Abstract
Inflammatory bowel diseases (IBD) are chronic, progressive diseases characterized by aberrant immune responses to environmental and gut microbial triggers in genetically susceptible hosts. Clinical, genetic, and experimental data support the role of gut microbes in causing and sustaining these diseases. Our understanding of IBD has changed dramatically as the result of advances in cultivation-independent approaches and computational platforms for the analysis of large data sets. However, investigations relevant to clinical observations and the natural history of the diseases will be essential for the development of microbial, genetic, and biological metrics that may be used to individualize assessment of risk and improve clinical outcomes in IBD.
The inflammatory bowel diseases (IBD) are a group of chronic diseases that result from a confluence of genetic and environmental triggers. IBD have two main clinical phenotypes, ulcerative colitis (UC) and Crohn’s disease (CD). UC is characterized by mucosal inflammation that is limited to the colon, beginning in the rectum and extending proximally in a continuous and circumferential fashion. CD, in contrast, can occur at any location in the gastrointestinal tract, from mouth to anus, though each patient’s CD typically involves a specific region, such as the small bowel or colon. CD is characterized by focal lesions that progress to ulcerations that can extend through the bowel wall and result in the development of fistulas, abscesses, and strictures.
A microbial basis for IBD has long been suspected, starting with the early descriptions of potential infectious agents associated with UC in the 19th century and CD in the early 20th century (1, 2). Despite these associations, no single agent has been proven to cause IBD. As a result, research and clinical management strategies shifted away from the notion that IBD are infectious diseases, and instead toward understanding and treating the underlying immune and inflammatory processes. However, the development of cultivation-independent approaches to characterize structural and functional profiles of complex microbial communities revealed many new insights into the relationships between host and microbes. At the same time, the pivotal role of indigenous gut microbes in causing IBD became apparent with the first descriptions of IBD susceptibility genes, as many were involved in mediating host responses to gut microbes.
Most experts now recognize that intestinal dysbiosis, or the imbalance in the structural and/or functional properties of the gut microbiota that can disrupt host-microbe homeostasis, is integral to the pathogenesis of IBD (3–5). However, the field remains descriptive, and some of the most basic questions about the role of the gut microbiota in IBD remain unanswered. Is the intestinal dysbiosis a cause or consequence of IBD? Are these diseases caused by the emergence of pathobionts and/or disappearance of symbionts, or merely due to an aberrant host immune response to commensal microbiota? Are the host-microbe disturbances constant or changing throughout the natural history of these diseases? What role do environmental and dietary factors play in determining the risk and course of IBD? Can IBD be prevented or treated by correcting the intestinal dysbiosis? Answering these questions will significantly advance the field and improve the understanding and management of human IBD. The clinical ramifications of the gut dysbiosis in IBD can only be sorted out by relating these findings to the natural history and complexities of these diseases.
Host factors affecting microbial assemblage and IBD etiopathogenesis
The assemblage of gut microbes throughout life is not stochastic, but determined by a combination of host, microbial (i.e., self-selective), and environmental factors. Genome-wide association studies (GWASs) provided some of the first insights into the host-microbe connection in IBD. Over 160 SNPs associated with increased risk for IBD have been identified (6). Of the genes and gene loci known to be associated with IBD susceptibility, many appear to be involved in processes critical for determining how the host responds to microbes, including barrier function, wound healing, autophagy, immune balance, and stress responses (6–8). Nucleotide-binding oligomerization domain–containing protein 2 (NOD2, also known as CARD15) was the first of these genes to be identified and encodes an intracellular receptor for the bacterial peptidoglycan muramyl dipeptide (MDP) (9, 10). This discovery and that of other IBD susceptibility genes (e.g., ATG16L1, IRGM, CARD9, and IL23R) shed light on fundamental relationships between host genetics with gut microbes, particularly with regard to innate and adaptive immune functions that affect the assemblage of the gut microbiome (8). Mutations in NOD2 and other genes that interact with it, for instance, are implicated in causing the intestinal dysbiosis associated with CD, possibly through altered states of innate and adaptive immune balance, impairment of autophagy, defective Paneth cell function, and perturbations of non-MDP–dependent functions (11, 12). Nod2-deficient mice have significantly increased Bacteroides, Firmicutes, and Bacillus bacterial loads in the terminal ileum, as well as decreased ability to clear a pathogenic bacterial species, Helicobacter hepaticus (13). Studies in human patients have also shown that the presence of a NOD2 risk allele (Leu1007fs, R702W, or G908R) is associated with shifts in microbial composition, such as decreased Clostridium Group XIVa and IV and increased Actinobacteria and Proteobacteria, in surgical specimens from CD and UC patients as compared with controls (14, 15). The luminal bacteria, in turn, appear to feed forward to affect Nod2 gene expression, because germ-free mice have less Nod2 expression in the terminal ileum. Conventionalization of these mice with gut microbiota restores Nod2 expression (13). On the other hand, the dysbiosis associated with Nod2-deficient mice appears to confer risk of colitis. Wild-type mice co-housed with Nod2-deficient mice developed greater inflammation in a model of chemically induced colitis and exhibited enhanced expression of genes related to apoptosis, necrosis, and tumorigenesis (16). The findings further raise the possibility that the maternal transmission of a disease-promoting microbiota to offspring might contribute to the risk of disease in familial forms of IBD.
