Abstract
We demonstrate the ability of pulsed dipolar electron spin resonance (ESR) spectroscopy (PDS) to report on the conformation of Cu-Zn superoxide dismutase (SOD1) through the sensitive measurement of dipolar interactions between inherent Cu2+ ions. Although the extent and the anisotropy of the Cu ESR spectrum provides challenges for PDS, Ku-band (17.3 GHz) double electron-electron resonance and double-quantum coherence variants of PDS coupled with distance reconstruction methods recover Cu-Cu distances in good agreement with crystal structures. Moreover, Cu-PDS measurements expose distinct differences between the conformational properties of wild-type SOD1 and a single-residue variant (I149T) that leads to the disease amyotrophic lateral sclerosis (ALS). The I149T protein displays a broader Cu-Cu distance distribution within the SOD1 dimer compared to wild-type. In a nitroxide (NO)-labeled sample, distance distributions obtained from Cu-Cu, Cu-NO, and NO-NO separations reveal increased structural heterogeneity within the protein and a tendency for mutant dimers to associate. In contrast, perturbations caused by the ALS mutation are completely masked in the crystal structure of I149T. Thus, PDS readily detects alterations in metalloenzyme solution properties not easily deciphered by other methods and in doing so supports the notion that increased range of motion and associations of SOD1 ALS variants contribute to disease progression.
Introduction
Only a few biophysical methods provide high-resolution long-range distance restraints for proteins in solution (1–4). One such technique is pulsed dipolar electron spin resonance (ESR) spectroscopy (PDS) (5–7), which when coupled with nitroxide (NO) radical spin labeling (8), can measure distances to within one Å accuracy and yield distance distributions within and between proteins that range up to 90 Å. However, despite the many advantages of PDS, nitroxide labels have certain limitations: their flexibility reduces effective distance resolution due to uncertainty in the positions of backbone atoms; their placement may perturb structure and/or function; multiple labels cannot be distinguished from each other (based, for example, on a difference in g-values); high labeling efficiency is necessary; and use is limited to environments where labels are stable; this usually necessitates biochemical reconstitution. Cu2+ ions offer an alternative spin probe (9) that for Cu-metalloproteins requires no perturbation of the sample (10). Moreover, when Cu2+ and nitroxide labels are combined in a single sample, triangulation of distance restraints (11,12) can provide a more global view of structure and dynamics (13). Here, we apply Cu-PDS to the enzyme Cu-Zn superoxide dismutase (SOD1, Fig. S1 in the Supporting Material) and demonstrate its ability to reveal aberrant conformational properties of SOD1 mutants linked to the disease amyotrophic lateral sclerosis (ALS). These differences are not well discriminated by other methods and their consequences speak to the mechanism of ALS.
Whereas mainstream use of PDS is based on nitroxide spin labels, recent work has shown the ability of PDS to provide distance distributions for separations between paramagnetic metal centers in peptides and proteins (13–18). PDS is mainly represented by double electron-electron resonance (DEER, aka PELDOR) and double-quantum coherence (DQC), with DEER currently implemented over a wide range of microwave frequencies (from cm to mm waves). Saxena et al. have developed techniques to measure Cu-Cu and Cu-nitroxide distances with DEER (17) and variants (13,15–17) of the generic DQC method (7,19,20) in both synthetic peptides (13,15,17), and proteins (17). In other studies, DEER was also used to determine distances between the Type 1 and 2 Cu centers of nitrite reductase (10). Similarly, DEER has been applied to Fe3+ and Mo4+ systems (14), and Gd3+ labels are increasingly employed in DEER measurements conducted at mm-wave frequencies (21–23). Here, we apply both DEER and DQC PDS experiments (see the Supporting Material) to characterize structural heterogeneity in the protein subunits of SOD1. DEER and DQC provide similar distance data when implemented in their most common 4-pulse and 6-pulse sequences, respectively (Fig. S2); although other variants have been developed (24–27). DEER provides an advantage for spin probes with widely separated ESR spectra and good isolation from nuclear coherence effects, whereas DQC has a wider distance range and often yields greater sensitivity. Our use of intense pulses with proportionally wide spectral excitation of ∼40 G or more significantly increases signal/noise ratio (SNR) for these Cu2+ studies (cf. Supporting Material).
