Abstract
Alternative splicing affects ~95% of eukaryotic genes, greatly expanding the coding capacity of complex genomes. Although our understanding of alternative splicing has increased rapidly, current knowledge of splicing regulation has largely been derived from studies of highly expressed mRNAs. Telomerase is a key example of a protein that is alternatively spliced, but it is expressed at very low levels, and although it is known that misregulation of telomerase splicing is a hallmark of nearly all cancers, the details of this process are unclear. Here we review work showing that hTERT expression is in part regulated by atypical alternative splicing, perhaps due to its exceptionally low expression level. We propose these differential regulatory mechanisms may be widely applicable to other genes and may provide new opportunities for development of cancer therapeutics.
Keywords: RNA splicing, regulation, low-abundance transcript, telomere, cancer therapy
Telomerase is an attractive yet challenging target for cancer therapeutics
Telomeres are dynamic DNA-protein structures at the end of chromosomes that prevent chromosome ends from being recognized as DNA damage [1]. Telomere repeats are bound by a shelterin protein complex composed of TRF1, TRF2, TIN2, RAP1, TPP1, and POT1 [2]. Together, the proteins of the shelterin complex recognize and bind to telomere repeats to promote the formation of a structure called a T-loop at telomeric ends by interacting with the 3′ guanine-rich termini of the telomere overhang, thereby concealing overhangs and preventing telomere degradation at DNA checkpoints [3, 4]. Initially, each human chromosome is capped by 15-20kb of telomeric TTAGGG repeats. Throughout the course of an organism's lifetime, these repeats slowly erode due to incomplete replication of the DNA lagging strand at the ends of the chromosomes (Figure 1). This process is called the end-replication problem [5, 6]. When a telomere becomes critically short, DNA damage signaling is induced, and cell growth is arrested, resulting in replicative senescence [7, 8]. The limited proliferative capacity of cells is widely accepted as an “aging time clock” mechanism in humans and most other large long-lived organisms. Such cells use this counting mechanism to prevent unlimited cell growth, which could lead to the accumulation of mutations over time and potentially progression to malignancy [7, 9, 10].
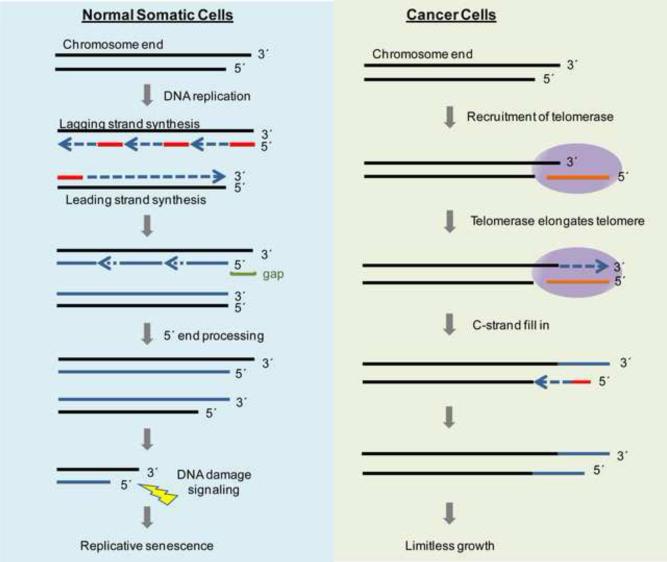
Figure 1.
Telomerase expression contributes to cancer cell immortality. In normal somatic cells, telomeres shorten with every cellular division due to the end-replication problem. During DNA replication, DNA polymerase makes complementary DNA using RNA primers (in red). The RNA primers are later removed. The newly synthesized fragments are used as primers for DNA polymerase to fill in the gaps. Although replication on the leading strand is complete, replication on the lagging strand is incomplete, resulting in telomere shortening with every cellular division. When a telomere reaches a critically short length, a DNA damage response is triggered and causes the cell to go into replicative senescence. In cancer cells, telomerase uses its RNA component (hTR, in orange) as a template for its catalytic component (hTERT) to elongate telomeres and evade cellular senescence, therefore cancer cells have unlimited proliferative capacity (e.g., become immortal), permitting additional alterations to occur to in more malignant tumors.
To overcome this brake on replicative aging, cancer cells almost universally up-regulate or re-express telomerase to re-elongate or maintain telomeres at lengths sufficient to avoid triggering DNA damage signaling [11]. Cancer cells have varying amounts of telomerase activity and almost all cancer cells have very short telomeres [9, 10, 12]. However, little is known about the regulation of this telomere maintenance program in either normal or cancer development. Telomerase is a ribonucleoprotein complex composed of a catalytic protein component with reverse transcriptase activity (hTERT) that uses a functional RNA component (hTR or hTERC) as a template to elongate telomeres [13]. Although telomerase is initially expressed in all cells during early fetal development, its expression is rapidly repressed to almost undetectable levels in somatic cells. Only a small subset of proliferating stem-like progenitor cells are capable of transient telomerase expression post-development [14].
