Abstract
Metastasis causes most cancer deaths, but is incompletely understood. MicroRNAs can regulate metastasis, but it is not known whether a single miRNA can regulate metastasis in primary cancer models in vivo. We compared the expression of miRNAs in metastatic and nonmetastatic primary mouse sarcomas and found that microRNA-182 (miR-182) was markedly overexpressed in some tumors that metastasized to the lungs. By utilizing genetically engineered mice with either deletion of or overexpression of miR-182 in primary sarcomas, we discovered that deletion of miR-182 substantially decreased, while overexpression of miR-182 considerably increased, the rate of lung metastasis after amputation of the tumor-bearing limb. Additionally, deletion of miR-182 decreased circulating tumor cells (CTCs), while overexpression of miR-182 increased CTCs, suggesting that miR-182 regulates intravasation of cancer cells into the circulation. We identified 4 miR-182 targets that inhibit either the migration of tumor cells or the degradation of the extracellular matrix. Notably, restoration of any of these targets in isolation did not alter the metastatic potential of sarcoma cells injected orthotopically, but the simultaneous restoration of all 4 targets together substantially decreased the number of metastases. These results demonstrate that a single miRNA can regulate metastasis of primary tumors in vivo by coordinated regulation of multiple genes.
Introduction
Metastatic disease remains the primary cause of mortality for cancer patients, despite decades of important advances in cancer research (1). To spread to distant sites, tumor cells must locally invade from the primary tumor through the extracellular matrix (ECM), intravasate into the circulation, extravasate into the secondary site, and finally colonize the tissue to form secondary tumors (2). Our understanding of pathways that define this multistep process is far from complete. Therefore, a better understanding of mechanisms underlying metastasis is not only important for identifying molecular markers that predict which patients will develop metastases, but also for developing novel therapeutic approaches.
miRNAs are small noncoding RNA molecules composed of 20 to 22 nucleotides that can inhibit translation of multiple mRNA targets (3, 4). Accumulating evidence suggests that miRNAs regulate tumor cell invasion and metastasis (5), and deregulation of miRNA expression has been linked to metastasis of human cancers. For example, the upregulation of certain miRNAs (miRNA-10b [miR-10b] and miR-21) has been associated with enhanced metastasis in breast cancer, while downregulation of certain miRNAs (miR-31, miR-126, miR-335, and miR-145) has been correlated with metastatic relapse in breast and colon cancer (6–9).
There is an emerging body of evidence from multiple in vitro and xenograft studies that suggests an important role for miRNAs in regulating metastasis, although, to our knowledge, a genetically engineered mouse model has not been used to determine whether a single miRNA can regulate metastasis from primary cancers in vivo. To address this question, we utilized a primary mouse model of soft tissue sarcoma (STS) in which approximately 40% of mice develop lung metastases following amputation of the tumor-bearing limb. Using an unbiased miRNA screen, we identified miR-182 as an overexpressed miRNA in a subset of tumors that metastasized to the lungs. To directly test the role of miR-182 in sarcoma metastasis, we genetically engineered mice that can conditionally delete or overexpress miR-182 in primary sarcomas in vivo. Deletion of miR-182 decreased the rate of lung metastasis after surgical removal of the primary tumor, while overexpression of miR-182 increased the rate of lung metastasis. Using fluorescently labeled reporter mice coupled with proteomic screening, we identified targets of miR-182 that regulate intravasation of tumor cells into the circulation. Taken together, our results demonstrate that miR-182 is a driver of tumor metastasis in vivo via the simultaneous repression of multiple proteins that control intravasation.
Results
miR-182 is elevated in a subset of metastatic sarcomas.
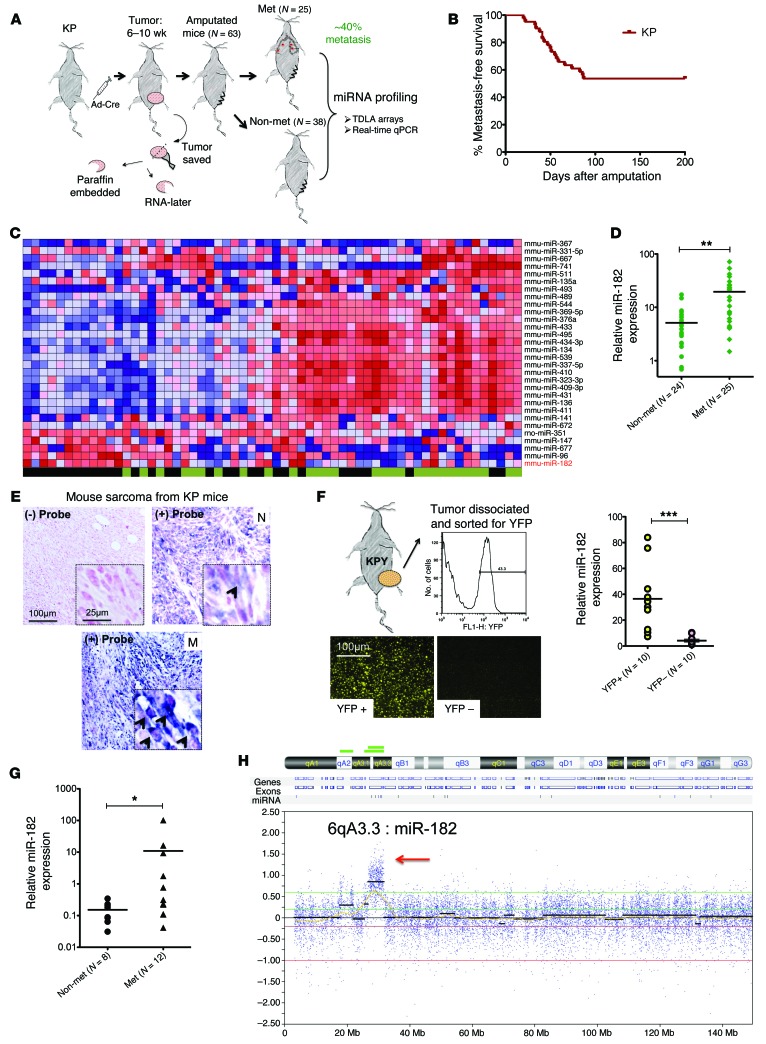
To study the development of lung metastasis in vivo, we utilized a spatially and temporally restricted mouse model of STS with conditional mutations in Kras and p53 (KP mice). Intramuscular injection of an adenovirus expressing Cre recombinase (Ad-Cre) into the hind limb activates oncogenic Kras and deletes p53, and high-grade spindle cell sarcomas develop at the site of injection in 2 to 4 months (10). We amputated the sarcoma-bearing limb and monitored the mice for the development of pulmonary metastases (Figure 1A). We observed that approximately 40% (n = 25/63) of KP mice developed lung metastases within 6 months after amputation (Figure 1B).
Figure 1. miR-182 is elevated in a subset of metastatic sarcomas.
(A) Schematic showing amputation strategy used to study metastasis in KP mice. (B) Kaplan-Meier curve showing metastasis-free survival in KP mice. (C) Heat map showing differential expression of miRNAs between nonmetastatic and metastatic primary mouse sarcomas using miRNA TLDA array (blue color, low expression; red color, high expression; green color, nonmetastatic [nonmet]; black color, metastatic [met]). (D) Validation of elevated miR-182 expression in primary mouse STS measured by real-time RT-PCR. (E) ISH detects miR-182 expression in primary metastatic (M), but not in nonmetastatic (N) sarcomas from KP mice (blue, miR-182–digoxigenin probe (arrows); pink, Nuclear Fast Red). (F) Primary tumors from KPY mice were dissociated and then sorted for YFP-positive and -negative cells. RNA was then extracted from those cells, and miR-182 expression was analyzed using qRT-PCR. miR-182 expression was specific to YFP-expressing sarcoma cells, but not to YFP-negative cells. (G) Elevated miR-182 expression in primary human STS measured by real time RT-PCR. (H) Comparative genomic hybridization shows amplification of miR-182 locus, 6qA3.3, in primary mouse sarcomas with known metastatic outcome. Two-tailed Student’s t test was used for statistical analysis in D–G. All data are mean ± SEM. Scale bars: 100 μm (E and F); 25 μm (E, insets). *P < 0.05; **P < 0.01; ***P < 0.005.
To investigate whether specific miRNAs were expressed in primary tumors that metastasized to the lungs, we performed a screen using miRNA-TaqMan low-density arrays (TLDAs) on the metastatic (n = 25) and the nonmetastatic (n = 38) primary mouse sarcomas. From the TLDA screen, the majority of mouse miRNAs were significantly downregulated in primary tumors that metastasized to the lungs compared with primary tumors that did not metastasize (Figure 1C and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI77116DS1). This is consistent with our prior study in KP mice in which deletion of 1 allele of the miRNA-processing protein Dicer promoted metastasis (11). However, we did identify few miRNAs that were upregulated in metastatic sarcomas, including miR-182. We focused on studying the role of miR-182 in lung metastasis because (a) miR-182 had been implicated in human melanoma metastasis (12) and (b) a genetic region spanning human miR-182 (7q32) is amplified in STS and correlates with poor prognosis (13, 14). We confirmed miR-182 upregulation in the metastatic primary sarcomas using TaqMan-based real-time reverse transcriptase–PCR (RT-PCR) and in situ hybridization (Figure 1, D and E, respectively). To track tumor cells using flow cytometry, we also crossed KP mice to mice expressing a Cre-activated yellow fluorescent protein (YFP) reporter ( hereafter called KPY mice), generating primary sarcomas with tumor-specific YFP expression. We found that miR-182 expression is specific to tumor cells (YFP+ cells) and is minimally expressed in the stroma (YFP– cells) (Figure 1F and Supplemental Methods).