While genetic risk is an important factor in IBD and can influence the assemblage of gut microbes in individuals, it is not likely to be sufficient in causing disease except in rare cases of aggressive, early-onset IBD such as the disease caused by deficiency of IL-10 or the IL-10 receptor (17). This partial role for genetics is highlighted by the concordance rate for CD between monozygotic twins, which is between 35% and 58%, while that for UC is only 16% to 18.5% (18–20). In both cases, these rates are much higher than that of the general population and dizygotic twins, which supports a genetic basis for IBD. However, the fact that the penetrance is not 100% strongly suggests that other, non-genetic factors are involved in determining disease, such as the gut microbiome, environmental factors, or epigenetic modifications acquired through life. These types of disease modifiers can potentially trigger the onset of disease and influence the phenotype and clinical course. For example, the T300A variant in ATG16L1 is associated with ileal CD, which may be related to disturbances in Paneth cell function and increased IL-1β production in response to MDP (21). However, many individuals in the general population carry this variant and remain healthy (21). In fact, the T300A variant increases disease risk by less than 2-fold, which suggests a role for other factors that interact with the gene to confer risk. Along these lines, mice hypomorphic for the Atg16l1 mutation exhibit Paneth cell abnormalities, such as abnormal lysozyme granule packaging (22). When these mice are infected with murine norovirus and then challenged with the colitis-inducing agent dextran sodium sulfate (DSS), the mice develop an intestinal pathology resembling CD. In contrast, Atg16l1-deficient mice infected with a nonreplicative norovirus develop far less severe inflammation.
Dietary and environmental factors affecting the gut microbiome
One of the most striking features of complex immune disorders such as IBD is the alarming increase in their incidence and prevalence over the past century, particularly in highly developed urban populations. A systematic review of population-based IBD data revealed that among studies showing a significant rise in incidence, the average annual percentage change ranged from 1.2% to 23.3% in CD and from 2.4% to 18.1% in UC over study periods from 1920 to 2010 (23). The rapid pace of these developments cannot be explained by genetic drift, but is more likely due to shifts in environmental factors and societal norms brought on by cultural westernization. Conditions that have been implicated in changing the gut microbiome of Western societies include the exposure to environmental pollutants and xenobiotics, wide and indiscriminate use of antibiotics by livestock producers and in medical practice, and lifestyle changes (e.g., diet, stress, and sleep disorders). Turnbaugh and colleagues have shown that short-term exposure to a specific medication, digoxin, elicited effects on the physiology, structure, and gene expression of the microbiome, which in turn altered the drug’s action on the host (24). The Blaser group studied the impact of sub-therapeutic antibiotic therapy (STAT) on gut microbes, a practice widely used in livestock to promote weight gain for food production (25). Mice exposed to STAT after weaning exhibited increased adiposity, changes in hepatic lipid metabolism, and altered immune balance that was associated with significant changes in gut microbial membership and function. In addition, STAT increased levels of colonic short-chain fatty acids (SCFAs), products of microbial fermentation that have many trophic biological effects on the host. The authors concluded from these findings that exposure of humans to antibiotics early in life could have long-lasting effects on metabolism and development. A study of Danish children from 1995 to 2003 provides further support for this notion, finding a 1.84-fold greater relative risk for IBD in children that received antibiotics as compared with those who did not (26). Caesarean sections are another common feature of Western medical practice that can affect the acquisition of the gut microbiome and possibly increase the risk of IBD. A Danish cohort study of subjects born from 1973 to 2008 found that Caesarean section was associated with a modest increased risk of IBD in children in the age range of 0 to 14 (27). Assuming causality, the authors estimated 3.2% of IBD cases diagnosed before the age of 15 were attributable to Caesarean section.