Telomerase is subject to transcriptional, post-transcriptional, and epigenetic levels of control, but there is no consensus on the precise mechanism(s) regulating telomerase repression during development and re-expression of telomerase in cancer progression. Regardless of whether a cell has telomerase activity or not, almost all cells have an excess amount of hTR (hTERC), the telomerase RNA template [15]. By contrast, hTERT can be detected at relatively low levels in stem cells, progenitor cells, and even in cancer cells. Recently it was demonstrated that both hTR and hTERT have a subpopulation in reserve that is not assembled into activate telomerase [16]. Although hTR is present in great excess over hTERT, only the assembled telomerase with both components can have telomere elongation activity. Therefore, only when the other component is in excess will more active telomerase be assembled, thereby making both hTR and hTERT limiting factors. The current best estimate for the number of catalytically active telomerase molecules per in vitro immortalized (telomerase-positive) cell or cancer cell is ~100-500 [17], produced from approximately 20 mRNA molecules per cell [15].
Because telomerase expression is restricted to cancer cells and some but not all proliferating stem cells, it offers a potentially highly specific target for cancer treatment. Moreover, telomeres are short in ~90% of primary cancers and cancer cell lines in comparison to the longer telomeres seen in the rarely dividing stem cells and actively dividing progenitor cells (Figure 2). Thus, inhibiting telomerase activity should result in telomere shortening leading to apoptosis of cancer cells while having little to no effect on quiescent stem cells. Significant efforts have been expended to develop cancer therapeutics targeting telomerase, yet the development of telomerase inhibitors has been largely unsuccessful. Although many telomerase-directed therapeutic approaches demonstrate inhibitory effects in in vitro systems, they rarely progress beyond early stage clinical trials due to lack of potency, low specificity, and/or increased toxicities (Box 1). A major challenge for developing an effective therapeutic agent against telomerase has been that telomerase is expressed at exceptionally low abundance even in cancer cells.
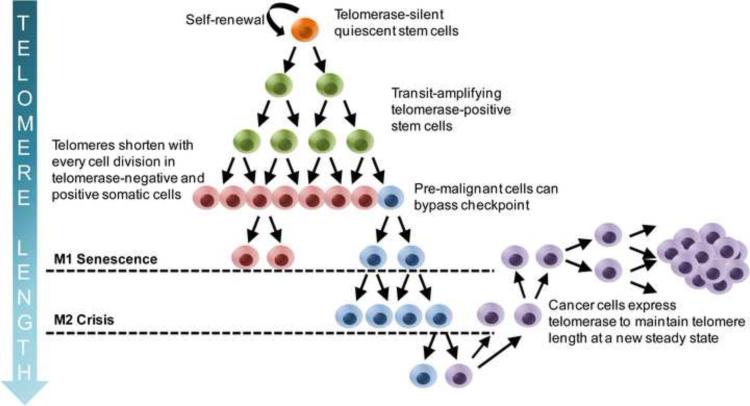
Figure 2.
Cancer cells have short telomeres compared to stem cells. Quiescent stem cells (orange) are telomerase-competent, but their telomerase activities remain silent most of the time and only transiently turn on during amplification (green). Because stem cells rarely divide, their telomere length remains long. Transit amplifying stem cells progressively lose telomere length even though they can transiently express telomerase and they eventually differentiate into tissues (red) that are telomerase-negative. All somatic cells including stem cells progressively lose telomeres with every cellular division and eventually go into replicative senescence when the telomeres become too short. Over time, a pre-malignant cell (blue) acquires sufficient alterations to become oncogenic and capable of bypassing cellular M1 senescence checkpoints. Eventually, the telomeres become so short that end-end fusions occur, leading to M2 crisis where there is a balance between cell growth and apoptosis. A very rare cancer (purple) capable of re-expressing telomerase can elongate its telomeres and maintain telomere length at a new steady state. Although both stem cells and cancer cells express telomerase, the difference in telomere length makes cancer cells more susceptible to telomerase inhibitor therapy.
Alternative splicing is a dynamic and highly regulated process
It is surprising that almost two decades after the cloning of hTERT it remains unclear how telomerase is regulated. During development it has been suggested that hTERT is in part regulated by alternative splicing [15, 18], which adds another layer of complexity to the problem for dissecting the mechanism(s) regulating telomerase. Mechanistically understanding hTERT alternative splicing offers the possibility of developing a novel anti-cancer agent that targets splicing to reduce telomerase activity in cancer. However, studying the alternative splicing of hTERT poses a new challenge because previous studies of splicing have largely been based on highly-expressed genes and the splicing of hTERT does not appear to conform to the established norm of alternative splicing regulation.
Alternative splicing affects about 95% of genes in multicellular eukaryotes, allowing for the generation of over 100,000 proteins from about 20,000 protein-coding sequences, thus greatly expanding the coding capacity of eukaryotic genomes [19]. RNA splicing, the joining of exon sequences via the removal of the noncoding intron sequences, occurs co-transcriptionally, and the splicing machinery, the spliceosome, is recruited and assembled around emerging splice sites as polymerase II transcribes the nascent pre-mRNA (Figure 3) [20, 21]. The spliceosome is a large and dynamic ribonucleoprotein that is composed of five small nuclear ribonucleic particles (snRNP) core components (U1, U2, U4, U5, and U6) and about 300 other proteins [22]. Spliceosome components assemble in an ordered and step-wise manner at each 5′ and 3′ splice site, branch point, and polypyrimidine tract in order to facilitate intron removal in the form of a lariat and subsequent exon ligation [23, 24].