SOD1 is a 32 kDa homodimer, with one Cu2+ ion tightly bound in each monomer (Fig. 1). The Cu2+ centers are 32 Å apart, which is well within the 10–90 Å range of PDS (Fig. 1, Fig. S1). Over 150 mutants of SOD1 have been implicated in the familial form of ALS (fALS), a late-onset terminal neurodegenerative disease (28). The connection between the SOD1 mutations and fALS is unclear because the mutant proteins have very similar x-ray crystal structures (29,30) and enzymatic activities (31) when compared to wild-type (WT) SOD1. Decreases in thermal stability versus unfolding have been observed in ALS mutants (32), and recently, dimer destabilization leading to aggregation has been proposed as a potential mechanism of disease progression (33–36). Here, we apply Cu-PDS to probe the structural properties of the fALS SOD1 variant I149T, which we find has one of the most aberrant Cu-Cu interactions of several fALS variants tested.
Figure 1.

Cu2+-Cu2+ separation in the SOD1 dimer detected by PDS. The I149T structure (yellow) is nearly identical to that of WT (blue). Inset: superposition of residue 149 with 2.38 Å resolution Fo-Fc omit electron density (3σ for Thr-149).
Materials and Methods
Protein expression and purification
Human SOD1 DNA in the pET28b expression vector was provided by the Getzoff lab at The Scripps Research Institute. Mutations were made using the QuikChange method (Stratagene, La Jolla, CA). Proteins were expressed in Escherichia coli BL21-DE3 cells. Cells were grown at 37°C until they reached an OD600 of 0.75, at which point they were induced with 0.3 mM IPTG and 0.25 mM CuSO4. Additional growth for 18 h at 25°C was followed by cell harvesting under centrifugation. Cells were lysed by sonication, insoluble debris was pelleted by centrifugation, and the supernatant was run over a nickel immobilized metal ion affinity chromatography column. After elution with excess imidazole (200 mM), the His-tag was cleaved with an overnight thrombin digest of 400 μg at room temperature. SOD1 was further purified on a Superdex 75 (Pharmacia/Pfizer, New York, NY) size-exclusion column. Exogenous metal was removed using EDTA at low pH as previously described (37) and copper and zinc were reconstituted by direct addition of 1–20 μM ZnSO4 (equimolar with protein) at 4°C overnight followed by the same concentration of CuSO4 at 4°C for 4 h. The protein was then concentrated to 350 μM. Spin labeling of Cys variants with (1-oxyl-2,2,5,5-tetamethylpyrroline-3-methyl)-methanethiossulfonate (MTSSL) was carried out on these purified, metallated samples, as previously described (38).
Upon purification, the metal content of the fALS variants (I149T, H48Q) was shown by inductively coupled plasma mass spectroscopy (University of Georgia, Department of Biochemistry (39)) not to vary significantly compared to that of the WT (all samples had 71–83% Cu2+ loading). Uniform metal content among the samples was also supported by only modest variations in ESR CW spectra and DEER echo modulation depth, the latter of which may in part be due to minor orientational effects (see the Supporting Material).
