Abstract
Advances in studies of microRNA (miRNA) expression and function in smooth muscles illustrate important effects of small noncoding RNAs on cell proliferation, hypertrophy and differentiation. An emerging theme in miRNA research in a variety of cell types including smooth muscles is that miRNAs regulate protein expression networks to fine tune phenotype. Some widely expressed miRNAs have been described in smooth muscles that regulate important processes in many cell types, such as miR-21 control of proliferation and cell survival. Other miRNAs that are prominent regulators of smooth muscle-restricted gene expression also have targets that control pluripotent cell differentiation. The miR-143~145 cluster which targets myocardin and Kruppel-like factor 4 (KLF4) is arguably the best-described miRNA family in smooth muscles with profound effects on gene expression networks that promote serum response factor (SRF)-dependent contractile and cytoskeletal protein expression and the mature contractile phenotype. Kruppel-family members KLF4 and KLF5 have multiple effects on cell differentiation and are targets for multiple miRNAs in smooth muscles (miR-145, miR-146a, miR-25). The feedback and feedforward loops being defined appear to contribute significantly to vascular and airway remodeling in cardiovascular and respiratory diseases. RNA interference approaches applied to animal models of vascular and respiratory diseases prove that miRNAs and RNA-induced silencing are valid targets for novel anti-remodeling therapies that alter pathological smooth muscle hyperplasia and hypertrophy.
Keywords: Asthma, Atherosclerosis, Hypertension, KLF4, Myocardin, Translation, Vascular Remodeling, Vascular injury
Introduction
Structural cells in the cardiovascular and respiratory systems adapt to permit changes in function during development and disease. Fibroblasts, myofibroblasts, smooth muscle, epithelial, endothelial and progenitor cells all undergo varying degrees of phenotypic modulation during organogenesis and in various diseases. In the cardiovascular system several clinically important conditions trigger adaptive and maladaptive blood vessel remodeling. Atherosclerosis, aneurysms, restenosis injury and ischemia are all conditions that elicit vessel remodeling. Remodeling can include cell hypertrophy, hyperplasia, matrix remodeling and secretion of numerous cell to cell signaling molecules. Structural cells of the respiratory tract also undergo significant remodeling in disease states. Asthma stimulates myofibroblasts to undergo a transition to a more contractile phenotype in both humans and animal models (1–3). Airway smooth muscle cells increase in number, sometimes increase in volume, and secrete signaling proteins thought to contribute to airway hyperactivity (reviewed by 4). In all smooth muscle tissues dynamic changes in gene expression and protein composition permit cells to respond to altered environmental conditions. Cellular plasticity is a fundamental characteristic of smooth muscle cells in vivo. Cellular plasticity is defined here as long lasting changes in the structure and function of a cell caused by altered gene expression and protein composition. Protein composition of smooth muscle cells, as in all mammalian cells, is determined by multiple parallel signaling pathways that regulate transcription, translation, mRNA half-life and protein catabolism. Several highly conserved protein kinase cascades (PKA, PKG, PKC, MAP kinases, JAK/Stat, Smad signaling and NFkB) regulate smooth muscle phenotype. Control of transcription by these pathways has been studied extensively in vascular and airway smooth muscle cells, but epigenetic mechanisms that modify smooth muscle phenotype are not as well described. Chromatin remodeling by histone modifications, DNA methylation and miRNA-induced gene silencing are not as well defined in smooth muscle cells as they are in other cell types such as cancer cells (5–7). This review will summarize the rapidly expanding knowledge of the function of the microRNA class of small, noncoding RNAs in determining smooth muscle cell phenotypes in normal and disease states. Emphasis will be placed on miRNAs with validated target genes in smooth muscles and on miRNAs that have demonstrated effects or high potential as druggable targets. Several examples of RNAi-based therapy of animal models of cardiovascular and respiratory diseases will be described that demonstrate proof of principle for RNAi therapy.
Smooth muscle cell phenotypes
Smooth muscle cells in vitro and in vivo are notable for their ability to adapt to the local milieu. In vitro, smooth muscle cells can be manipulated by altering culture conditions to induce a more contractile phenotype by culturing at high density at reduced serum concentrations in the presence of soluble factors that promote differentiation including retinoic acid, transforming growth factor beta 1 (TGF-β1) and insulin. The contractile phenotype will be defined here as cells that express smooth muscle-restricted contractile and cytoskeletal proteins and contract in response to neurotransmitters and autacoids. Verified smooth muscle-restricted contractile phenotype genes include: myosin II heavy chain, α and γ smooth-muscle actins, h-caldesmon, h1-calponin, smooth muscle tropomyosins, SM22 (transgelin) and smoothelin (8, 9). To promote the proliferative/migratory/secretory phenotype, smooth muscle cells are typically cultured in serum-containing medium with trophic growth factors epidermal growth factor and fibroblast growth factor. The proliferative/migratory phenotype is not as clearly defined as the contractile phenotype, but generally refers to cells in culture that proliferate in response to serum, migrate in response to stimuli including platelet derived growth factor (PDGF), and secrete a variety cytokines, chemokines and protein growth factors. The in vivo correlate of proliferating/migrating cells is inferred from the behavior of smooth muscle cells in culture and from studies of organogenesis of blood vessels and airways during fetal and neonatal development. Contractile and proliferating/migrating “phenotypes” are not necessarily stable, irreversible, or mutually exclusive. One view is that the phenotype of smooth muscle cells is more a graded than a binary phenomenon with cells in a particular tissue having a mosaic pattern of contractile protein gene expression (8–11). An alternate view is that smooth muscle cells can assume bistable states of gene expression in which the contractile and proliferative expression programs are mutually exclusive (9). Some combination of these models is also a formal possibility with gene expression programs being highly adaptable depending on tissue type, culture conditions or disease processes. Because smooth muscle cells adapt and remodel significantly in cardiovascular and lung diseases the role of epigenetic processes that shift cells from one state to another during development and disease is a very active area of investigation. Identifying miRNAs and the networks of target genes that participate in disease progression will have high impact on translational research aimed at identifying novel therapeutic targets for preventing or reversing pathological smooth muscle tissue remodeling.
