Abstract
Papillon-Lefèvre syndrome (PLS) results from mutations that inactivate cysteine protease cathepsin C (CTSC), which processes a variety of serine proteases considered essential for antimicrobial defense. Despite serine protease–deficient immune cell populations, PLS patients do not exhibit marked immunodeficiency. Here, we characterized a 24-year-old woman who had suffered from severe juvenile periodontal disease, but was otherwise healthy, and identified a homozygous missense mutation in CTSC indicative of PLS. Proteome analysis of patient neutrophil granules revealed that several proteins that normally localize to azurophil granules, including the major serine proteases, elastase, cathepsin G, and proteinase 3, were absent. Accordingly, neutrophils from this patient were incapable of producing neutrophil extracellular traps (NETs) in response to ROS and were unable to process endogenous cathelicidin hCAP-18 into the antibacterial peptide LL-37 in response to ionomycin. In immature myeloid cells from patient bone marrow, biosynthesis of CTSC and neutrophil serine proteases appeared normal along with initial processing and sorting to cellular storage. In contrast, these proteins were completely absent in mature neutrophils, indicating that CTSC mutation promotes protease degradation in more mature hematopoietic subsets, but does not affect protease production in progenitor cells. Together, these data indicate CTSC protects serine proteases from degradation in mature immune cells and suggest that neutrophil serine proteases are dispensable for human immunoprotection.
Introduction
Papillon-Lefèvre Syndrome (PLS) is a rare autosomal recessive disease caused by mutations in the gene encoding lysosomal cysteine protease cathepsin C (CTSC) (1–3), also known as dipeptidyl peptidase I (DPPI) (4). CTSC is expressed mainly in hematopoietic tissues. It is essential for posttranslational trimming of serine proteases stored in myeloid cells, in particular of the neutrophil granule-associated serine proteases: neutrophil elastase (NE), cathepsin G (CTSG), proteinase 3 (PR3) (5), and the recently described neutrophil serine protease 4 (NSP4) (6), but CTSC is also required for activation of granzymes A and B of cytotoxic lymphocytes (5, 7) and for activation of mast cell chymases (8). CTSC removes 2 N-terminal amino acids that block the active site of the proteases (5, 9). N-terminal trimming by CTSC is thus necessary for activation of these proteases, which are normally stored as active enzymes in granules. In neutrophils, the serine proteases localize to primary granules, also known as azurophil granules (9, 10).
Despite lack of functional serine proteases in neutrophils and cytotoxic T lymphocytes of patients with PLS, the associated immunodeficiency is remarkably mild (11, 12). This is also the case in the Ctsc-KO mouse (13), where lack of CTSC may even provide a survival advantage in experimental sepsis (14). The dominating clinical phenotype of PLS is a severe periodontal disease that results in loss of both primary and permanent teeth at an early age, and in most cases also in a skin condition known as keratosis palmoplantaris (15).
NE is a major serine protease of human neutrophils and is believed to play a key role in immunity both by virtue of its digestive capacity and due to direct antibacterial effects not related to its enzymatic activity (16). Mice with single deletion of Elane and Ctsg and with deletion of both serine proteases are susceptible to infections caused by Gram-negative bacteria, in particular Klebsiella pneumoniae (17). Neutrophils of mice differ substantially from human neutrophils, not least by lack of the antibacterial α-defensins. The mouse may thus be more dependent on NE and CTSG for antimicrobial activity, since these antibacterial proteases are not backed up by defensins, which in human neutrophils constitute 50% of the total protein of azurophil granules (18). It has, however, been noted that not only do neutrophils from PLS patients lack serine protease activity, as expected from the defective activating N-terminal trimming; the proteins themselves are also absent (12). While this has not been systematically investigated in patients, it seems to be at least partially recapitulated in the Ctsc-KO mouse (19).