Crystallization, x-ray diffraction data collection and structure solution
Crystals were grown by vapor diffusion with the hanging drop method. The well solution consisted of 100 mM NaCl, 100 mM Tris-HCl pH 7.6, 2.8 M (NH4)2SO4 and a protein concentration of 10 mg/mL in 2.25 mM potassium phosphate buffer, pH 7.0 and 160 mM NaCl. Crystals were soaked in a solution of 25% glycerol in mother liquor before flash-cooling in the cryostream. Data were collected at 100 K using 0.979 Å x-ray radiation at beamline A1 at the Cornell High Energy Synchrotron Source (CHESS). Data were reduced and scaled using HKL2000 (40). Initial phases were determined by molecular replacement in AutoMR (Phenix) (41) using WT SOD1 (PDB ID 2V0A (42)) as a model. Further refinement was carried out with XFIT (43), CNS (44), and Phenix (45) (Table S1). For comparing Cu ion positions in the WT and I149T SOD1, diffraction data were downloaded from the Protein Data Bank (PDB) and then limited to the same resolution as I149T. Cu ions were than randomly moved 0.5–1.0 Å and the structures refined in Phenix by conjugate gradient minimization. No restraints were placed on metal ion ligands. Variances of the Cu-Cu interdimer separations were calculated from the five unique SOD dimers in each asymmetric unit.
Pulsed ESR measurements
The 4-pulse DEER and the 6-pulse DQC PDS pulse sequences (Fig. S2) were used to measure distances between unpaired electron spins of catalytic Cu2+ ions in SOD1 and fALS mutants. DEER was also applied to measure distances between Cu2+ ions and MTSSL spin labels (R1) introduced at site V94, mutated to cysteine, as well as between the NO groups of R1 nitroxide side chains. Cu-Cu DEER experiments were conducted by flipping Cu spins and observing echo modulation at the field positions well outside of the nitroxide spectrum (Fig. S3). The field positions of pump and detection pulses, located at the Cu2+ gy maximum (∼5900–6000 G), were typically separated by 60 G. Experiments where pulse and pump positions were swapped and shifted by ∼200 G toward the NO spectrum in Cu-NO measurements produced no detectable difference in DEER modulation. NO-NO interactions were determined by confining both frequencies to the nitroxide spectrum (at ∼6200 G, Fig. S3) where Cu2+ absorption is low and will not contribute > ∼1% to the deep (∼50%) modulation of DEER signal when NO is pumped. Cu-NO interactions were measured by pumping the nitroxide spins between the pump and detection frequencies of the NO-NO case and detecting the Cu well outside of the NO envelope (Fig. S3). DQC experiments were carried out at the Cu spectrum maximum in the gy region. In general, the DQC method can easily circumvent the minimal 20 Å limit of standard DEER, as well as provide increased SNR. See the Supporting Material for more details on PDS implementation and results.
Multiangle light scattering coupled to size-exclusion chromatography (SEC)
SEC coupled with multiangle light scattering (MALS) was used to study the molar mass of WT SOD1 and the I149T mutant. Proteins (1–5 mg/mL) were run at room temperature on an SEC column (WTC050N5 - Wyatt) preequilibrated with 50 mM Tris-HCl pH 7.5, 150 mM NaCl buffer. Analysis and molecular weight determination was carried out with Wyatt technologies ASTRA. Bovine serum albumin (Sigma) was used as a control for data quality.
Results
Human SOD1 and fALS variants in the background of the stabilizing mutations C6A and C111S (46), (47) were expressed in E. coli cells and purified as described above. The C6A and C111S substitutions aid expression, but do not effect SOD1 structure, activity (46), (47) or PDS signals, and are henceforth referred to as WT (Supplemental Sec. A, Fig. S1). The resulting time-domain DEER traces of SOD1 (Fig. S4, Fig. S5) revealed sufficiently deep DEER modulation (6–10%) with pronounced oscillations, characteristic of a single dominant dipolar interaction well within the optimal DEER distance range (Fig. 2). Tikhonov regularization (48) and the maximum entropy method (49) produced a distance distribution function that peaks sharply at 32 Å, the Cu-Cu separation in the SOD1 dimer (Fig. 2 A). The standard orientation-free kernel was effective in reconstruction, despite substantial anisotropy of the SOD1 ESR spectrum (50) (see Supplemental Sec. B3 in the Supporting Material). DQC produced similar results compared to DEER, but after just an hour of data averaging (Fig. S6).
Figure 2.