The evolution of human dietary patterns over time and the recent popularization of the high-fat, high-sugar Western diet also represent major selective forces in microbiome assembly. African children on a high-fiber, plant-based diet exhibit a vastly different gut microbial community than their European counterparts whose dietary intake consists of a Western diet rich in sugar, fat, and protein (28). Such shifts in microbial assemblage have consequences for intestinal health, as was demonstrated in a study of genetically susceptible IL-10–deficient mice fed a diet rich in saturated milk fat. This diet promoted a bloom of Bilophila wadsworthia, which increased the incidence and severity of spontaneous colitis in these animals (29). Similarly, Turnbaugh and colleagues placed human subjects on either an animal-based diet of meats, eggs, and cheese, or a plant-based diet of grains, legumes, fruits, and vegetables, and compared microbial community structures in the subjects’ stool (30). The subjects on an animal-based diet had increased levels of bile-tolerant organisms such as Alistipes and B. wadsworthia and decreases in Firmicutes that metabolize plant polysaccharides, such as Roseburia, Eubacterium rectale, and Ruminococcus bromii.
While the highest rates of IBD have been reported in Europe and North America, the incidence has also risen in Asia over the past 50 years, particularly in Westernized nations (23, 31–33). One popular theory for this trend, dubbed the “hygiene hypothesis,” postulates that the adoption of hygienic practices has reduced exposure to important environmental and microbial cues necessary for proper immune development, resulting in perturbations that trigger aberrant immune responses in genetically susceptible hosts (34). For example, one Indian study found higher rates of IBD in homes with safe drinking water, whereas the presence of cattle in the home compound was negatively correlated with IBD (33). Kasper and colleagues proposed that the natural selection of host commensal microbes is necessary for the proper development of the intestinal immune system (35). This group colonized germ-free mice with mouse or human microbiota and found that the latter group exhibited fewer CD4+ and CD8+ T cells, proliferating T cells, and dendritic cells and lower antimicrobial peptide expression. Collectively, these studies suggest that driving factors associated with increased westernization perturb critical, evolutionarily determined host-microbe relationships, triggering diseases such as IBD in individuals unable to compensate for these changes.
Intestinal dysbiosis in IBD
Through rapid advances in cultivation-independent technologies and bioinformatics, the intestinal dysbiosis in active human IBD and experimental colitis has been fairly well described. Frank et al. found that CD and UC patients had depletion of members of Bacteroidetes and Lachnospiraceae (group IV and XIVa Clostridia) (4). Several other groups have described decreased biodiversity, with a lower proportion of Firmicutes and increased Gammaproteobacteria and Enterobacteriaceae in UC and CD patients (36–38). UC patients also have increased levels of sulfite-reducing Deltaproteobacteria (39). Faecalibacterium prausnitzii is a common intestinal microbial species that secretes metabolites that have antiinflammatory properties (40, 41). In one study, CD patients were observed to have lower levels of F. prausnitzii than healthy controls. Furthermore, depletion of this bacterium was predictive of postoperative disease recurrence in patients who had undergone ileal resection (42). A recent analysis of the mucosal-associated bacteria in ileal and rectal biopsies from treatment-naive, newly diagnosed pediatric CD patients has shown increased abundance in Enterobacteriaceae, Pasteurellaceae, Veillonellaceae, and Fusobacteriaceae and decreased abundance of Erysipelotrichales, Bacteroidales, and Clostridiales (43).
Bacteriophages far outnumber bacteria themselves and may represent another mechanism by which shifts in the bacterial community and dysbiosis occur. Lepage et al. reported on the abundance of virus-like particles (VLPs) from CD and healthy mucosal biopsy samples and found them to be quite abundant, on the order of 1.2 × 109 VLPs per mucosal biopsy (44). Each individual appeared to have one dominant phage type. In addition, VLPs appear to be more abundant in CD patients than in healthy controls, and localize more frequently to non-ulcerated areas of mucosa than to ulcerated regions. Another study of phages reported lower phage diversity and greater inter-individual variability among fecal samples from CD patients compared with healthy subjects (45). The functional significance of these findings remains unknown, due to the lack of reference genome inventories needed for identification and functional classification of bacteriophages.