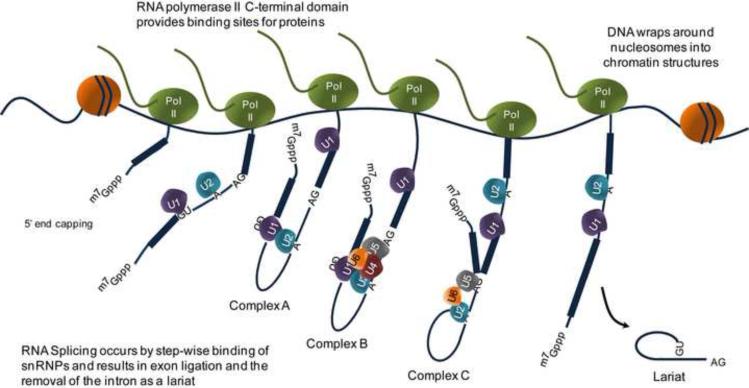
Figure 3.
RNA splicing occurs co-transcriptionally. DNA wraps around nucleosomes to form chromatin structures. During transcription, these chromatin structures need to partially disassemble and reassemble in order for RNA polymerase II (Pol II) to gain access to the DNA. The C-terminal domain (CTD) of Pol II acts as a docking site for a variety of proteins, facilitates pre-mRNA synthesis, and coordinates co-transcriptional processing events, including the initiation of transcription, including 5′ end capping of the RNA transcript, 3′ end formation by cleavage/polyadenylation, and RNA splicing. As nascent pre-mRNA is being transcribed, the spliceosome components (U1, U2, U4, U5, and U6) are recruited in a step wise manner onto the pre-mRNA for RNA splicing. U1 and U2 assemble at the 5′ splice site and branch point A respectively to form complex A. Recruitment of U4-U5-U6 snRNPs forms complex B, which goes through several rearrangements to form the catalytically active complex C. The intron is released as a lariat, the exons are ligated together, and the snRNPs disassemble.
An exon may be constitutive (always included in the mRNA of a gene) or alternative (may be included or excluded in the mRNA), giving rise to alternative splice variants. The usage of a splice site may be enhanced or suppressed by its proximity to local cis-regulatory sequences such as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs) [25]. The cis-regulatory sequences are in turn bound by trans-acting factors, or splicing factors. There are more than 500 splicing factors that can participate in alternative splicing.
The regulation of alternative splicing is intricately related to many other cellular processes. Splice site selection and pre-mRNA splicing are intimately connected to transcription, 5′-end capping, and 3′-end polyadenylation (Figure 3) [26, 27]. The coordinated control of splicing with other processes makes the regulation of alternative splicing an exciting and rapidly evolving field. Understanding the regulation of alternative splicing is especially important because deregulation of splicing factor expression has been linked to a variety of human disorders such as muscular dystrophies, premature aging disorders, and cancer [28-32].
Although our understanding of splicing regulation has greatly increased, most of what we know comes from studying the splicing of highly abundant genes. At present it is unknown if splicing regulation is similar for all genes or whether more unique forms of splicing regulation exist outside of the scope of the typical high abundant genes. Although there is little to no evidence for differential splicing regulation of low- versus high- abundant transcripts, based on recent studies of hTERT splicing, we speculate that low-abundant transcripts that are regulated by alternative splicing may require more specific mechanisms to fine-tune regulation and assist in recognition.
Low-abundance transcripts are often less conserved between species compared to high-abundant transcripts [33-35]. Although the coding sequence of hTERT is conserved among species, some of the intronic elements regulating hTERT splicing (discussed below) are only conserved amongst old-world primates [36]. In mouse, mTERT is constitutively expressed in many tissues, whereas expression of hTERT in humans is more tightly regulated [37]. This suggests that perhaps specific regulatory elements evolved with the need to fine-tune gene regulation ofhTERT and perhaps other low abundant transcripts with more specialized functions. Additionally, low-abundant transcripts have to compete with high-abundant transcripts for splicing machinery and the necessary splicing factors. Failure to recruit the necessary proteins for proper splicing is likely to be more detrimental to the low-abundant transcripts due to their inherent low transcript levels. Hence, it is feasible that cells might have evolved more specialized regulatory mechanisms for low-abundant transcripts to ensure proper splicing.