(A) Distance distributions for separation between Cu2+ centers of SOD1 WT and I149T variant. (B) Distance distributions for WT SOD1 and 3 fALS mutants at 350μM. The I149T P(r) is multiplied by 4 to aid comparison.
PDS measurements on fALS mutants H48Q and I149T produced Cu-Cu distance distributions decidedly different from WT (Fig. 2). H48Q substitutes a copper ligand in the active center and hence was expected to perturb the Cu-Cu separation (Fig. S1, Fig. S5). Indeed, the Cu-Cu distribution from H48Q is widened and peaks at ∼34 Å (Fig. 2 B). Remarkably, I149T, a substitution remote from the active center, shows a distribution that is severely broadened compared to WT (Fig. 2). Such behavior could arise from increased conformational variation within the protein itself, relocating the copper coordination site by ∼1–2 Å.
To further investigate the conformational properties of the I149T variant, we crystallized the protein and determined its structure to 2.4 Å resolution (Fig. 1, Table S1; PDB accession code 4OH2). The crystallographic electron density clearly reflects loss of the Ile-149 Cδ1 atom (Fig. 1), but otherwise, the I149T structure shows very little change compared to that of the WT. The 149 residue lies at the periphery of the dimer interface and the side chain faces into the hydrophobic core of the subunit β-barrel. The Ile→Thr substitution produces no change in β-sheet conformation or subunit association that can be observed among the five unique dimers of the crystal structure. Comparison of the Cu-Cu distances in the I149T and WT structures (determined from refinement at equivalent resolutions) shows a slightly broader distribution for the mutant dimers (σ = 0.07 vs. 0.02 Å), but the overall widening is extremely small (and likely insignificant) compared to that observed in the Cu-PDS experiment (FWHM = 5.3 vs. 1.8 Å). Note that the increased width in the distance distribution cannot be reasonably attributed to orientational correlation effects that differ in I149T (cf. Supplemental Sec. B3 in the Supporting Material).
To further investigate the source of the broad Cu-Cu distribution in the noncrystalline state of the I149T variant, we labeled the SOD1 dimer with a nitroxide at position V94 by mutating the residue to Cys and reacting it with methanethiosulfonate spin label to yield the spin-bearing side chain, commonly known as R1. The 94 position, which resides at an exposed β-turn, was chosen as a site that would generate unique Cu-NO, Cu-Cu, and NO-NO distances, but whose labeling would not interfere with dimerization or metal binding.
The combined set of three measurements for Cu and NO sites obtained from the V94R1 protein show four distinct distances that match very well with the expectations from the crystal structure (Fig. 3). However, the distance distributions derived from the I149T:V94R1 DEER data are much broader with all three types of spin interactions than those derived from V94R1 alone (Fig. 3). Thus, the I149T dimer is structurally heterogeneous and this likely reflects increased protein dynamics in the subunits, perhaps further superimposed on subunit rearrangements about the dimer interface. Indeed, the 24 Å intrasubunit Cu-NO separation in I149T is greatly diminished and merged with the longer Cu-NO intersubunit separation (Fig. 3 C). The small chemical change caused by the mutation (Fig. 1) thus increases the amplitude (and/or nature) of structural fluctuations within the entire protein, which nonetheless still center on the WT conformation. Crystallization of I149T must mask this behavior by stabilizing the WT state and damping any deviations from it. A similar pattern of behavior was recently reported for a membrane protein (51,52), for which a wide range of structural states were sampled by DEER but only two distinct states were captured by x-ray crystallography.
Figure 3.
