Abstract
Cell migration plays a pivotal role in many physiologically important processes such as embryogenesis, wound-healing, immune defense, and cancer metastasis. Although much effort has been directed toward motility of individual cells, the mechanisms underpinning collective cell migration remain poorly understood. Here we develop a collective motility model that incorporates cell mechanics and persistent random motions of individual cells to study coherent migratory motions in epithelial-like monolayers. This model, in absence of any external chemical signals, is able to explain coordinate rotational motion seen in systems ranging from two adherent cells to multicellular assemblies. We show that the competition between the active persistent force and random polarization fluctuation is responsible for the robust rotation. Passive mechanical coupling between cells is necessary but active chemical signaling between cells is not. The predicted angular motions also depend on the geometrical shape of the underlying substrate: cells exhibit collective rotation on circular substrates, but display linear back-and-forth motion on long and narrow substrates.
Introduction
Cell migration is a fundamental biological process in both physiological and pathological situations (1–3). In the canonical model of single-cell migration, four integrated steps are repeated: cell protrusion formation, formation of leading-edge adhesions, development of contractile forces, and detachment of tailing edge adhesions (4). These motions are governed by the polarization state of the cell, which controls the migration direction. In contrast, during collective migration, cells are physically and functionally connected through cell-cell junctions, and hence, cell motility is also coupled to the motions of neighbors (1,5–7). Collectively migrating cells have been shown to form intriguing ordered patterns (8–13), reminiscent of patterned motions seen in actively propelled systems such as bird flocks, fish schools, cytoskeletal bundles, and others (14–19). For example, when Madin-Darby canine kidney (MDCK) epithelial cells in monolayer sheet collectively invade their neighboring free space, the distribution of strain rate in the sheet behaves as a wavelike pattern propagated from the leading edge (20). Whether these collective motions are governed by mechanical or chemical signaling interactions or a combination of the two is not known. In addition, when collective cell movements are constrained by the environment, other novel features can appear. It has been observed that MDCK epithelial cells migrate with higher overall speed on narrow strips than on wide ones (21). This indicates that the geometry of the external environment also plays an important role in the coherent motion of cells (22).
Various experiments, some quite recent, found that clusters of cells (from two to three to many) on circular two-dimensional substrates can coherently rotate together in absence of any external cues (23–27). When two or three bovine capillary endothelial cells are confined on μm-scale, circular or squared islands coated with fibronectin, cells spontaneously break symmetry and rotate in a coordinated fashion. This is in contrast with isolated cells in open two-dimensional microenvironments where migration is random. The rotational motion depends on the geometrical shape of the substrate: coordinated rotation does not take place when the islands are long and rectangular (24,25), suggesting that cell-cell interaction and the geometry of confinement are critical for cell rotation. Cell-cell interaction and substrate geometry also influence the rotational motions seen in a larger number of cells. When MDCK cells with densities ranging from 800 to 10,000 cells per mm2 are seeded on circular fibronectin substrates, larger-scale rotational motions involving tens to hundreds of cells are also seen (26). Coherent rotational motions of cells are not only responsible for tissue morphogenesis but also related to the establishment of polarity of acini and assembly of basement membrane in three-dimensional tissues (25,28,29). Therefore, a mechanistic understanding that incorporates cell mechanics, cell geometry, and biochemical signaling is desirable. In 2013, a particle dynamics model was proposed to explain the rotation of two cells (27). However, a unified model that can be generalized to systems with different sizes and cell numbers is still unavailable.
In this article, we focus on developing a unified mechanical approach to describe collective cell rotation, as observed in previous experiments (24–26). We are especially interested in developing connections between macroscopic collective behavior with molecular-scale, single-cell behavior. Therefore, the model incorporates single-cell geometry and mechanics. We consider single-cell mechanics with cell elasticity, cell-cell adhesion, and cell contractility. Cell polarization dynamics is introduced in the form of an active persistence force and a random fluctuating force. Therefore, our approach falls in between the continuum collective motility approach of Lee and Wolgemuth (19) and the propelled particle models of Szabó et al. (15), Palsson and Othmer (30), and Basan et al. (31). This minimal geometric model is able to show that cells on confined substrates will rotate together in a coherent manner, agreeing with experiments. We discuss the roles of cell mechanical properties and the geometry of the confined boundary in the coherent rotation.
The Model
To capture essential shape and mechanical features of the cell, we use Dirichlet domains to describe the cell geometry in the monolayer (32). In this approach, a cell is represented by a convex polygon labeled by a center ri. The cell shape is defined by Voronoi tessellation (Fig. 1). Cells can exchange neighbors and change their shape in response to deforming forces without leaving gaps in the monolayer sheet. Because at cellular length scales and timescales, the viscosity is high, inertia can be neglected, therefore the motion of cells can be described by
| (1) |
where ri is the position of the ith cell, and η is the frictional coefficient (which may depend on the properties of substrate and the relative velocity between cells, but here is taken as a constant (Fig. 1)). Fi,p denotes the passive mechanical force on the ith cell arising from cell deformation and cell-cell adhesion interaction. Remaining two terms, Fi,a and FR are active forces generated by the cell, and model two important aspects of cell polarization dynamics.