miRNA silencing pathway
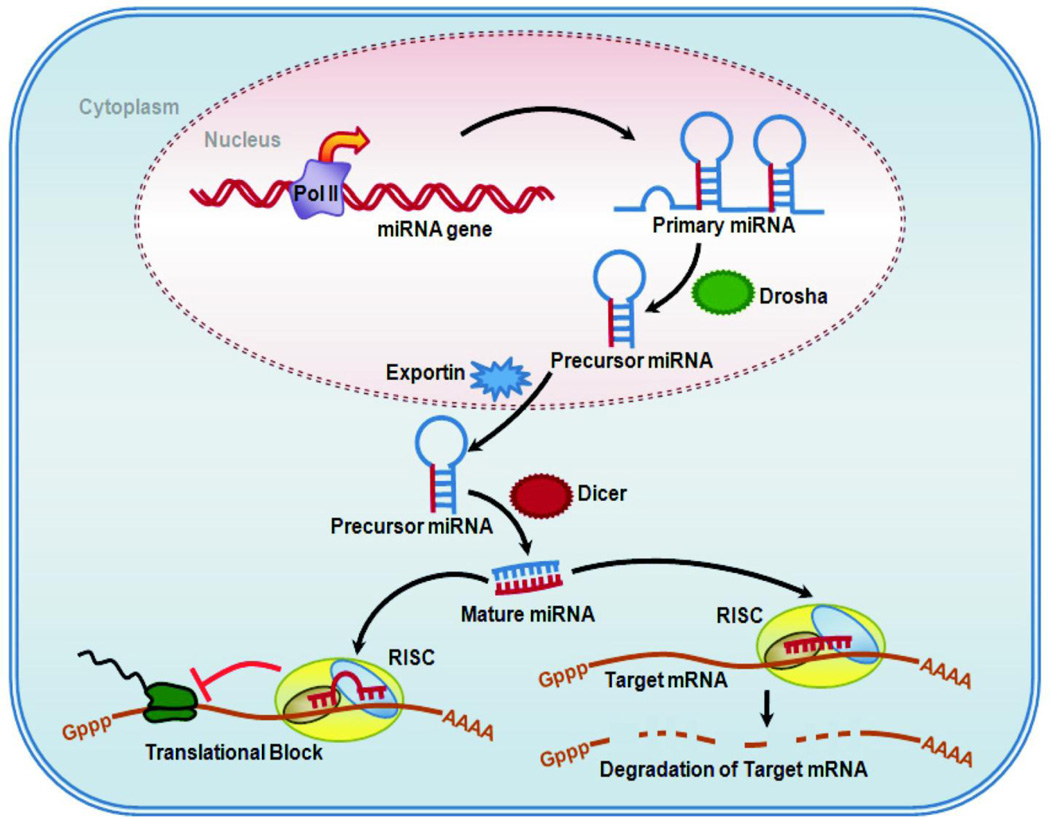
Basic features of miRNA biogenesis and RNA-induced gene silencing have been described in some detail within the past 10 years (reviewed by 12). This work is summarized in Figure 1. miRNA genes are present throughout mammalian genomes in introns, exons and intergenic regions with many miRNAs produced from clusters of coexpressed genes. Some miRNA genes are under control of the same Pol II promoters that drive expression of mRNAs and some have independent promoters. For example, intronic miRNA genes which comprise about half of known miRNA genes often depend on expression of the host gene. A few miRNA genes are also known to be transcribed by Pol III. Primary miRNA transcripts are processed by a nuclear ribonuclease RNase III (Drosha) and then exported to the cytoplasm where the mature miRNA is produced from ~70nt precursor miRNAs by RNase activity of Dicer (Figure 1). Dicer activity and the miRNA products are necessary for proper smooth muscle development, blood vessel formation and gastrointestinal development. Smooth-muscle restricted knockout of Dicer inhibits blood vessel maturation and intestinal tract development (13, 14). Mature miRNAs then complex with several proteins including Argonaute family members Ago-1 and Ago-2 in RNA-induced silencing complexes (RISC). RISCs mediate posttranscriptional silencing by several mechanisms. mRNA stability is reduced and/or translation is blocked depending on the degree of complementarity with the target sequence (Figure 1). mRNA is cleaved by the endonuclease activity of Ago-2 when complementarity is perfect, which is the mechanism of silencing by exogenous siRNAs. Further degradation of the cleaved transcript involves uridinylation, decapping and exonuclease activities. When complementarity is imperfect initiation can be blocked, premature termination and dissociation of ribosomes occurs followed by deadenylation, decapping and exonuclease degradation of mRNA. The net effect is RNA-induced gene silencing due to reduced translation of mRNA to proteins. In the sections below we review specific miRNAs for which some mechanistic information exists in smooth muscles. Our goal is to illustrate how miRNA-induced gene silencing might contribute to smooth muscle progenitor differentiation, smooth muscle restricted contractile protein expression, smooth muscle proliferation and proinflammatory mediator synthesis.
Figure 1. Biogenesis of micro RNA and Mechanism of Gene Silencing by micro RNA.
The outline of cell showing the transcription of primary micro RNA (Pri-miRNA) from miRNA gene by RNA polymerase II (Pol II), and its processing by Drosha (nuclear RNase III) in the nucleus. The Pri-miRNA is then exported to the cytoplasm by exportin via nuclear pore. In cytoplasm, Pri-miRNA is further processed by RNase activity of Dicer to mature micro RNA duplex. The duplex loads onto Ago in the RISC complex and separates. One of the mature miRNA strands (red strand) mediates small interfering RNA silencing by degrading the target mRNA or interfering with translational process. The outcome of RISC formation varies with the degree of complementarity of miRNA at 3’ untranslated regions (UTR) of the target mRNA.
MicroRNAs in smooth muscles
miRNAs in vascular smooth muscle plasticity
The literature on miRNA-mediated gene silencing in smooth muscles is expanding very rapidly with numerous recent studies of miRNAs in normal vascular development and in vascular pathologies. Table 1 summarizes factors regulating miRNA expression in smooth muscle, validated target proteins for those miRNAs and functions of the target proteins. These miRNAs are further classified into groups in table 2 to simplify the involvement of the miRNAs in determining the smooth muscle cell fate. Some of the earliest reports from Zhang and coworkers described the pro-proliferative and antiapoptotic effects of miR-21 in a carotid injury model in rats (15). miR-21 was the first miRNA shown to regulate vascular smooth muscle cell growth and survival by silencing expression of phosphatase and tensin homolog (PTEN) and increasing expression of B-cell leukemia/lymphoma 2 (BCL2) which increased proliferation and cell survival. Davis et al. (16) then described regulation of miR-21 processing by TGF-β1 and Smads in human pulmonary artery smooth muscle cells. Processing of the miR-21 primary transcript to the mature miRNA in pulmonary artery smooth muscle cells was enhanced by TGF-β1 and bone morphogenetic proteins (BMPs). BMP4 induced miR-21 which was then shown to upregulate smooth-muscle restricted contractile proteins by silencing programmed cell death 4 (PDCD4), a known tumor suppressor protein. This study is significant for two reasons. It was the first example of growth-factor regulation of miRNA processing in smooth muscle, and it suggested along with prior work on miR-21 by Zhang and coworkers (15) that the same miRNA (miR-21) can regulate features of the both contractile and proliferative phenotypes. Several other studies of miRNA modulation of the contractile phenotype followed in quick succession. Cordes et al. (17), in a landmark paper, described the master regulatory role of miR-143 and miR-145 in promoting contractile protein expression in vascular smooth muscle. There are now numerous studies confirming and extending the central regulatory role of the miR-143~145 cluster in vascular development (18–20), vascular damage response (21) and stem cell differentiation (22).
Table 1.
MiRNAs with Validated Targets in Smooth Muscles
| microRNA | Inducer / Regulator |
Target Proteins |
Cellular Functions of Target Proteins |
Physiology/ Pathology |
References |
|---|---|---|---|---|---|
| miR-1 | Myocardin | Pim-1 | Proliferation | Neointima | (45) |
| SRF | HDAC4 | Differentiation/myogenesis | formation | (79, 80) | |
| miR-10a | Retinoic acid | HDAC4 | Stem cell differentiation to smooth muscle |
Vascular development |
(50) |
| miR-21 | Vascular injury | PTEN, ↑Bcl2 |
Proliferation, apoptosis | Neointima formation |
(15) |
| TGF-β, BMPs | PDCD4 | Contractile protein synthesis |
(16) | ||
| miR-24 | PDGF-BB | Trb3 | Synthetic phenotype | Neointima formation |
(32) |
| miR-25 | IL-1β, TN-α, IFN-γ |
KLF4 | Contractile protein synthesis Matrix protein synthesis |
Airway remodeling |
(65) |
| miR-26a | Stretch C/EBPα |
GSK-3β | Hypertrophy | Airway remodeling |
(37) |
| Serum deprivation |
Smad1 Smad4 |
Proliferation | Aneurysm | (36) | |
| miR-133a | SRF | SRF | Suppress smooth muscle restricted gene expression |
(79, 81) | |
| Cyclin D2 | Myoblast proliferation | (80) | |||
| IL-13 | RhoA | Smooth Muscle Contraction |
Airway hyperreactivity |
(66, 82) | |
| miR-143 | p53 SRF/Myocardin |
PDGFR Elk-1 |
Podosome formation Differentiation |
Vascular remodeling |
(17, 18, 28) |
| miR- 143~145 |
SRF/Myocardin | Adducin3 SSh2 MRTF-B |
Cytoskeletal remodeling | (18) | |
| miR-145 | SRF/Myocardin | CamKII-δ KFL4 KLF5 |
Proliferation | Neointima formation |
(17) |
| PKC, Fascin |
Podosome formation | Remodeling | (28) | ||
| ACE | Contraction and proliferation |
Hypertension | (23) | ||
| Srgap2 | Cytoskeletal remodeling | (18) | |||
| OCT4 | KLF4 OCT4 SOX2 |
Stem cell differentiation to mesodermal cells |
Development | (22) | |
| p53 | c-Myc | Differentiation | Tumor suppression |
(40, 83) | |
| miR-146a | KLF5 | KLF4 | Proliferation | Neointima formation |
(27) |
| miR-204 | STAT3 | SHP2 | Proliferation | Pulmonary hypertension |
(56) |
| miR-221 | Injury, PDGF | p27, c-Kit | Proliferation | Neointima | (15, 33) |
Table 2.