Telomerase (hTERT) splicing is regulated by unique elements
The 42 kb telomerase (hTERT) gene on human chromosome 5p15.33 contains 16 exons and can be spliced into multiple isoforms [38]. To date, 22 isoforms of hTERT has been identified [39]. Besides the full length transcript with all 16 exons, none of the identified alternative spliced forms have reverse transcriptase activity and they cannot elongate telomeres [40, 41]. The alternatively spliced isoforms within the reverse transcriptase domain of hTERT include minus alpha, minus beta, or both (minus alpha beta) (Figure 4). The minus alpha splicing isoform uses an alternative 3′ splice acceptor site 36 bp into exon 6, resulting in an in-frame transcript that is translated into a dominant negative protein without reverse transcriptase activity [41, 42]. Overexpression of the minus alpha transcript inhibits telomerase activity in telomerase positive cell lines, resulting in either cell death or senescence [42]. It is not known if the alteration of levels of the minus alpha splicing isoform is part of telomerase activity regulation in cancer cells to fine tune telomere length. The minus beta splicing isoform skips exons 7 and 8, creating a frame-shift leading to a pre-mature stop codon in exon 10. The minus beta isoform is often the major alternatively spliced component of hTERT transcripts in cultured cancer cells [36], but hTERT splicing pattern varies greatly in cancerous patient samples with different tissue origins possibly due to tumor heterogeneity [43-44]. Although minus beta has a pre-mature stop codon and is therefore subject to non-sense mediated decay, its transcripts have been shown to be translated into protein, and overexpression of the minus beta protein has been reported to confer a growth advantage to breast cancer cells [45]. Thus, although the minus beta protein does not exhibit telomerase activity, it may have other potentially oncogenic functions. In addition, two hTERT splice variants with intron retentions (INS3 and INS4) have been shown to be expressed primarily in telomerase positive cells and act as dominant negative proteins that can bind DNA substrate but not the telomerase RNA component [46]. Although it is assumed that most variants encountering a premature stop codon would lead to degradation and unlikely be translated into proteins, whether the other isoforms of hTERT can be translated into proteins or not remains to be experimentally determined. The high amount of unspecific binding of the currently available TERT antibodies adds further difficulty to the task of identifying translated hTERT isoforms.
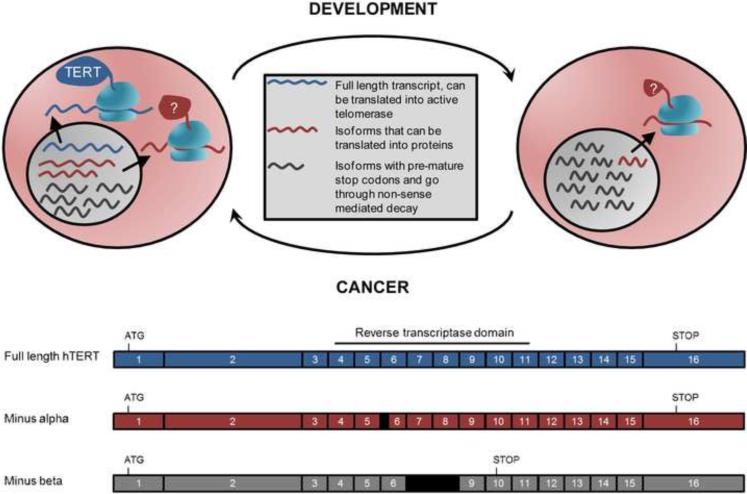
Figure 4.
Telomerase (hTERT) is partially regulated by alternative splicing. Early in development, hTERT pre-mRNAs can be spliced into 22 isoforms. The full length transcript (blue) contains all 16 exons that can make telomerase with reverse transcriptase activity. Some isoforms of hTERT have been shown to be translated into protein (red, with the minus alpha isoform as an example), although the functions of most isoforms remain to be determined. Many of the isoforms have a pre-mature stop codon (gray, with the minus beta isoform as an example) and are presumed to be degraded by non-sense mediated decay. During development the alternative splicing of hTERT changes, and for the most part, only short transcripts can be detected. As no full length transcripts are made, the cells are telomerase-negative. When telomerase activity is detected in transient amplifying stem cells and cancer cells, the pattern of hTERT splicing reverts back to its early developmental stage where some pre-mRNAs are spliced into full length hTERT and therefore can make functional telomerase.
During fetal development, telomerase activity disappears before transcripts do because of a dramatic shift in splicing pattern from full length hTERT to mostly minus beta and other isoforms without reverse transcriptase activity [18]. This shift in hTERT splicing is a highly regulated process that in humans is both tissue- and time- dependent. hTERT was traditionally believed to be transcriptionally silenced in somatic cells post-development because the common method of examining hTERT splicing uses primers that examine the inner reverse transcriptase region of the gene, leading to the false assumption that no transcription of hTERT occurs in somatic cells after development [39, 47]. Rather, a large proportion of the pre-mRNA in normal somatic cells is spliced into isoforms with the bulk of the middle portion of the transcript spliced out, rendering these isoforms undetectable using primers restricted within the reverse transcriptase domain [39]. This suggests that alternative splicing may have a key role in the regulation of telomerase activity during development.
In a wide variety of telomerase-positive embryonic stem cells, adult proliferating stem cells, and cancer cells examined, only a small fraction of the hTERT transcripts are spliced into the full length form that generates the catalytically active protein [18, 36]. The need to fine-tune the regulation to produce “just the right amount” of telomerase may be because too little telomerase would not be enough to maintain telomere length leading to increased genomic instability in cancer cells, but too much telomerase may lead to runaway elongation of telomeres and result in adverse effects including growth inhibition of the cancer cells. A working hypothesis to explain this observation is that the basal transcription machinery is unable to reduce transcription to a very low level that is optimal for telomerase function so the cell disposes of this excess transcription by alternatively splicing most transcripts into non-functional forms to produce the very low levels of protein needed to maintain telomere length in cancer cells [36]. Besides maintaining telomeres, TERT (full-length protein with reverse transcriptase activity) has been shown to have non-telomeric functions [48-50]. Reports have demonstrated non-telomeric functions for a few of the hTERT variants, such as the hTERT isoform with skipping of exons 4-13 functioning as an enhancer of Wnt signaling [39]. Potential functions for the other isoforms of hTERT are largely uncharacterized and may account for basal transcription of hTERT in telomerase-negative somatic cells. The precise amount of hTERT transcripts needed may be regulated by non-productive splicing, where most of the transcripts spliced are destined to be degraded. This may allow the cells to respond to stress and other cellular changes more rapidly by changing the splicing of pre-existing pre-mRNA. This has been demonstrated in clk-1 transcripts where a partially spliced clk-1 is retained in the nucleus and rapidly spliced into the mature clk-1 messenger RNA in response to stress [51].