We investigated a patient with severe juvenile periodontitis and found a homozygous missense mutation in the CTSC gene. Since the mutation was located in the propiece of CTSC and not previously described in PLS patients, we performed structural and functional analysis of the patient’s neutrophils to establish whether this novel mutation had functional consequences and could classify the patient as having PLS. Thus, neutrophils were isolated from peripheral blood and subjected to subcellular fractionation. Isolated granule subsets were subjected to proteome analysis to obtain an evaluation of the consequences of the genetic aberration. Bone marrow was aspirated to allow studies of synthesis of CTSC and the major serine proteases by radioactive pulse-chase.
The case is as follows: A 24-year-old female of European ethnicity was referred to one of the authors (N. Borregaard) from the Dental Science Center, University of Copenhagen, for diagnostic evaluation. She had lost her primary teeth at the age of 3 years and all her permanent teeth except 7 molars due to severe juvenile periodontal disease. Atopic dermatitis was diagnosed at 5 months of age, and allergy to grass pollen was documented. Several episodes of skin abscesses, mainly localized to the buttocks and many of which required surgical draining, had occurred between age 10 months and 2 years, but none had appeared since then. The patient had frequent styes that responded well to topical chloramphenicol. Bilateral tonsillectomy was performed at age 15 years due to repeated tonsillitis. Her physical and intellectual development had been normal, and no major illnesses had been recorded. The patient was without medication except for hormonal contraception.
On physical examination, the patient appeared normal except for dental prostheses. The patient was screened for immune deficiency by analyzing levels of immunoglobulins and subclasses of IgG, levels of mannan-binding lectin, complement, and complement activation through classical, lectin, and the alternative pathway, all of which were normal, as were T and B cell distribution and T cell proliferation in response to pokeweed mitogen. The number and differential counts of circulating white cells were normal, as were red cell number and morphology and platelet number. A screening for hemorrhagic diathesis was normal. Flow cytometry revealed normal amounts of β2 integrins on leukocytes. Comparative genomic hybridization was performed, but did not detect any abnormalities. These analyses were obtained as part of the laboratory service of the hospital by standard methods. The data are not shown but are kept in the patient’s record. DNA was extracted from peripheral blood and subjected to full exome sequencing using Illumina sequencing and Nimblegen SeqCap EZ Human Exome Library v 3.0. This revealed a homozygous missense mutation in the CTSC gene c.503A>G (exon 4). The mutation results in exchange of tyrosine 168 with cysteine.
CTSC has a unique structure in which the N-terminal 119 amino acids, which are part of the propiece (amino acids 1–206) and termed the “exclusion domain,” form a noncovalent association with the catalytically active mature protein after cleavage of the propiece. The mutated residue is located in the part of the propiece termed the “activation peptide” (amino acids 120–206), which is normally excised (20–22). Thus, this mutation is not predicted to induce stop of transcription or of translation. In fact, a mutation in this part of the propiece might go unnoticed. In order to evaluate the functional consequences, the studies reported below were carried out.
