Abstract
The transport and intracellular trafficking of heme biosynthesis intermediates are crucial for hemoglobin production, which is a critical process in developing red cells. Here, we profiled gene expression in terminally differentiating murine fetal liver-derived erythroid cells to identify regulators of heme metabolism. We determined that TMEM14C, an inner mitochondrial membrane protein that is enriched in vertebrate hematopoietic tissues, is essential for erythropoiesis and heme synthesis in vivo and in cultured erythroid cells. In mice, TMEM14C deficiency resulted in porphyrin accumulation in the fetal liver, erythroid maturation arrest, and embryonic lethality due to profound anemia. Protoporphyrin IX synthesis in TMEM14C-deficient erythroid cells was blocked, leading to an accumulation of porphyrin precursors. The heme synthesis defect in TMEM14C-deficient cells was ameliorated with a protoporphyrin IX analog, indicating that TMEM14C primarily functions in the terminal steps of the heme synthesis pathway. Together, our data demonstrate that TMEM14C facilitates the import of protoporphyrinogen IX into the mitochondrial matrix for heme synthesis and subsequent hemoglobin production. Furthermore, the identification of TMEM14C as a protoporphyrinogen IX importer provides a genetic tool for further exploring erythropoiesis and congenital anemias.
Introduction
Heme is a prosthetic group that plays a vital role in redox reactions involved in processes such as detoxification, oxygen transport, circadian rhythm, microRNA processing, regulation of transcription and translation, and apoptosis (1–4). The majority of heme is synthesized in red blood cells, whose main function is to transport oxygen via the heme-containing oxygen carrier protein, hemoglobin (5).
Despite extensive work on the regulation and mechanisms of heme synthetic enzymes, the mechanisms governing transport and intracellular trafficking of heme intermediates, which are crucial for heme synthesis, are poorly understood (6, 7). δ-Aminolevulinate (ALA), the first committed heme synthesis precursor, is synthesized in the mitochondria. ALA is exported from the mitochondria into the cytosol for subsequent processing by δ-aminolevulinic acid dehydratase (EC4.2.1.24), porphobilinogen dehydratase (EC2.5.1.61), uroporphyrinogen III (UROgenIII) synthase (EC4.2.1.75), and uroporphyrinogen decarboxylase (EC4.1.1.37) to form UROgenIII and coproporphyrinogen III (CPgenIII). CPgenIII is then transported back into the mitochondria to synthesize protoporphyrinogen IX (PPgenIX) by coproporphyrinogen oxidase (CPOX; EC1.3.3.3) and then oxidized to form protoporphyrin IX (PPIX) by protoporphyrinogen oxidase (PPOX; EC1.3.3.4). PPIX is ultimately metalated with the coordination of Fe(II) by ferrochelatase (FECH; EC4.99.1.1) to form heme. Hence, the transport and trafficking of these intermediates represent key regulatory points in the heme synthesis pathway (7–9). Dysregulation of heme intermediate transport can lead to cytotoxic accumulation of tetrapyrrolic synthetic intermediates, which are photoreactive and relatively insoluble when allowed to accumulate, as illustrated by porphyrias caused by deficiencies in heme synthesis enzymes (10). Anemia may also result from defects in porphyrin trafficking, as heme synthesis is ultimately impaired.
Genes for heme and globin synthesis are coordinately upregulated during erythroid differentiation (11, 12) by erythroid-specific transcription factors EKLF (also known as KLF1) (13–15) and GATA-1 (16–19). We hypothesized that proteins essential for transport of heme synthesis intermediates are also coregulated in differentiating erythroid cells. In this study, we identified genes that are upregulated in terminally differentiating erythroid cells present in the fetal liver, which synthesize large quantities of heme (20). We discovered that the expression of tmem14c, a gene coding for a transmembrane protein required for zebrafish embryonic erythropoiesis and heme synthesis (21), was upregulated in terminally differentiating, definitive primary murine erythroid cells.
To mechanistically dissect the functional role of Tmem14c in erythroid heme synthesis, we performed loss-of-function studies in the mouse, using cultured murine embryonic stem cells and embryoid bodies as well as cultured Friend murine erythroleukemia (MEL) cells (22). Our complementary studies, using biochemical, cell biology, pharmacologic and genetic methods, consistently demonstrate that TMEM14C plays a critical and conserved role in primitive and definitive erythropoiesis and is required for erythroid heme metabolism in vertebrate species. In particular, we show that TMEM14C functions to facilitate the import of PPgenIX into the mitochondria for terminal heme synthesis.
Results
TMEM14C expression is enriched in mammalian erythropoietic tissues.
Maturing erythroid cells synthesize large amounts of heme and acquire exogenous iron to keep pace with the high rate of hemoglobin synthesis during erythroid terminal differentiation (23, 24). To identify mitochondrial porphyrin transporters that are coregulated with the heme synthesis machinery during erythroid terminal differentiation, we performed RNA sequencing (RNAseq) analysis on murine fetal liver cells that were sorted into fractions corresponding to their differentiation stage (R1–R5) by their surface expression of TER119 and CD71 (20, 25). The expression of Tmem14c, which codes for a predicted mitochondrial transmembrane protein (21, 26), was upregulated in the terminally differentiating (R3–R5) subpopulation in parallel with heme synthetic enzymes relative to progenitor cells (R1–R2) (Figure 1A). The increase in Tmem14c expression during terminal erythroid differentiation was recapitulated in a MEL cell line (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI76979DS1). In contrast, expression of the related Tmem14a was not induced during erythroid differentiation (Figure 1A). The requirement of tmem14c for hemoglobinization in zebrafish morphants (21) and its coordinated expression with murine heme synthesis enzymes in fetal liver cells suggested that it could play a conserved role in vertebrate erythroid heme synthesis.
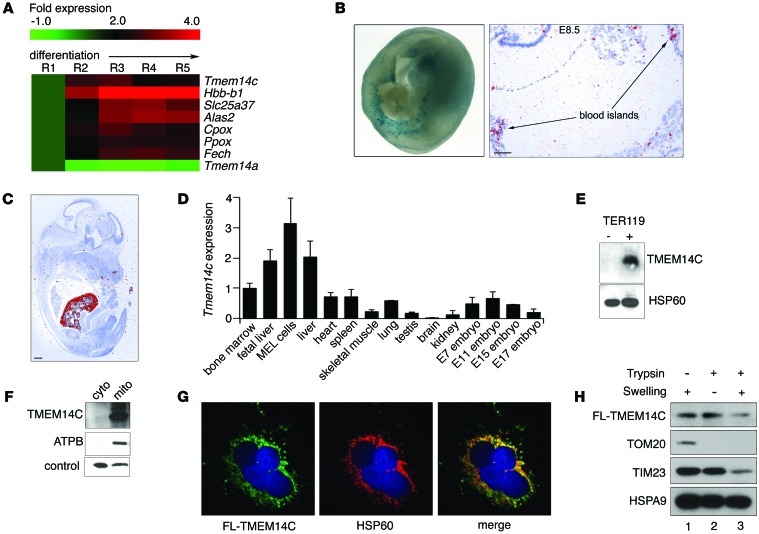
Figure 1. TMEM14C is enriched in differentiating murine erythroid cells and localizes to the inner mitochondrial membrane.