To gain insight into the clinical relevance of miR-182 in sarcoma metastasis, we performed real-time RT-PCR for miR-182 on patient sarcoma samples. We found that primary human undifferentiated peomorphic sarcomas (UPSs) sarcomas from patients with lung metastases had elevated levels of miR-182 (Figure 1G). As more than 50% of miRNA genes are located in fragile chromosomal sites (15), we performed array comparative genomic hybridization (CGH) on genomic DNA from mouse sarcomas to determine whether changes in DNA copy number could explain the upregulation of miR-182 in primary tumors that metastasized and that otherwise exhibited a general downregulation of miRNAs. Remarkably, in 4 of 9 mouse sarcomas that metastasized to the lungs, we detected a focal amplification of mouse locus 6qA3.3 (Figure 1H), suggesting a possible mechanism for the upregulation of miR-182, although, other members of this miRNA cluster (i.e., miR-96 and miR-183) were not upregulated (Supplemental Figure 1, A and B), presumably because miR-182 has an independent promoter (Supplemental Figure 1C). Interestingly, this region is syntenic to human locus 7q32, which includes human MIR182 and is amplified in human STS with poor outcome (13).
miR-182 regulates cell migration and invasion in primary mouse sarcoma cell lines.
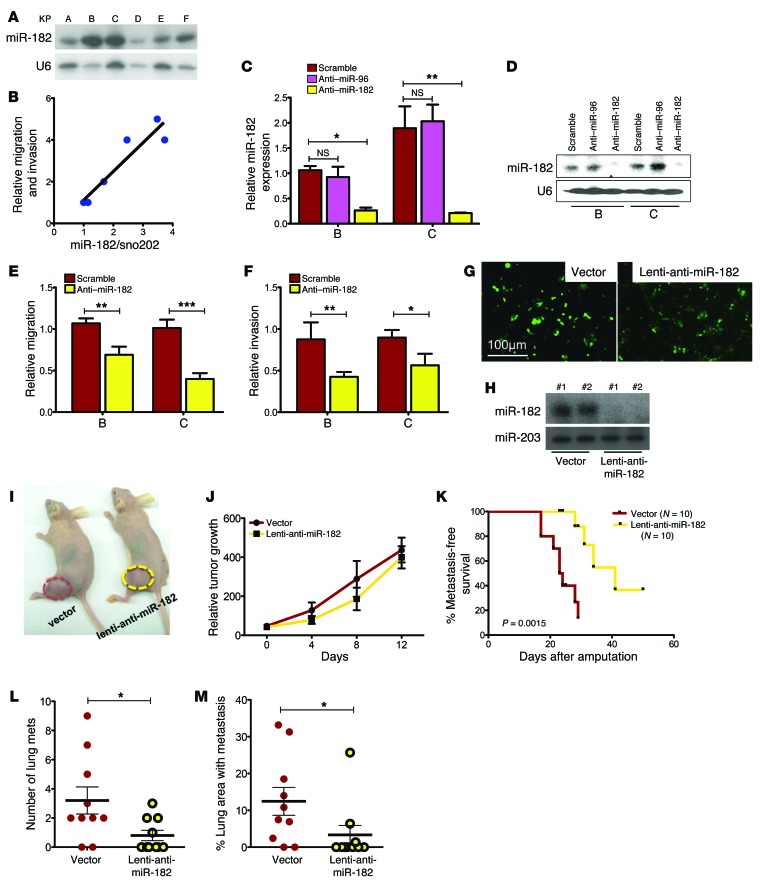
We examined miR-182 expression in primary sarcoma cell lines derived from KP mice using Northern blotting (Figure 2A). miR-182 was detected in all cell lines, and the level of miR-182 expression correlated with in vitro migration and invasion of KP cell lines (Figure 2B and Supplemental Methods; also see complete unedited blots in the supplemental material). Decreasing levels of miR-182 by knockdown (Figure 2, C and D) significantly decreased migration and invasion compared with a scrambled control (Figure 2, E and F). In contrast, stable overexpression of miR-182 in sarcoma cell lines A and B increased their migration (Supplemental Figure 2, A–C). Also, overexpression of miR-182 in human sarcoma cell lines derived from undifferentiated pleomorphic sarcomas (UPS) (STS-48 and STS-109) or pleomorphic rhabdomyosarcoma (STS-145) also increased migration and invasion in vitro (Supplemental Figure 2, D–G). A complementary in vivo experiment showed that orthotopic transplantation of sarcoma cell lines with stable knockdown of miR-182 (Figure 2, G and H) had no impact on primary tumor growth (Figure 2, I and J), but did improve metastasis-free survival (Figure 2, K–M).
Figure 2. Change in miR-182 levels modulates cell migration and invasion in primary mouse sarcoma cell lines.
(A) Northern blot detects miR-182 in multiple primary sarcoma cell lines from KP mice. (B) Levels of miR-182 in cells correlates with the relative migration and invasion ability of these KP cells. Average of migration and invasion index was plotted on the y axis. (C) Real-time RT-PCR and (D) Northern blot validates knockdown of miR-182 in KP cell lines B and C by an anti–miR-182 oligo. (E) Knockdown of miR-182 in B and C decreases migration and (F) invasion (n = 6 independent experiments). (G) Stably transduced cell line A with anti–miR-182 lentiviral construct. (H) Northern blot validates stable knockdown of miR-182 in 2 different clones. (I and J) Stable knockdown of miR-182 does not affect orthotopic tumor growth in nude mice, but prevents lung metastasis after surgical resection of the orthotopic tumor, as shown in a Kaplan-Meier plot (K) (Mantel-Cox log-rank test). (L) Quantification of the number of lung metastases and (M) percentage of lung area with metastasis shows significant difference between the 2 genotypes. One-way ANOVA (C, E, and F) and 2-tailed Student’s t test (L and M) were used for statistical analysis. All data are mean ± SEM. Scale bars: 100 μm. *P < 0.05; **P < 0.01; ***P < 0.005.
miR-182 deletion decreases the rate of sarcoma metastasis to the lungs.
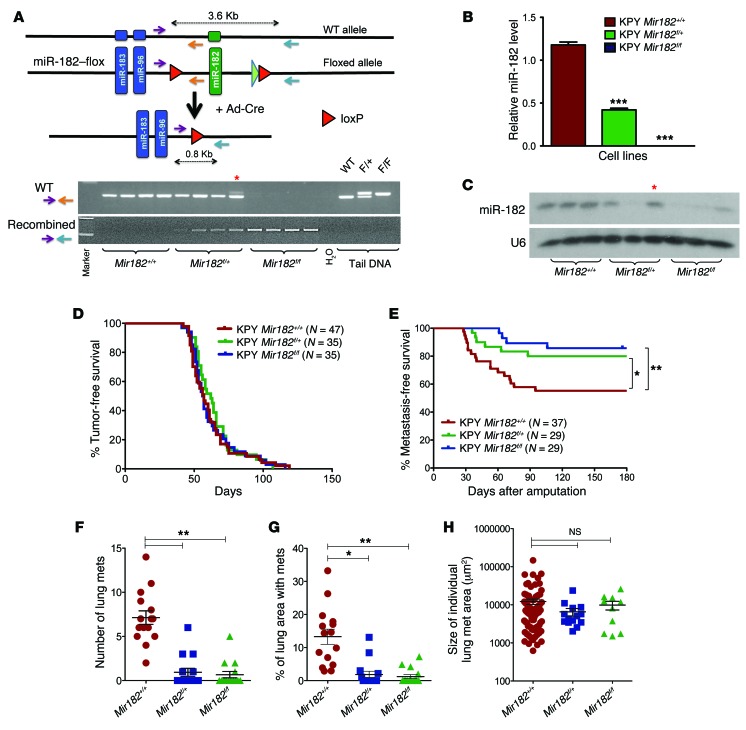
To our knowledge, no study has investigated whether altering a single miRNA can regulate metastasis in a primary tumor model system. To study how miR-182 affects metastasis in a primary model tumor system, we generated genetically engineered mice to delete miR-182 in primary sarcomas in vivo. In miR-182–flox (Mir182f/+) mice, loxP sites flank the miR-182 gene so that Cre recombinase deletes miR-182 (Figure 3A and Supplemental Figure 3, A and B). We then crossed these Mir182f/+ mice with KPY mice and injected Ad-Cre into the muscle of KPY Mir182f/+ mice, KPY Mir182f/f mice, and KPY littermate controls. No phenotypic differences were observed in primary sarcomas with or without miR-182 deletion. After successful isolation of primary cell lines from these sarcomas, we performed PCR analysis on genomic DNA, which showed that the primary sarcomas had efficiently recombined the miR-182–flox allele (Figure 3A). In addition, miR-182 expression was significantly decreased at the transcript level in Mir182f/+ and Mir182f/f mice by real-time RT-PCR and by Northern blot (Figure 3, B and C, respectively). We used these cell lines in Transwell migration chambers and Matrigel invasion chambers and found that deletion of either 1 or 2 alleles of miR-182 significantly suppressed both migration and invasion in vitro (Supplemental Figure 3, C and D).
Figure 3. miR-182 deletion decreases the rate of sarcoma metastasis to the lungs.