Figure 1.
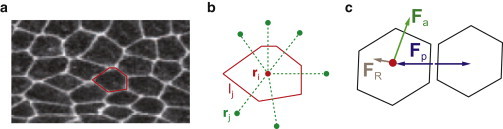
(a) Cell shape in a confluent monolayer sheet. (b) Dirichlet description of cells. ri is the position of the ith cell and rj denotes the positions of neighboring cells surrounding the ith cell. lj is the interface between the ith cell and its neighboring cell j. (c) Passive and active forces both drive collective cell migration. Fp is the passive force, which arises from cell volume elasticity and cell-cell adhesion; this force is cell-shape-dependent. Fa is the active force, which arises from cell protrusions and contractions in the polarization direction. FR is the random force, which describes fluctuations in the polarization direction. To see this figure in color, go online.
Passive force
For eukaryotic cells, cytoskeletal filaments such as actin, microtubules, and intermediate filaments provide mechanical rigidity to cells. In addition, cells control their volume by controlling their water content (33). To initiate this, an energy of the form Ei,v = 1/2Kv(Ai – A0)2 is introduced to account for cell volume/area elasticity, where Kv is a constant that describes the resistance of cells to area/volume changes, A0 denotes the preferred area of the cell, and Ai is the current area of the ith cell (34). In addition, in a cell monolayer, cell-cell adhesion energy contributes to the cell shape. Multiple factors can potentially influence cell-cell adhesion, for example, cell-cell junctions from cadherin bonds. In the two-dimensional case, the array of cells with minimal surface energy comprise the hexagonal lattice, inasmuch as that is where the total length of the cell boundaries is the shortest (34). Based on this, we introduce the adhesion energy Ei,s = 1/2Ks(ri – ri0)2 (35), where Ks is the constant describing the adhesion strength and ri0 is the geometric center of the cell. When the cell is completely symmetrical, i.e., ri = ri0, the adhesion energy is minimized. The total passive force can be derived using Fi,p = −∂(Ei,v + Ei,s)/∂ri, which gives
| (2) |
where lj is the length of the interface between cell i and its neighbor cell j. The value ni denotes the number of neighbors surrounding the ith cell (see the Supporting Material for details).
Active force
In addition to passive cell mechanics, cells also actively generate forces. Here we consider two kinds of active forces: First, the cell exerts a force, Fi,a, in the form of various types of protrusions in the polarization direction and contractile forces along the cell boundary. The polarization direction is equivalent to the velocity direction for isolated cells. (Note that during collective motility, the polarization direction is influenced by forces from cell neighbors, or potentially from contact biochemical signaling.) Second, the polarization direction of the cell, when there is no external chemotactic cues, also undergoes random orientational diffusion. FR is the random force that generates this polarization diffusion. It satisfies 〈FR〉 = 0 and
where σ is an adjustable parameter characterizing the magnitude of the polarization fluctuations. The values δ(t) and δα,β are, respectively, Dirac’s and Kronecker’s δ-functions. Because cells migrate in a directed fashion over a characteristic time required to disassemble and reassemble cytoskeletal networks necessary for motility, the active persistent force, Fi,a, is related to past velocities (polarization history) and the contractility of the cell,
| (3) |
where vi is dri/dt. The values α and β are the coefficients describing the strength of the persistent force and the memory decay rate of polarization history, respectively (36,37). The second term in Eq. 3 models forces that actively contract the cell cortical layer, which can be modeled from a contractile energy as
| (4) |
where Kc is the strength of cell contraction that minimizes the length of the cell boundary. This energy term written in terms of ri is different from previous models in which the contraction energy is expressed as Ei,c = ΔLim, with Δ being a contractility constant and Li as the circumferential length of the cell (38). However, they have similar effects because
is closely correlated with
as shown in Fig. 2. Therefore, Eq. 4 is closely related to the previous model for m = 2. It is worth noting that contractile forces in eukaryotic cells are also under potential chemical regulation through the Rho-ROCK signaling pathway (39). Therefore, Kc is also, in principle, a function of time. (Modeling the signaling pathway will not be discussed in this article.) Here we treat Kc as a constant. The definition of the active force given in Eq. 3 shows that the direction of the active force does not follow exactly the velocity direction. This is slightly different from the definition in the particle model of Basan et al. (31), where the active force has a tendency to align with the cell velocity direction.
Figure 2.
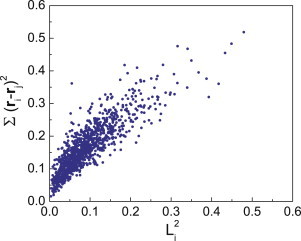
Comparison between the square of perimeter length and the sum of neighbor distance squared. Here 100 cells are simulated and sampled 10 times. We see that these quantities are linearly correlated, therefore approximate each other, except for a proportionality constant. To see this figure in color, go online.