Micro RNAs Regulating Smooth Muscle Cell Fate
| miRNA regulating SMC cell cycle | |||
|---|---|---|---|
| Proliferation | Apoptosis/Survival | Migration/Cytoskeletal | |
| miR-1 | miR-146a | miR-21 | miR-143~145 |
| miR-21 | miR-204 | ||
| miR-26a | miR-221 | ||
| miR-133a | |||
| miRNA regulating SMC phenotype | |||
| Contractile | Synthetic | Differentiation | |
| miR-1 | miR-24 | miR-10a | |
| miR-25 | miR-25 | miR-143~145 | |
| miR-133a | miR-26a | miR-155 | |
| miR-145 | |||
miRNAs in atherosclerosis and neointimal remodeling
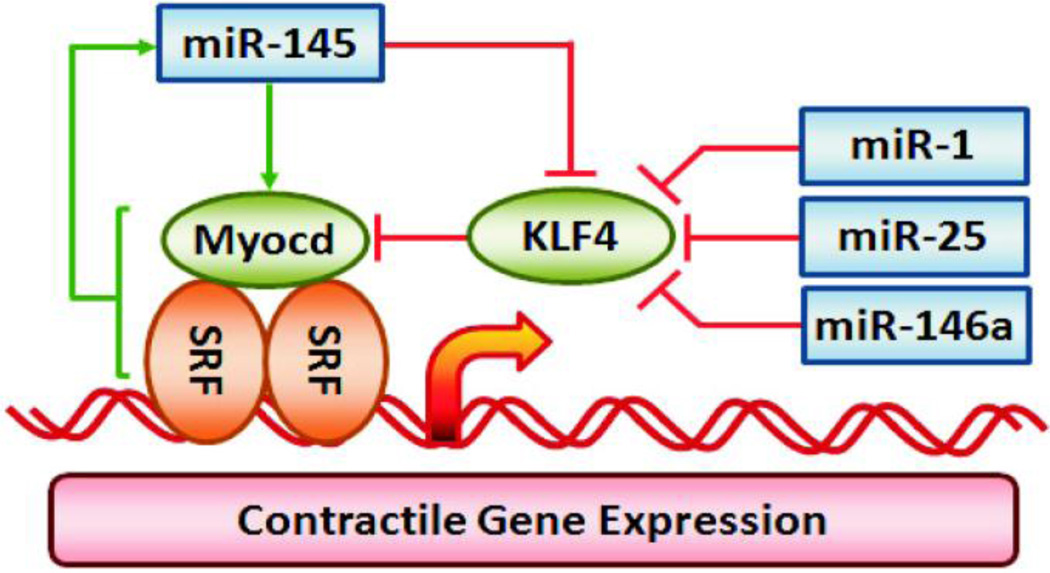
One of the earliest observations of changes in miRNA expression in blood vessels was that miR-145 is downregulated in neointimal lesions following vascular injury (21). The mechanism of miR-145 in regulating smooth muscle phenotype was subsequently defined by the elegant set of studies by Cordes et al. (17) mapping the pathway for reciprocal control of Kruppel-like factor 4 (KLF4) and myocardin expression by miR-145 as shown in Figure 2. Several knockout mouse studies have corroborated the initial observations and have verified the central role of miR-143~145 cluster in vascular smooth muscle contractile protein expression, vascular contractility and blood pressure regulation (18, 19, 23). The signaling scheme that is emerging includes a dominant effect of miR-145 to directly silence expression of KLF4 and an indirect upregulation of myocardin expression (Figure 2). Both events appear to contribute to TGF-β1 activation of serum response factor (SRF)-dependent smooth muscle restricted genes (24). The miR-143~145 cluster regulates a network of smooth muscle contractile, cytoskeletal and matrix protein genes with CArG boxes in the 5’ untranslated region (18, 25, 26). It is also clear that multiple miRNAs in addition to the miR-143~145 cluster can modulate KLF4 expression (Figure 2 ). miR-146a directly targets and silences KLF4 in vascular smooth muscle (27). A feedback loop was proposed that includes miR-146a silencing of KLF4 and KLF4 competing with KLF5 to reduce transcription of the miR-146a gene (Figure 3). This feedback loop is thought to be necessary for neointima formation by increasing KLF4 expression thus favoring smooth muscle proliferation and cell migration. The initial trigger for activating the miR-146a–KLF4/KLF5 pathway is not defined, but it would be an obvious target for reducing vascular remodeling during restenosis after angioplasty.
Figure 2. KLF4 and Myocardin Dependent Regulation of Smooth Muscle Contractile Gene Expression.
The signaling pathways illustrate miR-1, miR-25, miR-133a, miR-146a and miR-145 modulation of KLF and Myocardin dependent regulation of contractile gene expression. Red lines indicate silencing of protein expression or inhibition of miRNA expression by pathway components. Green arrows indicate activation or upregulation of the pathway component.
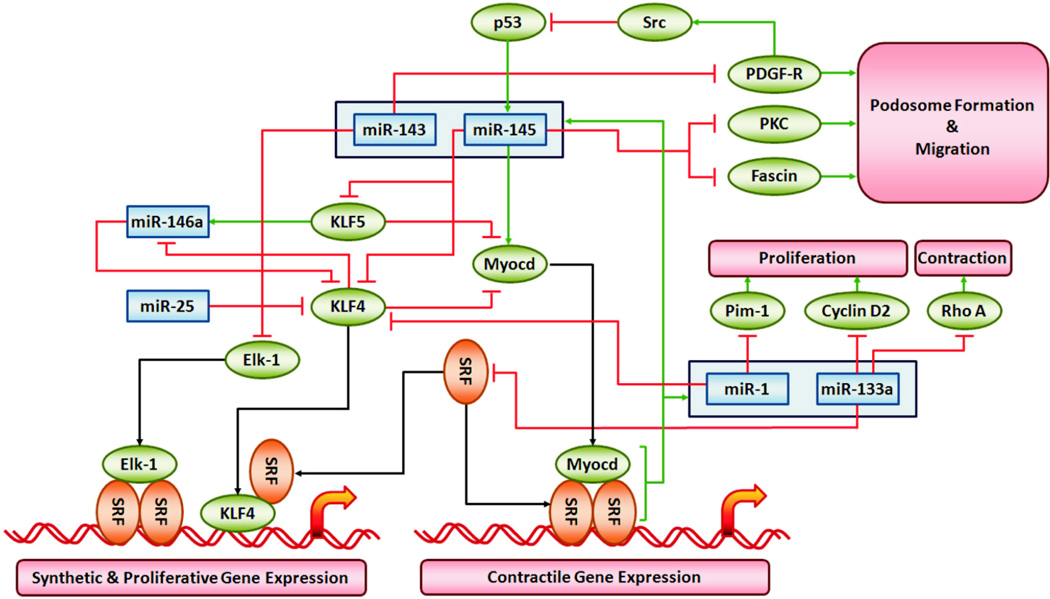
Figure 3. MiRNAs Regulating Smooth Muscle Phenotype via SRF-dependent Gene Expression.
The signaling pathways illustrate validated targets of miRNAs and miRNA families that control smooth muscle-restricted gene expression by serum response factor (SRF) and its co-regulators myocardin (Myocd), KLF4, KLF5 and Elk-1. See Table 1 for supporting references. Red lines indicate silencing of protein expression or inhibition of miRNA expression by pathway components. Green arrows indicate activation or upregulation of the pathway component. Black arrows illustrate known protein-protein interactions in the core SRF-dependent regulation of smooth muscle restricted gene expression program.
In addition to upregulating contractile protein expression in vascular smooth muscle the downregulation of miR-143~145 elicits upregulation of proteins important in podosome formation. Podosomes are local sites of matrix remodeling thought to be necessary for vascular wall remodeling. Quintavalle et al. (28) showed downregulation of miR-143~145 was sufficient to upregulate PDGF receptor, protein kinase C (PKC) epsilon and fascin, an actin bundling protein. The studies of miR-143~145 in atherosclerotic and neointimal vascular remodeling all point to a critical role of this miRNA cluster in repressing KLF4 expression, increasing myocardin expression and modulating a variety of other proteins that contribute to a more differentiated smooth muscle cell population (Figure 3).