(A) Schematic showing deletion of miR-182 by Ad-Cre (top panel) and PCR to show Cre-mediated excision of recombined allele from primary KP sarcomas (bottom panel). Asterisk denotes the only sample (1/25) with partial recombination of the miR-182–flox allele. (B) RT-PCR and (C) Northern blot validates deletion of miR-182 in primary sarcoma cells from KPY Mir182 mice. Asterisk denotes the only sample (1/25) with partial recombination of the miR-182–flox allele. (D) No significant change in tumor onset was observed. (E) Deleting miR-182 in primary sarcomas in mice decreases the rate of lung metastasis (Mantel-Cox log-rank test). (F) Quantification of the number of lung metastases, (G) percentage of lung area with metastasis, and (H) size of individual metastatic nodules among different genotypes. One-way Anova was used for statistical analysis. All data are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005.
After validating successful recombination of the miR-182–flox allele and impaired in vitro migration and invasion of miR-182–deleted cells, we generated a cohort of sarcomas with or without miR-182 deletion (Supplemental Figure 3E). We did not observe any significant changes in tumor onset or tumor growth kinetics after deletion of miR-182 in KP tumors (Figure 3D and Supplemental Figure 3F). Of note, immunostaining of sarcomas using Ki-67 showed no difference between these groups, suggesting that miR-182 deletion has no impact on the growth of sarcoma cells in vivo (Supplemental Figure 3, E and G). However, when primary tumors were resected and mice were followed for 6 months, we found that deletion of either 1 or both copies of miR-182 significantly decreased the rate of lung metastasis from 43% baseline to 20% and 13%, respectively (Figure 3E). Of note, we observed a significant decrease in the number of metastatic nodules and the metastatic burden in the lungs of the miR-182–deleted mice (Figure 3, F–H). These results demonstrate that the level of a single miRNA in primary cancers can regulate metastasis in vivo.
miR-182 overexpression increases the rate of sarcoma metastasis to the lungs.
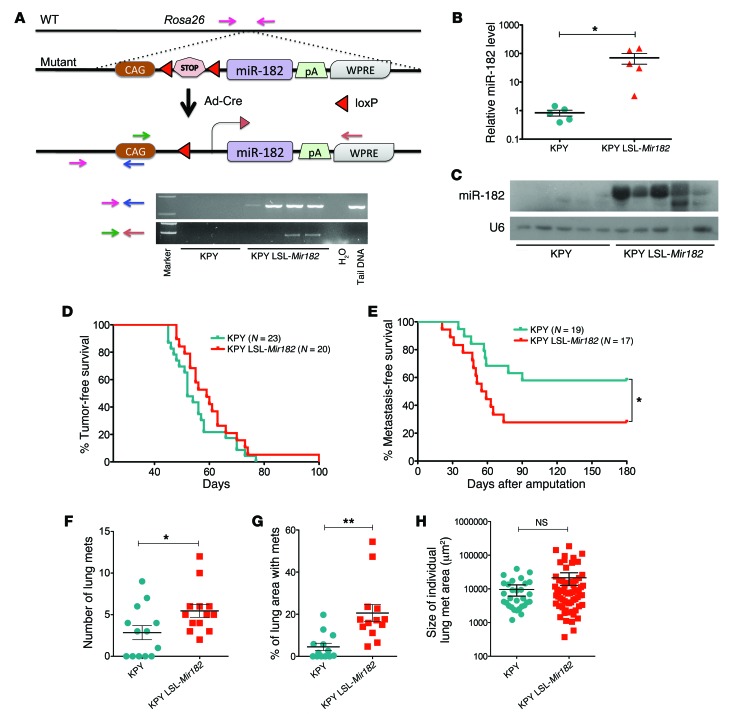
As a complementary approach, we also generated LSL–miR-182 mice (where LSL indicates LoxP-Stop-LoxP), in which Cre activates miR-182 overexpression. We generated primary sarcomas in KPY LSL-Mir182 mice (Figure 4A) and validated overexpression of miR-182 (Figure 4, B and C). We found that primary cell lines overexpressing miR-182 had enhanced in vitro migration and invasion (Supplemental Figure 4, A and B). miR-182 overexpression did not change the time for tumor onset (Figure 4D), growth of the tumors, or proliferation rate of the sarcomas (Supplemental Figure 4, C–E). However, overexpression of miR-182 significantly enhanced the rate of lung metastases from 40% in control mice to 75% (Figure 4E). In addition, miR-182 overexpression increased the number of metastatic nodules in the lungs (Figure 4, F–H). These findings complement the results of miR-182–knockout studies (Figure 3) and further demonstrate that the level of a single miRNA in primary cancers can regulate metastasis in vivo.
Figure 4. miR-182 overexpression increases the rate of sarcoma metastasis to the lungs.
(A) Schematic showing transgenic mice with conditional overexpression of miR-182 (top panel) and PCR to show Cre-mediated excision of the STOP cassette by Cre in primary KP sarcomas (bottom panel). (B) RT-PCR and (C) Northern blot validates significant overexpression of miR-182 in primary sarcoma cells from miR-182–overexpressor KP mice. (D) No change in tumor onset in primary sarcomas from KPY LSL-Mir182 mice. (E) Overexpression of miR-182 in primary sarcomas increases the rate of lung metastasis (Mantel-Cox log-rank test), *P < 0.05. (F) Quantification of the number of lung metastases, (G) percentage of lung area with metastasis, and (H) size of individual metastatic nodules between the 2 genotypes. Two-tailed Student’s t test is used for the statistical analysis. All data are mean ± SEM. *P < 0.05; **P < 0.01.
Deletion of miR-182 decreases intravasation of sarcoma cells into the circulation.
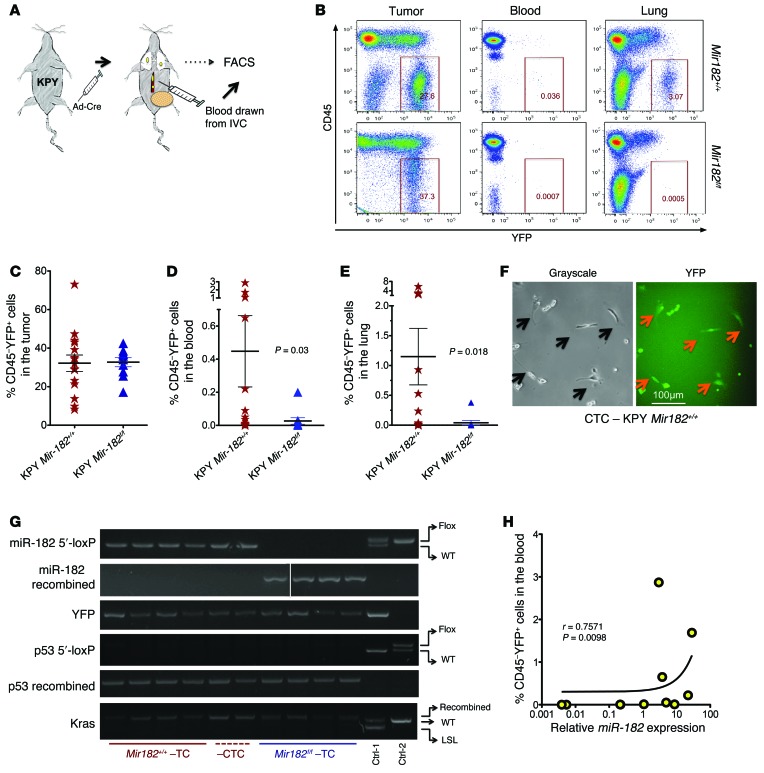
Metastasis has been correlated with the presence of circulating tumor cells (CTCs) in the vasculature (16), and we have shown that miR-182 levels within primary sarcomas correlate with the rate of lung metastasis. Because deletion of miR-182 promotes in vitro migration and invasion, we therefore hypothesized that loss of miR-182 would prevent sarcoma cells from entering into the circulation. We generated sarcomas in KPY Mir182f/f mice and KPY littermate controls to detect YFP-labeled (YFP+) cells in the circulation. Once tumors reached 250 mm3 in size, we drew blood from the inferior vena cava (IVC) to collect cells prior to filtration by the lungs. We used flow cytometry to analyze YFP+ cells in the primary tumors and in the circulation (Figure 5, A and B). We did not see any difference in the number of YFP+ cells in the primary tumors with or without miR-182 (Figure 5C). However, mice with miR-182–deficient sarcomas had a significant decrease in the number of YFP+ CTCs (Figure 5D). For example, approximately 90% of the control mice had detectable YFP+ CTCs in their blood, while YFP+ CTCs were detected in only 50% of mice with miR-182–deleted sarcomas (Supplemental Figure 5A). Furthermore, fluorescence microscopy of the sorted YFP+ CTCs confirmed that the CTCs retained a mesenchymal morphology (Figure 5F). In addition, flow cytometry further showed that YFP+ CTCs are positive for high expression of cell-surface vimentin similar to that of primary sarcoma cells (Supplemental Figure 5C), a finding that is in agreement with a recent study showing that vimentin might serve as an exclusive marker on sarcoma CTCs (17). Of importance, PCR analysis of genomic DNA from these YFP+ CTCs clearly demonstrated efficient recombination of KPY alleles with the presence of WT miR-182 in these cells (Figure 5G), indicating that the YFP+ CTCs are derived from KPY sarcomas. To investigate whether the number of YFP+ CTCs correlated with the level of miR-182 in primary sarcomas, we isolated RNA from primary sarcomas in KPY mice. Real-time quantitative PCR (qPCR) demonstrated a positive correlation between the number of YFP+ CTCs in the blood and expression of miR-182 within the primary tumor (Figure 5H), suggesting that miR-182 drives intravasation of the tumor cells. To further confirm the role of miR-182 in intravasation, we analyzed YFP+ cells in the primary tumor and the blood from miR-182 overexpressor mice (LSL–miR-182). As expected, we observed a higher number of YFP+ CTCs in the circulation in mice with miR-182–overexpressing sarcoma (Supplemental Figure 5, D–G). Interestingly, the percentage of YFP+ cells in primary tumors was found to be higher in miR-182–overexpressing mice compared with controls; however, no correlation between the percentages of YFP+ cells in primary tumors and the percentage of YFP+ CTCs was observed (Supplemental Figure 5J).