Connections with the persistent random walk model
Here we show that Eq. 1 is a multicellular generalization of the persistent random walk model (36,40,41). For a single isolated cell, the passive force in Eq. 2 and the contractile force modeled in Eq. 3 disappear. Only the persistent part of Eq. 3 and the random force FR remain. In this case, for the first term in Eq. 3, we can expand the velocity vector as
For large β or short persistent memory, it is permissible to drop higher order terms in τ − t. We have
| (5) |
where a and b are two new constants related to α and β. This form of the persistent force, in the absence of neighboring cells, is the so called persistent random walk model of cell motility (36,42). Without neighboring cells, Fi,p = 0 and the active contraction term disappears. The equation of motion for a single motile cell becomes
| (6) |
or
| (7) |
This model has been successfully used to describe single-cell motility for a variety of tissue cell types (36,40,41). Here we see that our model for the two-dimensional monolayer is nearly identical to the persistent random walk model if only a single cell is considered. For the two-dimensional layer, the only additional physics we introduce are passive cell-cell mechanical interactions and active contractility. Therefore, the coherent rotational motion observed in the model does not require additional signaling or communication between cells. The motion can arise if every cell generates protrusive forces and persistent motion as it would in isolation. Therefore, persistent motion plus cell-cell mechanical interaction can explain coherent cell rotations.
The persistent random walk model is also consistent with what we know about cell polarization dynamics. The polarization of the cell is correlated with the microtubule-organizing center position with respect to the cell nucleus (43). The cell nucleus undergoes random rotation, driven by microtubule motors. Therefore, the cell generates active protrusions in the direction of polarization, but the polarization undergoes slow diffusion. These molecular aspects support the persistent random walk model as a coarse-grained model for understanding motility over scales of hours.
Model setup
We scale the system by length scale , timescale , and force scale KvA03/2. The preferred area (A0) of each cell is set to be 1. Unless otherwise noted, the total area of the substrate is set to be N × A0 in all simulations, where N is the total number of cells in the system. In this fashion, the preferred density of cells is fixed as 1. For cells on fixed domains, cells cannot go beyond the boundary. For the cells near the fixed boundary, their shapes are determined by both neighboring cells and boundary. At the beginning of the simulation, cells are distributed randomly, followed by an equilibration phase. Order parameters are computed after the equilibration phase.
Results and Discussion
Clusters of two or three cells
Let us first consider simple configurations involving two or three cells constrained to circular islands (Fig. 3, a and b) (24,25). In our simulations, the interfaces between cells are straight lines, as shown in Fig. 3, c and d. Fig. 3, e and f, shows the trajectories of cells, which show consistent rotational motion. The motion can be either clockwise or counterclockwise, depending on initial conditions. We can examine the robustness of the observed rotation as a function of model parameters.
Figure 3.
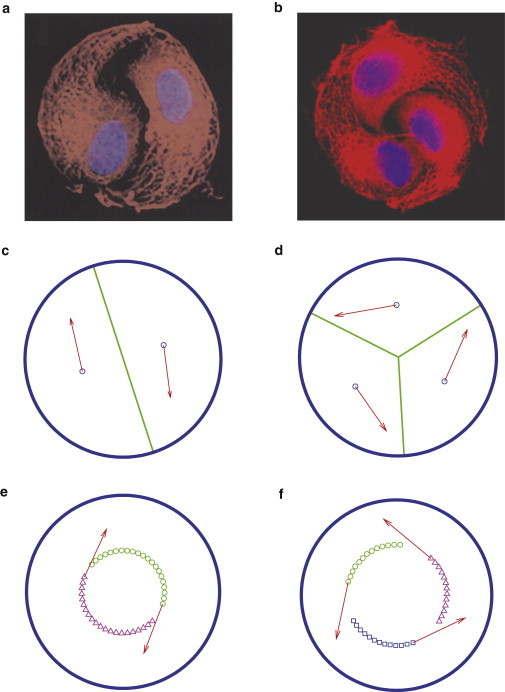
Two or three cells rotating on circular substrates. (a and b) Two and three bovine capillary endothelial cells migrating on micrometer-scale extracellular matrix island coated with a high-density of fibronectin (24,25). (c and d) Simulations by using Dirichlet domains. (e and f) Trajectory of cell migration. To see this figure in color, go online.
As expected, the velocity autocorrelation function, defined as CA(t) = 〈v(t) ⋅ v(0)〉/|v(0)|2, is oscillatory after the initial phase (see Fig. 4 a), rather than the exponential or double-exponential decays observed for the free movement of single cells (e.g., HaCaT cells and Dictyostelium discoideum) (36,40,44). The emergence of robust circular cell rotation can be captured by the order parameter g = 〈v(t) ⋅ τ(t)〉/|v(t)|, where τ denotes the tangential unit vector of cells. The value |g| approaching 1 implies full circular (clockwise or anticlockwise) rotation and |g| approaching 0 corresponds to cells migrating in a random manner. Because clockwise and counterclockwise rotations occur with equal probability, in the following, we take the tangential unit vector τ to point in the direction of cell rotation, so that we can treat clockwise and counterclockwise rotation equally. Our simulations with constant Kc show that after a short initial phase of stochastic motility, cells migrate in a circular fashion when σ(< α) is small (Fig. 4 b and see Movie S1 and Movie S2 in the Supporting Material). Increasing σ destroys the coherent motion. The phase diagrams of cell rotation are illustrated in Fig. 4, c and d. Here, robust rotation is defined by the order parameter g. The rotation is robust when g > 0.8. In Fig. 4, we computed 10 trajectories for each parameter combination (α,σ) to obtain the order parameter g. The plotted phase boundary corresponds to (α,σ), where |g – 0.8| < 0.001.