Results
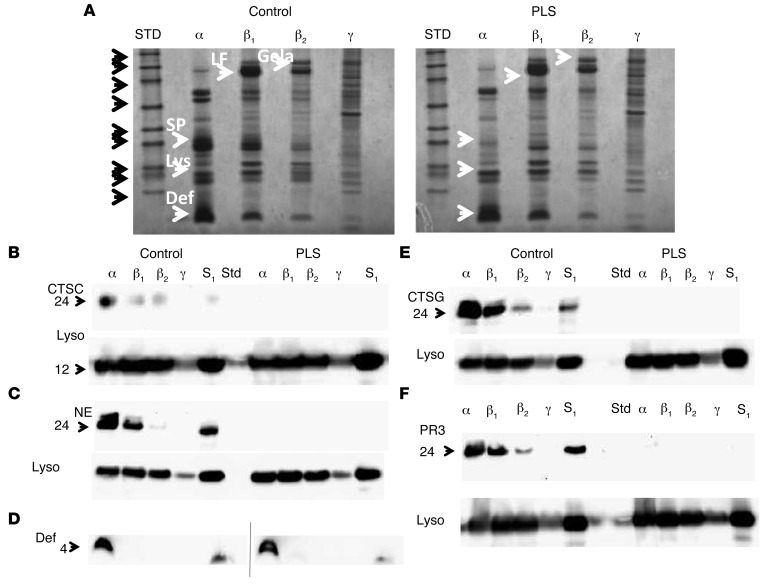
Subcellular fractionation was performed on isolated neutrophils from the PLS patient and a normal control. The distribution of marker proteins used for pooling of fractions containing azurophil granules (α band), specific granules (β1 band), gelatinase granules (β2 band), and light membranes including plasma membranes and secretory vesicles (γ band) is given in Supplemental Figure 1 (supplemental material available online with this article; doi:10.1172/JCI76009DS1). The protein profiles of these isolated fractions (α, β1, β2, and γ) are given in Figure 1. It is readily observed that the major band around 28 to 31 kD in azurophil granules seen in the control sample is lacking in the patient neutrophils. Since there is a minor contamination of azurophil granules in the β1 band, which otherwise contains the bulk of specific granules, there is a similar lack of the much less intense bands in the lane containing the material from the β1 band. Notably, a normal amount of protein around 4 to 5 kD corresponding to the localization of defensins (23) is present in the patient neutrophils and confirmed to react with anti-defensin antibodies. Western blotting, however, showed absence of CTSG, NE, PR3, and CTSC both in lysates of neutrophils (S1) and in subcellular fractions (Figure 1, B–F). Lysozyme, a constituent of all neutrophil granule subsets (24), was present and used as a loading control. The absence of serine proteases observed on the Coomassie-stained gel and by Western blots corresponds with the proteome data mentioned below and thus allows us to conclude that the serine proteases as well as CTSC are absent from the patient neutrophils (Figure 1). The band at 62 kD present in azurophil granules from control and largely absent from the patient was identified as eosinophil peroxidase by proteome analysis (Supplemental Table 1) and by Western blotting (not shown).
Figure 1. Deficiency of cathepsin C and serine proteases in PLS neutrophils.
(A) Coommassie-stained 4%–12% SDS-PAGE–separated proteins from isolated azurophil granules (α), specific granules (β1), gelatinase granules (β2), and light membranes (γ) from PLS neutrophils and neutrophils from a normal control. Molecular weight (MW) standards are indicated with black arrows: from top, 225; 102; 76; 52; 38; 31; 24; 17; 12 kDa. Selected proteins are indicated by white arrows. SP, serine proteases; Lys, lysozyme; Def, defensins; LF, lactoferrin; Gela, gelatinase. (B–F) Western blotting for proteases of the isolated fractions α, β1, β2, and γ and of the postnuclear supernatant (S1), which is the material loaded onto the density gradient. (B) CTSC and lysozyme (Lyso); (C) NE and lysozyme; (D) defensin (control and patient samples were run on 2 separate gels that had a slightly curved run); (E) CTSG and lysozyme; (F) PR3 and lysozyme. Blotting was done first for serine proteases. The blots were then stripped and reprobed with antibody against lysozyme as a loading control.