Differentiated smooth muscle cells in the fibrous cap of atherosclerotic plaques provide important structural integrity necessary for plaque stability. Smooth muscle cells in plaques are thought to be heterogeneous, expressing a range of phenotypes (29). Smooth muscle cells in plaque secrete extracellular matrix (ECM) proteins. Plaque stability can be achieved not only by increasing the ECM production but also by decreasing the ECM degradation. ECM synthesis by smooth muscle cells is tightly regulated by various factors in the cellular environment, such as growth factors, cytokines, nitric oxide and surrounding ECM. Also, smooth muscle cell proliferation increases plaque stability, while smooth muscle cell apoptosis is thought to decrease plaque stability especially in the shoulder of the plaque. miRNA regulation of vascular smooth muscle phenotype may therefore be vital for plaque stability. A survey of circulating miRNAs in patients with stable coronary artery disease showed decreased levels of circulating miR-145 (30). Loss of miR-143~145 function leading to reduced contractile protein expression might reasonably contribute to the vascular damage response and possibly contribute to plaque instability (31). Further studies of RNA and protein expression of stable and unstable plaques would be needed to critically test this hypothesis. A predicted corollary would be delivery of miR-145 mimics should reduce plaque complexity, enhance plaque stability and reduce the incidence of acute cardiovascular events due to plaque rupture.
In addition to regulation of smooth muscle cell phenotype by the miR143~145 cluster, there are a variety of miRNAs that determine smooth muscle cell fate following injury and neointima formation. PDGF and TGF-β1 are important signaling proteins that contribute to the injury response. They do so in part by altering primary miRNA transcript expression and processing. PDGF-BB promotes the proliferative/migratory/ secretory phenotype. In contrast, TGF-β family proteins usually promote the contractile phenotype via Smad-dependent signaling. PDGF-BB was found to induce expression of miR-24 in human pulmonary artery smooth muscle cells (32). miR-24 was shown to directly bind to the 3’ untranslated region (UTR) of Tribbles-like protein 3 (Trb3) and to downregulate Trb3 expression. Downregulation of Trb3 decreased Smad1 levels, thus inhibiting TGF-β1 and BMP signaling. Forced expression of miR-24 reduced Smad2 and Smad3 as well as TGF-β-mediated activation of Smad2. Thus, miR-24 is a novel regulator of smooth muscle plasticity that mediates the well-known functional antagonism of PDGF-BB and the TGF-β family in determining vascular smooth muscle phenotype.
Activation of the PDGF signaling pathway in vascular smooth muscle also leads to upregulation of miR-221 which may contribute to neointimal proliferation (33). miR-221 upregulation has been implicated in a variety of cancers and is known to silence expression of the cell cycle inhibitor protein p27Kip1 during skeletal muscle differentiation (34). In cultured vascular smooth muscle cells miR-221 also downregulates expression of p27Kip1 thus increasing proliferation (33). miR-221 also downregulates expression of c-Kit, which was shown to be a positive regulator of myocardin and contractile protein expression. Regulation of cell cycle control proteins in smooth muscles by miR-221 was corroborated by Liu et al. (35) who reported that both miR-221 and miR-222 were induced by PDGF in a dose and time dependent manner which decreased p27Kip1 and p57Kip2 expression. miR-221 and miR-222, much like miR-21, are examples of miRNAs that are conserved in many cells and have consistent effects on expression of conserved components of cell cycle control machinery in vascular smooth muscle cells.
miRNAs in vascular development and smooth muscle differentiation
miR-26a
A survey of miRNA expression during differentiation of vascular smooth muscle cells identified several miRNAs that were upregulated following serum withdrawal (36). Pathway analysis of targets of 31 regulated miRNAs suggested mitogen activated protein (MAP) kinase signaling, actin cytoskeleton and focal adhesions, Wnt signaling and TGF-β signaling were all targets of multiple upregulated miRNAs. Gain-of-function and loss of function approaches showed miR-26a had a dedifferentiation effect mediated by silencing of Smad1 and by inhibiting TGF-β signaling. These results are paradoxical in that a previous study in airway smooth muscle found miR-26a was induced by stretch, that it silenced glycogen synthase kinase 3 and promoted airway smooth muscle hypertrophy in culture (37). These apparently disparate observations in vascular and airway smooth muscle might point up important tissue-specific differences in miRNA functions, or important differences in experimental conditions that result in opposing effects on differentiation. The apparent paradox is not unprecedented. miR-21 was reported by several groups to be pro-proliferative and anti-apoptic in vascular smooth muscles (15, 38), yet miR-21 can also promote TGF-β-family induction of contractile protein expression by silencing PDCD4 expression (16). Further studies of miRNAs in multiple smooth muscles under growth conditions vs differentiation conditions is warranted to explore this interesting paradox.
miR-143~145 cluster
During muscle development progenitor cells typically differentiate from a pluripotent state to a more differentiated state. The miRNAs that modify the various differentiation events in cardiac, skeletal and smooth muscles are the subject of intense interest because of the fundamental biological significance and the potential for identifying novel targets to manipulate muscle tissue remodeling. One of the key miRNAs in smooth muscle development and differentiation, miR-145, also has an important role in cell fate determination early in embryonic development. miR-145 triggers fate decision in pluripotent stem cells by silencing several key transcription factors and transcriptional coregulators including c-Myc, Sox2, Oct4 and KLF4 (39, 40). In mature tissues miR-145 frequently acts as a tumor suppressor. Downregulation of expression promotes the most common solid tumors (breast, bladder, lung and colon). Therefore, in addition to promoting the contractile phenotype of smooth muscles, miR-145 also promotes stem cell differentiation and suppresses tumor formation by silencing gene expression networks in many cell types. It is important to note KLF4 is probably a major effector molecule for the differentiation and tumor suppressive properties of miR-145 (Figure 3). KLF4 is a validated target of miR-145 with significant effects on gene expression profiles in stem cells and in tumor cells. For these reasons miR-145 is the subject of intense investigation in a variety of cardiovascular disorders, lung diseases gastrointestinal disorders and neoplastic diseases. Of the miRNAs discussed in this review the miR-143~145 cluster could be considered master regulators of smooth muscle differentiation.
miR-155
Differentiation of precursor cells into mature smooth muscle cells is a fundamental process during organ development that also contributes to development of vascular diseases. Much of the recent interest in miRNAs in smooth muscle is stimulated by insufficient information about how precursor cells differentiate to mature smooth muscle cells. Some of the earliest work on this issue was a study of miR-155 on angiotensin receptor (AT1R) expression and signaling (41). A polymorphism in the 3’UTR of the human AT1R gene, which is clearly linked to cardiovascular disease, disrupts miR-155 silencing of the AT1R receptor. The resulting upregulation of AT1R signaling is thought to contribute to development of hypertension, cardiac hypertrophy and myocardial infarction. Regulation of smooth muscle function by miR-155 may be common to multiple smooth muscle tissues because Martin et al. (41) found miR-155 expressed in both vascular and airway smooth muscle by in situ hybridization. In addition to regulating AT1R expression miR-155 has been shown regulate genes necessary for differentiation of stem cells to smooth muscles. Using two independent protocols for smooth muscle cell differentiation Danielson et al. (42) showed differentiation of mature smooth muscle cells from bone-marrow derived mesenchymal stem cells depended on mature miRNA expression. They identified sets of miRNAs that increased or decreased monotonically during mesenchymal stem cell to smooth muscle cell differentiation. miR-155 was downregulated during differentiation, which was necessary to generate differentiated smooth muscle cells. Exogenous overexpression of miR-155 inhibited expression of smooth muscle myosin II heavy chain and prevented maturation of differentiated smooth muscle cells. Additionally, Zheng et al. (43) reported that miR-155 regulates the differentiation of aortic adventitial fibroblasts to myofibroblasts. Overexpression of miR-155 inhibited AT1R signaling, reduced smooth muscle α-actin expression and inhibited differentiation consistent with the earlier report of Martin et al. (41). Altogether these results suggested that downregulation of miR-155 expression might contribute significantly to cardiovascular diseases by permitting increased AT1R signaling in fibroblasts and smooth muscle cells in vivo.