Figure 4.
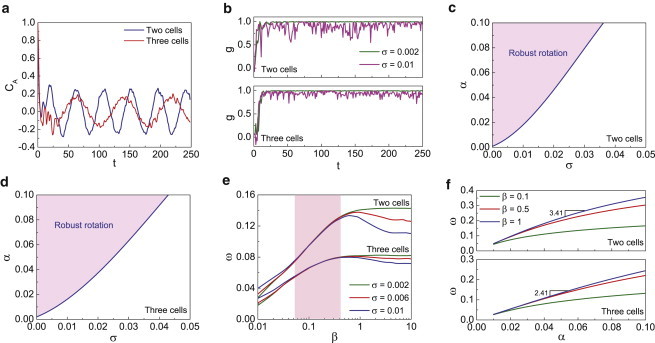
Simulations of two and three cells rotating on circular substrates. (a) Autocorrelation function of cell velocity. (b) Robustness of cell rotation is influenced by the polarization fluctuation σ. (c and d) Phase diagrams of cell rotation for two and three cell systems, respectively. (e) The rotation frequency ω versus the persistent memory decay rate, β. There is an insensitive region (pink) in which the rotation does not depend on the variance of σ. (f) Dependence of ω on the strength of persistent force α. For large β the relationship is linear, and the slope is explained by Eq. 9. To see this figure in color, go online.
We find that the persistent force is the dominant factor controlling rotational motion. The value β, which describes the rate of decay of the polarization memory, strongly affects the rotation frequency ω. There exists a peak angular frequency when the memory decay rate is increased, depending on the random force (Fig. 4 e). There is also a regime where ω is insensitive to the random force, indicating robust rotation. The angular frequency (ω) is enhanced by increasing the strength (α) of the persistent force, as shown in Fig. 4 f. During the rotation, a frictional torque (Tf ) arises due to the friction between the cell monolayer and the substrate. Tf can be defined as (26)
| (8) |
where R is the radius of the substrate and ηs is the friction coefficient between the cell monolayer and its substrate. When the system reaches a uniform rotation state, the cell cluster rotates like a solid disk, and in this case the relative movements between cells can be ignored. Therefore, the active torque is balanced by Tf (26). For short memory, i.e., large β, the direction of the persistent force approximately coincides with the tangent direction. In this case, the active torque generated by each cell is given by αh, where h is the distance from geometrical center of the island (see the Supporting Material). For two cells, h = 4R/(3π), and for three cells, . Hence,
| (9) |
where N denotes the number of cells. This α−ω relationship is shown in Fig. 4 f, where the observed slope is consistent with our predictions. This gives a simple estimate of the total cell force when the persistent memory is short.
Large clusters
A cluster of many cells migrating on a confined substrate also forms rotational patterns (Fig. 5 a). This phenomenon can be reproduced by our model, as shown in Fig. 4, b and c (and see Movie S3 and Movie S4). Fig. 5 d plots the order parameter g and the velocity autocorrelation function CA, which demonstrates that after initial random motility, the cells arrive at a robust rotation state. Fig. 5 e shows that the magnitude of velocity is proportional to the distance from the substrate center, indicating that the system rotates like a solid disk, as evidenced in experiments (26). The slope of this proportional relationship is the angular frequency of rotation, which correlates with the persistent force. The coordinated rotation predicted here is different from spirals seen in collective dynamics of polar gels and bacterial suspensions (45,46), where hydrodynamics play a significant role in the pattern formation. A relationship similar to Eq. 9 is derived as α ≈ 3/4 ηsA0Rω (see the Supporting Material for details). The velocity relationship between cells can be quantitatively described by a spatial velocity correlation function (Fig. 5 f), which is defined as
| (10) |
where denotes the deviation of the velocity from the mean velocity (21). It reveals that the correlation length (the distance where Cs reaches 0) depends on the number of cells in the system. Systems with more cells have longer correlation lengths, consistent with previous experiments (26). Fig. 5 g shows the area of cells in the monolayer sheet. The cells expand ((Ai – A0)/A0 > 0) in the central region, whereas they are compressed ((Ai – A0)/A0 < 0) near the edge due to the confinement of the boundary, again consistent with experiments (Fig. 5 a). Furthermore, we find that the rotation frequency ω is related to the cell number and average density (Fig. 5 h). Here, the average density is defined as ρ = N/S, with S being the area of the substrate. For a multicellular system, increasing cell density increases the rotation frequency, also agreeing with experimental observations (26). When the persistent memory is short, the linear α−ω relationship can be recast as ω = kN−1/2, where . This shows that a system with more cells favors lower ω.
Figure 5.