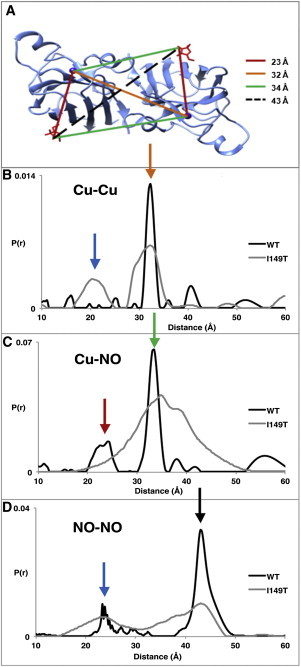
Cu-NO PDS experiment reveals conformational differences of the I149T variant. (A) Distances based on the I149T crystal structure (modeled nitroxides shown in red). (B) Cu-Cu, (C) Cu-NO, and (D) NO-NO distance distributions from WT (C6A:V94C:C111S) and with addition of I149T shown with colored arrows designating separations in (A). Short distances can be explained by dimer association (blue). The I149T trace in (C) has been multiplied by 3 to aid comparison. Sharp 23 Å peak in NO-NO WT due to D2O modulation is not present in protiated buffer. WT measurements were made at 150 μM, I149T at 350 μM.
Of importance, new short distances appear for I149T in both the Cu-Cu and NO-NO data (Figs. S7–S9), a fact that can only be explained by higher order associations of SOD1 dimers (Fig. 3). In particular, the strong ∼25 Å NO-NO signal requires an association of SOD1 subunits through an interface distinct from that of the WT SOD1 dimer. Differences in the high SNR time-domain data between WT and I149T are also in full support of this interpretation (Figs. S7–S9). Although short distance components indicative of aggregation clearly manifest in both I149T and I149T:V94R1, they are more pronounced when the spin label is present. Thus, V94R1 further destabilizes the I149T variant (Fig. S10), even though V94R1 alone produces no conformational destabilization or aggregation (Fig. 3 and Fig. S9). Aggregation of I149T is also supported by the increase in DEER modulation depth in all cases (cf. Figs. S7–S9 in Supporting Material) and by MALS experiments that show a concentration-dependent increase in the average molecular weight of the I149T variant, in contrast to the WT, which remains at the dimeric molecular weight over the same concentration range (Fig. S11). Hence, at least this particular ALS mutation appears to promote increased oligomerization of the protein. The plausibility of a tetrameric state containing short spin separations is supported by a dimer-to-dimer contact in the I149T crystal structure that produces close Cu-Cu and NO-NO separations consistent with those observed by PDS (Fig. S12, Fig. S13). Due to the moderate intensity of the component derived from a dimer-of-dimers, even less probable, higher order aggregates are not expected to significantly change the signal. Indeed, a long-distance component in the range of the SOD1 dimer size (∼60 Å), which would result from abundant multimeric aggregates, was not observed. However, characterization of large protein aggregates currently presents a difficult task for PDS and is often only evident by an increase in the baseline slope (7,38). This issue could be explored further in the future.
Discussion
The Cu-PDS experiments on SOD1 I149T indicate that the disease-causing residue substitution increases conformational heterogeneity in the overall protein fold. These data along with MALS experiments further show that the increase in heterogeneity leads to aggregation. Previous studies on the solution properties of SOD1 fALS mutants demonstrated that the residue substitutions caused local destabilization (53) or produced metal loss, but that such changes were only detected in denaturing conditions (54,55). An exception is an ALS mutational site in contact with I149 (I113T). The I113T substitution also did not affect the SOD1 crystal structure, yet the solution structure was considerably altered, as judged by small-angle x-ray scattering (30). Our method is the first, to our knowledge, to reveal structurally defined conformational differences between WT SOD and an ALS mutant that promote aggregation under native solution conditions. We have found that substitutions at the G93 position of SOD1 (a hot spot for fALS mutations) also give perturbed Cu-Cu PDS spectra, albeit to a lesser extent than I149T (Fig. 2 and Fig. S5). Thus, an altered Cu-Cu PDS distribution, which appears as a consistent feature of fALS variants tested thus far, may prove a useful diagnostic tool for identifying mutations in SOD1 that lead to ALS. In conclusion, Cu-PDS, when combined with orthogonal (e.g., nitroxide) labeling, enables a straightforward assessment of metalloprotein conformation and association state relative to the active center metal ions. This information fills an important gap between low-resolution methods such as electron microscopy, small-angle x-ray scattering, fluorescence resonance energy transfer, and high-resolution techniques such as NMR and x-ray crystallography.