(A) RNAseq analysis of murine fetal liver cells sorted into 5 progressively differentiated erythroid subpopulations (R1–R5) shows that Tmem14c is upregulated during erythroid differentiation. (B) Tmem14c mRNA is expressed in hematopoietic organs, as shown by β-galactosidase staining (blue) of Tmem14c LacZ reporter expression in an E10.5 murine yolk sac (original magnification, ×63) and in situ hybridization of an E8.5 yolk sac (scale bar: 100 μm) and (C) fetal liver at E15.5 (pseudo-red; scale bar: 500 μm). (D) qRT-PCR shows Tmem14c mRNA is highly expressed in erythropoietic tissues and a MEL cell line. Tmem14c expression was normalized to Hprt levels. (E) Western blot analysis shows specific expression of TMEM14C protein in differentiating TER119+ murine fetal liver erythroid cells. HSP60 serves as a loading control. (F) Western blot analysis of fractionated Tmem14c-transfected HEK293T cells demonstrates localization of TMEM14C to the mitochondria. The control band indicates a protein that nonspecifically cross-reacts with MFRN1 antibody and migrates at a different molecular weight. (G) Confocal immunofluorescence microscopy (original magnification, ×63) shows that most of the transiently transfected FLAG-TMEM14C (fluorescein) colocalizes (merged, yellow) with HSP60 (rhodamine), a mitochondrial resident protein; nuclei were stained with DAPI (blue). (H) Transiently transfected FLAG-TMEM14C localizes to the inner mitochondrial membrane. TMEM14C, like TIM23, an inner mitochondrial protein, is sensitive to trypsin digestion when the outer mitochondrial membrane is disrupted by hypotonic swelling. The residual FLAG-TMEM14C that is trypsin resistant reflects mitochondria that are resistant to osmotic shock.
Analysis of cDNA from murine tissue and in situ hybridization of E8.5–E14.5 mouse embryos revealed that Tmem14c expression is enriched in hematopoietic organs, such as the yolk sac, bone marrow, fetal liver, and spleen (Figure 1, B–D, and Supplemental Figure 1A). β-Galactosidase staining of murine embryos carrying a Tmem14c gene trap cassette expressing LacZ under the control of the endogenous Tmem14c promoter confirmed that Tmem14c was highly expressed in the yolk sac blood cells in the vasculature at E8.5–E10.5 (Figure 1B). Consistent with the RNAseq experiments (Figure 1A), expression of the endogenous TMEM14C protein was specifically enriched in the TER119+ maturing erythroid population of the murine fetal liver (Figure 1E). The enrichment of TMEM14C expression in differentiating erythroid cells suggested that it played an important role in terminal erythroid maturation.
TMEM14C is localized to the inner mitochondrial membrane.
To determine the mechanism of TMEM14C function, we characterized its subcellular localization. Based on proteomic studies, TMEM14C was predicted to be a mitochondrial protein (21, 26). Western blot analysis of mitochondrial and cytosolic fractions from HEK293T cells that were transiently transfected with TMEM14C confirmed colocalization of TMEM14C with the β-subunit of ATP synthase in the mitochondria (Figure 1F). In addition, confocal immunofluorescence detection of transiently transfected FLAG-tagged TMEM14C in COS-7 cells showed colocalization of TMEM14C with HSP60, a mitochondrial resident protein, with a Mander’s overlap coefficient greater than 0.7 and a Pearson’s overlap coefficient of 0.67 (Figure 1G).
To assess the submitochondrial localization of TMEM14C, we isolated mitochondria from transiently transfected HeLa cells expressing murine FLAG-TMEM14C and disrupted their outer membrane by hypotonic swelling to create mitoplasts. Hypotonic swelling did not disrupt inner mitochondrial and intermitochondrial membrane proteins (Figure 1H, lane 1) but liberated proteins in the intermembrane space. Hypotonic swelling also rendered proteins in the intermembrane space and outer mitochondrial membrane accessible to trypsin digestion (Figure 1H, lane 3). The presence of TMEM14C in the mitochondria after hypotonic swelling showed that it is not an intermembrane space protein (Figure 1H, lane 1). Trypsin treatment of mitochondria degraded outer membrane proteins, such as TOM20, but not inner membrane proteins, like TIM23, and did not affect the presence of TMEM14C in mitochondria, thereby demonstrating that TMEM14C is not an outer membrane protein (Figure 1H, lane 2). In contrast, a large proportion of TMEM14C was degraded when mitochondria were subjected to both hypotonic swelling and trypsin treatment, which exposes inner membrane proteins, such as TIM23, to trypsin degradation, while mitochondrial matrix proteins, such as HSPA9, are resistant. These data demonstrate that the majority of TMEM14C resides in the inner mitochondrial membrane (Figure 1H, lane 3).
TMEM14C is specifically required for terminal erythropoiesis.
To confirm that TMEM14C is required for mammalian erythropoiesis, we examined hematopoiesis in embryoid bodies derived from a Tmem14c gene trap murine embryonic stem cell line from the German Gene Trap Consortium (Figure 2A). The original embryonic stem gene trap line (gt/+), E295C12, was grown under high puromycin step-up selection to target the remaining wild-type allele by gene conversion, thus generating a null (gt/gt) clone (refs. 27–29 and Figure 2B). Quantitative RT-PCR (qRT-PCR) analysis (Figure 2C) showed absence of Tmem14c mRNA in the null Tmem14cgt/gt clone compared with that in wild-type and heterozygous control embryonic stem cell clones.
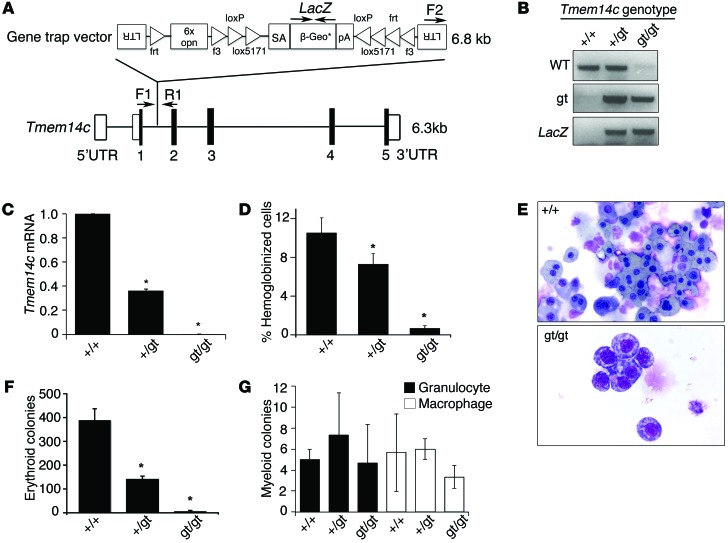
Figure 2. Tmem14c is specifically required for maturation of the primitive erythroid lineage.
(A) Design of Tmem14c gene trap construct (E295C12) used to disrupt expression of Tmem14c. LTR, long term repeat; 6x opn, 6x osteopontin enhancer element; SA, splice acceptor sequence; β-Geo*, β-galactosidase gene fused with the neomycin resistance gene; pA, polyadenylation sequence; UTR, untranslated region. (B) Genomic PCR verifies disruption of Tmem14c locus. (C) Expression of Tmem14c mRNA is abrogated in a Tmem14cgt/gt murine embryonic stem line. (D) The number of hemoglobinized cells is significantly reduced in Tmem14cgt/gt cells, suggesting a defect of erythroid differentiation. (E) Primitive erythroid cells derived from Tmem14cgt/gt embryoid bodies are developmentally arrested at the proerythroblast stage (original magnification, ×60). (F) The number of erythroid colonies formed is decreased in Tmem14cgt/gt derived embryoid bodies. (G) Myeloid lineages derived from embryoid bodies are not affected by Tmem14c deficiency. *P < 0.05.