Figure 5. Deletion of miR-182 decreases intravasation of sarcoma cells into the circulation.
(A) Schematic showing the study design for isolation and analysis of YFP+ CTCs in the blood. (B) Flow cytometry reveals significantly lower number of YFP+ cells in both the blood and lung after miR-182 deletion. (C) No change in YFP+ cells in primary tumors between the 2 genotypes. (D and E) Quantification of CD45–YFP+ cells within the blood and the lungs between the 2 genotypes. (F) Images show the morphology of isolated YFP+ sarcoma cells from the blood. (G) PCR demonstrates that isolated CD45–YFP+ cells have successfully recombined LSL-Kras, deleted p53, and recombined LSL-YFP. TC, tumor cells. (H) Number of YFP+ cells in the blood correlates with the level of miR-182 in the primary sarcoma in KPY mice. Two-tailed Student’s t test and Fisher’s exact test were used for statistical analysis. All data are mean ± SEM. Scale bars: 100 μm.
We next asked whether YFP+ CTCs are competent to extravasate from the blood vessels in the lungs. Therefore, we analyzed YFP+ cells in homogenized lungs from the same mice using flow cytometry. We found a lower percentage of YFP+ cells in the lungs of mice with sarcomas deleted for miR-182 compared with mice with control sarcomas (Figure 5E and Supplemental Figure 5B). In contrast, a higher percentage of YFP+ cells in the lungs of mice with miR-182–overexpressing sarcomas were observed compared with mice with control sarcomas. Notably, all mice with miR-182–overexpressing sarcomas had YFP+ cells in their lungs (Supplemental Figure 5E). Taken together, our findings indicate that miR-182 enhances the ability of tumor cells from primary sarcomas to enter into the circulation and reach the lungs.
miR-182 does not affect lung colonization in an experimental metastasis assay.
To test whether miR-182 also promotes lung colonization, in addition to promoting intravasation, we used an experimental metastasis model to test whether miR-182 modulates metastasis once intravasation is bypassed. We injected early passage primary mouse sarcoma cell lines either deficient for Mir182 (Mir182-KO), overexpressing Mir182 (Mir182-OE), or from their respective controls directly into the circulation via the tail vein in nude mice. Then all lungs were harvested after 2 weeks. As shown in Supplemental Figure 6, A–D, among the different genotypes we did not observe any significant difference, either in the number of lung colonies formed or in the metastatic burden in the lung. In addition, we did not see any difference in the number of lung metastases following tail-vein injection of a KP sarcoma cell line with stable knockdown of miR-182 compared with its vector control (Supplemental Figure 6, E–G), a finding which is inconsistent with other published reports that show miR-182 regulate metastasis in breast and gall bladder cancer (18, 19). Since tail-vein injection bypasses the requirement for invasion in metastasis, these results suggest that miR-182 regulates sarcoma metastasis by controlling intravasation rather than extravasation and colonization.
miR-182 target genes that regulate metastasis.
To dissect the molecular mechanism by which miR-182 mediates sarcoma metastasis, we utilized 3 independent, unbiased approaches to search for potential miR-182 targets: in silico prediction, luciferase reporter assay, and proteomics. First, in silico programs, including Targetscan and Miranda, identified 5 genes implicated in metastasis with a miR-182–binding motif: metastasis suppressor protein-1 (Mtss1, also called missing-in-metastasis [MIM]), Ras suppressor protein-1 (Rsu1), L1 cell adhesion molecule (L1cam), RAS p21 protein activator (Rasa1), and Ras association domain family 1 isoform A (Rassf1a). We followed this analysis with a 3′ UTR luciferase reporter screen (Figure 6A) to investigate whether miR-182 targets these 5 genes because seed pairing alone may not be sufficient for miRNA-target interactions in cells (20). In fact, results from the luciferase assay revealed that miR-182 suppresses luciferase activity only for the 3′ UTRs of Mtss1 and Rsu1. Moreover, this suppression was abolished in these constructs where the miR-182–binding site was mutated (Supplemental Figure 7, A–E). In contrast, the 3′ UTR of other genes (including L1cam, Rasa1, and Rassf1a) failed to regulate luciferase activity in a miR-182–dependent manner (Supplemental Figure 7). Of note, both Rsu1 and Mtss1 have been implicated in remodeling the cytoskeleton and cell migration (21, 22).
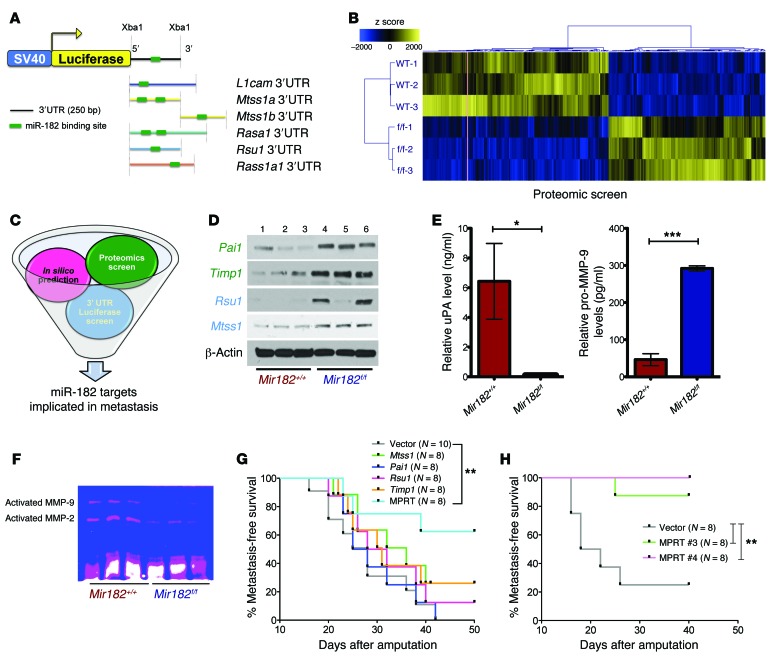
Figure 6. miR-182 target genes cooperate to regulate metastasis.
(A) In silico/3′ UTR luciferase screen used to predict miR-182 targets (top panel) and number of putative miR-182–binding sites on respective 3′ UTRs of the different genes. (B) Heat map shows differential expression of approximately 400 proteins between miR-182 WT and miR-182 deleted primary sarcoma cells from a proteomic screen. (C) Schematic shows the different approaches used to identify miR-182 target genes implicated in metastasis. (D) Western blot validates upregulation of Pai1, Timp1, Rsu1, and Mtss1 in miR-182–deleted KP cells (1–3, different KPY Mir182 WT; 4–6, different KPY Mir182f/f cell lines). (E) ELISA validates decreased uPA and increased unprocessed pro–MMP-9 in the cell-culture media from miR-182–deleted primary sarcoma cells. (F) Zymography confirms decreased activation of MMP-2 and MMP-9 in the cell-culture media from miR-182–deleted KP cells. (G and H) Kaplan-Meier curves show that ectopic expression of all 4 miR-182 targets together (H), but not individual targets alone (G) are required to rescue miR-182–mediated metastasis in an orthotopic metastasis assay. Mice were euthanized 40 days following amputation. Five mice had microscopic lung nodules despite having no clinical evidence of metastasis (Supplemental Figure 13G). MPRT, cell line expressing all 4 miR-182 targets, i.e., Mtss1, Pai1, Rsu1, and Timp1. Two-tailed Student’s t test and 1-way ANOVA were used for statistical analysis. All data are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005.
To search for additional metastasis-related miR-182 targets, we performed a liquid chromatography–mass spectrometry (LC-MS) proteomic screen of cell lines derived from primary sarcomas with or without miR-182 deletion. The proteomic screen revealed that more than 400 proteins were differentially regulated at least 2-fold between the miR-182–expressing and –deleted sarcoma cells. This included 225 upregulated and 175 downregulated proteins (Figure 6B and Supplemental Table 2, A and B). Out of those 225 upregulated proteins, only 8 had 1 or more putative miR-182–binding motifs in their mRNA, as determined using in silico analysis, suggesting that most differentially expressed proteins are indirectly regulated by miR-182 (Supplemental Table 3). We subsequently focused on proteins implicated in metastasis and/or with putative binding sites for miR-182 (Figure 6C). With these criteria, plasminogen activator inhibitor-1 (Pai1, upregulated 12.9-fold), tissue inhibitor of metalloproteases-1 (Timp1, upregulated 6.3-fold), and BCL2/adenovirus E1B 19 kDa protein-interacting protein-3 (Bnip3, upregulated 3.5-fold) stood out as potential targets for miR-182 (Supplemental Table 2). Western blot analysis validated that Rsu1, Mtss1, Pai1, and Timp1 (Figure 6D), but not Bnip3 (Supplemental Figure 8B and Supplemental Methods), were elevated in miR-182–deleted sarcoma cells. Moreover, Rsu1, Mtss1, Pai1, and Timp1 were all upregulated in miR-182–deleted sarcoma cell lines and downregulated in miR-182–overexpressing cell lines (Supplemental Figure 9A). Furthermore, a luciferase assay indicated that (in addition to Rsu1 and Mtss1) both Pai1 and Timp1 were also direct targets of miR-182. As shown in Supplemental Figure 8, C–H, miR-182 suppressed luciferase activity of the WT 3′ UTR of Pai1 and Timp1, whereas this suppression was abolished when the miR-182–binding site was mutated. Consistent with the data from mouse sarcoma cell lines, overexpression of miR-182 in human sarcoma cells also caused downregulation of PAI1, TIMP1, and MTSS1 (Supplemental Figure 9B).