miR-1~133a cluster
The miR-1~miR-133 family is another group of miRNAs of great interest in smooth muscle differentiation and hypertrophy. These miRNAs have been studied primarily in cardiac and skeletal muscle development as silencers of smooth muscle-restricted gene expression. Recently, Jiang et al. (44) found that overexpression of myocardin in human aortic smooth muscle cell increased both smooth muscle cell contractility and the expression of miR-1. However, exogenous miR-1 mimetic inhibited smooth muscle contractility and expression of smooth muscle contractile proteins (SM22 and smooth muscle α-actin) basally and in response to myocardin expression. Antisense inhibition of endogenous miR-1 enhanced contractility and increased contractile protein expression. miR-1 expression was found to have no effect on either myocardin or SRF. In a subsequent study, the same group reported that overexpression of myocardin in human aortic smooth muscle cell increased expression of miR-1 and decreased smooth muscle cell proliferation (45). Overexpression of myocardin decreased the proliferation of smooth muscle cells which was reversed by an antisense miR-1 inhibitor. Exogenous miR-1 mimetic inhibited proliferation and negatively regulated expression of a serine/threonine kinase, Pim-1, but not other miR-1 target genes [histone deacetylase 4 (HDAC4), heart and neural crest derivatives expressed 2 (Hand2), and Ras homolog enriched in brain (Rheb)]. Neointimal lesions following carotid artery ligation showed decreased expression of myocardin and miR-1 and upregulation of Pim-1 suggesting reduced miR-1 expression is a contributing factor in vascular remodeling after injury. In addition to regulating the phenotype of adult smooth muscle cells a recent study of embryonic stem cell differentiation showed miR-1 promotes smooth muscle cell differentiation by directly targeting KLF4 3’UTR, silencing KLF4 protein expression and enhancing expression of smooth muscle-restricted contractile proteins (46). Current data suggests a signaling loop in which myocardin enhances contractile protein expression and miR-1 expression. miR-1 feeds back to represses KLF4 thus reducing proliferation, increasing contractile protein expression and promoting smooth muscle cell differentiation (Figure 3). Reduced miR-1 expression during vascular injury may promote smooth muscle cell proliferation and neointimal thickening.
miR-10a
A variety of multipotent cells, including embryonic stem cells, can differentiate to smooth muscle cells in culture (47–49). One of the factors that induces multipotent progenitors and pluripotent stem cells to express smooth-muscle restricted proteins is all-trans retinoic acid, a vitamin A metabolite important in differentiation and development of a variety of tissues and organs. In addition to regulating a wide range of protein coding genes retinoic acid also regulates expression of miRNAs that influence smooth muscle differentiation. Huang et al. (50) found that expression of miR-10a was upregulated during retinoic acid-mediated differentiation of mouse embryonic stem cells (ESC). miR-10a negatively regulated HDAC4 which was shown by others to regulate expression of smooth muscle restricted genes (51) and to mediate PDGF-induced proliferation (52). The role of miR-10a in silencing HDAC4 expression in the setting of mouse ESC differentiation appears to be consistent with reduced HDAC4 levels resulting in derepression of smooth muscle contractile proteins and differentiation to a contractile phenotype (51). The role of miRNAs in stem cell and vascular wall progenitor cell differentiation has profound implications for pathogenesis of atherosclerosis, the response to vascular injury and vascular remodeling in hypertension syndromes.
miRNA and pulmonary hypertension
Several of the miRNAs with conserved functions described in smooth muscle cells (eg. miR-21 and miR-221) have also been assigned roles in differentiation, proliferation and survival of endothelial cells and other vascular mural cells (53). miRNAs that participate in vascular remodeling have recently become subjects of intense interest. The initial studies of miRNAs in the cardiovascular system suggested several obvious targets for RNAi-based antagonism of remodeling including miR-21, miR-145 and miR-221 (54). However, until very recently it was not known which miRNAs were relevant to pulmonary arterial hypertension (PAH) where arterial muscularization occurs and irreversible occlusive lesions develop. These remodeling events are thought to contribute to lack of response to vasodilators and inevitable right heart failure and death. Caruso et al. (55) surveyed miRNA expression in total lung extracts from rat models (chronic hypoxia and monocrotaline model) of PAH and found downregulation of miR-21 to be prominent in both models. Courboulin et al. (56) later found miR-204 was also downregulated in human as well as rat models of PAH, and that delivery of miR-204 to rat lungs would reduce the severity of the disease. In addition, they found that in mononuclear cells isolated from the buffy coat of blood from PAH patients the expression of miR-204 were similarly downregulated compared to controls. This suggests miR-204 in peripheral blood mononuclear cells may be as a useful biomarker of PAH pathogenesis. Potential targets for miR-204 were investigated because Stat3 activation was increased upon attenuation of miR-204 expression. It was found that miR-204 directly regulates SHP2 by targeting its 3’UTR. Therefore, decreased miR-204 increases expression of SHP2, which by activating Src increases Stat3 activation contributing to smooth muscle proliferation and pulmonary vessel wall thickening. In addition, ROCK1 was shown to be silenced by miR-204, probably by an indirect effect although the exact mechanism was not defined nor was it the primary point of the study. The study by Courboulin et al. (56) provides solid proof of principle that “rescue” of low miRNA expression can prevent progression of established PAH. It also supports prior suggestions that RNAi-based therapy might be useful in treating several diseases involving vascular remodeling including atherosclerosis and restenosis injuries (57–59).
The study by Courboulin et al. (56) is unique in that miR-204 has not been implicated previously in vascular disease or myogenesis. Given the significant reversal of pulmonary vascular remodeling in vivo with miR-204 mimetic therapy, it is tempting to speculate that expression of miR-204 promotes differentiated vascular smooth muscle cells and downregulation of miR-204 might accompany other diseases involving vascular remodeling including atherosclerosis and restenosis. In addition, several targets of miR-204 previously validated in other cell types have profound roles in smooth muscle cell physiology and pathophysiology. Examples include: TGF-β receptor 2 (60), epidermal growth factor (EGF) receptor signaling (61), forkhead box C1 (FOXC1) (62), and runt-related transcription factor 2 (Runx2) (63). The regulation of smooth muscle phenotype by miR-204 should be explored further to establish the significance of these putative target proteins in smooth muscle development and disease.
miRNA modulation of pro-inflammatory signaling cascades in airway smooth muscle
Airway smooth muscle shares with vascular smooth muscle the ability to adapt to pathological mechanical and soluble signals by undergoing hyperplasia, hypertrophy, cell migration and increased synthetic activity. These processes may be especially important in severe asthmatics where the airway wall thickens markedly and is thought to be a factor in airway hyperreactivity (64). Inflammation is a major feature of asthma and a number of pro-inflammatory signaling cascades are involved in regulating the pro-inflammatory gene expression as well as miRNA gene expression (Figure 4). Some of the key pro-inflammatory mediators that contribute to asthma are interleukin-13 (IL-13), interferon-γ (IFN-γ), tumor necrosis factor-a (TNF-α) and interleukin-1β (IL-1β). Recent studies of miRNAs regulated by these inflammatory mediators and by mechanical signaling illustrate an important role of miRNAs in phenotypic plasticity of airway smooth muscle. Singer and associates first described a general inhibition of miRNA expression in cultured human airway smooth muscle cells (ASMC) when treated with a cytokine cocktail consisting of TNFα, IL-1β, and IFN-γ (65). The results suggest the well-known enhancement of gene expression in airway smooth muscle by inflammation may be due in part to a general inhibition of normal gene silencing mechanisms. Another novel finding was that the rarely reported miR-25 was shown to enhance smooth muscle contractile protein expression (65). Informatics analysis suggested, and biochemical studies confirmed, this uncommon miRNA directly targets the 3’UTR of KLF4. Transfection of miR-25 mimic reduced expression of KLF4 which was then verified as a negative regulator of myosin II heavy chain expression in airway smooth muscle (Figure 2 and 3).