The role of fungi and Archaea in IBD is less well understood. The intestines of several mammalian species, including humans, also harbor fungal communities that interact with the host immune system via receptors such as Dectin-1 (46). Dectin-1 recognizes the β1,3 glucans found in the fungal cell wall and is necessary for phagocytosis and killing of fungi by myeloid phagocytes. Several lines of evidence suggest fungi may be relevant to IBD. Dectin-1 signals through CARD9, a gene with many UC-associated variants. A SNP for CLEC7A (the gene encoding Dectin-1) appears to be associated with medically refractory UC. Moreover, mice lacking Dectin-1 are more susceptible to DSS-induced colitis. Thus, several plausible links exist between the ability of the host to respond immunologically to intestinal fungi and susceptibility to inflammation. A few studies in humans have also observed increased fungal diversity in patients with CD, particularly in inflamed areas, based on 18S ribosomal gene sequencing (47). While few studies have looked at the Archaea in the intestine, data suggest that CD and UC patients have fewer methanogens as compared with healthy controls (48). Methanogens remove end products of fermentation, and a decrease in their membership could be associated with metabolic dysfunction.
While general trends in gut dysbiosis are observed in IBD, the question now becomes how these changes affect host immune states and responses. The risk of onset or relapse of IBD can also be triggered by decreases in function and/or representation of symbionts of the gut microbiota, such as microorganisms with immunomodulatory cell wall agents such as polysaccharide A (PSA), or that express quorum-sensing molecules such as competence sporulation factor (CSF). Mazmanian et al. showed that the symbiont Bacteroides fragilis can protect mice from H. hepaticus–induced inflammation in an experimental colitis model through the action of PSA (49). PSA is able to suppress proinflammatory cytokine IL-17 production and provide protection from colitis by inducing protective IL-10 production. Bacillus subtilis–derived quorum-sensing pentapeptide CSF is taken up by the membrane transporter OCTN2 to activate key survival pathways such as heat shock proteins in intestinal epithelial cells (50). Studies of probiotic mechanisms have also shown that soluble bacterial components can modulate the host immune system (51–53). These data illustrate that many bacterial components contribute to the maintenance of intestinal and immune homeostasis.
Bacterial fermentation products such as SCFAs can also affect the host immune system. A decrease in the bacterial populations that produce SCFAs from the metabolism of fiber is a common observed trend in IBD. Atarashi et al. isolated a group of SCFA producing bacterial strains within Clostridia cluster IV, XIVa, XVII from human stool and found that these strains induced colonic Treg differentiation, accumulation, and function (54). These strains also reduced the severity of trinitrobenzene sulphonic acid–induced (TNBS-induced) colitis in mice, an action possibly mediated by the increased bioavailability of SCFAs to elicit TGF-β and promote the differentiation and expansion of Tregs. Others have similarly reported decreased SCFAs and Treg function in human IBD and other models of experimental colitis (4, 5, 55). Garrett and colleagues has shown that SCFAs regulate the size and function of the colonic Treg pool (56). In Western diets low in plant fiber, decreased microbial production of SCFAs potentially reduces colonic Treg development, shifting immune homeostasis to a more proinflammatory state (56). The effects of SCFAs on Treg expression appeared to be in part mediated by the G protein–coupled receptor GPR43 (gene symbol Ffar2), which then promotes epigenetic modifications that may involve the inhibition of histone deacetylase (56, 57).
The loss of beneficial microbes and their products can also provide opportunity for the emergence of disease-promoting microbes that produce microbial metabolites with negative effects on the intestine under inflammatory conditions. Patients with UC have increased representation of sulfur-reducing, bile-tolerant bacteria in their gut microbiota, with concomitant increases in production of toxic, proinflammatory hydrogen sulfide (39, 58). Many have strong antioxidant capabilities, allowing them to survive the hostile host-induced inflammatory environment (39). In non-transformed colonic epithelial cells, H2S can be genotoxic at concentrations found in the colon. H2S has also been shown to modulate expression of genes involved in cell cycle progression, trigger immune and inflammatory responses, as well as impair DNA repair (59). H2S also inhibits SCFA oxidation and utilization properties (60). One of the two enzymes responsible for detoxification of H2S, thiosulfate sulfur transferase, has recently been described as significantly decreased in patients with active UC and can be restored upon successful medical treatment (61). Enterobacteriaceae have also been implicated in IBD, as the relative abundance of these facultative anaerobes is increased in active disease (36–38, 40). Two Enterobacteriaceae species, Klebsiella pneumoniae and Proteus mirabilis, isolated from TRUC mice (mice deficient in innate and adaptive immunity that develop spontaneous UC-like inflammation) can elicit colitis in wild-type mice in the presence of the endogenous microbiota (62, 63). A recent study showed that Enterobacteriaceae such as E. coli are able to utilize nitrate generated as a by-product of the inflamed gut, and can therefore outcompete bacteria that require fermentation substrates (64). These microorganisms may therefore contribute to the inflammatory process by suppressing commensal microbiota seen in health and inciting mechanisms that activate immune and inflammatory processes.