In embryoid body cultures, Tmem14c deficiency specifically resulted in a decrease in the percentage of hemoglobinized cells and erythroid cells (Figure 2, D–F), while myelopoiesis was unaffected (Figure 2G). Erythroid cells derived from the Tmem14cgt/gt embryoid bodies were developmentally arrested at an early erythroblast stage (Figure 2E). These data show that TMEM14C is specifically required for erythroid terminal maturation or hemoglobin synthesis rather than hematopoietic stem cell biology or lineage determination.
We confirmed that TMEM14C is required for erythropoiesis by silencing Tmem14c in differentiating E14.5 mouse fetal liver cells with shRNA hairpin constructs (30). Fetal liver cells transduced with shRNA to Tmem14c exhibited decreased hemoglobin synthesis (Figure 3A), indicating that TMEM14C is continuously required for heme synthesis in the primary definitive erythroid lineage.
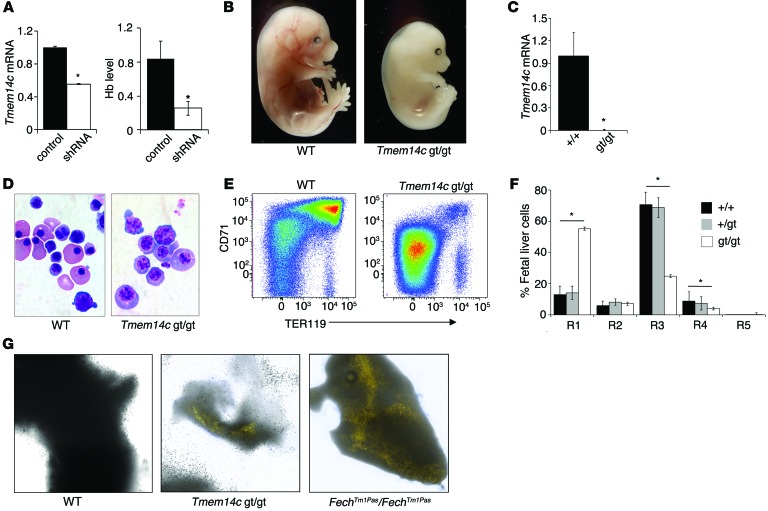
Figure 3. TMEM14C is specifically required for terminal differentiation of primary definitive murine erythroid cells.
(A) Primary murine fetal liver cells transduced with shRNA lentivirus, silencing the expression of Tmem14c mRNA, have significantly decreased hemoglobinization, compared with control cells. (B) E15.5 Tmem14cgt/gt mice are profoundly anemic; their livers are deficient in hemoglobinization compared with wild-type littermates (original magnification, ×20). (C) Tmem14c mRNA expression is abrogated in fetal livers derived from E12.5 Tmem14cgt/gt embryos. (D) Erythroid cells (original magnification, ×100) from E12.5 Tmem14cgt/gt embryos are developmentally arrested. (E) Erythroid maturation arrest of E12.5 Tmem14cgt/gt fetal liver population was confirmed by flow cytometry analysis of CD71 and TER119 expression. (F) Quantification of the R1–R5 subpopulations. Tmem14c+/gt fetal liver erythropoiesis is similar to that in wild-type fetal livers. (G) Autofluorescent erythrocytes are visualized in Tmem14cgt/gt and FechTm1Pas FechTm1Pas fetal livers under fluorescent light (original magnification, ×630) (λ,405 nm excitation and λ,620 nm emission; Supplemental Methods). Autofluorescent erythrocytes are not present in wild-type fetal livers. *P < 0.05.
We further interrogated the Tmem14c-deficient phenotype in vivo by generating Tmem14cgt/+ mice. A cross between heterozygous animals did not yield any viable homozygous pups, suggesting embryonic mortality of Tmem14cgt/gt mice. Tmem14cgt/gt mice died in utero by E13.5 (Supplemental Table 1), a developmental stage at which definitive fetal liver erythropoiesis becomes the main source of red cells (31). Tmem14cgt/gt embryos had pale livers compared with livers of wild-type mouse embryos and were visibly anemic (Figure 3B). Erythroid cells from Tmem14cgt/gt fetal livers, which do not express Tmem14c mRNA (Figure 3C), were developmentally arrested (Figure 3D). Flow cytometry analyses revealed a decrease in the number of terminally differentiating TER119+ erythroid cells (R3–R5) in Tmem14c-deficient fetal livers, whereas the numbers of erythroid progenitors (R1–R2) remained unchanged (Figure 3, E and F). Other hematopoietic lineages were unaffected in the absence of Tmem14c (Supplemental Figure 2), highlighting its erythroid-restricted role. Erythroid defects were only present in Tmem14cgt/gt embryos; heterozygous mice were viable and fertile and did not exhibit any hematopoietic lineage defects (Figure 3, E and F; Supplemental Table 2; and Supplemental Figure 2).
Since TMEM14C is a mitochondrial protein that is required for hemoglobinization and terminal erythroid maturation, we considered whether it could play a role in heme metabolism. Haploinsufficiency of heme synthesis enzymes, such as FECH (32–34) or UROgenIII synthase (35), results in the accumulation of photoreactive tetrapyrrole biosynthetic intermediates, particularly in erythroid and hepatic tissue (36). E12.5 Tmem14cgt/gt fetal livers autofluoresced under fluorescence illumination, similar to fetal livers from Fech mutant mice (FechTm1Pas FechTm1Pas mice), while wild-type fetal livers did not, indicating an accumulation of heme intermediates, which autofluoresce due to their highly conjugated tetrapyrrole ring structures (Figure 3G). We therefore concluded that TMEM14C is involved in mitochondrial heme metabolism.
TMEM14C is required for mitochondrial porphyrin metabolism.