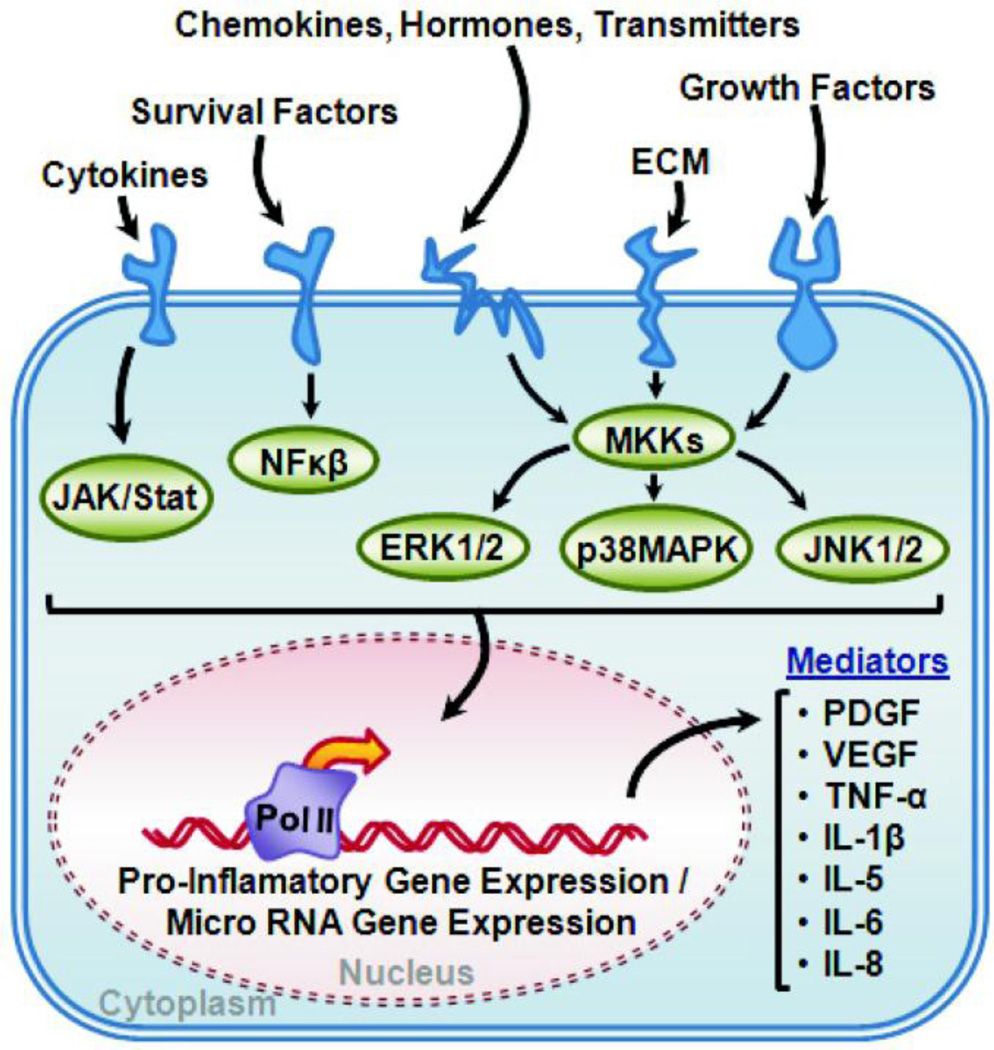
Figure 4. Pro-Inflammatory Signaling Pathway.
The schematic shows common upstream signaling mediators and the major signaling pathways transducing pro-inflammatory signals in smooth muscles Conserved kinase cascades (JAK/Stat, NFκβ, ERK1/2, p38MAPK, and JNK1/2) have been described in all smooth muscles that regulate pro-inflammatory as well as miRNA gene expression. Some important pro-inflammatory gene products are listed as autocrine and paracrine mediators of vascular and airway inflammation and remodeling.
Cytokine treatment was also found to alter expression of miR-133a in airway smooth muscle by Chiba et al. (66). miR-133a expression was reduced by treatment of human ASMCs with IL-13, and this coincided with an increase in RhoA gene expression (66). IL-13 appeared in bronchoalveolar lavage fluid of ovalbumin challenged mice and miR-133a levels decreased in bronchial smooth muscle coincident with increased RhoA mRNA (66). Isolated bronchial smooth muscle from naive mice exhibited enhanced contractility when treated with IL-13 in organ culture, possibly due to downregulation of miR-133a and increased RhoA signaling which is known to enhance calcium sensitivity of airway smooth muscle. miR-133a may normally repress expression of RhoA in airway smooth muscle, and perhaps in other types of smooth muscles. This repression may be disrupted by elevated IL-13 levels in allergic asthma leading to increased smooth muscle contraction and airways hyperreactivity.
The targets and functions of miR-145 have been extensively characterized in vascular smooth muscle, but its functions in the airway have largely been extrapolated from the cardiovascular literature. In a study utilizing a house dust mite model of acute allergic asthma in mice, Foster and coworkers observed that miR-145 levels increased in the larger airways (minus parenchyma) after repeated challenge (67). In a separate study using a chronic ovalbumin model, Foster and coworkers also observed an increased in miR-145 that peaked by the second week of challenge (68). miR-145 was also observed to be increased by Colige and co-workers in mice using a chronic ovalbumin model (69). Through an unknown mechanism, inhibition of miR-145 with a 2’-O-methyl phosphoroamidite modified antagomir prevented the development of allergic airways disease after house dust mite sensitization. Multiple aspects of the asthmatic phenotype were inhibited including TH2 cytokine production, mucus hypersecretion, and airway hyperresponsiveness (67). The anti-inflammatory efficacy of the miR-145 antagomir was found to be equivalent to the effects of dexamethasone. Part of the asthmatic allergic response is also likely to be due to production of interferons. A recent study has identified that both interferon-γ and interferon-β increased miR-145 and a-actin expression in human airway smooth muscle cells (70). Altogether these studies provide a plausible link between cytokine and interferon upregulation in allergic asthma, induction of miR-145 in airway smooth muscle and enhanced contractility.
In future, studies investigating the effects of miR-145 inhibition and airway remodeling are warranted due to the current lack of anti-remodeling drugs. A recent study showing that repeated bronchoconstriction alone without inflammation is sufficient to elicit airway remodeling in humans (71) suggests that miRNA or siRNA targeting contractile and cytoskeletal proteins may be useful for antagonizing mechanical signals that stimulate ASMC hypertrophy. RNAi-based treatments that inhibit both smooth muscle contraction and smooth muscle hypertrophy are very appealing, especially for therapy of severe asthmatics who are corticosteroid-resistant.
Another appealing therapeutic strategy is to inhibit synthesis of inflammatory mediators in multiple cell types in the lung, a strategy that is usually successful when inhaled glucocorticoids are used to treat asthma. miR-146a is a well-defined anti-inflammatory miRNA with profound effects on blunting the innate immune response. Since airway smooth muscle cells secrete many protein mediators involved in innate immunity a role of miR-146a was investigated by Lindsay and coworkers in airway smooth muscle (72). Based upon their previous work with miR-146a in alveolar epithelial cells and the published data on antagonism of Toll-receptor signaling it was reasonable to surmise miR-146a might inhibit expression of cytokine and chemokine synthesis in airway smooth muscle cells. IL-1β treatment of airway smooth muscle induced the expression of miR-146a, but this effect differed in magnitude from observations in other cell types (72). Pri-miR-146a post-transcriptional processing was found to be regulated by MEK-1/2 and JNK-1/2 and expression of Pri-miR-146a was activated by NFκβ, which was a novel observation in regulation of miRNA biogenesis. miR-146a negatively regulated IL-6 and IL-8 release in human airway smooth muscle cells in culture, and this was confirmed using miR-146a mimics at concentrations that achieved 3000-fold more miR-146a compared to the 20–50 fold increase that was observed with IL-1β stimulation (72). The investigators attributed this effect on secretion to be a false positive that occurred due to “supra-maximal levels” of miR-146a. Both the miR-146a inhibitor and the nonsilencing control inhibitor caused a decrease in IL-6 secretion at high concentrations, which the authors dismissed as non-specific disruption of miRNA silencing. This interpretation is supported by a previous report showing concentration- and time-dependent saturation of RISC complexes occur when exogenous small RNAs are transfected into cells (73). Also, the false positive effect on secretion was not due to the down regulation of interleukin-1 receptor-associated kinase 1 (IRAK-1) and TNF receptor-associated factor 6 (TRAF-6) expression by miR-146a. All of these findings led to the conclusion that the mechanism of miR-146a function is cell-type dependent. The study also illustrates an important limitation of gain-of-function studies using high concentrations of siRNA or miRNA mimics.