In addition to transcriptional changes, hTERT splicing in cancer cells largely reverts back to the splicing pattern seen during development in which the pre-mRNA is often spliced into non-functional isoforms with only a smaller proportion of full length transcripts that can be translated into functional reverse transcriptase. Consistent with the common theme in cancer, the alternative splicing of hTERT appears to be reprogrammed in cancer cells to return to its embryonic/fetal status.
Instead of being regulated by elements near the splice sites, hTERT alternative splicing is regulated by long range interactions using elements that are >1 kb away from the exon/intron junction within introns 6 and 8. An unusual 1.1 kb region of 38 bp variable numbers of tandem repeats (VNTR) that is conserved among old world primates was determined to be essential for the exclusion of exons 7 and 8 to produce minus beta splicing [36]. Within a minigene context, the VNTR described may use RNA:RNA pairing as a mechanism to regulate splicing of hTERT [52]. Altogether, the use of a VNTR that is far from the exon/intron junctions along with the potential usage of RNA:RNA pairing as the mechanism to promote exon skipping makes the regulation of hTERT splicing atypical of those previously described, which may have evolved in large and long-lived organisms in need of regulating TERT expression more precisely [37].
The splicing of low abundant transcripts may provide unique targets for cancer therapeutics
Cancers arise as a result of the inherent instability of the cancer genome leading to the accumulation of hundreds of genetic aberrations over time which provide the collective advantage for subsets of cancer cells to proliferate and survive [53, 54]. Whole-genome sequencing and whole-gene expression analyses have identified a large number of mutations and aberrant gene expression seen in cancer cells, but such analyses are often biased for highly abundant genes while lowly abundant genes such as hTERT are often lost or undetectable, leaving the cancer genome picture grossly incomplete.
Low-abundance genes play important roles in fundamental biological processes such as cellular differentiation, metabolism, and development, and account for an estimated 20%-40% of the mRNA mass [55]. The function and the regulation of low-abundance genes have been understudied due to the technical difficulties in isolating these transcripts [56, 57]. Yet, low-abundance transcripts may have more specialized functions compared to high-abundance transcripts (which tend to have housekeeping functions). It has been suggested that a better understanding of low-abundance transcripts could lead to the identification of more suitable cancer-specific therapeutic targets [56]. A new appreciation for the importance of low-abundance genes in cancer biology has recently gained momentum due to technical advances that enable their examination systematically. RNA-seq can provide an unbiased snapshot of the transcriptome, including both high- and low- abundance genes [58, 59]. However, more sensitive approaches need to be developed to detect extremely low-abundance genes, such as hTERT. In addition, several groups have used PCR-based suppressive subtractive hybridization to determine the differentially regulated low-abundance and rare transcripts by comparing normal and cancer cells in a variety of cancer cells types [60-63]. Using this method, several previously unexamined low-abundance transcripts were identified as new biomarkers and potential targets for cancer therapy. Importantly, at the present time very little is known about how these genes are regulated.
Due to the direct correlation of telomerase activity with cancer cell immortality, telomerase is one of the very few low-abundance genes that have received wide-spread attention in the cancer field. Although some aspects of the precise mechanism leading up to aberrant hTERT splicing in cancer cells remain to be determined, the altered splicing of hTERT appears to be a necessary event as hTERT is often spliced into a similar pattern in telomerase-positive cancer cells [36]. In addition to changes in hTERT splicing, many oncogenes and tumor suppressor genes are aberrantly spliced in cancer cells [64, 65]. Recent exon expression arrays have shown differential inclusion/exclusion of exons when comparing cancer cells with normal cells [66]. It is possible that the newly identified low-abundance cancer-related transcripts may be subject to regulation by alternative splicing. Genome-wide studies have shown that changes in splicing factor expression may activate splicing program changes in many cancer-related genes that subsequently promote cancer growth [31, 67, 68]. This poses a new avenue for cancer therapeutics by developing approaches to alter alternative splicing. Any drug targeting splicing, however, will need to be restricted to a splicing factor that regulates a limited number of splicing events. Because most splicing factors participate in the splicing of a wide variety of genes, targeting such global splicing factors would be predicted to result in increased toxicity to both cancer and normal cells. Overcoming this challenge, however, may not be out of the realm of possibility. As the first of its kind, hTERT alternative splicing has been shown to be regulated differently from more abundant housekeeping transcripts, perhaps due to its low abundance. This suggests that examining the alternative splicing of other low-abundance cancer genes may provide unique opportunities for drug development.
Is hTERT splicing regulation a new paradigm or an isolated incident?