Because PAI1 and TIMP1 can suppress metastasis by preventing the activation of proteases (23, 24), we next analyzed the expression of proteases in the media secreted from sarcoma cells by ELISA (Supplemental Methods). In sarcoma cells lacking miR-182, we found reduced activation of urokinase (uPA) and MMP-9 (Figure 6E), which are downstream targets of Pai1 and Timp1, respectively. These results link the activation of uPA and MMP-9, which control ECM degradation, to miR-182. Consistent with the results from the ELISA, gelatin zymography revealed reduced gelatinolytic activity of both MMP-2 and MMP-9 in the media from the miR-182–deleted sarcoma cells, showing that miR-182 regulates the activity of these extracellular proteases (Figure 6F).
To investigate whether miR-182 regulates all 4 of the validated miR-182 target genes in vivo, we analyzed an independent cohort of primary mouse sarcoma samples with known metastatic outcome and correlated the level of miR-182 by real-time RT-PCR with protein expression of the miR-182 target genes by Western blot. Similar to the initial cohort of primary sarcomas (Figure 1D), this independent cohort of 30 sarcomas showed elevated miR-182 in about 70% of sarcomas that metastasized to the lungs (Supplemental Figure 10C). As anticipated, Western blotting of lysates from these tumors showed decreased protein expression for all 4 target genes in metastatic sarcomas, consistent with regulation by miR-182 in vivo (Supplemental Figure 10, A and B). Also, ELISA measurements of uPA and pro–MMP-9 demonstrated that in primary sarcomas that metastasized to the lungs, uPA levels were increased, while pro–MMP-9 levels trended downwards (Supplemental Figure 10). Of note, the strength of these interactions was further demonstrated by 1 sample that did not metastasize to the lungs, but nevertheless had an elevated level of miR-182 and retained the same pattern of expression of the miR-182 target genes (Supplemental Figure 10, D–J). Although the expression of miR-182 target genes is lower in metastatic sarcomas taken as a group, there was considerable variation in expression among individual tumors (Supplemental Figure 11).
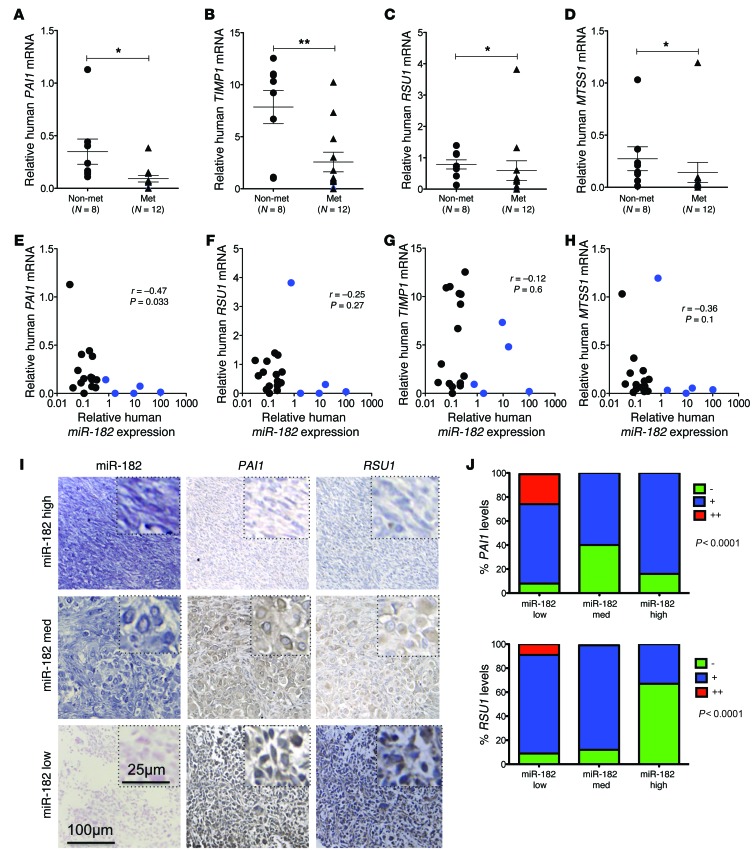
In addition, real time RT-PCR demonstrated downregulation of RSU1, MTSS1, PAI1, and TIMP1 in a subset of primary human metastatic sarcomas (Figure 7, A–D) that trended with elevated miR-182 expression (Figure 7, E–H). Finally, a strong negative correlation between miR-182 and its target genes, PAI1 and RSU1, was observed in matched sarcoma specimens on a tissue microarray (TMA) (Figure 7, I and J, and Supplemental Table 6).
Figure 7. Decreased expression of miR-182 target genes in primary human metastatic sarcomas.
(A–D) Decreased PAI1, TIMP1, RSU1, and MTSS1 mRNA expression in primary human STS measured by real-time RT-PCR. (E–H) miR-182 levels negatively correlate with mRNA levels of PAI1, TIMP1, RSU1, and MTSS1 in primary human sarcomas. The metastatic human sarcomas with high miR-182 expression are denoted in blue circles, and these colored samples are followed through parts A–D to correlate miR-182 expression with the level of its targets. (I) Expression of miR-182, PAI1 and RSU1 in matched human sarcoma specimens. (No immunostaining was observed for MTSS1 and TIMP1 on human TMAs). (J) Quantification of immunostaining. Mann-Whitney test was used for statistical analysis (A–D). Spearman R correlation test was used for statistical analysis (E and F). Fisher’s exact test was used for statistical analysis (J). All data are mean ± SEM. Scale bars: 100 μm (I); 25 μm (I, insets). *P < 0.05; **P < 0.01.
miR-182 must target multiple genes to control metastasis.
To test whether miR-182 required multiple genes to control metastasis, we performed rescue experiments with the KP LSL-Mir182 sarcoma cell line, which expresses approximately 20-fold higher miR-182 expression than a KP LSL-control cell line (Supplemental Figure 12A). We stably overexpressed each target alone or all 4 miR-182 gene targets in different combinations. After validating stable overexpression of the target genes at both the RNA and protein levels (Supplemental Figure 12, B–D), we subjected these cells to Transwell and Matrigel assays. Whereas overexpression of each individual gene modestly decreased invasion and migration without reaching statistical significance, overexpression of all 4 miR-182 targets together (Mtss1, Pai1, Rsu1, Timp1 [MPRT]) significantly suppressed both migration and invasion in vitro (Supplemental Figure 12, F and G). When these cells were injected into the muscle of lower hind limb and the subsequent tumors were amputated, improved metastasis-free survival was observed only with MPRT cells (Figure 6G and Supplemental Figure 12, I and J). A further improvement in metastasis-free survival was seen when the same in vivo experiment was repeated with 2 separate monoclonal MPRT clones, no. 3 and no. 4 (Figure 6H), which were isolated as single cells by flow cytometry and subsequently validated for overexpression of all 4 miR-182 targets (Supplemental Figure 13). In addition, both uPA and activated MMP-9 levels were found decreased in the supernatant of MPRT-expressing cells (Supplemental Figure 13). Therefore, our results show that miR-182 regulates multiple genes that function at different steps (migration and invasion) of the metastatic process. Taken together, we have demonstrated that miR-182 targets multiple genes to enhance the migration of sarcoma cells (Rsu1 and Mtss1) and ECM invasion (Pai1 and Timp1) via enhanced activation of extracellular proteases, including uPA and MMP-9. Moreover, all of these targets appear to be required for miR-182 to promote metastasis in vivo.
Discussion
Metastasis is a complex process that can be regulated at several different steps. Here, we demonstrated that levels of miR-182 correlated with metastasis in primary STS in mice. To better understand whether miR-182 is a driver of metastasis or only a passenger in the metastasis process, we generated genetically engineered mice to either delete or overexpress miR-182 in primary sarcomas in mice. By deleting miR-182 in primary tumors, the rate of metastasis was decreased (43% to 13%), while overexpressing miR-182 increased the rate of metastasis (40% to 75%). Therefore, miR-182 is a driver of metastasis in primary STS in mice. By using both loss-of-function and gain-of-function approaches, we show for the first time, to our knowledge, that a single miRNA can regulate metastasis in a primary cancer model. Also, we found that miR-182 was enriched in a subset of human metastatic sarcomas. Consistent with our findings, other studies have correlated a high level of miR-182 to cancer progression in a number of other human cancers. For example, miR-182 expression was found to be elevated in metastatic melanoma compared with benign nevi (12), in gliomas compared with adjacent normal brain tissue (25), and in follicular carcinomas compared with normal thyroid tissue (26). In invasive bladder cancer, elevated miR-182 was correlated with aggressiveness and overall survival (27). In addition, in high-grade prostate cancer, miR-182 was elevated more than any other miRNA analyzed (28). Therefore, our finding that miR-182 drives metastasis in primary mouse sarcomas is relevant for a large number of human cancers.