Mechanotransduction Signaling in Airway Smooth Muscle
Because smooth muscle cells are mechanosensitive cells it is reasonable to predict expression and function of miRNAs in these cells might respond to mechanical signals and modify cell and tissue mechanics. In a survey of human airway smooth muscle cells a small number of miRNAs were found to be mechanosensitive (37). Cyclic stretch of cultured human airway smooth muscle cells identified miR-16, miR-26a, and miR-140 as mechanosensitive molecules. These three miRNA were investigated further as possible contributors to stretch-induced hypertrophy and hyperplasia in airway smooth muscle. Only miR-26a was identified as contributing to hypertrophy and none of the miRNAs contributed to hyperplasia. Stretch was found to directly increase miR-26a expression through the transcription factor, CCAAT enhancer-binding protein α(C/EBPα). miR-26a silenced the expression of glycogen synthase kinse-3β (GSK-3β), which can promote both inflammation and hypertrophy by pleiotropic effects on substrates that regulate metabolism, structural proteins and transcription. Overexpression of pre-miR-26a induced hypertrophy independent of stretch. Antagonism of miR-26a with antagomirs or knockdown of C/EBPα with siRNA prevented the development of stretch induced hypertrophy (37). Overexpression of miR-26a was also sufficient to increase the expression of α-actin, SM22, and myosin II heavy chain, but this effect could be an indirect result of a global increase in translation. This pioneering study is the first to our knowledge that investigates stretch-mediated activation of miRNA expression in smooth muscle and it clearly demonstrates the link between stretch, miRNAs and hypertrophy in a mechanically active tissue. It does raise an interesting question of cell-type or culture-condition dependence of the action of miR-26a, which as discussed above has anti-differentiation effects in vascular smooth muscle (36) (see Table 1). It will be important to test for mechanosensitivity of miR-26a in vascular and visceral smooth muscles to test for tissue-dependent differences in miRNA function.
Expression Surveys in Visceral and Urogenital Smooth Muscles
Several recent surveys of miRNA expression in nonvascular smooth muscle cells suggest miRs that are functionally significant in vascular smooth muscles are also necessary for normal development and function in visceral smooth muscles. In gastrointestinal smooth muscles a Dicer knockout mouse model demonstrated that, as during vascular development (13), proper synthesis of miRNA is required for normal intestinal smooth muscle development (14). A deep sequencing analysis of miRNAs expressed in differentiated intestinal smooth muscle compared to a smooth muscle cell line (PAC1) revealed many of the same critical miRNAs described above in vascular and airway smooth muscle phenotype determination e.g., miR-1, miR-133a, miR-24, miR-26a and the miR-143~145 cluster. Park et al. (14) described a novel role of miR-199a and miR-214 in promoting smooth muscle proliferation that has not yet been reported in other smooth muscles. This may be due to a unique role for these miRNAs in intestinal smooth muscle phenotype.
Uterine smooth muscle is a highly dynamic muscle that remodels dramatically during gestation, immediately prior to partuition and post-delivery. It is not surprising that dynamic changes in miRNA expression would occur in uterine smooth muscle cells and tissues in response to changes in hormone status. Estrogen reduces expression of miR-21 in myometrial cells (74), which may reduce proliferation and enhance survival by silencing PTEN and upregulating BCL2 (15), or it may modulate contractile protein expression by silencing PDCD4, a transcriptional co-regulator of SRF gene expression (33). These competing possibilities, which have profoundly different functional outcomes, point out the challenge of defining the functional significance of individual miRNAs in a dynamically remodeling tissue like the uterus. Significantly more functional analysis of miRNAs in intestinal and uterine smooth muscles is required before a clear set of principles or tissue-restricted patterns of regulation will become apparent. The results of such studies will be important for translational research in organ and tissue-selective RNAi therapies of intestinal and uterine motility disorders and leiomyomas arising in these organs.
Conclusion and Future Directions
Conducting miRNA expression surveys in disease models involving remodeling of smooth muscles is an approach that has yielded important insights into disease mechanisms (summarized in Table 1). It has also identified some novel targets for future drug therapy. Several conserved miRNAs and functions have been described in smooth muscles that were previously described in the cancer literature–for example miR-21 and miR-221/222 in proliferation and cell survival. Other miRNAs are expressed in striated and smooth muscles and have important effects on muscle cell differentiation (eg. miR-1 and miR-133a). In contrast, the anti-inflammatory miR-146a appears to not have the same profound silencing effect on Toll receptor signaling that has in immune cells and airway epithelial cells. Some miRNAs appear to have special significance in smooth muscle and some novel insights into differentiation mechanisms were driven by investigations of vascular and airway remodeling in disease models and in humans. The central role of the miR-143~145 cluster in smooth muscle development and differentiation is a good example, as is the role of miR-25 in control of contractile protein expression in airway smooth muscle. There is need for further investigation of how smooth-muscle regulating miRNAs can control a set of highly smooth-muscle restricted genes and yet in other settings act as tumor suppressors and regulators of pluripotency.
Several important questions arise from the current state of understanding of miRNAs in smooth muscles. Do smooth muscle cells possess a particular set of epigenetic conditions or factors such as DNA methylation patterns in promoters or histone modifications that prime them to respond to expression of the miR-143~145 cluster in the characteristic manner of differentiated smooth muscles? Would additional measurements of miRNA expression and miRNA processing during development and disease pathogenesis in more smooth muscle tissues identify new candidate molecules for inhibiting pathological smooth muscle remodeling? Would RNA mimics or antagonists be effective in vivo? To answer the latter question novel delivery methods of RNA-based drugs in humans would need to be developed.
In principle RNAi, either antisense oligonucleotides or modified miRNA, is effective “therapy” in animal models and is on the verge of use in humans. One of the earliest examples of RNAi therapy was intranasal delivery of antisense oligonucleotides against a viral protein to the lungs of mice inhibited respiratory virus replication (75, 76). RNAi therapy can be scaled up to primates as shown by use of locked nucleic acid modified miR-122 administered intravenous to green monkeys to inhibit cholesterol synthesis (77). Recently RNAi therapies targeting smooth muscle remodeling have been found to be effective in several diseases. Pulmonary hypertension and asthma in animal models are both responsive to lung-restricted delivery of RNAi drugs that rescue (56), or antagonize (68) miRNAs altered by the disease. There is also hope that atherosclerotic plaque stability might be susceptible to manipulation via systemic delivery of RNAi-based drugs (31, 58, 78). Although there are still major issues of drug delivery, metabolism and bioavailability to be addressed, defining the function of specific miRNAs in smooth muscle cell phenotypes is an important first step towards identifying novel targets and developing novel treatments to inhibit pathological smooth muscle remodeling.
Acknowledgements
Supported in part by NIH grants HL077726 and HL092270 to WTG. This article has an overlap with prior review by Joshi et al (84). Figures 1 & 2 and Table 2 are reproduced from a previous review of miRNAs in pulmonary hypertension (84); authors own copyright, permission to reproduce was not required.
