleases are another means to improve T cells for adoptive immunotherapy. Zinc-finger nucleases (ZFNs)[154], transcription activator-like (TAL) effector nucleases (TALENs)[72], and CRISPR/Cas9[73]–[75],[155] introduce double-stranded DNA breaks at specific sites, leading to repair by non-homologous end joining, which may result in gene inactivation. These technologies display potential for modulating the immune response to generate a favorable outcome in cancer immunotherapy. Artificial nucleases, such as CTLA-4 and programmed cell death-1 (PD-1), can be used to remove negative regulators of the anticancer response[156]–[158]. However, the removal of negative regulators of T-cell function must be approached with caution. Deaths have resulted following the infusion of CAR+ T cells[95],[109], one of which was attributed to the rapid and systemic release of inflammatory proteins[109]. Furthermore, artificial nucleases could be used to remove the TCR from the αβ T cells used for immunotherapy to eliminate the HLA alloreactivity of the infused T cells to reduce the risk of GVHD and to generate an off-the-shelf T-cell product[159].
Conclusions
The promise of T cells for the killing of cancer cells has been demonstrated for decades. Recent advances in gene therapy have facilitated the successful ex vivo re-programming of primary human T cells to express engineered CARs to achieve therapeutic effects in vivo. Both virus- and transposon-mediated technologies enable the re-specification of T cells to target TAAs. We believe that the SB system displays particular promise to help democratize T-cell therapy and personalize the genetically modified products to meet the needs of a patient with cancer. In the future, gene transfer will be augmented by genome editing to further increase the effectiveness of adoptive T-cell immunotherapy.
References
- 1.Mathe G, Amiel JL, Schwarzenberg L, et al. Successful allogenic bone marrow transplantation in man: chimerism, induced specific tolerance and possible anti-leukemic effects. Blood. 1965;25:179–196. [PubMed] [Google Scholar]
- 2.Weiden PL, Flournoy N, Thomas ED, et al. Antileukemic effect of graft-versus-host disease in human recipients of allogeneic-marrow grafts. N Engl J Med. 1979;300:1068–1073. doi: 10.1056/NEJM197905103001902. [DOI] [PubMed] [Google Scholar]
- 3.Weiden PL, Sullivan KM, Flournoy N, et al. Anti-leukemic effect of chronic graft versus host-disease: contribution to improved survival after allogeneic marrow transplantation. N Engl J Med. 1981;304:1529–1533. doi: 10.1056/NEJM198106183042507. [DOI] [PubMed] [Google Scholar]
- 4.Marmont AM, Horowitz MM, Gale RP, et al. T-cell depletion of HLA-identical transplants in leukemia. Blood. 1991;78:2120–2130. [PubMed] [Google Scholar]
- 5.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 6.Apperley JF, Mauro FR, Goldman JM, et al. Bone marrow transplantation for chronic myeloid leukemia in first chronic phase: importance of a graft-versus-leukemia effect. Br J Haematol. 1988;69:239–245. doi: 10.1111/j.1365-2141.1988.tb07628.x. [DOI] [PubMed] [Google Scholar]
- 7.Kolb HJ, Mittermuller J, Clemm C, et al. Donor leukocyte transfusions for treatment of recurrent chronic myelogenous leukemia in marrow transplant patients. Blood. 1990;76:2462–2465. [PubMed] [Google Scholar]
- 8.Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood. 2008;112:4371–4383. doi: 10.1182/blood-2008-03-077974. [DOI] [PubMed] [Google Scholar]
- 9.Collins RH, Shpilberg O, Drobyski WR, et al. Donor leukocyte infusions in 140 patients with relapsed malignancy after allogeneic bone marrow transplantation. J Clin Oncol. 1997;15:433–444. doi: 10.1200/JCO.1997.15.2.433. [DOI] [PubMed] [Google Scholar]
- 10.Kolb HJ, Schattenberg A, Goldman JM, et al. Graft-versus-leukemia effect of donor lymphocyte transfusions in marrow grafted patients. Blood. 1995;86:2041–2050. [PubMed] [Google Scholar]
- 11.Shlomchik WD, Couzens MS, Tang CB, et al. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 1999;285:412–415. doi: 10.1126/science.285.5426.412. [DOI] [PubMed] [Google Scholar]
- 12.Teshima T, Ordemann R, Reddy P, et al. Acute graft-versus-host disease does not require alloantigen expression on host epithelium. Nat Med. 2002;8:575–581. doi: 10.1038/nm0602-575. [DOI] [PubMed] [Google Scholar]