Assessing the communal function of the gut microbiota has been a challenge. Several functional metagenomic analyses of the inflammation-associated gut microbiota have been reported, although these data have been limited by incomplete inventories of annotated functional gene sets (65). Taking a different approach, Morgan et al. analyzed fecal specimens and intestinal biopsies from individuals with IBD and from healthy subjects to develop inferred microbial functions based on reference genomes of well-represented taxa, with confirmation of some samples via shotgun metagenomic sequencing (66). Twelve percent of total metabolic modules were differentially abundant between patients with IBD and controls, while only 2% of genus-level clades reached this significance threshold. Changes in microbial function therefore appeared more consistent and informative than changes in community structure. Notably, increases in glycolysis and carbohydrate transport/metabolism and decreases in lipid metabolism and catabolism modules were observed in ileal CD, suggesting perturbations in overall microbial energy metabolism. In addition, increased representation of genes involved in secretion systems associated with cell wall degradation and exotoxins as well as microbial adherence/invasion were observed in ileal CD. These data suggest a complex interplay between the host and microbiota in which microbial functions that allow microbes to survive in and promote the inflammatory environment are selected. Greenblum et al. utilized a shotgun metagenomic data set from patients with IBD and subjects without IBD, integrating this data with an in silico systems analysis of metabolic networks (67). The microbial enzyme level variation that was associated with IBD appeared to be related to the way in which the microbiome interacted with the host gut environment, rather than with core metabolic processes. Gevers et al. analyzed treatment-naive pediatric patients with CD; they used the PICRUSt algorithm to estimate the functional changes in the CD patient mucosal-associated microbial communities, and predicted the switch toward auxotrophy with increases in aerobic or aerotolerant taxa such as Proteobacteria and Pasteurellaceae (43). Shotgun metagenomic sequencing of a subset of stool samples from this cohort suggests that the bacterial species that are increased in CD patients contribute components of the glycerophospholipid and lipopolysaccharide metabolism pathways thought to initiate inflammation and also contribute phosphonoacetate hydrolase, which provides access to novel carbon and phosphate sources.
Studies of individual bacterial species’ transcriptomes have also revealed interesting insights into the effects of inflammation on microbial function and the ability to survive in the inflammatory milieu. Analysis of E. coli NC101, a non-pathogenic murine strain isolated from Il10–/– mice with colitis, demonstrated upregulation of the stress response operon, including the small heat shock proteins IbpA and IbpB, which participate in protection from oxidative stress (68). Induction of these bacterial stress response proteins in the inflamed intestine may serve to reduce inflammation, as expression of IbpA and IbpB reduced histologic signs of inflammation, secretion of IL-12/23p40 from colon tissue explants, and secretion of interferon-γ by stimulated mesenteric lymph node cells in vitro. IBD patients also have increased numbers of adherent-invasive E. coli (AIEC) and increased expression of CEACAM6, to which the FimH component of type I AIEC pili specifically adheres. FimH is subject to amino acid substitutions, and specific FimH mutations have been associated with CD and UC patients and may be a mechanism by which AIEC proliferates (69, 70). Thus, the inflammatory environment may select for expression of certain bacterial proteins that affect microbial survival and the perpetuation of the host’s inflammation.
In summary, despite the large amount of information that describes the gut dysbiosis of IBD, many questions remain about the microbial role and mechanisms that are involved in human IBD. As studies become more focused on the functional consequences of microbial shifts and investigators appreciate the subtleties of host age, medications, diet, and environment on the microbiota, there is every reason to believe that solutions to overcome these challenges will be found.