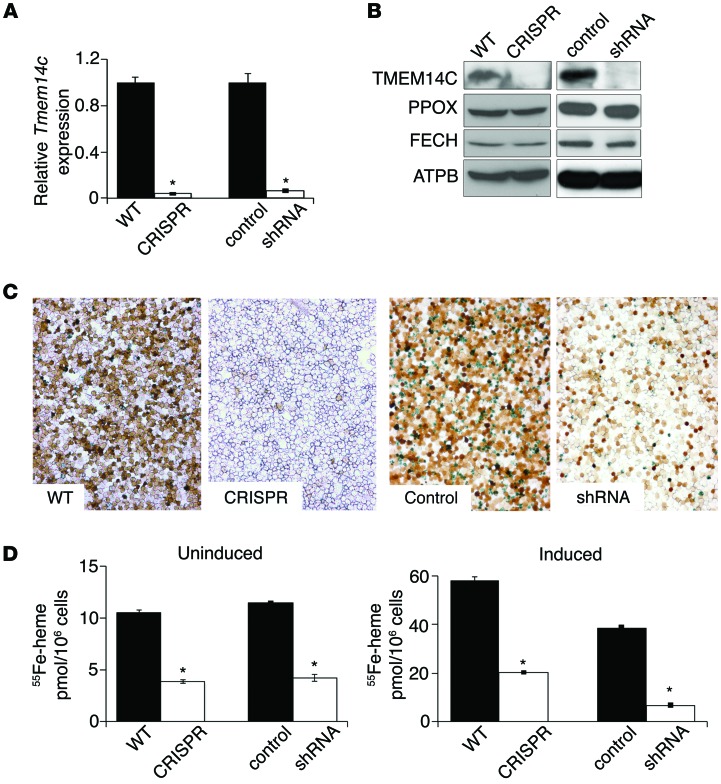
To further characterize the role of TMEM14C in erythroid heme and iron metabolism, we depleted Tmem14c in MEL cells using two methods. First, we used CRISPR/Cas-mediated genomic editing to generate stable compound heterozygote knockout (CRISPR) cells (Supplemental Figure 3 and refs. 37, 38). Second, we generated stable Tmem14c knockdown MEL clones using shRNA silencing (shRNA-silenced clones are referred to herein as shRNA). Both cell lines expressed negligible steady-state levels of Tmem14c mRNA and protein (Figure 4, A and B). TMEM14C-deficient and control mitochondria contained similar amounts of PPOX and FECH proteins, showing that TMEM14C did not regulate protein levels of the heme synthetic enzymes (Figure 4B). Consistent with observations gathered from tmem14c morphant zebrafish embryos (21), differentiating murine embryonic stem–derived erythroid cells (Figure 2), and murine fetal livers (Figure 3), Tmem14c-silenced cells exhibited decreased heme synthesis upon induction of terminal differentiation with DMSO in comparison with that of control cells, as shown by o-dianisidine staining for hemoglobinized cells (Figure 4C). Quantitation of heme synthesis rate by 55Fe labeling indicated that TMEM14C was required for heme synthesis both basally and during terminal erythroid differentiation in MEL cells (Figure 4D). This indicated that the heme synthesis defect in Tmem14c-deficient cells was not secondary to an erythroid differentiation defect but rather that TMEM14C was directly involved in the heme homeostasis.
Figure 4. TMEM14C is required for heme synthesis in murine erythroid cells both basally and during terminal differentiation.
(A) qRT-PCR analysis shows a significant decrease in Tmem14c mRNA in both CRISPR and shRNA-silenced MEL cells compared with their respective controls. (B) Western blot analysis show deficiency of TMEM14C protein, but not the other heme synthesis enzymes, in CRISPR cells and shRNA-silenced MEL cells. (C) o-dianisidine staining shows a heme synthesis defect in CRISPR cells and shRNA-silenced cells (original magnification, ×20). (D) 55Fe-Tf metabolic labeling demonstrates that basal heme synthesis (left) and heme synthesis in differentiating Tmem14c-deficient erythroid cells (right) are significantly decreased. *P < 0.05.
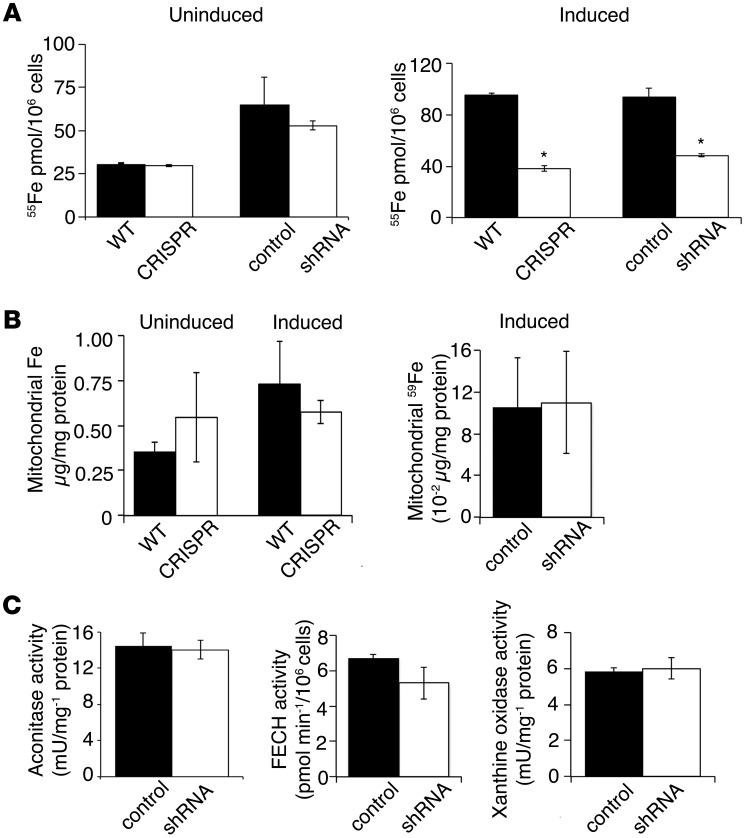
Because TMEM14C is a mitochondrial protein required for heme synthesis, we assayed for biomarkers reflective of iron homeostasis (39). Tmem14c deficiency did not alter basal cellular 55Fe uptake, although the amount of 55Fe uptake was decreased in differentiating Tmem14c-deficient cells (Figure 5A). The decrease in cellular 55Fe in Tmem14c-deficient MEL cells may reflect the block in erythroid maturation (Figure 4C), with a decreased demand for iron in heme production. Significantly, mitochondrial iron levels, as measured by inductively coupled plasma mass spectrometry and 59Fe labeling, were similar in controls and Tmem14c-deficient cells, both basally and during terminal differentiation (Figure 5B). These data indicated that TMEM14C does not regulate heme synthesis via the control of mitochondrial iron import. In Tmem14c-deficient cells, mitochondrial aconitase (EC4.2.1.3), FECH, and cytosolic xanthine oxidase (EC1.17.3.2) exhibited normal activity, indicating normal [2Fe-2S] cluster assembly in both the mitochondria and cytosol, further excluding a role for TMEM14C in mitochondrial iron metabolism per se (Figure 5C).
Figure 5. TMEM14C is not required for mitochondrial iron homeostasis.
(A) Quantitation of cellular iron status by 55Fe metabolic labeling shows that basal cellular iron import is not decreased in Tmem14c-deficient MEL cells. However, cellular iron content is decreased in Tmem14c-deficient erythroid cells during differentiation. (B) Deficiency in heme synthesis in Tmem14c-silenced cells is not due to a defect in mitochondrial iron content. Inductively coupled plasma analysis of mitochondrial iron shows that Tmem14c deficiency does not cause a defect in mitochondrial iron content either basally or during erythroid differentiation (left). This was confirmed by 59Fe metabolic labeling and quantitation of mitochondrial iron from differentiating Tmem14c-silenced cells (right). (C) Normal activities for [2Fe-2S] cofactor–dependent mitochondrial aconitase, FECH, and cytosolic xanthine oxidase in Tmem14c-silenced cells exclude defects in [2Fe-2S] cluster assembly. *P < 0.05.