Footnotes
Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Richter A, Puddicombe SM, Lordan JL, et al. The contribution of interleukin (IL)-4 and IL-13 to the epithelial-mesenchymal trophic unit in asthma. Am J Respir Cell Mol Biol. 2001;25:385–391. doi: 10.1165/ajrcmb.25.3.4437. [DOI] [PubMed] [Google Scholar]
- 2.Wicks J, Haitchi HM, Holgate ST, Davies DE, Powell RM. Enhanced upregulation of smooth muscle related transcripts by TGF beta2 in asthmatic (myo) fibroblasts. Thorax. 2006;61:313–319. doi: 10.1136/thx.2005.050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finotto S, Hausding M, Doganci A, et al. Asthmatic changes in mice lacking T-bet are mediated by IL-13. Int Immunol. 2005;17:993–1007. doi: 10.1093/intimm/dxh281. [DOI] [PubMed] [Google Scholar]
- 4.Bentley JK, Hershenson MB. Airway smooth muscle growth in asthma: proliferation, hypertrophy, and migration. Proc Am Thorac Soc. 2008;5:89–96. doi: 10.1513/pats.200705-063VS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 6.Majesky MW, Dong XR, Regan JN, Hoglund VJ. Vascular smooth muscle progenitor cells: building and repairing blood vessels. Circ Res. 2011;108:365–377. doi: 10.1161/CIRCRESAHA.110.223800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clifford RL, Coward WR, Knox AJ, John AE. Transcriptional regulation of inflammatory genes associated with severe asthma. Curr Pharm Des. 2011;17:653–666. doi: 10.2174/138161211795429000. [DOI] [PubMed] [Google Scholar]
- 8.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 9.Larsson E, McLean SE, Mecham RP, Lindahl P, Nelander S. Do two mutually exclusive gene modules define the phenotypic diversity of mammalian smooth muscle? Mol Genet Genomics. 2008;280:127–137. doi: 10.1007/s00438-008-0349-y. [DOI] [PubMed] [Google Scholar]
- 10.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 11.Eddinger TJ, Meer DP. Myosin II isoforms in smooth muscle: heterogeneity and function. Am J Physiol Cell Physiol. 2007;293:C493–C508. doi: 10.1152/ajpcell.00131.2007. [DOI] [PubMed] [Google Scholar]
- 12.Ku G, McManus MT. Behind the scenes of a small RNA gene-silencing pathway. Hum Gene Ther. 2008;19:17–26. doi: 10.1089/hum.2007.1226. [DOI] [PubMed] [Google Scholar]
- 13.Albinsson S, Skoura A, Yu J, et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One. 2011;6:e18869. doi: 10.1371/journal.pone.0018869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park C, Yan W, Ward SM, et al. MicroRNAs dynamically remodel gastrointestinal smooth muscle cells. PLoS One. 2011;6:e18628. doi: 10.1371/journal.pone.0018628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 16.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia L, Quintavalle M, Zhang J, et al. The knockout of miR-143 and −145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucher JM, Peterson SM, Urs S, Zhang C, Liaw L. The mir-143/145 cluster is a novel transcriptional target of jagged-1/notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286:28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Liu X, Yang J, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 23.Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis-Dusenbery BN, Chan MC, Reno KE, et al. Downregulation of KLF4 by MIR-143/145 is critical for modulation of vascular smooth muscle cell phenotype by TGF-{beta} and BMP. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C, Hennig GW, Sanders KM, et al. SRF-Dependent microRNAs Regulate Gastrointestinal Smooth Muscle Cell Phenotypes. Gastroenterology. 2011;141:164–175. doi: 10.1053/j.gastro.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long X, Miano JM. TGF{beta}1 utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286:30119–30129. doi: 10.1074/jbc.M111.258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun SG, Zheng B, Han M, et al. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J Cell Biol. 2010;189:13–22. doi: 10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao H, Gabbiani G, Bochaton-Piallat ML. Arterial smooth muscle cell heterogeneity: implications for atherosclerosis and restenosis development. Arterioscler Thromb Vasc Biol. 2003;23:1510–1520. doi: 10.1161/01.ATV.0000090130.85752.ED. [DOI] [PubMed] [Google Scholar]
- 30.Fichtlscherer S, De RS, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 31.O'Sullivan JF, Martin K, Caplice NM. Microribonucleic acids for prevention of plaque rupture and in-stent restenosis: “a finger in the dam”. J Am Coll Cardiol. 2011;57:383–389. doi: 10.1016/j.jacc.2010.09.029. [DOI] [PubMed] [Google Scholar]
- 32.Chan MC, Hilyard AC, Wu C, et al. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardinali B, Castellani L, Fasanaro P, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leeper NJ, Raiesdana A, Kojima Y, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J Biol Chem. 2010;285:29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299:L861–L871. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin MM, Buckenberger JA, Jiang J, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Danielson LS, Menendez S, Attolini CS, et al. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol. 2010;177:908–917. doi: 10.2353/ajpath.2010.091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L, Xu CC, Chen WD, et al. MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem Biophys Res Commun. 2010;400:483–488. doi: 10.1016/j.bbrc.2010.08.067. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y, Yin H, Zheng XL. MicroRNA-1 inhibits myocardin-induced contractility of human vascular smooth muscle cells. J Cell Physiol. 2010;225:506–511. doi: 10.1002/jcp.22230. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Yin H, Jiang Y, et al. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol. 2011;31:368–375. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie C, Huang H, Sun X, et al. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20:205–210. doi: 10.1089/scd.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drab M, Haller H, Bychkov R, et al. From totipotent embryonic stem cells to spontaneously contracting smooth muscle cells: a retinoic acid and db-cAMP in vitro differentiation model. FASEB J. 1997;11:905–915. doi: 10.1096/fasebj.11.11.9285489. [DOI] [PubMed] [Google Scholar]
- 48.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 49.Xie CQ, Huang H, Wei S, et al. A comparison of murine smooth muscle cells generated from embryonic versus induced pluripotent stem cells. Stem Cells Dev. 2009;18:741–748. doi: 10.1089/scd.2008.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 2010;285:9383–9389. doi: 10.1074/jbc.M109.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellis JJ, Valencia TG, Zeng H, Roberts LD, Deaton RA, Grant SR. CaM kinase IIdeltaC phosphorylation of 14–3–3beta in vascular smooth muscle cells: activation of class II HDAC repression. Mol Cell Biochem. 2003;242:153–161. [PubMed] [Google Scholar]
- 52.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. Am J Physiol Cell Physiol. 2007;292:C886–C895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 53.Ohtani K, Dimmeler S. Control of cardiovascular differentiation by microRNAs. Basic Res Cardiol. 2011;106:5–11. doi: 10.1007/s00395-010-0139-7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang C. MicroRNA-145 in vascular smooth muscle cell biology: a new therapeutic target for vascular disease. Cell Cycle. 2009;8:3469–3473. doi: 10.4161/cc.8.21.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caruso P, MacLean MR, Khanin R, et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 56.Courboulin A, Paulin R, Giguere NJ, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albinsson S, Sessa WC. Can microRNAs control vascular smooth muscle phenotypic modulation and the response to injury? Physiol Genomics. 2011;43:529–533. doi: 10.1152/physiolgenomics.00146.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C. MicroRNA and vascular smooth muscle cell phenotype: new therapy for atherosclerosis? Genome Med. 2009;1:85. doi: 10.1186/gm85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19(3):205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang FE, Zhang C, Maminishkis A, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2011;130:1036–1045. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosse Y, Pare PD, Seow CY. Airway wall remodeling in asthma: from the epithelial layer to the adventitia. Curr Allergy Asthma Rep. 2008;8:357–366. doi: 10.1007/s11882-008-0056-0. [DOI] [PubMed] [Google Scholar]
- 65.Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–513. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- 67.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128:160–176. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 68.Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garbacki N, Di VE, Huynh-Thu VA, et al. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One. 2011;6:e16509. doi: 10.1371/journal.pone.0016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goncharova EA, Lim PN, Chisolm A, et al. Interferons modulate mitogen-induced protein synthesis in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2010;299:L25–L35. doi: 10.1152/ajplung.00228.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grainge CL, Lau LC, Ward JA, et al. Effect of bronchoconstriction on airway remodeling in asthma. N Engl J Med. 2011;364:2006–2015. doi: 10.1056/NEJMoa1014350. [DOI] [PubMed] [Google Scholar]
- 72.Larner-Svensson HM, Williams AE, Tsitsiou E, et al. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respir Res. 2010;11:68. doi: 10.1186/1465-9921-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12:227–240. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 76.Bitko V, Barik S. Nasal delivery of siRNA. Methods Mol Biol. 2008;442:75–82. doi: 10.1007/978-1-59745-191-8_6. [DOI] [PubMed] [Google Scholar]
- 77.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 78.Jamaluddin MS, Weakley SM, Zhang L, et al. miRNAs: roles and clinical applications in vascular disease. Expert Rev Mol Diagn. 2011;11:79–89. doi: 10.1586/erm.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X, Azhar G, Helms SA, Wei JY. Regulation of cardiac microRNAs by serum response factor. J Biomed Sci. 2011;18:15. doi: 10.1186/1423-0127-18-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiba Y, Misawa M. MicroRNAs and their therapeutic potential for human diseases: MiR-133a and bronchial smooth muscle hyperresponsiveness in asthma. J Pharmacol Sci. 2010;114:264–268. doi: 10.1254/jphs.10r10fm. [DOI] [PubMed] [Google Scholar]
- 83.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joshi SR, McLendon JM, Comer BS, Gerthoffer WT. MicroRNAs-control of essential genes: Implications for pulmonary vascular disease. Pulm Circ. 2011;1:357–364. doi: 10.4103/2045-8932.87301. [DOI] [PMC free article] [PubMed] [Google Scholar]