Closing the gap between clinical and microbial investigations
Clinical investigators who study IBD are often unfamiliar with microbial and ecological concepts, while most microbial ecologists are adept at large-scale surveys but unaware of the nuances of IBD. Thus, bringing the two sides together to leverage their respective expertise represents one of the major challenges at hand. The next phase of research investigation of the gut microbiome in IBD should be guided by specific biological questions relevant to the clinical aspects and natural history of the disease. For instance, the intestinal dysbiosis associated with active IBD was initially felt to be irrelevant to what causes these diseases, i.e., merely a consequence of the aberrant immune and inflammatory process. However, these changes may in fact be important in sustaining these diseases, promoting the development of long-term complications, and contributing to poor responses to certain therapeutic approaches.
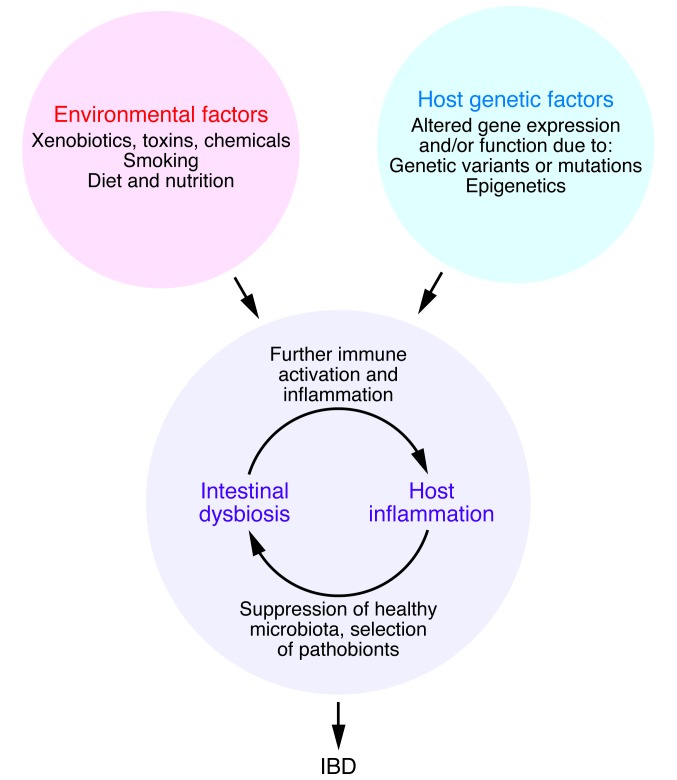
The chronicity of many diseases arises from a series of self-reinforcing events that create a new steady state that is difficult to interrupt (Figure 1). In IBD, the inflammatory milieu created by altered immune states selects against commensal microbes associated with health in favor of those that can survive and flourish in this hostile environment. Many of these microbes are also highly proinflammatory (pathobionts), a property that confers a selective advantage over the return of commensal microbes by maintaining the host inflammatory state. In IBD, immune homeostasis cannot be easily restored, because of inherent defects or inability to shut down the immune activation and correct the proinflammatory intestinal dysbiosis. Therefore, interventions aimed at correcting only one facet of this vicious cycle (e.g., solely targeting the host or only targeting gut microbes) are not likely to be successful. Broad-spectrum antibiotics, for instance, have not been useful in IBD as primary therapy because they suppress the return of a diverse microbiota and do little in mitigating the host inflammatory/immune derangements (71–74). Probiotics and prebiotics are also of limited value because the former cannot survive in an inflammatory environment and the latter require a diverse microbiota associated with health that is not present in the inflammatory state to mediate their actions. Fecal microbiota transplant (FMT) or interventions with defined, complex communities of microbiota are theoretically better than probiotics because the microbial populations are assembled and self-selected under healthy conditions. However, they too are not likely to be effective in moderate or severe cases of IBD in absence of other therapies directed against the host inflammatory and immune dysregulation (75). Even then, the question remains how long the membership, diversity, and function of the transferred microbiota can be sustained under different set of conditions presented by the new host’s genetic, environmental, and physiological factors. The correct strategy in any scenario will likely call for a personalized, multi-pronged approach aimed at halting the chain of events that maintains the disease-sustaining steady state.
Figure 1. IBD are caused by a confluence of factors that perturb the balance between the environment, gut microbes, and host.
IBD are believed to arise from a “perfect storm” that involves the confluence of environmental and genetic factors tipping the balance between host immune and gut microbial factors and thus triggering disease in genetically susceptible individuals. IBD are maintained by pathophysiological host processes and events that feed forward to promote a dysbiotic microbiota that can in turn worsen inflammation and create a vicious loop.