Flow cytometry analysis of MitoTracker Red–stained cells (see Supplemental Methods) revealed no differences between the CRISPR cells and wild-type cells in the number of MitoTracker–positive cells (Supplemental Figure 4A) or mean cellular MitoTracker fluorescence (Supplemental Figure 4B), excluding a role for TMEM14C in the regulation of mitochondrial number, apoptosis, or mitochondrial membrane potential. As cellular iron availability is a requirement for normal mitochondrial biogenesis (40), we analyzed HSP60 levels in lysates of uninduced and induced CRISPR and wild-type MEL cells, as a surrogate for mitochondrial protein content. Wild-type and CRISPR cells contained similar levels of HSP60 and control cytosolic marker, GAPDH, under both uninduced and induced conditions (Supplemental Figure 4C). These data confirmed that the absence of TMEM14C did not affect mitochondrial biogenesis or function and excluded the possibility that the heme synthetic defect was secondary to a general defect in mitochondrial physiology.
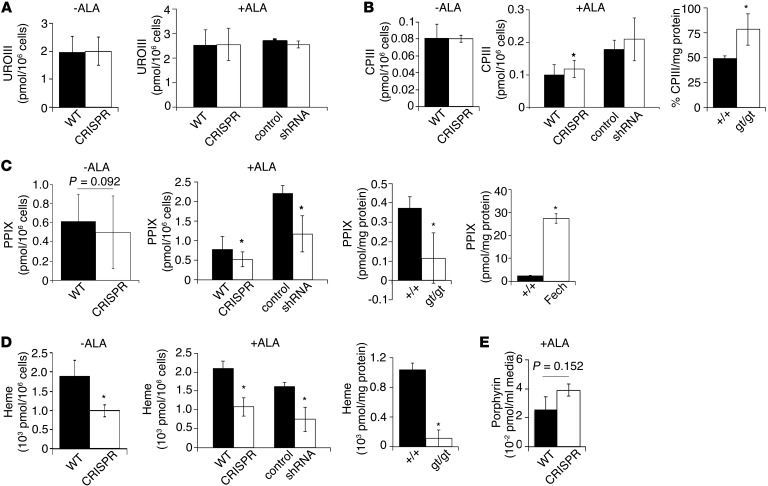
We then considered the possibility that TMEM14C might play a regulatory role in mitochondrial heme synthesis. To quantitate the levels of porphyrin intermediates, we performed HPLC analysis on MEL cells differentiated in the presence of DMSO. Whether or not cells were treated with exogenous ALA, an early synthetic precursor in heme synthesis, Tmem14c-silenced cells contained similar levels of intracellular uroporphyrin III as control cells (Figure 6A). ALA-treated CRISPR cells and Tmem14cgt/gt fetal liver cells contained significantly more coproporphyrin III than wild-type controls (Figure 6B). In comparison, Tmem14c-deficient cells and Tmem14cgt/gt fetal liver cells contained significantly less PPIX than wild-type controls (Figure 6C). This contrasted with high levels of PPIX accumulation in the fetal FechTm1Pas FechTm1Pas mouse liver (Figure 6C). Consistent with decreased PPIX production in Tmem14c-deficient cells and Tmem14cgt/gt fetal liver, heme levels were decreased (Figure 6D). The media of ALA-treated CRISPR cells exhibited a trend of elevated total porphyrin levels (Figure 6E), indicative of increased extracellular excretion of cellular porphyrin, consistent with porphyrin accumulation observed in Tmem14cgt/gt fetal liver tissues (Figure 3F and Figure 6B). While we were unable to directly assay reduced porphyrinogens in our samples due to their spontaneous oxidation, these data demonstrate that TMEM14C is required for the formation of PPIX from CPgenIII in the mitochondria. In the absence of TMEM14C, this pathway is blocked, causing an accumulation of upstream porphyrins.
Figure 6. TMEM14C is required for synthesis of PPIX.
HPLC analysis of porphyrin intermediates and heme in wild-type, CRISPR, control, and silenced cell lines and E12.5 fetal liver tissue. Porphyrinogens were oxidized to their corresponding porphyrins during isolation; porphyrin levels are a surrogate for porphyrinogen levels in vivo. (A) Uroporphyrin III (UROIII) levels are normal in differentiating Tmem14c-deficient cells in the absence and presence of exogenous ALA. (B) Coproporphyrin III (CPIII) levels are normal in the absence of exogenous ALA but are mildly elevated in ALA-supplemented CRISPR cells. Coproporphyrin III is significantly increased in Tmem14cgt/gt fetal liver. (C) PPIX levels are reduced in Tmem14c-deficient cells in the absence and presence of exogenous ALA. PPIX levels are significantly decreased in Tmem14cgt/gt fetal liver. As a control for our assay, PPIX is significantly elevated in FechTm1Pas FechTm1Pas (Fech) fetal liver. (D) Heme levels are significantly reduced in Tmem14c-deficient cells both in the absence and presence of exogenous ALA as well as in Tmem14cgt/gt fetal liver tissue. (E) There is an increase in the level of total porphyrin excreted into the cell culture media of CRISPR cells. *P < 0.05.
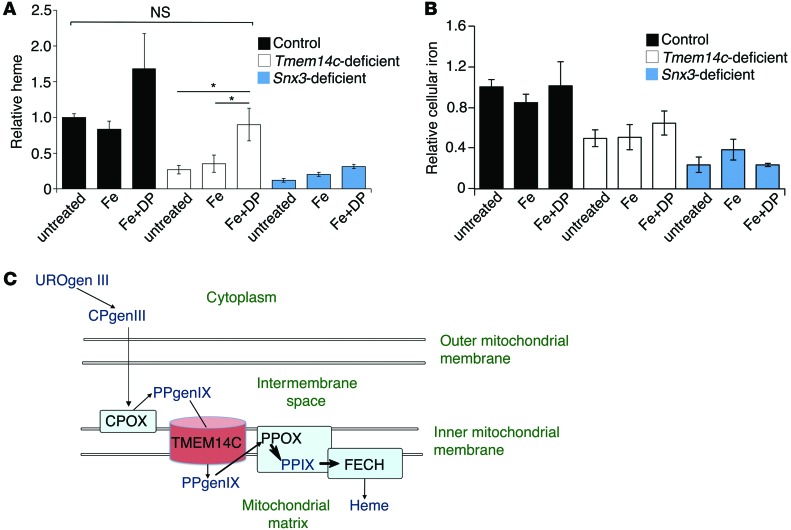
To confirm that TMEM14C plays a role in porphyrin metabolism, we used deuteroporphyrin IX (DP), a synthetic analog of PPIX in which the 2,4 vinyl groups are replaced by hydrogen atoms, to chemically complement the heme synthesis defect in shRNA and CRISPR cells. We differentiated control, Tmem14c-deficient, and Snx3-silenced cells, which have a primary iron uptake defect (41), and concurrently treated them with either 5 μM Fe-dextran alone or in combination with 5 μM DP. We omitted treatment with DP alone, as DP is inherently cytotoxic in the absence of supplemental iron and inhibited MEL cell differentiation (data not shown). Cellular heme synthesis was assayed by 55Fe-heme incorporation. Consistent with previous data, 55Fe-heme incorporation confirmed that Fe-dextran with DP, but not Fe-dextran alone, rescued heme synthesis in Tmem14c-deficient cells to a level similar to that of differentiated control cells. In contrast, DP could not complement the heme synthetic defect in Snx3-silenced cells, demonstrating the specificity of the heme synthesis rescue by DP (Figure 7A). The increase in heme synthesis in response to Fe-dextran and DP was not due to an increase in cellular iron content in the Tmem14c-deficient cells (Figure 7B). In combination with the HPLC data, these chemical complementation results demonstrate that TMEM14C is specifically required for the terminal steps in the heme synthesis pathway. The presence of TMEM14C in the mitochondrial inner membrane, and the active site orientation of heme synthesis enzymes in its proximity (42), defines a role for TMEM14C in the transport of PPgenIX within the mitochondrial intermembrane space into the mitochondrial matrix for the synthesis of PPIX (Figure 7C). The final step in heme synthesis, the metalation of PPIX by FECH, occurs within the mitochondrial matrix.