- 13.Mapara MY, Kim YM, Wang SP, et al. Donor lymphocyte infusions mediate superior graft-versus-leukemia effects in mixed compared to fully allogeneic chimeras: a critical role for host antigen-presenting cells. Blood. 2002;100:1903–1909. doi: 10.1182/blood-2002-01-0023. [DOI] [PubMed] [Google Scholar]
- 14.Matte CC, Liu JL, Cormier J, et al. Donor APCs are required for maximal GVHD but not for GVL. Nat Med. 2004;10:987–992. doi: 10.1038/nm1089. [DOI] [PubMed] [Google Scholar]
- 15.Chakraverty R, Eom HS, Sachs J, et al. Host MHC class II+ antigen-presentirig cells and CD4 cells are required for CD8-mediated graft-versus-leukemia responses following delayed donor leukocyte infusions. Blood. 2006;108:2106–2113. doi: 10.1182/blood-2006-03-007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storb R, Gyurkocza B, Storer BE, et al. Graft-versus-host disease and graft-versus-tumor effects after allogeneic hematopoietic cell transplantation. J Clin Oncol. 2013;31:1530–1538. doi: 10.1200/JCO.2012.45.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong HD, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 18.Wojtowicz-Praga S. Reversal of tumor-induced immunosuppression by TGF-beta inhibitors. Invest New Drug. 2003;21:21–32. doi: 10.1023/a:1022951824806. [DOI] [PubMed] [Google Scholar]
- 19.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7:1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 20.Ahmadzadeh M, Rosenberg SA. TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol. 2005;174:5215–5223. doi: 10.4049/jimmunol.174.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natali PG, Nicotra MR, Bigotti A, et al. Selective changes in expression of HLA class-I polymorphic determinants in human solid tumors. Proc Natl Acad Sci U S A. 1989;86:6719–6723. doi: 10.1073/pnas.86.17.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Cao Y, Albino AP, et al. Lack of HLA class I antigen expression by melanoma-cells SK-MEL-33 caused by a reading frameshift in beta-2-microglobulin messenger-RNA. J Clin Invest. 1993;91:684–692. doi: 10.1172/JCI116249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrera T, Fernandez MA, Sierra A, et al. High frequency of altered HLA class I phenotypes in invasive breast carcinomas. Hum Immunol. 1996;50:127–134. doi: 10.1016/0198-8859(96)00145-0. [DOI] [PubMed] [Google Scholar]
- 24.Wu XL, Li Y, Crise B, et al. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 25.Schroder ARW, Shinn P, Chen HM, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 26.Hacein-Bey-Abina S, Hauer J, Lim A, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 28.Gaspar HB, Cooray S, Gilmour KC, et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 29.Newrzela S, Cornils K, Li ZX, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 30.Cattoglio C, Maruggi G, Bartholomae C, et al. High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS One. 2010;5:e15688. doi: 10.1371/journal.pone.0015688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholler J, Brady TL, Binder-Scholl G, et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med. 2012;4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8:S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balciunas D, Wangensteen KJ, Wilber A, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:1715–1724. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabundzija I, Irgang M, Mates L, et al. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SCY, Meir YJJ, Coates CJ, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2 and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding S, Wu XH, Li G, et al. Efficient transposition of the piggyBac resource (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Fraser MJ, Coszczon T, Elick T, et al. Precise excision of TTAA-specific lepidopteran transposons piggyBac(IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 39.Wilson MH, Coates CJ, George AL. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 40.Galvan DL, Nakazawa Y, Kaja A, et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivics Z, Hackett PB, Plasterk RH, et al. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 42.Geurts AM, Yang Y, Clark KJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 43.Mates L, Chuah MKL, Belay E, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 44.Zayed H, Izsvak Z, Walisko O, et al. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Vigdal TJ, Kaufman CD, Izsvak Z, et al. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 46.Yant SR, Wu XL, Huang Y, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berry C, Hannenhalli S, Leipzig J, et al. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:1450–1462. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karsi A, Moav B, Hackett P, et al. Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol. 2001;3:241–245. doi: 10.1007/s101260000072. [DOI] [PubMed] [Google Scholar]