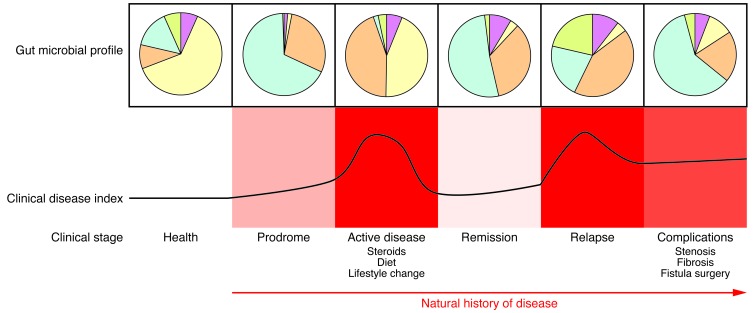
IBD are progressive diseases that arise from a wide variety of pathophysiological processes (Figure 2). Changes in gut microbial composition and function are likely to evolve through the course (natural history) of IBD, reflecting transitions in host-microbe relationships arising from disease-intrinsic and confounding factors. Microbial factors that trigger the onset of disease may be quite different from those that sustain the inflammatory process or are a consequence of long-term complications and interventions. The interpretation of gut microbial data in absence of this contextual information can be limited and potentially misleading.
Figure 2. IBD are chronic, progressive diseases.
The composition and function of the gut microbiota likely change through the course (natural history) of IBD, reflecting transitions in host-microbe relationships that arise from disease-intrinsic and confounding factors. Microbial factors that trigger the onset of disease may be quite different from those that sustain the inflammatory process or result from the consequences of long-term complications and interventions. The interpretation of gut microbial data in the absence of this contextual information can be limited and potentially misleading. The pie charts in this figure illustrate the concept of general shifts in microbial composition and/or function over time and are not meant to indicate any quantifiable information.
Along the same lines, the same treatment or intervention is unlikely to work for all cases and stages of IBD. Our interventional goals should therefore be guided by a combination of microbial, genetic, and biological metrics that assess risk and states of health of patients on an individual basis. As we approach the era of personalized medicine, this strategy is most likely to yield the best and most sustainable clinical outcomes.
Acknowledgments
The authors received funding from the NIH grants P30 DK42086, R37 DK47722, R01 DK097268, and T32 DK07074 (to E.B. Chang) and F32 DK09025 (to S.R. Dalal), the Leona M. and Harry B. Helmsley Charitable Trust, and the Gastro-Intestinal Research Foundation of Chicago. Also special thanks to Kyle Dolan, Vanessa Leone, and Jeannette Messer for their help in editing the manuscript. This manuscript is dedicated to the memory of Joseph B. Kirsner, a pioneer in the study of IBD and a tireless advocate for his IBD patients, and to Lloyd Mayer, who will always be remembered for his contributions to the field, collegiality, and steadfast friendship.
Footnotes
Conflict of interest: Eugene B. Chang received research funding support from Tsumura & Co. to study natural products.
Reference information:J Clin Invest. 2014;124(10):4190–4196. doi:10.1172/JCI72330.