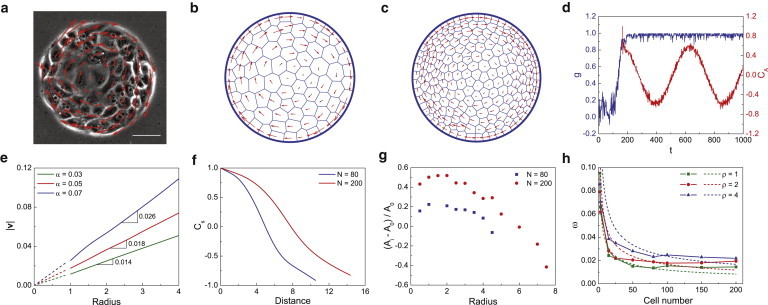
Many cells migrating on circular substrates. (a) Experiment of MDCK cells rotating on a fibronectin-coated substrate (26). (b and c) Simulations of cell rotation with the cell number N = 80 and N = 200. (d) The order parameter and autocorrelation function, where N = 80 and α = 0.03. (e) The absolute velocity as a function of the position radius (N = 80). The slopes of curves give the angular frequency of cell rotation. (f) The spatial correlation function of cell movement. (g) The area of cells with the position radius, where Ai is the actual area of cell. (h) The dependence of rotation frequency on the cell number under different average density ρ. (Symbol and line) Simulation. (Dashed line) ω = kN−1/2. In panels b–d and f–h, we take α = 0.03, β = 0.1, and σ = 0.002. To see this figure in color, go online.
For even larger systems with many cells, our simulations show that a larger persistent force is required to drive robust rotation. If the persistent force is not sufficiently large, the rotation center can deviate from the geometric center of the substrate, as shown in Fig. 6 a. In this case, the rotation center may execute eccentric motion. Some local rotational patterns may appear, as demonstrated in previous experiments (Fig. 6 b and see Movie S5) (26). This observation indicates that the correlation length also depends on the strength of the active persistent force. For substrates that are larger than the correlation length, other hydrodynamics modes can appear.
Figure 6.
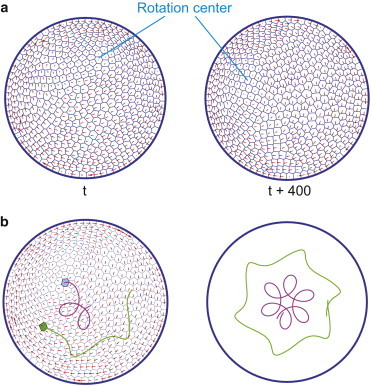
Eccentric rotation in a system with a large number of cells. Coherent rotations are still observed; however, the rotation center is dynamic. (a) The rotation center moves throughout the cluster over time. (b) Trajectories of two representative cells. The rotation shows higher order structure, representative of additional modes in this system. To see this figure in color, go online.
Rotation driven by a portion of cells
We also find that one needs only a few cells with persistent force to generate coherent rotation of all cells. A few cells that persistently migrate while the rest of the colony do not actively exert forces can still lead to coherent rotation of the whole system, as shown in Fig. 7 and Movie S6. Systems with a smaller number of persistent cells require a larger persistence force (α) to drive rotation. These observations are consistent with the idea that some cells within epithelial sheets are so-called “leader” cells (5,47–49). Migratory motion can arise from the action of leader cells. We also found that persistent cells initially located at the substrate rim are more effective in driving robust rotation than those initially located at the center. This is because cells at the rim exert a larger torque on the system.
Figure 7.
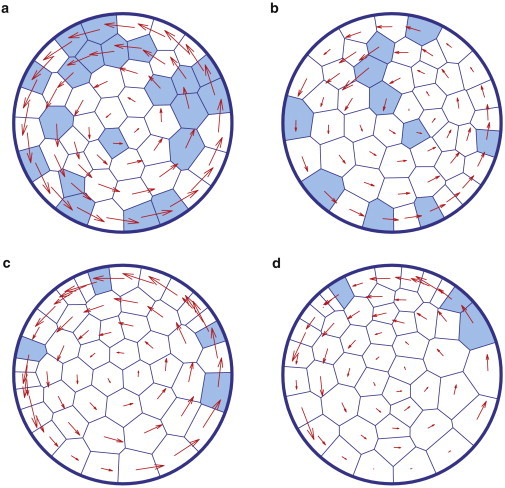
Collective cell rotation driven by a portion of cells with persistence force. Here, colored cells can exert persistent force whereas the remaining cells are passive. (a) Number of cells with persistent force is M = 20. The persistent force magnitude is α = 0.1. (b) M = 10 and α = 0.15. (c) M = 4 and α = 0.25. (d) M = 3 and α = 0.25. The total number of cells in the system is N = 50. In these simulations, we take β = 0.1 and σ = 0.002. To see this figure in color, go online.