- 49.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 50.Liu G, Aronovich EL, Cui Z, et al. Excision of Sleeping Beauty transposons: parameters and applications to gene therapy. J Gene Med. 2004;6:574–583. doi: 10.1002/jgm.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell JB, Aronovich EL, Schreifels JM, et al. Duration of expression and activity of Sleeping Beauty transposase in mouse liver following hydrodynamic DNA delivery. Mol Ther. 2010;18:1796–1802. doi: 10.1038/mt.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hackett PB, Largaespada DA, Switzer KC, et al. Evaluating risks of insertional mutagenesis by DNA transposons in gene therapy. Transl Res. 2013;161:265–283. doi: 10.1016/j.trsl.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilber A, Frandsen JL, Geurts JL, et al. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg SA, Spiess P, Lafreniere R. A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science. 1986;233:1318–1321. doi: 10.1126/science.3489291. [DOI] [PubMed] [Google Scholar]
- 55.Crowley NJ, Slingluff CL, Jr, Darrow TL, et al. Generation of human autologous melanoma-specific cytotoxic T-cells using HLA-A2-matched allogeneic melanomas. Cancer Res. 1990;50:492–498. [PubMed] [Google Scholar]
- 56.Lee S, Margolin K. Tumor-infiltrating lymphocytes in melanoma. Curr Oncol Rep. 2012;14:468–474. doi: 10.1007/s11912-012-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisegang M, Engels B, Meyerhuber P, et al. Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J Mol Med. 2008;86:573–583. doi: 10.1007/s00109-008-0317-3. [DOI] [PubMed] [Google Scholar]
- 58.Mizuguchi H, Xu Z, Ishii-Watabe A, et al. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 59.Hughes MS, Yu YY, Dudley ME, et al. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gnjatic S, Jager E, Chen W, et al. CD8(+) T cell responses against a dominant cryptic HLA-A2 epitope after NY-ESO-1 peptide immunization of cancer patients. Proc Natl Acad Sci U S A. 2002;99:11813–11818. doi: 10.1073/pnas.142417699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y, Zheng Z, Robbins PF, et al. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;174:4415–4423. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen CJ, Zheng Z, Bray R, et al. Recognition of fresh human tumor by human peripheral blood lymphocytes transduced with a bicistronic retroviral vector encoding a murine anti-p53 TCR. J Immunol. 2005;175:5799–5808. doi: 10.4049/jimmunol.175.9.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis JL, Theoret MR, Zheng Z, et al. Development of human anti-murine T-cell receptor antibodies in both responding and nonresponding patients enrolled in TCR gene therapy trials. Clin Cancer Res. 2010;16:5852–5861. doi: 10.1158/1078-0432.CCR-10-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmitz M, Diestelkoetter P, Weigle B, et al. Generation of survivin-specific CD8+ T effector cells by dendritic cells pulsed with protein or selected peptides. Cancer Res. 2000;60:4845–4849. [PubMed] [Google Scholar]
- 66.Robbins PF, Morgan RA, Feldman SA, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Moysey R, Molloy PE, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 68.Chervin AS, Aggen DH, Raseman JM, et al. Engineering higher affinity T cell receptors using a T cell display system. J Immunol Methods. 2008;339:175–184. doi: 10.1016/j.jim.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cameron BJ, Gerry AB, Dukes J, et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bendle GM, Linnemann C, Hooijkaas AI, et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med. 2010;16:565–570. doi: 10.1038/nm.2128. [DOI] [PubMed] [Google Scholar]