Recently, several studies have shown that CTCs can be used to predict disease progression and metastasis (29, 30). For example, a study utilizing serial PCR-based analysis of CTCs was able to identify a subgroup of melanoma patients at higher risk of disease recurrence (31). Similarly, other studies have correlated the presence of CTCs with metastasis in a variety of cancers, including prostate, breast, and colon (32–35). Recently, investigators have also shown that CTCs can be detected in mice bearing orthotopic xenografts of breast cancers (36). Also, a recent study showed that CTCs could be detected even before the primary tumor in a genetically engineered mouse model of pancreatic cancer (37). We detected CTCs (YFP-labeled cells) in the blood of mice bearing primary sarcomas and observed a decrease in the number of YFP+ CTCs in mice with miR-182–deficient sarcomas compared with littermate controls. This finding suggests that miR-182 regulates intravasation of cancer cells into the circulation.
Several studies have suggested that miRNA-associated phenotypes can be mediated by a single miRNA target (12, 38, 39). However, each miRNA is known to regulate the expression of hundreds of genes. Therefore, a single miRNA target may not be sufficient to coordinate a multistep process, such as metastasis. Utilizing 3 unbiased approaches to search for miR-182 targets, we discovered 4 novel targets of miR-182 implicated in metastasis: Rsu1, Mtss1, Pai1, and Timp1. Both Pai1 and Timp1 regulate ECM proteases and decrease invasion of sarcoma cells in vitro. Pai1 is a serine protease inhibitor (Serpin) that inhibits uPA (40, 41); uPA in turn blocks the activation of plasmin, a protease that has been shown to promote ECM degradation and cell migration and invasion (42–46). Therefore, by preventing the activation of both uPA and plasmin, Pai1 reduces tumor cell migration and invasion (43, 45, 46). Although other groups have reported prometastatic properties of Pai1 in other tumor types (47, 48), our data support a metastasis suppressor role for Pai1 in STS.
Another miR-182 target identified through a proteomic screen, Timp1 is known to inhibit 2 MMPs, i.e., MMP-2 and MMP-9 (49, 50). Timp1 has been shown to inhibit tumor cell invasion and metastasis via MMP inhibitory activity in experimental models of hepatocellular carcinomas, osteosarcoma, and ovarian cancer (51–54). Interestingly, a clinical study implicates TIMP1 in STS in human patients. Analyzing the expression of MMPs and TIMP1 in 69 biopsies from patients with high-grade STS (including 28 UPS), TIMP1 expression and plasma levels were significantly lower in STS patients compared with healthy controls (n = 56) (55). Our study extends this observation by finding that TIMP1 expression is lower in both mouse and human metastatic sarcomas. Furthermore, in the mouse sarcoma, Timp1 is inversely related to MMP-9 levels and negatively correlates with the level of miR-182; overexpression of Timp1 is sufficient to reduce invasion of sarcoma cells in vitro.
Rsu1 and Mtss1 were validated as 2 additional miR-182 targets in our study.
Rsu1 has been shown to inhibit tumorigenesis and metastasis in multiple tumor types (56, 57). Rsu1 has been shown to interact with the Lim5 domain of Pinch1 that inhibits cell migration by stabilizing the IPP ILK, pinch, parvin (IPP) focal adhesion complex (58). Furthermore, a recent study demonstrated that Ras activation perturbs this inhibitory function of Rsu1 by promoting Rsu1 splicing and inhibiting its association with the IPP complex leading to Rac activation (21). Therefore, in addition to the previously reported mechanism of Ras regulating Rsu1, we established a novel mechanism of Rsu1 regulation by miR-182, which increases the migration of sarcoma cells. Mtss1 is also a gene that has previously been implicated in metastasis (59, 60). Mtss1 plays an important role in cytoskeleton remodeling both by activation of Rac and induction of actin-rich protrusions (61, 62). Therefore, Mtss1 is important in regulating cell motility, and its expression has been reported to be lost during metastasis in a variety of cancers, including bladder, colon, and prostate cancer (63–65). Consistent with a role for MTSS1 in regulating metastasis, we detected lower MTSS1 expression in both mouse and human metastatic sarcomas. Interestingly, a recent study by Wang et al. showed a similar negative regulation between MTSS1 and miR-182 in hepatocellular carcinoma (66) that further strengthens our findings.
To further understand how miR-182 acts through multiple targets, we performed rescue experiments in sarcoma cells, with overexpression of each target independently or overexpression of all 4 miR-182 gene targets in different combinations. We found that independent overexpression of any one of the 4 genes failed to suppress metastasis in vivo. Conversely, overexpression of all 4 genes together not only suppressed migration and invasion in vitro, but also metastasis in vivo in an orthotopic model. Taken together, these results indicate that the regulation of metastasis by a miRNA extends beyond the “1 miRNA–1 target” model, as a single miRNA can target multiple genes to regulate the multi-step process of metastasis. For example, we propose that miR-182 targets Rsu1 and Mtss1 to enhance the migration of sarcoma cells, while miR-182 targets Pai1 and Timp1 to increase invasion of cells through the ECM via enhanced activation of extracellular proteases, including uPA and MMP-9. Despite the fact that miR-182 regulates multiple genes, we observed that there was considerable variation in the expression of miR-182 target genes among individual sarcomas in both mouse and human. For instance, some sarcomas with elevated miR-182 suppressed all 4 miR-182 target genes, while others had less suppression of a subset of miR-182 targets. This suggests that other factors in addition to miR-182 contribute to regulation of these genes in vivo. Also, the changes observed in the proposed target gene mRNA levels in humans were not as impressive as the changes observed in the protein levels in both mice and human. We observed that, except for MTSS1, Targetscan does not identify a conserved binding site for miR-182 in other human targets. Nevertheless, in human sarcoma cells miR-182 can downregulate PAI1 and TIMP1 in addition to MTSS1 (Supplemental Figure 10B). Furthermore, a negative correlation between miR-182 and PAI1 and RSU1 was observed at the protein level in a human STS TMA, as shown in Figure 7, I and J. Since miRNAs can cause translation suppression without causing mRNA degradation, it is possible that miR-182 regulates its targets via translational repression. Furthermore, nonconserved miR-182 binding sites were identified using an alternative prediction program (RNAHybrid). Therefore, it is possible that miR-182 regulates these targets in humans either through nonconserved binding motifs or via some indirect mechanisms. This observation emphasizes the complexity of target regulation by miRNAs in vivo and underscores the value of testing the role of miRNAs in genetically engineered mouse models.
In conclusion, we found that miR-182 is elevated in a subset of primary mouse and human sarcomas that metastasized to the lungs. By overexpressing or deleting miR-182 in primary sarcomas, we demonstrate that the level of a single miRNA can drive metastasis in vivo. In addition, we identified targets of miR-182 that regulate different stages of metastasis to control intravasation into the circulation. These studies establish that miR-182 is a driver of metastasis that must target multiple genes to promote metastasis in vivo. We anticipate that other miRNAs can similarly target numerous genes at multiple steps to drive metastasis of primary cancers.
Methods
Animals and in vivo metastatic study.
Primary STSs were generated in LSL-KrasG12D and p53f/f mice. LSL-KrasG12D mice were provided by T. Jacks (Massachusetts Institute of Technology, Cambridge, Massachusetts, USA) and p53f/f mice by A. Berns (The Netherlands Cancer Institute, Amsterdam, The Netherlands) (67, 68). KP mice were crossed to mice expressing a Cre-activated YFP (LSL-YFPf/f) reporter to get KPY mice. KPY mice were then crossed to mice that conditionally delete miR-182 (miR-182–flox) or overexpress miR-182 (LSL–miR-182). Tumors were inducted by injecting adenovirus expressing Cre recombinase into the hind limb. For the orthotopic metastasis assay, athymic nude (nu/nu) mice (5 to 6 weeks old) were purchased from Taconic labs (NCRNU-M) and maintained in Duke University’s accredited animal facility. In brief, 5 × 104 exponentially growing KP-vector or KP–anti–miR-182 cells were injected into the hind limb of the nude mice. When the tumor size reached about 250 to 300 mm3, the tumor-bearing limb was amputated and mice were followed for the development of lung metastases. Similarly, in experiments with primary sarcomas in mice, the tumors were amputated when they were approximately 250 mm3. Mice were examined about 3 times per week following amputation until they developed signs of systemic illness, such as hunched posture, lethargy, decreased weight, or ruffled fur. For those mice that developed lung metastases, this often occurred at 1 to 2 months following amputation. At this point, the mice were euthanized and the lungs analyzed for metastasis. Most of the mice that developed lung metastases did so by 120 days after amputation. We sacrificed surviving mice at 180 days after amputation so that a sarcoma pathologist (D.M. Cardona) could examine the lungs to confirm the absence of lung metastases.
TLDAs.
64 tumors annotated for metastatic outcome were identified (metastatic, n = 25, nonmetastatic, n = 39). Samples were macrodissected, stored in RNAlater, and homogenized in liquid nitrogen; total RNA was isolated using TRIzol reagent (Invitrogen) per the manufacturer’s recommendation. TaqMan arrays were performed as described previously (11). Samples were compared using the ΔΔCT method by normalizing to SnoRNA202 expression and the median array. Samples were clustered using sparse hierarchical clustering (69) with average linkage. The heat map was made using differentially expressed miRNAs based on a 2-tailed t test (P < 0.05) between primary tumors that did or did not give rise to distant metastases.
Human sarcoma samples.
Human STS samples were acquired from MD Anderson under a protocol approved by the Duke University and MD Anderson Institutional Review Boards. The human sarcomas included in the study are UPS. Total RNA was extracted from frozen tumor samples using TRIzol, and cDNA synthesis was performed for mature miR-182 using the TaqMan MicroRNAs Reverse Transcription Kit, followed by quantitative PCR with its respective probe, per the manufacturer’s suggestion (Applied Biosystems). miR-182 expression was calculated using the ΔΔCT method (70) after normalization to SnoRNA234 expression. A sarcoma TMA (TMA 23) was acquired from the Radiation Therapy Oncology Group (RTOG). In situ hybridization for miR-182 and immunohistochemistry (IHC) for PAI1 and RSU1 were performed on matched sections from the TMA, which were then analyzed and scored by a pathologist (D.M. Cardona) in a blinded-fashion.