Influence of confined boundary on cell rotation
An interesting question is how substrate geometry can influence the coherent motion. We perform numerical experiments where cells migrate on substrates with different geometrical shapes. We introduce the shape factor ϕ = 4πS/L2s, with Ls being the perimeter of the substrate. The value ϕ approaches 1 for a circular substrate and approaches 0 for a narrow, stripelike substrate. Results on elliptic substrates with varying aspect ratios are shown in Fig. 8 a. Note that in these simulations, we vary the substrate geometry but the total number of cells is fixed, therefore the total persistent force exerted by cells is a constant. We find that the order parameter g is correlated with ϕ: circular substrates favor robust circular rotation (g > 0.8) whereas on narrow or stripelike substrates (ϕ < 0.8), cells prefer unidirectional migration (21) or back-and-forth migration (insets in Fig. 8 a). Cells can also collectively rotate on annulus substrates and the width of the annulus plays a role in the rotation frequency: larger width of the annulus is correlated with increasing ω (Fig. 8 b). However, for small H/R, our simulations show that the averaged migration speed is enhanced by the increased H/R. This is in contrast with experiments in Vedula et al. (21), which showed an opposite dependence. This disparity arises possibly from the varying total number of cells in the experiments; or the presence of a free edge in experiments, which can change the surface tension; or possibly signaling-dependent force generation in cells.
Figure 8.
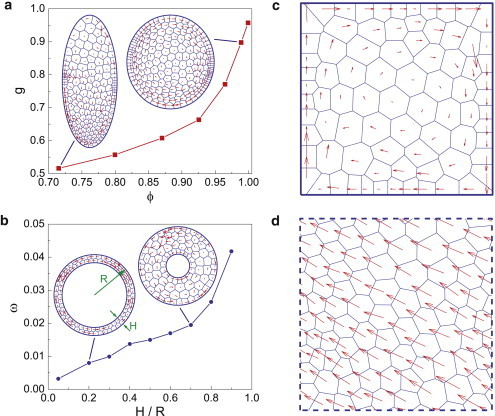
Collective cell migration on substrate with varying geometries. (a) On elliptic substrate with N = 210. (b) On annulus substrate with fixed N = 100, the rotation frequency ω is positively correlated with the width-radius ratio H/R. (c and d) On squared substrate with fixed boundary and periodic boundary (N = 100), respectively. In all these simulations, we take α = 0.03, β = 0.1, and σ = 0.002. To see this figure in color, go online.
Collective rotation can also take place in a confined, squared geometry (Fig. 8 c and see Movie S7) (15), but is less likely on a narrow rectangular substrate. Again, this confirms the important role of the aspect ratio in cell rotation. Our results show that the rotation hinges on the geometrical confinement. For a space without confinement, robust cell rotation does not occur and cells prefer to migrate unidirectionally, as shown in the squared system with a periodic boundary (Fig. 8 d and see Movie S8).
Discussion and Conclusions
In this article, we demonstrated that a persistent random walk model together with mechanical coupling can explain coherent rotational motion in two-dimensional epithelial monolayers. Cells propel themselves as they would in isolation; additional chemical signaling between cells is not necessary to explain rotation. Robust rotation depends on the geometrical shapes of the substrates. In addition, material properties of the substrates, and hence the mechanical response of the substrates, may also play a role in this collective cell migration (50,51). In our model, this is manifested in the parameter η, which affects rotation and migration speed. However, substrate properties may also affect cell elasticity and the magnitude of active contraction and adhesion. Therefore, further dissection of this problem is required.
Our model retains simple geometric information of individual cells in the cluster, therefore the model is able to capture mechanics of cells with reasonable detail. However, as of this writing, our model does not allow for cells that are not geometrically convex. This limitation suggests that some types of collectively motility, such as the “bursty” motions seen in highly deformed cancer cells, cannot be easily captured within this model (52). Nevertheless, the basic framework of the model remains valid. Moreover, because we use the Dirichlet domains to describe the cell shape, the cell monolayer covers the entire space without leaving any gaps or overlaps. Therefore, this model is unable to examine cell-cell collisions, which may be another mechanism that can orient and align cells during collective migration in sparse monolayers (53,54)
Mechanical interactions and propulsive motion generating coherent rotations is not a new notion. For instance, continuum models utilizing these features can also generate rotary motions (19). However, our approach is unique in that it allows for further investigation of the interplay between cell signaling and collective motility. Within our framework, cell signaling will change parameters such as Kc, which describes the strength of contractility, and Ks, which describes the strength of adhesions. As the biochemical state of the cell changes, these parameters will change with time. Therefore, quantitative studies on collective motility must quantitatively distinguish passive versus active forces and contributions from signaling changes in the cell. This presents a serious difficulty, inasmuch as measurable quantities such as velocity correlation functions and detailed velocity fields cannot easily disentangle these contributions. Further experiments and modeling are needed to fully resolve these issues.
Our model is an example of rotational pattern formation in systems with a director field. Other examples include liquid crystals (55), cytoskeletal systems (14), and flocking (14). Along with these other topics, studies on actively propelled cells will further our understanding on collective phenomena prominent in diverse fields such as cell biology, tissue morphogenesis, and even social organization processes.
Acknowledgments
This work has been supported by the National Institutes of Health grants No. 1U54CA143868-01 and No. 1R01GM075305, and National Science Foundation grant No. PHY-1205795.
Supporting Material
Supporting Citations
Reference (56) appears in the Supporting Material.