Figure 7. The heme defect in Tmem14c-deficient MEL cells is complemented by the addition of DP.
(A) 55Fe-metabolic labeling of differentiating MEL cells that were treated with either Fe-dextran (Fe) or Fe-dextran with DP (Fe+DP). Fe-dextran with DP supplementation, but not Fe-dextran alone, significantly increased heme synthesis in Tmem14c-deficient (CRISPR and shRNA) cells to similar quantities as untreated control cells. In contrast, Fe-dextran with DP does not complement the heme synthesis defect in Snx3-deficient cells. *P < 0.05. (B) Quantitation of total cellular iron by 55Fe metabolic labeling. Fe-dextran and Fe-dextran with DP supplementation did not significantly alter total cellular iron content in control, Tmem14c-deficient, or Snx3-deficient cells. (C) Proposed model for the function of TMEM14C (brown cylinder) as a PPgenIX transporter, enabling access to PPOX and facilitating synthesis of PPIX and eventually heme in the mitochondria of developing erythroblasts. Heme synthesis enzymes are represented by light blue boxes, while heme and porphyrin intermediates are indicated in dark blue text.
Discussion
Although porphyrias are currently understood to result from defects in heme synthetic pathway enzymes, the low genetic penetrance of mutations in these genes suggests the requirement for additional genetic modifiers that predispose an individual to heme synthesis defects and porphyrin accumulation (43). Thus far, genetic interactions associated with congenital porphyrias have comprised mutations in multiple components of the heme synthetic pathway (43, 44). Of note, mutations in Mfrn1 (also known as Slc25a37), required for mitochondrial iron import (28), do not by themselves cause porphyria (27). However, they predispose vertebrates to develop porphyria when cellular ALA levels are increased by gain-of-function mutations in ALAS2 (45) or dietary intake (46).
Our study reveals the potential for defects in porphyrin transport to cause or exacerbate porphyria. We identified TMEM14C as a protein that facilitates the transport of terminal heme synthesis intermediates, in particular PPgenIX. TMEM14C is a member of the uncharacterized TMEM14 superfamily of transmembrane proteins and has 4 predicted helical domains that are conserved in vertebrate species (26), 3 of which span the mitochondrial membrane (47). Development of other organs and nonerythroid hematopoietic lineages is largely normal in tmem14c-deficient zebrafish morphants (21) and mouse embryos (Supplemental Figure 2), indicating that these structurally similar proteins may play analogous, compensatory roles to those of TMEM14C in nonerythroid heme synthesis. Consistent with the proposed role of Tmem14c in erythroid heme synthesis, it is not present in Caenorhabditis elegans, a heme auxotroph devoid of heme synthesis genes (8).
Tmem14c-deficient primary murine fetal liver tissue and erythroid cell lines exhibited defective PPIX synthesis and accumulated coproporphyrin III (Figure 6, B and C) or excreted excess porphyrin into the cell culture media (Figure 6E). The porphyrin accumulation in Tmem14cgt/gt fetal liver tissue caused autofluorescence characteristic of murine porphyria models (Figure 3G and refs. 34–36). The apparent absence of autofluorescence in tmem14c morphant zebrafish embryos (21) could be reconciled by the inherent chemical instability and photoreactivity of CPgenIII and PPgenIX, excretion of these intermediates into the water and yolk sac, or a combination of these factors. In addition, other structurally similar TMEM14 family proteins like TMEM14A may partially compensate for the loss of TMEM14C function (47), mitigating porphyrin accumulation in tmem14c morphant embryos.
Due to the localization of TMEM14C in the inner mitochondrial membrane, we considered and excluded several possibilities for the function of TMEM14C in mitochondrial heme metabolism. We excluded the possibility of a role for TMEM14C in regulating the levels of mitochondrial heme synthesis enzymes (Figure 4B), mitochondrial iron (Figure 5B), and [Fe-S]-cluster assembly (Figure 5C). The observed decrease in cellular iron content in Tmem14c-deficient MEL cells was a secondary effect of differentiation defects in these cells, which require less iron for heme production (Figure 5A). Even so, it is likely that normal levels of mitochondrial iron in Tmem14c-deficient cells are maintained by direct import of iron from transferrin-containing endosomes, and the imported iron is preferentially used to carry out essential cellular functions (48).
The requirement for TMEM14C in housekeeping erythroid heme synthesis suggests that TMEM14C plays a broader role in erythroid heme homeostasis, beyond hemoglobin synthesis. We speculate that, in addition to TMEM14C, other TMEM14 family proteins play a major role in maintenance of housekeeping heme synthesis, a process critical for the function of mitochondrial respiratory proteins and cell survival (49, 50). The redundant functions of other mitochondrial proteins are underscored by the observation that Tmem14c-deficient cells have survival rates and mitochondrial function comparable to that of wild-type cells (Supplemental Figure 4).
Tmem14c-deficient cells synthesized uroporphyrin III and coproporphyrin III at levels similar to control cells (Figure 6, A and B). Furthermore, ALA supplementation of cells used in metabolic iron radiolabeling experiments (Figure 4D) and porphyrin HPLC analysis (Figure 6, A–D) did not complement heme synthesis in Tmem14c-deficient cells. However, DP, a synthetic protoporphyrin analog, but not iron, dramatically complemented the heme defect in Tmem14c CRISPR cells (Figure 7A). The specificity of our complementation assay is maintained by the marginal rescue of heme in Snx3-deficient cells. These data demonstrate that TMEM14C primarily and directly facilitates the terminal steps of mitochondrial heme synthesis.
Proteomic mapping of proteins in the mitochondria reassigned PPOX to the mitochondrial matrix and confirmed the localization of the CPOX and FECH active sites in the intermembrane space and mitochondrial matrix, respectively (42). ABCB6 (EC3.6.3), a mitochondrial outer membrane protein, has been implicated as a putative mitochondrial importer of CPgenIII, enabling the conversion of CPgenIII to PPgenIX by inner membrane–bound CPOX in the intermembrane space (51–54); although, more recent work suggests that it is dispensable for heme synthesis and specifies the blood group Langereis (55). Based on the colocalization of TMEM14C and CPOX, and the restricted role of TMEM14C in mitochondrial heme synthesis, we propose that TMEM14C directly accepts PPgenIX from CPOX and transports PPgenIX into the mitochondrial matrix (Figure 7C). The proposed role of TMEM14C as PPgenIX importer is consistent with its tightly packed helical structure that is typical of transmembrane transport proteins (47, 56). However, due to spontaneous oxidation of porphyrinogens to porphyrins under our porphyrin isolation conditions, we could not directly measure UROgenIII, CPgenIII, or PPgenIX levels to demonstrate this.