Generation of miR-182–flox and LSL–miR-182 mice.
To generate conditional miR-182 alleles, targeting constructs were generated from a 129Sv BAC by recombineering in E. coli (71). A mobile recombineering system (pSIM vector; ref. 72) was introduced into a BAC clone bMQ340c02 (obtained from Source BioScience LifeSciences) containing 120 kb of mouse chromosome 6 harboring miR-182. LoxP sites and a neomycin cassette flanked by frt sites were recombineered into the BAC, followed by recombineering out into the targeting vector. To obtain a conditional allele of LSL–miR-182 in the Rosa-26 locus, the fluorescent reporter in the Rosa targeting vector Ai9 (73) was replaced with the miR-182 sequence. Linearized targeting vectors were electroporated into 129Sv ES cells, and homologous recombination events were detected by PCR (74). After verification of correct rearrangement by Southern blot analysis, clones were injected into C57BL/6 blastocysts following standard procedures. Chimeric males were mated to WT C57BL/6 females to generate heterozygotes. These mice were subsequently crossed to a flpO deleter strain to excise the frt-Neo-frt cassette to generate miR-182–flox mice and R26-LSL–miR-182 mice. Primer sequences are provided in Supplemental Table 4.
Cell culture.
All KP cell lines, i.e., A–F, were derived from primary sarcomas in KP mice. In brief, sarcomas between 250 and 300 cm3 were harvested in a cell-culture hood and cut into tiny pieces. Then, they were digested using collagenase, dispase, and trypsin for a minimum of 1 hour. The digested mixture/slurry was then filtered through a 70-micron filter, followed by rbc lysis using ACK buffer (Lonza) according to the manufacturer’s protocol. Cells were finally filtered using a 40-micron sieve and cultured 5 to 8 passages before being used for experiments. Cells were cultured in DMEM medium supplemented with 10% FBS and incubated at 37°C with 5% CO2 in a humidified cell-culture incubator. Additional KP cell lines with miR-182 deletion or overexpression were also derived using a strategy similar to that described above. Human sarcoma cell lines STS-48 and STS-109 (UPS), and STS-145 (pleomorphic rhabdomyosarcoma) were cultured in DMEM/F12 medium supplemented with 10% FBS and incubated at 37°C with 5% CO2 in a humidified cell-culture incubator.
Plasmids.
The plasmids expressing miR-182 and miR-96 were cloned into the lentiviral vector pCDH-CMV-MCS-EF1-copGFP (System Biosciences). Pre–miR-182 and Pre–miR-96 were first PCR amplified using mouse genomic DNA as a template and Platinum Taq polymerase enzyme (Invitrogen) with corresponding specific primers. Primer sequences are shown in Supplemental Table 4. The amplified fragment was then cloned into a lentiviral vector (pCDH-CMV-MCS-EF1-copGFP from System Biosciences) at EcoR1 and Not1 sites using the Choo-Choo Cloning Kit per the manufacturer’s protocol (MCLAB). The Lentiviral anti–miR-182 plasmid was purchased from System Biosciences. The Luciferase reporter constructs were generated by introducing the 3′ UTR of a specific gene carrying a putative miR-182–binding site into pGL3 control vector (Promega). We first amplified the 3′ UTR sequence by PCR using gene-specific primers (Supplemental Table 4) and directly cloned them into the Xba1 site of the pGL3 using the Choo-Choo Cloning Kit per the manufacturer’s protocol (MCLAB). Site-directed mutagenesis of the miR-182–binding site in the Mtss1, Pai1, Rsu1, and Timp1 3′ UTR was carried out by the 2-step PCR approach as described previously (75). All PCR products were verified by DNA sequencing (Primer sequences are provided in Supplemental Table 4).
LC-MS (proteomics).
Proteomic analysis was performed at the Duke Proteomics Core Facility (DPCF). Three cell pellets from either miR-182 WT or Mir182f/f cells were washed with 50 mM ammonium bicarbonate and solubilized by sonication in 200 μl of 0.2 % Rapigest-SF (w/v). Protein yield was then determined using a Bradford assay (Bio-Rad), and 30 μg of each sample was normalized to approximately 0.5 μg/μl in ammonium bicarbonate. Samples were reduced with 10 mM DTT at 80°C for 15 minutes and alkylated with iodoacetamide at room temperature in the dark for 30 minutes. Samples were then subjected to trypsin digestion overnight at 37°C at an enzyme to protein ratio (w/w) of 1:50. After digestion, all samples were acidified with 1% TFA and heated at 60°C for 2 hours to hydrolyze RapiGest and centrifuged at 25,000 g for 5 minutes; supernatants were lyophilized. Samples were resuspended at 1 μg/μl in 200 mM ammonium formate (pH 10) containing 25 fmol/μl trypsinized yeast alcohol dehydrogenase 1 (MassPREP; Waters Corp.) as an internal standard. Quantitative 2D LC/LC-MS/MS was performed on 3 μg of protein digest per sample in singlicate, and the pool was analyzed in triplicate with 3-μg injections (once each at the beginning, middle, and end of the queue). The method uses 2D LC in a high-low pH reversed phase/reversed phase configuration on a nanoAcquity UPLC system (Waters Corp.) coupled to a Synapt G2 HDMS high-resolution accurate mass tandem mass spectrometer (Waters Corp.) with nanoelectrospray ionization in a manner similar to that previously described (76, 77). The total analysis cycle time for each sample injection was approximately 6 hours. Statistical significance was determined by a 2-tailed Student’s t test on log2-transformed data.
Gelatin zymography.
Cell media was collected from cells cultured overnight, and protein was measured using the BCA assay (Bio-Rad). 1 μg of the protein (approx. 10 μl of the media) was separated by 10% SDS-PAGE with 0.1% gelatin purchased commercially from Bio-Rad (Hercules). Gels were then washed in 2.5% Triton X-100 and then incubated for 30 minutes in development buffer. After Coomassie blue staining for 1 hour, gels were destained and analyzed for MMP-2 and MMP-9 activity by densitometry.
IHC.
Primary sarcomas or lungs were fixed in 10% formalin and paraffin embedded. 5-Micron sections were cut and subjected to H&E staining. All images were taken with a ×10 objective using a Leica DM5500B microscope, with insets images taken with a ×40 objective. IHC was used to detect Ki67 (BD) in paraffin-embedded sarcomas using the Vectastain Elite ABC Reagent (Vector Labs). All the information on primary and secondary antibodies is provided in Supplemental Table 5.
Statistics.
Statistical significance of the studies was analyzed by 2-tailed Student’s t test, Mann-Whitney test, 1-way ANOVA, or 2-way ANOVA. P values of less than 0.05 are considered significant.
Study approval.
Human STS samples were acquired from MD Anderson under a protocol approved by the Duke University and MD Anderson Institutional Review Boards. All animal studies were performed in accordance with protocols approved by the Duke University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Tyler Jacks for providing the LSL-KrasG12D mice and Anton Berns for providing the p53f mice. We thank Q. Liu and the NeuroTransgenic Lab (NTL) at Duke University for blastocyst injections of ES cells. NTL is supported in part with funding from NIH-NINDS Center Core grant 5P30NS061789. We thank RTOG for kindly providing human sarcoma TMA. This project was supported by RTOG grant U10 CA21661 from the National Cancer Institute (NCI). We also thank Arthur Moseley, Brenna M. Richardson, and Matthew W. Foster from the Duek Proteomics Core Facility. This work was supported by a QuadW-AACR Fellowship for Clinical/Translational Sarcoma Research (to M. Sachdeva), R01 CA138265 (to D.G. Kirsch), and was partially supported by Duke Cancer Center support grant 5P30-CA14236-38 from the NCI.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(10):4305–4319. doi:10.1172/JCI77116.
Jeffrey K. Mito’s present address is: Department of Pathology, Brigham and Women’s Hospital, Boston, Massachusetts USA.
Zhizhong Li’s present address is: Department of Discovery Oncology, Novartis Institutes for Biomedical Research, San Diego, California, USA.