References
- 1.Kirsner JB. Historical aspects of inflammatory bowel disease. J Clin Gastroenterol. 1988;10(3):286–297. doi: 10.1097/00004836-198806000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Dalziel TK. Chronic interstitial enteritis. Br Med J [Clin Res]. 1913;2:1068–1070. [Google Scholar]
- 3.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 10.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 11.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaser A, Blumberg RS. Autophagy, microbial sensing, endoplasmic reticulum stress, and epithelial function in inflammatory bowel disease. Gastroenterology. 2011;140(6):1738–1747. doi: 10.1053/j.gastro.2011.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petnicki-Ocwieja T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106(37):15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(1):179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li E, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7(6):e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123(2):700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah N, Kammermeier J, Elawad M, Glocker E-O. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Curr Allergy Asthma Rep. 2012;12(5):373–379. doi: 10.1007/s11882-012-0286-z. [DOI] [PubMed] [Google Scholar]
- 18.Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14(7):968–976. doi: 10.1002/ibd.20380. [DOI] [PubMed] [Google Scholar]
- 19.Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35(10):1075–1081. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 20.Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124(7):1767–1773. doi: 10.1016/S0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 21.Prescott NJ, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132(5):1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152(1–2):39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60(1):49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- 27.Bager P, Simonsen J, Nielsen NM, Frisch M. Cesarean section and offspring’s risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis. 2012;18(5):857–862. doi: 10.1002/ibd.21805. [DOI] [PubMed] [Google Scholar]
- 28.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10–/– mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thia KT, Loftus EVJ, Sandborn WJ, Yang S-K. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008;103(12):3167–3182. doi: 10.1111/j.1572-0241.2008.02158.x. [DOI] [PubMed] [Google Scholar]
- 32.Zheng JJ, Zhu XS, Huangfu Z, Shi XH, Guo ZR. Prevalence and incidence rates of Crohn’s disease in mainland China: a meta-analysis of 55 years of research. J Dig Dis. 2010;11(3):161–166. doi: 10.1111/j.1751-2980.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 33.Pugazhendhi S, Sahu MK, Subramanian V, Pulimood A, Ramakrishna BS. Environmental factors associated with Crohn’s disease in India. Indian J Gastroenterol. 2011;30(6):264–269. doi: 10.1007/s12664-011-0145-1. [DOI] [PubMed] [Google Scholar]
- 34.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 35.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giaffer MH, Holdsworth CD, Duerden BI. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J Med Microbiol. 1991;35(4):238–243. doi: 10.1099/00222615-35-4-238. [DOI] [PubMed] [Google Scholar]
- 37.Seksik P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52(2):237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44(11):4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roediger WE, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci. 1997;42(8):1571–1579. doi: 10.1023/A:1018851723920. [DOI] [PubMed] [Google Scholar]
- 40.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 41.Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol. 2010;26(4):327–331. doi: 10.1097/MOG.0b013e328339536b. [DOI] [PubMed] [Google Scholar]
- 42.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepage P, Colombet J, Marteau P, Sime-Ngando T, Dore J, Leclerc M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57(3):424–425. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Brocal V, et al. Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin Transl Gastroenterol. 2013;4:e36. doi: 10.1038/ctg.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott SJ, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43(7):831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 48.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 50.Fujiya M, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1(4):299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Yan F, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121(6):2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, et al. Probiotic yeast inhibits VEGFR signaling and angiogenesis in intestinal inflammation. PLoS One. 2013;8(5):e64227. doi: 10.1371/journal.pone.0064227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrof EO, et al. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127(5):1474–1487. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 55.Vital M, et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 2013;1(1):8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine J, Ellis CJ, Furne JK, Springfield J, Levitt MD. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93(1):83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- 59.Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen. 2010;51(4):304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 60.Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Preter V, et al. Decreased mucosal sulfide detoxification is related to an impaired butyrate oxidation in ulcerative colitis. Inflamm Bowel Dis. 2012;18(12):2371–2380. doi: 10.1002/ibd.22949. [DOI] [PubMed] [Google Scholar]
- 62.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8(3):292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339(6120):708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3(6):417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109(2):594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patwa LG, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011;141(5):1842–1851. doi: 10.1053/j.gastro.2011.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dreux N, et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9(1):e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iebba V, et al. Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease. Infect Immun. 2012;80(4):1408–1417. doi: 10.1128/IAI.06181-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scribano ML, Prantera C. Use of antibiotics in the treatment of Crohn’s disease. World J Gastroenterol. 2013;19(5):648–653. doi: 10.3748/wjg.v19.i5.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borgaonkar MR, MacIntosh DG, Fardy JM. A meta-analysis of antimycobacterial therapy for Crohn’s disease. Am J Gastroenterol. 2000;95(3):725–729. doi: 10.1111/j.1572-0241.2000.01842.x. [DOI] [PubMed] [Google Scholar]
- 73.Colombel JF, et al. A controlled trial comparing ciprofloxacin with mesalazine for the treatment of active Crohn’s disease. Groupe d’Etudes Thérapeutiques des Affections Inflammatoires Digestives (GETAID). Am J Gastroenterol. 1999;94(3):674–678. doi: 10.1111/j.1572-0241.1999.935_q.x. [DOI] [PubMed] [Google Scholar]
- 74.Arnold GL, Beaves MR, Pryjdun VO, Mook WJ. Preliminary study of ciprofloxacin in active Crohn’s disease. Inflamm Bowel Dis. 2002;8(1):10–15. doi: 10.1097/00054725-200201000-00002. [DOI] [PubMed] [Google Scholar]
- 75.Angelberger S, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol. 2013;108(10):1620–1630. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]