In vertebrates, the expression of Tmem14c mRNA is enriched in embryonic (Figure 1B and ref. 21), fetal, and adult hematopoietic tissues (Figure 1, C and D). Tmem14c expression is most pronounced in terminally differentiating erythroid cells (Figure 1E and Supplemental Figure 1B). Consistent with this observation, the Tmem14c promoter is occupied by the erythroid transcription factor GATA-1 (19). As many genes regulating heme and iron homeostasis are regulated by iron regulatory proteins and cellular iron status (57, 58), we searched for iron response elements, which are bound by iron regulatory proteins, in the Tmem14c mRNA sequence (http://ccbg.imppc.org/sires/index.html) and found no evidence of iron response elements. We also treated MEL cells with desferrioxamine and Fe-citrate to examine the effects of iron depletion or supplementation on Tmem14c mRNA expression. Neither treatment had an effect on Tmem14c mRNA levels (Supplemental Figure 1C), indicating that cellular iron status does not directly regulate steady-state Tmem14c mRNA levels.
Our current data extend the findings that transport proteins play a critical role in heme homeostasis (52, 59–63). Although the National Human Genome Research Institute human genome-wide association studies database (64) and genome-wide association studies have yet to link mutations in TMEM14C to hematologic disease (65–67), the profound anemic and mild porphyric phenotypes in Tmem14cgt/gt mice indicate that TMEM14C could function as a genetic modifier for the severity of anemia and porphyria in humans. We predict that further genetic sequencing studies will uncover TMEM14C hypomorphic mutations in individuals suffering from anemias or porphyrias of unknown etiology. Our identification of Tmem14c as an essential regulator of heme synthesis thus provides a novel genetic tool for further studies on normal vertebrate erythropoiesis and pathological states, such as anemia and porphyria.
Methods
Cell lines.
DS19 MEL cells were obtained from Arthur Skoultchi (Albert Einstein College of Medicine, New York, New York, USA). Gene trap mouse embryonic stem cells for TMEM14C (E295C12) were obtained from the German Gene Trap Consortium.
Knockdown of Tmem14c by shRNA hairpins in mouse cells.
Tmem14c (GenBank NM_025387) stable knockdown MEL clones were obtained by stable transfection of an shRNA hairpin (TRCN0000009763, Sigma-Aldrich). Electroporation of the DS19 MEL cells and stable selection of clones was carried out as previously described (37). Knockdown efficiency was assessed by qRT-PCR (Tmem14c probe: Mm00481276_m1, Invitrogen; Hprt probe: Mm01545399_m1, Invitrogen) and Western blot analyses (Figure 4, A and B).
Fe radiolabeling and radio Fe-heme measurements.
59FeCl3 (specific activity: 1 Ci/mmol) and 55FeCl3 (specific activity: 1.28 Ci/mmol) (Perkin Elmer) were loaded onto transferrin as described previously (68). Metabolic labeling was carried out as described previously (28).
Chemical complementation assays.
MEL cells were differentiated chemically with 1.5% DMSO for 72 hours. Concurrently, cells were treated with 5 μM Fe-dextran (Sigma-Aldrich) alone or in combination with 5 μM DP (Frontier Scientific). Heme synthesis was assayed by labeling with 55Fe (Supplemental Methods).
Statistics.
Statistical analysis was carried out using 2-tailed paired or unpaired Student’s t test. Significance was set at P < 0.05. All graphs depict the mean ± SEM.
Study approval.
The present studies in animals were reviewed and approved by The Jackson Laboratory Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank James Bieker, Roslyn Orkin, David Williams, and Leonard Zon for critical feedback on the manuscript. We thank Caiyong Chen, Jacky Chung, and Amy Medlock for technical advice and helpful scientific discussions. We thank Mahnaz Paktinat for training on the use of the BD FACSCanto cell sorter, as mentioned in the Supplemental Methods, and Marie McConkey, Esther Obeng, and Benjamin Ebert for providing mouse tissues. We also thank Jonathan Thon for his assistance in analysis of confocal images. The DS19 MEL clones were obtained from Arthur Skoultchi (Albert Einstein College of Medicine). As detailed in the Supplemental Methods, fluorescence confocal microscopy was performed at the Harvard Digestive Disease Center Imaging Facility (Boston Children’s Hospital, supported by NIH center grant P30 DK34854) with assistance from Ramiro Massol. This work was supported by grants from the American Heart Association (to J.D. Cooney), Cooley’s Anemia Foundation (to D.I. Shah), the March of Dimes Foundation (6-FY09-289, to B.H. Paw), the NWO Dutch National Science Fund (to I.J. Schultz), the American Physiological Society (to E.M. Keenan), and the NIH (K01 DK085217, to D.I. Shah; K08 DK076848, to S.M. Hattangadi; T32 HL007574 and F32 DK098866, to Y.Y. Yien; K08 DK093705, to D.E. Bauer; P01 HD032062, to S. DiMauro; R01 DK09361, to J.D. Phillips; DK020503 and U54 DK083909, to J.D. Phillips; R01 DK096051, to H.A. Dailey; R01 GM61721, to C.M. Koehler; R01 DK052830, to J. Kaplan and D.M. Ward; R01 HL088468, to L.L. Peters; R01 DK070838, to B.H. Paw; and P01 HL032262, to A.B. Cantor, H.F. Lodish, S.H. Orkin, and B.H. Paw).
Iman J. Schultz’s present address is: InteRNA Technologies, Utrecht, The Netherlands.
Gordon J. Hildick-Smith’s present address is: Weill Cornell Medical College, New York, New York, USA.
Jeffrey D. Cooney’s present address is: University of Texas Health Science Center, San Antonio, Texas, USA.
Gloria Yi’s present address is: MGH Institute of Health Professions, Boston, Massachusetts, USA.
Eric L. Pierce’s present address is: Georgia Institute of Technology, Atlanta, Georgia, USA.
Non Miyata’s present address is: Kyushu University, Fukuoka, Japan.
Wen Chen’s present address is: Texas A&M Health Science Center, Temple, Texas, USA.
Shilpa M. Hattangadi’s present address is: Yale University Medical School, New Haven, Connecticut, USA.
Hui Huang’s present address is: Columbia University Medical Center, New York, New York, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(10):4294–4304. doi:10.1172/JCI76979.