Acknowledgments
The authors thank Ria Sircar for her crystallographic assistance.
This research was supported by National Institutes of Health (NIH) grants GM066775 GM079679 (B.R.C.) P41GM103521 (J.H.F.) 2R01GM039345 (E.D.G) and Molecular Biophysics Training grant T32GM008267 (G.E.M.). For access to data collection facilities the authors thank the Cornell High-Energy Synchrotron (supported by NIH grant GM103485 and National Science Foundation (NSF) grant DMR-0936384).
Contributor Information
Jack H. Freed, Email: jhf3@cornell.edu.
Brian R. Crane, Email: bc69@cornell.edu.
Supporting Material
Supporting Citations
References (56–59) appear in the Supporting Material.
References
- 1.Jain D., Lamour V. Computational tools in protein crystallography. Methods Mol. Biol. 2010;673:129–156. doi: 10.1007/978-1-60761-842-3_8. [DOI] [PubMed] [Google Scholar]
- 2.Clore G.M., Gronenborn A.M. NMR structure determination of proteins and protein complexes larger than 20 kDa. Curr. Opin. Chem. Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 3.Moerner W.E. Single molecule spectroscopy of autofluorescent proteins. J. Chem. Phys. 2002;117:10925–10937. [Google Scholar]
- 4.Klare J.P., Steinhoff H.J. Spin labeling EPR. Photosynth. Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 5.Borbat P.P., Freed J.H. Measuring distances by pulsed dipolar ESR spectroscopy: spin-labeled histidine kinases. Methods Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 6.Jeschke G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 7.Borbat P.P., Freed J.H. Pulse dipolar ESR: distance measurements. In: Timmel C.R., Harmer J.R., editors. Vol. 152. Springer-Verlag; 2014. pp. 1–82. (Structure and Bonding). [Google Scholar]
- 8.Hubbell W.L., Gross A., Lietzow M.A. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 9.Ruthstein S., Stone K.M., Saxena S. Pulsed electron spin resonance resolves the coordination site of Cu²(+) ions in α1-glycine receptor. Biophys. J. 2010;99:2497–2506. doi: 10.1016/j.bpj.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Wonderen J.H., Kostrz D.N., MacMillan F. Refined distances between paramagnetic centers of a multi-copper nitrite reductase determined by pulsed EPR (iDEER) spectroscopy. Angew. Chem. Int. Ed. Engl. 2013;52:1990–1993. doi: 10.1002/anie.201208166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaffney B.J., Bradshaw M.D., Borbat P. Locating a lipid at the portal to the lipoxygenase active site. Biophys. J. 2012;103:2134–2144. doi: 10.1016/j.bpj.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Kurpiewski M.R., Saxena S. ESR spectroscopy identifies inhibitory Cu2+ sites in a DNA-modifying enzyme to reveal determinants of catalytic specificity. Proc. Natl. Acad. Sci. USA. 2012;109:E993–E1000. doi: 10.1073/pnas.1200733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarver J., Silva K.I., Saxena S. Measuring Cu2+-nitroxide distances using double electron-electron resonance and saturation recovery. Appl. Magn. Reson. 2013;44:583–594. [Google Scholar]
- 14.Astashkin A.V., Rajapakshe A., Enemark J.H. Determination of the distance between the Mo(V) and Fe(III) heme centers of wild type human sulfite oxidase by pulsed EPR spectroscopy. J. Phys. Chem. B. 2012;116:1942–1950. doi: 10.1021/jp210578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jun S., Becker J.S., Saxena S. Unfolding of alanine-based peptides using electron spin resonance distance measurements. Biochemistry. 2006;45:11666–11673. doi: 10.1021/bi061195b. [DOI] [PubMed] [Google Scholar]
- 16.Ruthstein S., Ji M., Saxena S. Sensitive Cu2+-Cu2+ distance measurements in a protein-DNA complex by double-quantum coherence ESR. J. Phys. Chem. B. 2013;117:6227–6230. doi: 10.1021/jp4037149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., Kise D., Saxena S. An approach towards the measurement of nanometer range distances based on Cu2+ ions and ESR. J. Phys. Chem. B. 2010;114:6165–6174. doi: 10.1021/jp911637s. [DOI] [PubMed] [Google Scholar]