Although there is little current evidence for other low-abundance genes being regulated by alternative splicing, we believe that the unique mechanisms regulating telomerase (hTERT) splicing are not an isolated example. In 2011, it was reported that low-abundance human Acyl-CoA binding protein (ACBP) is regulated by alternative splicing and alternative promoter usage to achieve its multiple regulatory functions in the cell [69]. With advanced tools, the alternative splicing of low-abundance genes can now be more thoroughly examined, and additional examples such as ACBP may be found.
The discovery that hTERT alternative splicing is regulated by a VNTR that is far from the exon/intron junctions along with the potential use of RNA:RNA pairing as the mechanism to promote exon skipping is both provoking and puzzling. It begs the question whether hTERT alternative splicing regulation is one of a kind or may be widely applicable to other genes. If so, could this more specialized mode of splicing regulation have evolved for the fine-tuning of low-abundance genes with specialized functions?
Bioinformatics analyses have discovered many potential splicing factor binding sites including VNTRs far away from the intron/exon junctions in a number of low abundant genes besides hTERT that await validation as to whether those sites are bound by splicing factors that can affect alternative splicing from a distance [70]. VNTRs have been reported throughout the human genome with various functions identified [71]. The repeat sequence varies in length from 5bp (microsatellites) to longer repeat sequences (minisatellites), such as observed in the block of repeats in intron 6 in hTERT (38 bp). It is likely that other VNTRs exist, participating in alternative splicing regulation in a similar manner as hTERT, especially if it is highly conserved. In addition to the block of repeats in intron 6 of hTERT that was characterized, there are three additional blocks of repeats within the hTERT pre-mRNA. A second block of repeats in intron 6 is composed of repeats 36 bp long. Two blocks of repeats are in intron 2 with repeats of 61 bp and 42 bp. The consensus sequences of each of the four blocks of repeats are independent of one another. Although functioning differently, a 60 bp VNTR within exon 2 of MUC1 (a transmembrane mucin) makes up part of the extracellular domain (or cytoplasmic tail), which is important for cell-cell and cell-matrix interactions [72, 73]. The VNTR of MUC1 exon 2 can be recognized as an intron and is often spliced out in cancer cells but rarely in normal cells, which may result in changes in the interactions with the extracellular environment and support tumor growth [74].
RNA:RNA pairing is a splicing mechanism first observed for mutually exclusive exon choice in a few insects (such as Dscam in D. melanogaster and related species) [75], and until very recently it had not been seen in mammals [76]. It has now been demonstrated that the human splicing factor (SF1, also termed zinc finger protein 162:ZFM162) uses RNA:RNA pairing. hTERT may be the first examples of a VNTR participating in RNA:RNA pairing to regulate splicing in a mammalian gene. A caveat of the study is that it was performed using the hTERT minigene system. The precise RNA secondary structure the block of repeats forms in vivo is expected to be different than the one predicted since only selected portions of the hTERT sequence are included in the minigene. Furthermore, in vivo RNA folding is a dynamic interplay between the RNA binding proteins coating the RNA and the adaptation of the more stable secondary structure as nascent RNA is being transcribed. Confirmation of whether RNA:RNA pairing is the mechanism governing minus beta splicing in endogenous hTERT will require determination of the RNA secondary structure of hTERT in vivo, which at present is not known.
However, the unconventional ways hTERT alternative splicing is regulated appears to have been hinted at for other genes by large-scale genome-wide studies. The precise regulatory mechanisms for each gene, especially for low-abundance cancer-relevant transcripts, warrant careful examination to determine if alternative splicing manipulations are potential approaches for cancer therapeutics. Overall, hTERT alternative splicing regulation may pose a new paradigm for exploration in different types of splicing regulation.
Concluding Remarks
With the advance in technology, the depth of our understanding of alternative splicing has increased tremendously. The unconventional ways telomerase (hTERT) splicing is regulated shed new light on different mechanisms of splicing regulation. It remains to be determined how prevalent this type of splicing regulation is and whether a distinct mechanism has evolved to specifically regulate splicing of low-abundance transcripts with more specialized functions in the cell. A better understanding of the unique features of hTERT splicing in cancer cells may serve as a springboard for the wider study of cancer-specific low-abundance, alternatively spliced transcripts, and ultimately, the development of a novel class of highly specific anticancer therapeutics.
Box 1. The search for an ideal telomerase inhibitor.
Telomerase expression is almost entirely specific to cancer cells with the exception of proliferating stem cell progenitors. Due to the increased number of alterations that must occur in pre-cancerous cell evolution, the vast majority of pre-neoplastic and cancer cells have very short telomeres in comparison to the longer telomeres seen in the rarely dividing stems cells. Thus, inhibiting telomerase activity should in theory result in telomere shortening leading to apoptosis of cancer cells while having minimal effects on stem cells. For these reasons, telomerase has been well recognized as an attractive and almost universal target for cancer therapeutics.
Significant efforts have been expended to develop cancer therapeutics targeting telomerase, yet the development of telomerase inhibitors has been largely unsuccessful. A variety of approaches have been undertaken to inhibit telomerase activity, such as antisense oligonucleotides, ribozymes, G-quadruplex stabilizers, natural compounds, small molecule inhibitors (BIBR1532), and RNA interference [11, 77]. Although many of these telomerase inhibitor approaches have encouraging effects in in vitro systems, they rarely progress into clinical trials due to lack of potency, low specificity, and/or high toxicity.