References
- 1.Friedl P., Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Theveneau E., Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell. Mol. Life Sci. 2013;70:3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaritsky A., Natan S., Tsarfaty I. Emergence of HGF/SF-induced coordinated cellular motility. PLoS ONE. 2012;7:e44671. doi: 10.1371/journal.pone.0044671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauffenburger D.A., Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 5.Poujade M., Grasland-Mongrain E., Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. USA. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czirók A., Varga K., Szabó A. Collective cell streams in epithelial monolayers depend on cell adhesion. New J. Phys. 2013;15:075006. doi: 10.1088/1367-2630/15/7/075006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepúlveda N., Petitjean L., Hakim V. Collective cell motion in an epithelial sheet can be quantitatively described by a stochastic interacting particle model. PLOS Comput. Biol. 2013;9:e1002944. doi: 10.1371/journal.pcbi.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riedel I.H., Kruse K., Howard J. A self-organized vortex array of hydrodynamically entrained sperm cells. Science. 2005;309:300–303. doi: 10.1126/science.1110329. [DOI] [PubMed] [Google Scholar]
- 9.Trepat X., Wasserman M.R., Fredberg J.J. Physical forces during collective cell migration. Nat. Phys. 2009;5:426–430. [Google Scholar]
- 10.Petitjean L., Reffay M., Silberzan P. Velocity fields in a collectively migrating epithelium. Biophys. J. 2010;98:1790–1800. doi: 10.1016/j.bpj.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelini T.E., Hannezo E., Weitz D.A. Glass-like dynamics of collective cell migration. Proc. Natl. Acad. Sci. USA. 2011;108:4714–4719. doi: 10.1073/pnas.1010059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J.H., Serra-Picamal X., Fredberg J.J. Propulsion and navigation within the advancing monolayer sheet. Nat. Mater. 2013;12:856–863. doi: 10.1038/nmat3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deforet M., Hakim V., Silberzan P. Emergence of collective modes and tri-dimensional structures from epithelial confinement. Nat. Commun. 2014;5:3747. doi: 10.1038/ncomms4747. [DOI] [PubMed] [Google Scholar]
- 14.Toner J., Tu Y. Long-range order in a two-dimensional dynamical XY model: how birds fly together. Phys. Rev. Lett. 1995;75:4326–4329. doi: 10.1103/PhysRevLett.75.4326. [DOI] [PubMed] [Google Scholar]
- 15.Szabó B., Szöllösi G.J., Vicsek T. Phase transition in the collective migration of tissue cells: experiment and model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- 16.Schaller V., Weber C., Bausch A.R. Polar patterns of driven filaments. Nature. 2010;467:73–77. doi: 10.1038/nature09312. [DOI] [PubMed] [Google Scholar]
- 17.Vicsek T., Zafeiris A. Collective motion. Phys. Rep. 2012;517:71–140. [Google Scholar]
- 18.Marchetti M.C., Joanny J.F., Simha R.A. Hydrodynamics of soft active matter. Rev. Mod. Phys. 2013;85:1143–1189. [Google Scholar]
- 19.Lee P., Wolgemuth C.W. Crawling cells can close wounds without purse strings or signaling. PLOS Comput. Biol. 2011;7:e1002007. doi: 10.1371/journal.pcbi.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serra-Picamal X., Conte V., Trepat X. Mechanical waves during tissue expansion. Nat. Phys. 2012;8:628–634. [Google Scholar]
- 21.Vedula S.R.K., Leong M.C., Ladoux B. Emerging modes of collective cell migration induced by geometrical constraints. Proc. Natl. Acad. Sci. USA. 2012;109:12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolli C.G., Nakayama H., Nakanishi J. Switchable adhesive substrates: revealing geometry dependence in collective cell behavior. Biomaterials. 2012;33:2409–2418. doi: 10.1016/j.biomaterials.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Rappel W.-J., Nicol A., Loomis W.F. Self-organized vortex state in two-dimensional Dictyostelium dynamics. Phys. Rev. Lett. 1999;83:1247–1250. [Google Scholar]
- 24.Brangwynne C., Huang S., Ostuni E. Symmetry breaking in cultured mammalian cells. In Vitro Cell. Dev. Biol. Anim. 2000;36:563–565. doi: 10.1007/BF02577523. [DOI] [PubMed] [Google Scholar]
- 25.Huang S., Brangwynne C.P., Ingber D.E. Symmetry-breaking in mammalian cell cohort migration during tissue pattern formation: role of random-walk persistence. Cell Motil. Cytoskeleton. 2005;61:201–213. doi: 10.1002/cm.20077. [DOI] [PubMed] [Google Scholar]
- 26.Doxzen K., Vedula S.R.K., Lim C.T. Guidance of collective cell migration by substrate geometry. Integr Biol (Camb) 2013;5:1026–1035. doi: 10.1039/c3ib40054a. [DOI] [PubMed] [Google Scholar]