- 18.Ezhevskaya M., Bordignon E., van Doorslaer S. Distance determination between low-spin ferric haem and nitroxide spin label using DEER: the neuroglobin case. Mol. Phys. 2013;111:2855–2864. [Google Scholar]
- 19.Borbat P.P., Freed J.H. Distance measurements in biological systems by EPR. In: Berliner L.J., Eaton G.R., Eaton S.S., editors. Vol. 19. Academic/Plenum Publishers; New York: 2000. pp. 385–459. (Biological Magnetic Resonance). [Google Scholar]
- 20.Borbat P.P., Freed J.H. Multiple-quantum ESR and distance measurements. Chem. Phys. Lett. 1999;313:145–154. [Google Scholar]
- 21.Yagi H., Banerjee D., Otting G. Gadolinium tagging for high-precision measurements of 6 nm distances in protein assemblies by EPR. J. Am. Chem. Soc. 2011;133:10418–10421. doi: 10.1021/ja204415w. [DOI] [PubMed] [Google Scholar]
- 22.Song Y., Meade T.J., Raitsimring A. Pulsed dipolar spectroscopy distance measurements in biomacromolecules labeled with Gd(III) markers. J. Magn. Reson. 2011;210:59–68. doi: 10.1016/j.jmr.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garbuio L., Bordignon E., Yulikov M. Orthogonal spin labeling and Gd(III)-nitroxide distance measurements on bacteriophage T4-lysozyme. J. Phys. Chem. B. 2013;117:3145–3153. doi: 10.1021/jp401806g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovett J.E., Lovett B.W., Harmer J. DEER-Stitch: combining three- and four-pulse DEER measurements for high sensitivity, deadtime free data. J. Magn. Reson. 2012;223:98–106. doi: 10.1016/j.jmr.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Borbat P.P., Georgieva E.R., Freed J.H. Improved sensitivity for long-distance measurements in biomolecules: five-pulse double electron-electron resonance. J Phys Chem Lett. 2013;4:170–175. doi: 10.1021/jz301788n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borbat P.P., Davis J.H., Freed J.H. Measurement of large distances in biomolecules using double-quantum filtered refocused electron spin-echoes. J. Am. Chem. Soc. 2004;126:7746–7747. doi: 10.1021/ja049372o. [DOI] [PubMed] [Google Scholar]
- 27.Jeschke G., Bender A., Godt A. Sensitivity enhancement in pulse EPR distance measurements. J. Magn. Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 28.Pratt A.J., Getzoff E.D., Perry J.J. Amyotrophic lateral sclerosis: update and new developments. Degener Neurol Neuromuscul Dis. 2012;2012:1–14. doi: 10.2147/DNND.S19803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galaleldeen A., Strange R.W., Hart P.J. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch. Biochem. Biophys. 2009;492:40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hough M.A., Grossmann J.G., Hasnain S.S. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc. Natl. Acad. Sci. USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borchelt D.R., Lee M.K., Cleveland D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez J.A., Valentine J.S., Hayward L.J. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J. Biol. Chem. 2002;277:15932–15937. doi: 10.1074/jbc.M112088200. [DOI] [PubMed] [Google Scholar]
- 33.DiDonato M., Craig L., Tainer J.A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 34.Bruijn L.I., Houseweart M.K., Cleveland D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 35.Stathopulos P.B., Rumfeldt J.A., Meiering E.M. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elam J.S., Taylor A.B., Hart P.J. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 37.Lyons T.J., Nersissian A., Valentine J.S. Metal ion reconstitution studies of yeast copper-zinc superoxide dismutase: the “phantom” subunit and the possible role of Lys7p. JBIC. 1998;3:650–662. [Google Scholar]