- 71.Okamoto S, Mineno J, Ikeda H, et al. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer Res. 2009;69:9003–9011. doi: 10.1158/0008-5472.CAN-09-1450. [DOI] [PubMed] [Google Scholar]
- 72.Miller JC, Tan SY, Qiao GJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–U149. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 73.Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jena B, Moyes JS, Huls H, et al. Driving CAR-based T-cell therapy to success. Curr Hematol Malig Rep. 2014;9:50–56. doi: 10.1007/s11899-013-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown CE, Starr R, Aguilar B, et al. Stem-like tumor-initiating cells isolated from IL13Rα2 expressing gliomas are targeted and killed by IL13-zetakine-redirected T cells. Clin Cancer Res. 2012;18:2199–2209. doi: 10.1158/1078-0432.CCR-11-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumaresan PR, Manuri PR, Albert ND, et al. Bioengineering T cells to target carbohydrate to treat opportunistic fungal infection. Proc Natl Acad Sci U S A. 2014;111:10660–10665. doi: 10.1073/pnas.1312789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shaffer DR, Savoldo B, Yi Z, et al. T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood. 2011;117:4304–4314. doi: 10.1182/blood-2010-04-278218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.James SE, Greenberg PD, Jensen MC, et al. Antigen sensitivity of CD22-specific chimeric TCR is modulated by target epitope distance from the cell membrane. J Immunol. 2008;180:7028–7038. doi: 10.4049/jimmunol.180.10.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hudecek M, Lupo-Stanghellini MT, Kosasih PL, et al. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res. 2013;19:3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hombach A, Hombach AA, Abken H. Adoptive immunotherapy with genetically engineered T cells: modification of the IgG1 Fc ‘spacer’ domain in the extracellular moiety of chimeric antigen receptors avoids ‘off-target’ activation and unintended initiation of an innate immune response. Gene Ther. 2010;17:1206–1213. doi: 10.1038/gt.2010.91. [DOI] [PubMed] [Google Scholar]
- 83.Bridgeman JS, Hawkins RE, Bagley S, et al. The optimal antigen response of chimeric antigen receptors harboring the CD3zeta transmembrane domain is dependent upon incorporation of the receptor into the endogenous TCR/CD3 complex. J Immunol. 2010;184:6938–6949. doi: 10.4049/jimmunol.0901766. [DOI] [PubMed] [Google Scholar]
- 84.Eshhar Z, Waks T, Gross G, et al. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma-subunit or zeta-subunit of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci U S A. 1993;90:720–724. doi: 10.1073/pnas.90.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sadelain M, Brentjens R, Riviere I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savoldo B, Ramos CA, Liu E, et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J Clin Invest. 2011;12:1822–1826. doi: 10.1172/JCI46110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen MC, Popplewell L, Cooper LJ, et al. Antitransgene rejection responses contribute to attenuated persistence of adoptively transferred CD20/CD19-specific chimeric antigen receptor redirected T cells in humans. Biol Blood Marrow Transplant. 2010;16:1245–1256. doi: 10.1016/j.bbmt.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh H, Figliola MJ, Dawson MJ, et al. Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS One. 2013;8:e64138. doi: 10.1371/journal.pone.0064138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maiti SN, Huls H, Singh H, et al. Sleeping beauty system to redirect T-cell specificity for human applications. J Immunother. 2013;36:112–123. doi: 10.1097/CJI.0b013e3182811ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh H, Huls H, Kebriaei P, et al. A new approach to gene therapy using sleeping beauty to genetically modify clinical-grade T cells to target CD19. Immunol Rev. 2014;257:181–190. doi: 10.1111/imr.