A major challenge to developing an effective therapeutic against telomerase is that telomerase is expressed at low abundance in cancer cells. Therefore, therapeutic approaches against telomerase at the splicing level may be more advantageous by preventing the translation of functional TERT protein. An ideal telomerase-specific anticancer agent would be a small molecule that targets very few transcripts in addition to hTERT (minimizing toxicity) and one that alters splicing from functional hTERT to non-functional splice variants, thereby inhibiting the production of sufficient telomerase required to maintain telomere function.
Highlights.
Regulation of alternative splicing in abundant housekeeping genes is well-studied
Low-abundance gene regulation, including splicing, is understudied but may play key roles in cancer
TERT, a low-abundance transcript, is alternatively spliced via unique regulatory mechanisms
Understanding TERT splicing may provide new cancer therapy approaches
Acknowledgements
This work was supported by a pre-doctoral fellowship (BC100756 to M.S.W.); CA154805 (W.E.W.); the Simmons Cancer Center Support Grant (5P30 CA 142543-03), and the Southland Financial Corporation Distinguished Chair in Geriatric Research (W.E.W. and J.W.S.). This work was performed in laboratories constructed with support from National Institute of Health grant C06 RR30414
Glossary
- Alternative splicing
During pre-mRNA processing, particular exons of a gene maybe in included or excluded from the final messenger RNA. This process allows for the generation of multiple proteins with potentially multiple functions from a single gene in the genome.
- End-replication problem
During DNA replication, the lagging strand cannot be completely copied because no polymerase can fill in the resulting gap after the last RNA primer is removed. As a result, each round of replication generates shorter and shorter telomeres.
- Replicative senescence
Also known as the Hayflick limit. After so many cellular divisions when telomere ends are too short to for the stable T-loop structure, a DNA damage signal is induced and cells cease to divide.
- Splicing factor
Proteins that affect splice site selection by either directly binding to the pre-mRNA or directly through binding to other proteins.
- Variable number of tandem repeat (VNTR)
Short tandem repeats of a repetitive nucleotide sequence are dispersed throughout the genome. The repeat sequence varies in length and copy number. An array of functions has been assigned to VNTRs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Griffith JK, et al. Reduced telomere DNA content is correlated with genomic instability and metastasis in invasive human breast carcinoma. Breast cancer research and treatment. 1999;54:59–64. doi: 10.1023/a:1006128228761. [DOI] [PubMed] [Google Scholar]
- 4.Stansel RM, et al. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. The EMBO journal. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy MZ, et al. Telomere end-replication problem and cell aging. Journal of molecular biology. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 6.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. Journal of theoretical biology. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 7.Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 8.Wright WE, et al. Experimental elongation of telomeres extends the lifespan of immortal x normal cell hybrids. The EMBO journal. 1996;15:1734–1741. [PMC free article] [PubMed] [Google Scholar]
- 9.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. European journal of cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 10.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 11.Shay JW, Wright WE. The reactivation of telomerase activity in cancer progression. Trends in genetics : TIG. 1996;12:129–131. doi: 10.1016/0168-9525(96)30018-8. [DOI] [PubMed] [Google Scholar]
- 12.Shay JW, et al. Telomerase and cancer. Human molecular genetics. 2001;10:677–685. doi: 10.1093/hmg/10.7.677. [DOI] [PubMed] [Google Scholar]
- 13.Greider CW. Telomeres, telomerase and senescence. BioEssays : news and reviews in molecular, cellular and developmental biology. 1990;12:363–369. doi: 10.1002/bies.950120803. [DOI] [PubMed] [Google Scholar]
- 14.Wright WE, et al. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Yi X, et al. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic acids research. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xi L, et al. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic acids research. 2014 doi: 10.1093/nar/gku560. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen SB, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 18.Ulaner GA, et al. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. International journal of cancer. Journal international du cancer. 2001;91:644–649. [PubMed] [Google Scholar]
- 19.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt U, et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. The Journal of cell biology. 2011;193:819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstrohm AC, et al. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/s0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- 22.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Molecular cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 23.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & development. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 24.Tennyson CN, et al. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature genetics. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 25.Roca X, et al. Pick one, but be quick: 5' splice sites and the problems of too many choices. Genes & development. 2013;27:129–144. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graveley BR. Coordinated control of splicing and translation. Nature structural & molecular biology. 2005;12:1022–1023. doi: 10.1038/nsmb1205-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 28.Bonnal S, et al. The spliceosome as a target of novel antitumour drugs. Nature reviews. Drug discovery. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 29.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature reviews. Genetics. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 30.Tazi J, et al. Alternative splicing and disease. Biochimica et biophysica acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukong KE, et al. RNA-binding proteins in human genetic disease. Trends in genetics : TIG. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Padgett RA. New connections between splicing and human disease. Trends in genetics : TIG. 2012;28:147–154. doi: 10.1016/j.tig.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee A, et al. Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS genetics. 2008;4:e1000299. doi: 10.1371/journal.pgen.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature genetics. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 36.Wong MS, et al. Regulation of telomerase alternative splicing: a target for chemotherapy. Cell reports. 