References
- 1.Martinkova M, Kitanishi K, Shimizu T. Heme-based globin-coupled oxygen sensors: linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J Biol Chem. 2005;288(39):27702–27711. doi: 10.1074/jbc.R113.473249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763(7):723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2010;47(2):158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girvan HM, Munro AW. Heme sensor proteins. J Biol Chem. 2013;288(19):13194–13203. doi: 10.1074/jbc.R112.422642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Paw BH. Cellular and mitochondrial iron homeostasis in vertebrates. Biochim Biophys Acta. 2012;1823(9):1459–1467. doi: 10.1016/j.bbamcr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung J, Chen C, Paw BH. Heme metabolism and erythropoiesis. Curr Opin Hematol. 2012;19(3):156–162. doi: 10.1097/MOH.0b013e328351c48b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yien YY, Paw BH. Iron and heme transport and trafficking. In: Culotta VC, Scott RS, eds. Metals in Cells. Chichester, United Kingdom: John Wiley and Sons; 2013:113–130. [Google Scholar]
- 8.Hamza I, Dailey HA. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta. 2012;1823(9):1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz IJ, Chen C, Paw BH, Hamza I. Iron and porphyrin trafficking in heme biogenesis. J Biol Chem. 2010;285(35):26753–26759. doi: 10.1074/jbc.R110.119503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassa S. Modern diagnosis and management of the porphyrias. Br J Haematol. 2006;135(3):281–292. doi: 10.1111/j.1365-2141.2006.06289.x. [DOI] [PubMed] [Google Scholar]
- 11.Li L, et al. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121(22):4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wontakal SN, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci U S A. 2012;109(10):3822–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yien YY, Bieker JJ. EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol Cell Biol. 2013;33(1):4–13. doi: 10.1128/MCB.01058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilon AM, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Krüppel-like factor during erythrocyte differentiation. Blood. 2011;118(17):e139–e148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tallack MR, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20(8):1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amigo JD, et al. Identification of distal cis-regulatory elements at mouse mitoferrin loci using zebrafish transgenesis. Mol Cell Biol. 2011;7(7):1344–1356. doi: 10.1128/MCB.01010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19(12):2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36(4):682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102(12):3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson R, et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 2009;10(2):119–130. doi: 10.1016/j.cmet.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ney PA, D’Andrea AD. Friend erythroleukemia revisited. Blood. 2000;96(12):3675–3680. [PubMed] [Google Scholar]
- 23.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105(5):1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 24.Richardson DR, et al. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A. 2010;107(24):10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118(16):e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung J, et al. Iron regulatory protein-1 protects against mitoferrin1-deficient porphyria. J Biol Chem. 2014;289(11):7835–7843. doi: 10.1074/jbc.M114.547778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440(7080):96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 29.Woo AJ, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol. 2008;28(8):2675–2689. doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattangadi SM, Burke KA, Lodish HF. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood. 2010;115(23):4853–4861. doi: 10.1182/blood-2009-07-235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron MH, Isern J, Fraser ST. The embryonic origins of erythropoiesis in mammals. Blood. 2012;119(21):4828–4837. doi: 10.1182/blood-2012-01-153486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Childs S, Weinstein BM, Mohideen MA, Donohue S, Bonkovsky H, Fishman MC. Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr Biol. 2000;10(16):1001–1004. doi: 10.1016/S0960-9822(00)00653-9. [DOI] [PubMed] [Google Scholar]
- 33. Kiupel M, Montagutelli X, Sundberg JP. The ferrochelatase deficiency (Fechtm1Pas) mouse mutation: a mouse model for erythropoietic protoporphyria. JAX Communication. 1999;1:1–7. http://www.porfyrier.dk/pdf_filer/communication01.pdf. [Google Scholar]
- 34.Tutois S, et al. Erythropoietic protoporphyria in the house mouse. A recessive inherited ferrochelatase deficiency with anemia, photosensitivity, and liver disease. J Clin Invest. 1991;88(5):1730–1736. doi: 10.1172/JCI115491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop DF, et al. Uroporphyrinogen III synthase knock-in mice have the human congenital erythropoietic porphyria phenotype, including the characteristic light-induced cutaneous lesions. Am J Hum Genet. 2006;78(4):645–658. doi: 10.1086/502667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop DF, Clavero S, Mohandas N, Desnick RJ. Congenital erythropoietic porphyria: characterization of murine models of the severe common (C73R/C73R) and later-onset genotypes. Mol Med. 2011;17(7):748–756. doi: 10.2119/molmed.2010.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289(31):21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Barrientos T, Andrews NC. Iron and copper in mitochondrial diseases. Cell Metab. 2013;17(3):319–328. doi: 10.1016/j.cmet.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rensvold JW, Ong SE, Jeevananthan A, Carr SA, Mootha VK, Pagliarini DJ. Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep. 2013;3(1):237–245. doi: 10.1016/j.celrep.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17(3):343–352. doi: 10.1016/j.cmet.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balwani M, Desnick RJ. The porphyrias: advances in diagnosis and treatment. Blood. 2012;120(23):4496–4504. doi: 10.1182/blood-2012-05-423186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.To-Figueras J, et al. ALAS2 acts as a modifier gene in patients with congenital erythropoietic porphyria. Blood. 2011;118(6):1443–1451. doi: 10.1182/blood-2011-03-342873. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, et al. Abnormal mitoferrin-1 expression in patients with erythropoietic protoporphyria. Exp Hematol. 2011;39(7):784–794. doi: 10.1016/j.exphem.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Troadec MB, et al. Targeted deletion of the mouse Mitoferrin1 gene: from anemia to protoporphyria. Blood. 2011;117(20):5494–5502. doi: 10.1182/blood-2010-11-319483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klammt C, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9(8):834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 2007;110(1):125–132. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 49.Kim HJ, Khalimonchuk O, Smith PM, Winge DR. Structure, function and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 2012;1823(9):1604–1616. doi: 10.1016/j.bbamcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homedan C, et al. Acute intermittent porphyria causes hepatic mitochondrial energetic failure in a mouse model. Int J Biochem Cell Biol. 2014;51:93–101. doi: 10.1016/j.biocel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 51.Chavan H, Khan MM, Tegos G, Krishnamurthy P. Efficient purification and reconstitution of ATP binding cassette transporter B6 (ABCB6) for functional structural studies. J Biol Chem. 2013;288(31):22658–22669. doi: 10.1074/jbc.M113.485284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnamurthy PC, et al. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443(7111):586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- 53.Krishnamurthy P, Schuetz JD. The role of ABCG2 and ABCB6 in porphyrin metabolism cell survival. Curr Pharm Biotechnol. 2011;12(4):647–655. doi: 10.2174/138920111795163995. [DOI] [PubMed] [Google Scholar]
- 54.Ulrich DL, et al. ATP-dependent mitochondrial porphyrin importer ABCB6 protects against phenylhydrazine toxicity. J Biol Chem. 2012;287(16):12676–12690. doi: 10.1074/jbc.M111.336180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helias V, et al. ABCB6 is dispensable for erythropoiesis specifies the new blood group system Langereis. Nat Genet. 2012;44(2):170–173. doi: 10.1038/ng.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maslennikov I, et al. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci U S A. 2010;107(24):10902–10907. doi: 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson CP, Shen M, Eisenstein RS, Leibold EA. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim Biophys Acta. 2012;1823(9):1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casey JL, et al. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240(4854):924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 59.Chiabrando D, et al. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J Clin Invest. 2012;122(12):4569–4579. doi: 10.1172/JCI62422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desuzinges-Mandon E, Arnaud O, Martinez L, Huche F, Di Pietro A, Falson P. ABCG2 transports transfers heme to albumin through its large extracellular loop. J Biol Chem. 2010;285(43):33123–33133. doi: 10.1074/jbc.M110.139170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A. Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004;23(8):1720–1728. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogino T, et al. Serum-dependent export of protoporphyrin IX by ATP-binding cassette transporter G2 in T24 cells. Mol Cell Biochem. 2011;358(1):297–307. doi: 10.1007/s11010-011-0980-5. [DOI] [PubMed] [Google Scholar]
- 63.Proulx KL, Woodard SI, Dailey HA. In situ conversion of coproporphyingen III to heme by murine mitochondria: terminal steps of the heme biosynthetic pathway. Protein Sci. 1993;2(7):1092–1098. doi: 10.1002/pro.5560020703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganesh SK, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41(11):1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem. 1999;274(13):9022–9028. doi: 10.1074/jbc.274.13.9022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.