- 27.Leong F.Y. Physical explanation of coupled cell-cell rotational behavior and interfacial morphology: a particle dynamics model. Biophys. J. 2013;105:2301–2311. doi: 10.1016/j.bpj.2013.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner K., Mori H., Bissell M.J. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc. Natl. Acad. Sci. USA. 2012;109:1973–1978. doi: 10.1073/pnas.1119578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Lacoche S., Muthuswamy S.K. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc. Natl. Acad. Sci. USA. 2013;110:163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palsson E., Othmer H.G. A model for individual and collective cell movement in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 2000;97:10448–10453. doi: 10.1073/pnas.97.19.10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basan M., Elgeti J., Levine H. Alignment of cellular motility forces with tissue flow as a mechanism for efficient wound healing. Proc. Natl. Acad. Sci. USA. 2013;110:2452–2459. doi: 10.1073/pnas.1219937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda H. Description of cellular patterns by Dirichlet domains: the two-dimensional case. J. Theor. Biol. 1978;72:523–543. doi: 10.1016/0022-5193(78)90315-6. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H., Sun S.X. Cellular pressure and volume regulation and implications for cell mechanics. Biophys. J. 2013;105:609–619. doi: 10.1016/j.bpj.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai T., Honda H. A dynamic cell model for the formation of epithelial tissures. Philos. Mag. B. 2001;81:699–719. [Google Scholar]
- 35.Barrio R.A., Romero-Arias J.R., Álvarez-Buylla E.R. Cell patterns emerge from coupled chemical and physical fields with cell proliferation dynamics: the Arabidopsis thaliana root as a study system. PLOS Comput. Biol. 2013;9:e1003026. doi: 10.1371/journal.pcbi.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campos D., Méndez V., Llopis I. Persistent random motion: uncovering cell migration dynamics. J. Theor. Biol. 2010;267:526–534. doi: 10.1016/j.jtbi.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Kabla A.J. Collective cell migration: leadership, invasion and segregation. J. R. Soc. Interface. 2012;9:3268–3278. doi: 10.1098/rsif.2012.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farhadifar R., Röper J.-C., Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 39.Amano M., Chihara K., Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275:1308–1311. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- 40.Selmeczi D., Mosler S., Flyvbjerg H. Cell motility as persistent random motion: theories from experiments. Biophys. J. 2005;89:912–931. doi: 10.1529/biophysj.105.061150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L., Nørrelykke S.F., Cox E.C. Persistent cell motion in the absence of external signals: a search strategy for eukaryotic cells. PLoS ONE. 2008;3:e2093. doi: 10.1371/journal.pone.0002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu P.H., Giri A., Wirtz D. Three-dimensional cell migration does not follow a random walk. Proc. Natl. Acad. Sci. USA. 2014;111:3949–3954. doi: 10.1073/pnas.1318967111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.S.H., Chang M.I., Wirtz D. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol. Biol. Cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi H., Sato M.J., Ueda M. Functional analysis of spontaneous cell movement under different physiological conditions. PLoS ONE. 2008;3:e2648. doi: 10.1371/journal.pone.0002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruse K., Joanny J.F., Sekimoto K. Asters, vortices, and rotating spirals in active gels of polar filaments. Phys. Rev. Lett. 2004;92:078101. doi: 10.1103/PhysRevLett.92.078101. [DOI] [PubMed] [Google Scholar]
- 46.Wioland H., Woodhouse F.G., Goldstein R.E. Confinement stabilizes a bacterial suspension into a spiral vortex. Phys. Rev. Lett. 2013;110:268102. doi: 10.1103/PhysRevLett.110.268102. [DOI] [PubMed] [Google Scholar]
- 47.Gov N.S. Collective cell migration patterns: follow the leader. Proc. Natl. Acad. Sci. USA. 2007;104:15970–15971. doi: 10.1073/pnas.0708037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rørth P. Collective cell migration. Annu. Rev. Cell Dev. Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 49.Mark S., Shlomovitz R., Silberzan P. Physical model of the dynamic instability in an expanding cell culture. Biophys. J. 2010;98:361–370. doi: 10.1016/j.bpj.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelini T.E., Hannezo E., Weitz D.A. Cell migration driven by cooperative substrate deformation patterns. Phys. Rev. Lett. 2010;104:168104. doi: 10.1103/PhysRevLett.104.168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng M.R., Besser A., Brugge J.S. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 2012;199:545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee M.H., Wu P.H., Wirtz D. Mismatch in mechanical and adhesive properties induces pulsating cancer cell migration in epithelial monolayer. Biophys. J. 2012;102:2731–2741. doi: 10.1016/j.bpj.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bindschadler M., McGrath J.L. Sheet migration by wounded monolayers as an emergent property of single-cell dynamics. J. Cell Sci. 2007;120:876–884. doi: 10.1242/jcs.03395. [DOI] [PubMed] [Google Scholar]
- 54.Bricard A., Caussin J.-B., Bartolo D. Emergence of macroscopic directed motion in populations of motile colloids. Nature. 2013;503:95–98. doi: 10.1038/nature12673. [DOI] [PubMed] [Google Scholar]
- 55.de Gennes P.G., Prost J. Oxford University Press; New York: 1993. The Physics of Liquid Crystals. [Google Scholar]
- 56.Marinari E., Mehonic A., Baum B. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature. 2012;484:542–545. doi: 10.1038/nature10984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