2013;3:1028–1035. doi: 10.1016/j.celrep.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes NM, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilian A, et al. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Human molecular genetics. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 39.Hrdlickova R, et al. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Molecular and cellular biology. 2012;32:4283–4296. doi: 10.1128/MCB.00550-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeboe-Larssen S, et al. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol. 2006;7:26. doi: 10.1186/1471-2199-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi X, et al. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colgin LM, et al. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia. 2000;2:426–432. doi: 10.1038/sj.neo.7900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaffaroni N, et al. Transcription and alternative splicing of telomerase reverse transcriptase in benign and malignant breast tumours and in adjacent mammary glandular tissues: implications for telomerase activity. J Pathol. 2002;198:37–46. doi: 10.1002/path.1178. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, et al. Differentiating alternative splice variant patterns of human telomerase reverse transcriptase in thyroid neoplasms. Thyroid. 2008;18(10):1055–64. doi: 10.1089/thy.2008.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Listerman I, et al. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer research. 2013;73:2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu S, et al. Inactive C-terminal telomerase reverse transcriptase insertion splicing variants are dominant-negative inhibitors of telomerase. Biochimie. 2014;101:93–103. doi: 10.1016/j.biochi.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 47.Hu BT, Insel RA. Up-regulation of telomerase in human B lymphocytes occurs independently of cellular proliferation and with expression of the telomerase catalytic subunit. Eur J Immunol. 1999;29:3745–3753. doi: 10.1002/(SICI)1521-4141(199911)29:11<3745::AID-IMMU3745>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 48.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman R, et al. hTERT antagonizes p53-induced apoptosis independently of telomerase activity. Oncogene. 2005;24:1320–1327. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 50.Cao Y, et al. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene. 2002;21:3130–3138. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- 51.Ninomiya K, et al. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. The Journal of cell biology. 2011;195:27–40. doi: 10.1083/jcb.201107093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong MS, et al. Regulation of human telomerase splicing by RNA:RNA pairing. Nature communications. 2014;5:3306. doi: 10.1038/ncomms4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 54.Merlo LM, et al. Cancer as an evolutionary and ecological process. Nature reviews. Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 55.Carninci P, et al. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome research. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim YC, et al. Pan-genome isolation of low abundance transcripts using SAGE tag. FEBS letters. 2006;580:6721–6729. doi: 10.1016/j.febslet.2006.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, et al. Detecting novel low-abundant transcripts in Drosophila. Rna. 2005;11:939–946. doi: 10.1261/rna.7239605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilad Y, et al. Characterizing natural variation using next-generation sequencing technologies. Trends in genetics : TIG. 2009;25:463–471. doi: 10.1016/j.tig.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diatchenko L, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bizama C, et al. The low-abundance transcriptome reveals novel biomarkers, specific intracellular pathways and targetable genes associated with advanced gastric cancer. International journal of cancer. Journal international du cancer. 2014;134:755–764. doi: 10.1002/ijc.28405. [DOI] [PubMed] [Google Scholar]
- 62.Liu BH, et al. Identification of unique and common low abundance tumour-specific transcripts by suppression subtractive hybridization and oligonucleotide probe array analysis. Oncogene. 2008;27:4128–4136. doi: 10.1038/onc.2008.50. [DOI] [PubMed] [Google Scholar]
- 63.Barraclough DL, et al. Microarray analysis of suppression subtracted hybridisation libraries identifies genes associated with breast cancer progression. Cellular oncology : the official journal of the International Society for Cellular Oncology. 2010;32:87–99. doi: 10.3233/CLO-2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. Journal of cell science. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 65.Venables JP. Unbalanced alternative splicing and its significance in cancer. BioEssays : news and reviews in molecular, cellular and developmental biology. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 66.Brosseau JP, et al. Tumor microenvironment-associated modifications of alternative splicing. Rna. 2014;20:189–201. doi: 10.1261/rna.042168.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie W, et al. Entropy measures quantify global splicing disorders in cancer. PLoS computational biology. 2008;4:e1000011. doi: 10.1371/journal.pcbi.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim E, et al. Insights into the connection between cancer and alternative splicing. Trends in genetics : TIG. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Nitz I, et al. Specific regulation of low-abundance transcript variants encoding human Acyl-CoA binding protein (ACBP) isoforms. Journal of cellular and molecular medicine. 2011;15:909–927. doi: 10.1111/j.1582-4934.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gemayel R, et al. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual review of genetics. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 72.Hilkens J, et al. Cell membrane-associated mucins and their adhesion-modulating property. Trends in biochemical sciences. 1992;17:359–363. doi: 10.1016/0968-0004(92)90315-z. [DOI] [PubMed] [Google Scholar]
- 73.Fukuda M. Roles of mucin-type O-glycans in cell adhesion. Biochimica et biophysica acta. 2002;1573:394–405. doi: 10.1016/s0304-4165(02)00409-9. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, et al. Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms. Cancer immunology, immunotherapy : CII. 2013;62:423–435. doi: 10.1007/s00262-012-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pervouchine DD, et al. Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. Rna. 2012;18:1–15. doi: 10.1261/rna.029249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White LK, et al. Telomerase inhibitors. Trends in biotechnology. 2001;19:114–120. doi: 10.1016/s0167-7799(00)01541-9. [DOI] [PubMed] [Google Scholar]