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang M, Maiti S, Bernatchez C, et al. A new approach to simultaneously quantify both TCR alpha- and beta-chain diversity after adoptive immunotherapy. Clin Cancer Res. 2012;18:4733–4742. doi: 10.1158/1078-0432.CCR-11-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kochenderfer JN, Wilson WH, Janik JE, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116:4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kochenderfer JN, Dudley ME, Feldman SA, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119:2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brentjens RJ, Riviere I, Park JH, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118:4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brentjens R, Yeh R, Bernal Y, et al. Treatment of chronic lympho-cytic leukemia with genetically targeted autologous T cells: case report of an unforeseen adverse event in a phase I clinical trial. Mol Ther. 2010;18:666–668. doi: 10.1038/mt.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davila ML, Riviere I, Wang X, et al. Efficacy and toxicity manage-ment of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Medicine. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Porter DL, Levine BL, Kalos M, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kochenderfer JN, Dudley ME, Carpenter RO, et al. Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood. 2013;122:4129–4139. doi: 10.1182/blood-2013-08-519413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Till BG, Jensen MC, Wang J, et al. Adoptive immunotherapy for indolent non-Hodgkin lymphoma and mantle cell lymphoma using genetically modified autologous CD20-specific T cells. Blood. 2008;112:2261–2271. doi: 10.1182/blood-2007-12-128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Till BG, Jensen MC, Wang J, et al. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012;119:3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kershaw MH, Westwood JA, Parker LL, et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin Cancer Res. 2006;12:6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lamers CH, Willemsen R, van Elzakker P, et al. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood. 2011;117:72–82. doi: 10.1182/blood-2010-07-294520. [DOI] [PubMed] [Google Scholar]
- 106.Lamers CH, Sleijfer S, van Steenbergen S, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013;21:904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park JR, Digiusto DL, Slovak M, et al. Adoptive transfer of chimeric antigen receptor re-directed cytolytic T lymphocyte clones in patients with neuroblastoma. Mol Ther. 2007;15:825–833. doi: 10.1038/sj.mt.6300104. [DOI] [PubMed] [Google Scholar]
- 108.Louis CU, Savoldo B, Dotti G, et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood. 2011;118:6050–6056. doi: 10.1182/blood-2011-05-354449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morgan RA, Yang JC, Kitano M, et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vera J, Savoldo B, Vigouroux S, et al. T lymphocytes redirected against the kappa light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood. 2006;108:3890–3897. doi: 10.1182/blood-2006-04-017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jensen M, Tan G, Forman S, et al. CD20 is a molecular target for scFvFc:zeta receptor redirected T cells: implications for cellular immunotherapy of CD20+ malignancy. Biol Blood Marrow Transplant. 1998;4:75–83. doi: 10.1053/bbmt.1998.v4.pm9763110. [DOI] [PubMed] [Google Scholar]
- 112.Haso W, Lee DW, Shah NN, et al. Anti-CD22-chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia. Blood. 2013;121:1165–1174. doi: 10.1182/blood-2012-06-438002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hinrichs CS, Restifo NP. Reassessing target antigens for adoptive T-cell therapy. Nat Biotechnol. 2013;31:999–1008. doi: 10.1038/nbt.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fukuda T, Chen L, Endo T, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008;105:3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hudecek M, Schmitt TM, Baskar S, et al. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood. 2010;116:4532–4541. doi: 10.1182/blood-2010-05-283309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang S, Chen L, Wang-Rodriguez J, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181:1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kloss CC, Condomines M, Cartellieri M, et al. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol. 2013;31:71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fedorov VD, Themeli M, Sadelain M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci Transl Med. 2013;5:215ra172. doi: 10.1126/scitranslmed.3006597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spencer DM, Wandless TJ, Schreiber SL, et al. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 120.Clackson T, Yang W, Rozamus LW, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Straathof KC, Pule MA, Yotnda P, et al. An inducible caspase 9 safety switch for T-cell therapy. Blood. 2005;105:4247–4254. doi: 10.1182/blood-2004-11-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Di Stasi A, Tey SK, Dotti G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bonini C, Ferrari G, Verzeletti S, et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science. 1997;276:1719–1724. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 124.Marin V, Cribioli E, Philip B, et al. Comparison of different suicide-gene strategies for the safety improvement of genetically manipulated T cells. Hum Gene Ther Methods. 2012;23:376–386. doi: 10.1089/hgtb.2012.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Introna M, Barbui AM, Bambacioni F, et al. Genetic modification of human T cells with CD20: a strategy to purify and lyse transduced cells with anti-CD20 antibodies. Human Gene Ther. 2000;11:611–620. doi: 10.1089/10430340050015798. [DOI] [PubMed] [Google Scholar]
- 126.Serafini M, Manganini M, Borleri G, et al. Characterization of CD20-transduced T lymphocytes as an alternative suicide gene therapy approach for the treatment of graft-versus-host disease. Hum Gene Ther. 2004;15:63–76. doi: 10.1089/10430340460732463. [DOI] [PubMed] [Google Scholar]
- 127.Paulos CM, Carpenito C, Plesa G, et al. The inducible costimulator (ICOS) is critical for the development of human T(H)17 cells. Sci Transl Med. 2010;2:55ra78. doi: 10.1126/scitranslmed.3000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alshaker HA, Matalka KZ. IFN-γ, IL-17 and TGF-β involvement in shaping the tumor microenvironment: the significance of modulating such cytokines in treating malignant solid tumors. Cancer Cell Int. 2011;11:33. doi: 10.1186/1475-2867-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hiraga J, Tomita A, Sugimoto T, et al. Down-regulation of CD20 expression in B-cell lymphoma cells after treatment with rituximab-containing combination chemotherapies: its prevalence and clinical significance. Blood. 2009;113:4885–4893. doi: 10.1182/blood-2008-08-175208. [DOI] [PubMed] [Google Scholar]
- 130.Terui Y, Mishima Y, Sugimura N, et al. Identification of CD20 C-terminal deletion mutations associated with loss of CD20 expression in non-Hodgkin's lymphoma. Clin Cancer Res. 2009;15:2523–2530. doi: 10.1158/1078-0432.CCR-08-1403. [DOI] [PubMed] [Google Scholar]
- 131.Grada Z, Hegde M, Byrd T, et al. TanCAR: a novel bispecific chimeric antigen receptor for cancer immunotherapy. Mol Ther Nucleic Acids. 2013;2:e105. doi: 10.1038/mtna.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu B, Pizarro JC, Holmes MA, et al. Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA. Proc Natl Acad Sci U S A. 2011;108:2414–2419. doi: 10.1073/pnas.1015433108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Groh V, Rhinehart R, Secrist H, et al. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O'Brien RL, Born W. Heat shock proteins as antigens for gamma delta T cells. Semin Immunol. 1991;3:81–87. [PubMed] [Google Scholar]
- 135.Kong Y, Cao W, Xi X, et al. The NKG2D ligand ULBP4 binds to TCRgamma9/delta2 and induces cytotoxicity to tumor cells through both TCRgammadelta and NKG2D. Blood. 2009;114:310–317. doi: 10.1182/blood-2008-12-196287. [DOI] [PubMed] [Google Scholar]
- 136.Sturm E, Braakman E, Fisch P, et al. Human V gamma 9-V delta 2 T cell receptor-gamma delta lymphocytes show specificity to Daudi Burkitt's lymphoma cells. J Immunol. 1990;145:3202–3208. [PubMed] [Google Scholar]
- 137.Fisch P, Malkovsky M, Kovats S, et al. Recognition by human V gamma 9/V delta 2 T cells of a GroEL homolog on Daudi Burkitt's lymphoma cells. Science. 1990;250:1269–1273. doi: 10.1126/science.1978758. [DOI] [PubMed] [Google Scholar]
- 138.Kaur I, Voss SD, Gupta RS, et al. Human peripheral gamma delta T cells recognize hsp60 molecules on Daudi Burkitt's lymphoma cells. J Immunol. 1993;150:2046–2055. [PubMed] [Google Scholar]