- 38.Bhatnagar J., Borbat P.P., Crane B.R. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaccaro B., Menon A.L., Adams M.W.W. Vol. 4. John Wiley & Sons; 2009. Metallomics using inductively coupled plasma mass spectrometry; pp. 249–274. (Current Protocols in Chemical Biology). [Google Scholar]
- 40.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr., editor. Academic Press; San Diego, CA: 1997. pp. 307–326. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 41.Adams P.D., Afonine P.V., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strange R.W., Yong C.W., Hasnain S.S. Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA. 2007;104:10040–10044. doi: 10.1073/pnas.0703857104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McRee D.E. XtalView: A visual protein crystallographic software system for X11/Xview. J. Mol. Graph. 1992;10:44–47. [Google Scholar]
- 44.Brünger A.T., Adams P.D., Warren G.L. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Afonine P.V., Grosse-Kunstleve R.W., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hallewell R.A., Imlay K.C., Cousens L.S. Thermostabilization of recombinant human and bovine CuZn superoxide dismutases by replacement of free cysteines. Biochem. Biophys. Res. Commun. 1991;181:474–480. doi: 10.1016/s0006-291x(05)81443-3. [DOI] [PubMed] [Google Scholar]
- 47.Lepock J.R., Frey H.E., Hallewell R.A. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J. Biol. Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 48.Chiang Y.W., Borbat P.P., Freed J.H. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Chiang Y.W., Borbat P.P., Freed J.H. Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J. Magn. Reson. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 50.Lieberman R.A., Sands R.H., Fee J.A. A study of the electron paramagnetic resonance properties of single monoclinic crystals of bovine superoxide dismutase. J. Biol. Chem. 1982;257:336–344. [PubMed] [Google Scholar]
- 51.Georgieva E.R., Borbat P.P., Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 2013;20:215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hänelt I., Wunnicke D., Slotboom D.J. Conformational heterogeneity of the aspartate transporter Glt(Ph) Nat. Struct. Mol. Biol. 2013;20:210–214. doi: 10.1038/nsmb.2471. [DOI] [PubMed] [Google Scholar]
- 53.Museth A.K., Brorsson A.C., Jonsson B.H. The ALS-associated mutation G93A in human copper-zinc superoxide dismutase selectively destabilizes the remote metal binding region. Biochemistry. 2009;48:8817–8829. doi: 10.1021/bi900703v. [DOI] [PubMed] [Google Scholar]
- 54.Kitamura F., Fujimaki N., Takeuchi H. Structural instability and Cu-dependent pro-oxidant activity acquired by the apo form of mutant SOD1 associated with amyotrophic lateral sclerosis. Biochemistry. 2011;50:4242–4250. doi: 10.1021/bi200338h. [DOI] [PubMed] [Google Scholar]
- 55.Rumfeldt J.A., Lepock J.R., Meiering E.M. Unfolding and folding kinetics of amyotrophic lateral sclerosis-associated mutant Cu,Zn superoxide dismutases. J. Mol. Biol. 2009;385:278–298. doi: 10.1016/j.jmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Borbat P.P., Crepeau R.H., Freed J.H. Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J. Magn. Reson. 1997;127:155–167. doi: 10.1006/jmre.1997.1201. [DOI] [PubMed] [Google Scholar]
- 57.Marko A., Margraf D., Prisner T. Molecular orientation studies by pulsed electron-electron double resonance experiments. J. Chem. Phys. 2009;130:064102. doi: 10.1063/1.3073040. [DOI] [PubMed] [Google Scholar]
- 58.Becker J.S., Saxena S. Double quantum coherence electron spin resonance on coupled Cu(II)-Cu(II) electron spins. Chem. Phys. Lett. 2005;414:248–252. [Google Scholar]
- 59.Polyhach Y., Bordignon E., Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