References
- 1.Lyman GH, Djulbegovic B. The challenge of systematic reviews of diagnostic and staging studies in cancer. Cancer Treat Rev. 2005;31(8):628–639. doi: 10.1016/j.ctrv.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.White NM, Fatoohi E, Metias M, Jung K, Stephan C, Yousef GM. Metastamirs: a stepping stone towards improved cancer management. Nat Rev Clin Oncol. 2011;8(2):75–84. doi: 10.1038/nrclinonc.2010.173. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 7.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 10.Kirsch DG, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13(8):992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 11.Mito JK, et al. Oncogene-dependent control of miRNA biogenesis and metastatic progression in a model of undifferentiated pleomorphic sarcoma. J Pathol. 2013;229(1):132–140. doi: 10.1002/path.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segura MF, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larramendy ML, et al. Comparative genomic hybridization of malignant fibrous histiocytoma reveals a novel prognostic marker. Am J Pathol. 1997;151(4):1153–1161. [PMC free article] [PubMed] [Google Scholar]
- 14.Parente F, Grosgeorge J, Coindre JM, Terrier P, Vilain O, Turc-Carel C. Comparative genomic hybridization reveals novel chromosome deletions in 90 primary soft tissue tumors. Cancer Genet Cytogenet. 1999;115(2):89–95. doi: 10.1016/S0165-4608(99)00082-5. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59(1):110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 17.Satelli A, et al. Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer Res. 2014;74(6):1645–1650. doi: 10.1158/0008-5472.CAN-13-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu Y, et al. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Mol Biosyst. 2014;10(3):679–685. doi: 10.1039/c3mb70479c. [DOI] [PubMed] [Google Scholar]
- 19.Lei R, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33(10):1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 20.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13(9):849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 21.Dougherty GW, Jose C, Gimona M, Cutler ML. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol. 2008;87(8):721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Xu MR, Wang T, Li T, Zhu J. MTSS1 overexpression correlates with poor prognosis in colorectal cancer. J Gastrointest Surg. 2011;15(7):1205–1212. doi: 10.1007/s11605-011-1546-2. [DOI] [PubMed] [Google Scholar]
- 23.Ulisse S, Baldini E, Sorrenti S, D’Armiento M. The urokinase plasminogen activator system: a target for anti-cancer therapy. Curr Cancer Drug Targets. 2009;9(1):32–71. doi: 10.2174/156800909787314002. [DOI] [PubMed] [Google Scholar]
- 24.Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477(1):267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, et al. miR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dettmer M, et al. MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J Clin Endocrinol Metab. 2013;98(1):E1–E7. doi: 10.1210/jc.2012-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pignot G, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132(11):2479–2491. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchiyama K, et al. Expression of MicroRNAs associated with Gleason grading system in prostate cancer: miR-182-5p is a useful marker for high grade prostate cancer. Prostate. 2013;73(8):827–834. doi: 10.1002/pros.22626. [DOI] [PubMed] [Google Scholar]
- 29.Ghossein RA, Bhattacharya S, Rosai J. Molecular detection of micrometastases and circulating tumor cells in solid tumors. Clin Cancer Res. 1999;5(8):1950–1960. [PubMed] [Google Scholar]
- 30.Ghossein RA, Bhattacharya S. Molecular detection and characterization of circulating tumor cells and micrometastases in prostatic, urothelial, and renal cell carcinomas. Semin Surg Oncol. 2001;20(4):304–311. doi: 10.1002/ssu.1048. [DOI] [PubMed] [Google Scholar]
- 31.Mocellin S, et al. Molecular detection of circulating tumor cells is an independent prognostic factor in patients with high-risk cutaneous melanoma. Int J Cancer. 2004;111(5):741–745. doi: 10.1002/ijc.20347. [DOI] [PubMed] [Google Scholar]
- 32.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 33.Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 34.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 35.Moreno JG, et al. Changes in circulating carcinoma cells in patients with metastatic prostate cancer correlate with disease status. Urology. 2001;58(3):386–392. doi: 10.1016/S0090-4295(01)01191-8. [DOI] [PubMed] [Google Scholar]
- 36.Eliane JP, et al. Monitoring serial changes in circulating human breast cancer cells in murine xenograft models. Cancer Res. 2008;68(14):5529–5532. doi: 10.1158/0008-5472.CAN-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei R, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33(10):1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 39.Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem. 2013;114(7):1464–1473. doi: 10.1002/jcb.24488. [DOI] [PubMed] [Google Scholar]
- 40.Cubellis MV, Wun TC, Blasi F. Receptor-mediated internalization and degradation of urokinase is caused by its specific inhibitor PAI-1. EMBO J. 1990;9(4):1079–1085. doi: 10.1002/j.1460-2075.1990.tb08213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis V, Wun TC, Behrendt N, Ronne E, Dano K. Inhibition of receptor-bound urokinase by plasminogen-activator inhibitors. J Biol Chem. 1990;265(17):9904–9908. [PubMed] [Google Scholar]
- 42.Fisher JL, Mackie PS, Howard ML, Zhou H, Choong PF. The expression of the urokinase plasminogen activator system in metastatic murine osteosarcoma: an in vivo mouse model. Clin Cancer Res. 2001;7(6):1654–1660. [PubMed] [Google Scholar]
- 43.Kramer MD, Reinartz J, Brunner G, Schirrmacher V. Plasmin in pericellular proteolysis and cellular invasion. Invasion Metastasis. 1994;14(1):210–222. [PubMed] [Google Scholar]
- 44.Setyono-Han B, et al. Suppression of rat breast cancer metastasis and reduction of primary tumour growth by the small synthetic urokinase inhibitor WX-UK1. Thromb Haemost. 2005;93(4):779–786. doi: 10.1160/TH04-11-0712. [DOI] [PubMed] [Google Scholar]
- 45.Sidenius N, Blasi F. The urokinase plasminogen activator system in cancer: recent advances and implication for prognosis and therapy. Cancer Metastasis Rev. 2003;22(2):205–222. doi: 10.1023/a:1023099415940. [DOI] [PubMed] [Google Scholar]
- 46.Henneke I, et al. Inhibition of urokinase activity reduces primary tumor growth and metastasis formation in a murine lung carcinoma model. Am J Respir Crit Care Med. 2010;181(6):611–619. doi: 10.1164/rccm.200903-0342OC. [DOI] [PubMed] [Google Scholar]
- 47.Kotzsch M, et al. Combined mRNA expression levels of members of the urokinase plasminogen activator (uPA) system correlate with disease-associated survival of soft-tissue sarcoma patients. BMC Cancer. 2011;11:273. doi: 10.1186/1471-2407-11-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vial D, McKeown-Longo PJ. PAI1 stimulates assembly of the fibronectin matrix in osteosarcoma cells through crosstalk between the αvβ5 and α5β1 integrins. J Cell Sci. 2008;121(pt 10):1661–1670. doi: 10.1242/jcs.020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74(2):111–122. [PubMed] [Google Scholar]
- 50.Yasumitsu H, Wakabayashi E, Kitajima H, Sakamoto Y, Miyazaki K. [TIMPs (tissue inhibitors of metalloproteinases): their species, functions and effects on tumor metastasis]. Nihon Rinsho. 1995;53(7):1798–1804. [PubMed] [Google Scholar]
- 51.Miyagi E, et al. Characterization of matrix-degrading proteinases and their inhibitors secreted by human gynecological carcinoma cells. Jpn J Cancer Res. 1995;86(6):568–576. doi: 10.1111/j.1349-7006.1995.tb02436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang N, Zhu M, Tsao SW, Man K, Zhang Z, Feng Y. Up-regulation of TIMP-1 by genipin inhibits MMP-2 activities and suppresses the metastatic potential of human hepatocellular carcinoma. PLoS One. 2012;7(9):e46318. doi: 10.1371/journal.pone.0046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka K, et al. Cyclic AMP-regulated synthesis of the tissue inhibitors of metalloproteinases suppresses the invasive potential of the human fibrosarcoma cell line HT1080. Cancer Res. 1995;55(13):2927–2935. [PubMed] [Google Scholar]
- 54.Xin ZF, Shen CC, Tao LJ, Yan SG, Wu HB. Gambogic acid inhibits invasion of osteosarcoma via upregulation of TIMP-1. Int J Mol Med. 2013;31(1):105–112. doi: 10.3892/ijmm.2012.1192. [DOI] [PubMed] [Google Scholar]
- 55.Benassi MS, et al. Tissue and serum loss of metalloproteinase inhibitors in high grade soft tissue sarcomas. Histol Histopathol. 2003;18(4):1035–1040. doi: 10.14670/HH-18.1035. [DOI] [PubMed] [Google Scholar]
- 56.Cutler ML, Bassin RH, Zanoni L, Talbot N. Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol Cell Biol. 1992;12(9):3750–3756. doi: 10.1128/mcb.12.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasaturo F, Dougherty GW, Cutler ML. Ectopic expression of Rsu-1 results in elevation of p21CIP and inhibits anchorage-independent growth of MCF7 breast cancer cells. Breast Cancer Res Treat. 2000;61(1):69–78. doi: 10.1023/A:1006462323260. [DOI] [PubMed] [Google Scholar]
- 58.Dougherty GW, Chopp T, Qi SM, Cutler ML. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp Cell Res. 2005;306(1):168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 59.Machesky LM, Johnston SA. MIM: a multifunctional scaffold protein. J Mol Med. 2007;85(6):569–576. doi: 10.1007/s00109-007-0207-0. [DOI] [PubMed] [Google Scholar]
- 60.Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia. 2002;4(4):291–294. doi: 10.1038/sj.neo.7900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120(pt 9):1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 62.Dawson JC, Bruche S, Spence HJ, Braga VM, Machesky LM. Mtss1 promotes cell-cell junction assembly and stability through the small GTPase Rac1. PLoS One. 2012;7(3):e31141. doi: 10.1371/journal.pone.0031141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Callahan CA, et al. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004;18(22):2724–2729. doi: 10.1101/gad.1221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu K, Wang G, Ding H, Chen Y, Yu G, Wang J. Downregulation of metastasis suppressor 1 (MTSS1) is associated with nodal metastasis and poor outcome in Chinese patients with gastric cancer. BMC Cancer. 2010;10:428. doi: 10.1186/1471-2407-10-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parr C, Jiang WG. Metastasis suppressor 1 (MTSS1) demonstrates prognostic value and anti-metastatic properties in breast cancer. Eur J Cancer. 2009;45(9):1673–1683. doi: 10.1016/j.ejca.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 69.Witten DM, Tibshirani R. A framework for feature selection in clustering. J Am Stat Assoc. 2010;105(490):713–726. doi: 10.1198/jasa.2010.tm09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 72.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 73.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90(18):8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sachdeva M, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28(14):1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 77.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77(19):6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.