- 139.Wrobel P, Shojaei H, Schittek B, et al. Lysis of a broad range of epithelial tumour cells by human gamma delta T cells: involvement of NKG2D ligands and T-cell receptor versus NKG2D-dependent recognition. Scand J Immunol. 2007;66:320–328. doi: 10.1111/j.1365-3083.2007.01963.x. [DOI] [PubMed] [Google Scholar]
- 140.Schild H, Mavaddat N, Litzenberger C, et al. The nature of major histocompatibility complex recognition by gamma delta T cells. Cell. 1994;76:29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 141.Morita CT, Beckman EM, Bukowski JF, et al. Direct presentation of nonpeptide prenyl pyrophosphate antigens to human gamma delta T cells. Immunity. 1995;3:495–507. doi: 10.1016/1074-7613(95)90178-7. [DOI] [PubMed] [Google Scholar]
- 142.Lanier LL, Ruitenberg J, Bolhuis RLH, et al. Structural and serological heterogeneity of gamma/delta-T-cell antigen receptor expression in thymus and peripheral blood. Eur J Immunol. 1988;18:1985–1992. doi: 10.1002/eji.1830181218. [DOI] [PubMed] [Google Scholar]
- 143.Kato Y, Tanaka Y, Miyagawa F, et al. Targeting of tumor cells for human gamma delta T cells by nonpeptide antigens. J Immunol. 2001;167:5092–5098. doi: 10.4049/jimmunol.167.9.5092. [DOI] [PubMed] [Google Scholar]
- 144.Kunzmann V, Bauer E, Feurle J, et al. Stimulation of gamma delta T cells by aminobisphosphonates and induction of antiplasma cell activity in multiple myeloma. Blood. 2000;96:384–392. [PubMed] [Google Scholar]
- 145.Dieli F, Gebbia N, Poccia F, et al. Induction of gamma delta T-lymphocyte effector functions by bisphosphonate zoledronic acid in cancer patients in vivo. Blood. 2003;102:2310–2311. doi: 10.1182/blood-2003-05-1655. [DOI] [PubMed] [Google Scholar]
- 146.Sicard H, Ingoure S, Luciani B, et al. In vivo immunomanipulation of V gamma 9V delta 2 T cells with a synthetic phosphoantigen in a preclinical nonhuman primate model. J Immunol. 2005;175:5471–5480. doi: 10.4049/jimmunol.175.8.5471. [DOI] [PubMed] [Google Scholar]
- 147.Casetti R, Perretta G, Taglioni A, et al. Drug-induced expansion and differentiation of V gamma 9V delta 2 T cells in vivo: the role of exogenous IL-2. J Immunol. 2005;175:1593–1598. doi: 10.4049/jimmunol.175.3.1593. [DOI] [PubMed] [Google Scholar]
- 148.Kondo M, Sakuta K, Noguchi A, et al. Zoledronate facilitates large-scale ex vivo expansion of functional gamma delta T cells from cancer patients for use in adoptive immunotherapy. Cytotherapy. 2008;10:842–856. doi: 10.1080/14653240802419328. [DOI] [PubMed] [Google Scholar]
- 149.Nicol AJ, Tokuyama H, Mattarollo SR, et al. Clinical evaluation of autologous gamma delta T cell-based immunotherapy for metastatic solid tumours. Br J Cancer. 2011;105:778–786. doi: 10.1038/bjc.2011.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kobayashi H, Tanaka Y, Yagi J, et al. Safety profile and anti-tumor effects of adoptive immunotherapy using gamma-delta T cells against advanced renal cell carcinoma: a pilot study. Cancer Immunol Immun. 2007;56:469–476. doi: 10.1007/s00262-006-0199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bennouna J, Bompas E, Neidhardt EM, et al. Phase-I study of innacell gammadelta, an autologous cell-therapy product highly enriched in gamma9delta2 T lymphocytes, in combination with IL-2, in patients with metastatic renal cell carcinoma. Cancer Immunol Immun. 2008;57:1599–1609. doi: 10.1007/s00262-008-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Deniger DC, Switzer K, Mi TJ, et al. Bispecific T-cells expressing polyclonal repertoire of endogenous gamma delta T-cell receptors and introduced CD19-specific chimeric antigen receptor. Mol Ther. 2013;21:638–647. doi: 10.1038/mt.2012.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Deniger DC, Maiti S, Mi T, et al. Activating and propagating polyclonal gamma delta T cells with broad specificity for malignancies. Clin Cancer Res. 2014 May 15; doi: 10.1158/1078-0432.CCR-13-3451. pii: clincanres. 3451.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Phan GQ, Yang JC, Sherry RM, et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc Natl Acad Sci U S A. 2003;100:8372–8377. doi: 10.1073/pnas.1533209100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Torikai H, Reik A, Liu PQ, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]