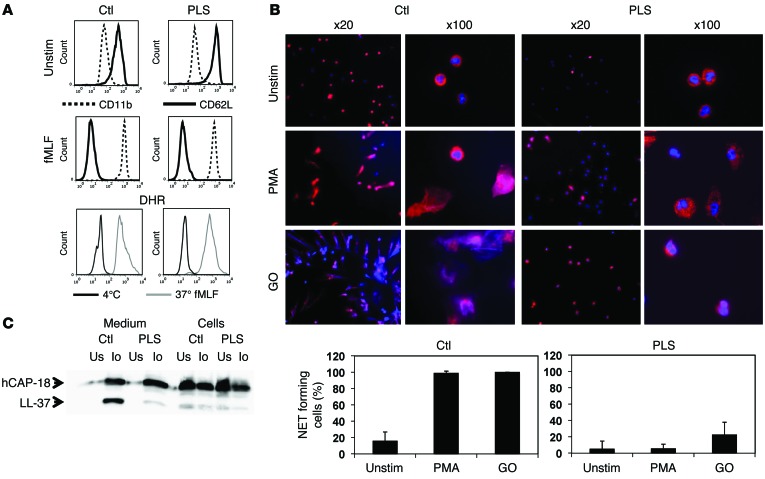
To test the functional consequences, neutrophils were stimulated with formyl-methionyl-leucyl-phenylalanine (fMLF) to see whether PLS neutrophils responded normally to stimulation. Patient neutrophils upregulated CD11b in response to fMLF as well as control neutrophils, indicating that stimulus-response coupling was normal in patient neutrophils. Shedding of CD62L by patient neutrophils, which is executed by the membrane-bound proteinase TNF-α–converting enzyme (TACE), also known as ADAM17 (25), was also normal (Figure 2A). The ability to mount a respiratory burst was evaluated by flow cytometry (Figure 2A) and showed activities in PLS neutrophils similar to those of control neutrophils.
Figure 2. Functional studies on PLS neutrophils.
(A) Flow cytometry of nonstimulated and fMLF-stimulated (10–8 M) neutrophils from PLS and a normal control demonstrating equal disappearance of CD62L and upregulation of CD11b in response to stimulation. Respiratory burst was quantified as DHR-positive cells by flow cytometry of isolated neutrophils stimulated by fMLF (10–8 M). (B) NETosis. Neutrophils were stimulated with PMA or glucose oxidase (GO) to undergo NETosis and stained with DAPI and antibodies against MPO. NETosis was quantified as an increase in MPO-positive areas normally caused by formation of NETs with bound MPO. While some PLS neutrophils had increased MPO-positive areas after stimulation with glucose oxidase, these cells did not form NETs. The increased MPO-positive areas in these cells were caused by the presence of MPO in decondensated nuclei, as seen in the panels with ×100 magnification. Error bars for the PLS neutrophils demonstrate SD between the 2 experiments performed. One representative experiment out of 3 is shown for quantification of NETosis from the normal controls, and here error bars indicate SD between the 20 images used for quantification. (C) Processing of hCAP-18 by ionomycin-stimulated neutrophils as described in Methods. Cells and medium from ionomycin-stimulated (Io) or unstimulated neutrophils (Us) were blotted with a rabbit antibody against hCAP-18 (34). These functional studies (except the respiratory burst experiment, which was performed only once) were performed on 2 independent occasions 3 months apart with similar results.
Neutrophil extracellular traps (NETs) (26) have recently come to be regarded as important in the defense against dissemination of invading microorganisms (27, 28). NET formation depends on the presence of NE, myeloperoxidase (MPO), and the ability to mount a respiratory burst (26, 29). We suspected PLS neutrophils to be incapable of forming NETs due to lack of elastase. This was confirmed (Figure 2B).
We have previously demonstrated that the ability of neutrophils to cleave proantibacterial human cathelicidin antimicrobial protein at 18 kD (hCAP-18) and generate free antibacterial C-terminal peptides, also known as LL-37, is due to proteolysis mediated by PR3 (30). Thus, we suspected that this cleavage would be deficient in patient neutrophils. This was tested by subjecting patient and control neutrophils to stimulation with ionomycin, a known and potent activator of degranulation of neutrophil granules (30). Figure 2C shows a major reduction in generation of LL-37 from hCAP-18 in patient neutrophils. The fact that major amounts of hCAP-18 are released to the supernatant of ionomycin-stimulated cells testifies that PLS neutrophils are capable of degranulating in response to stimulation.
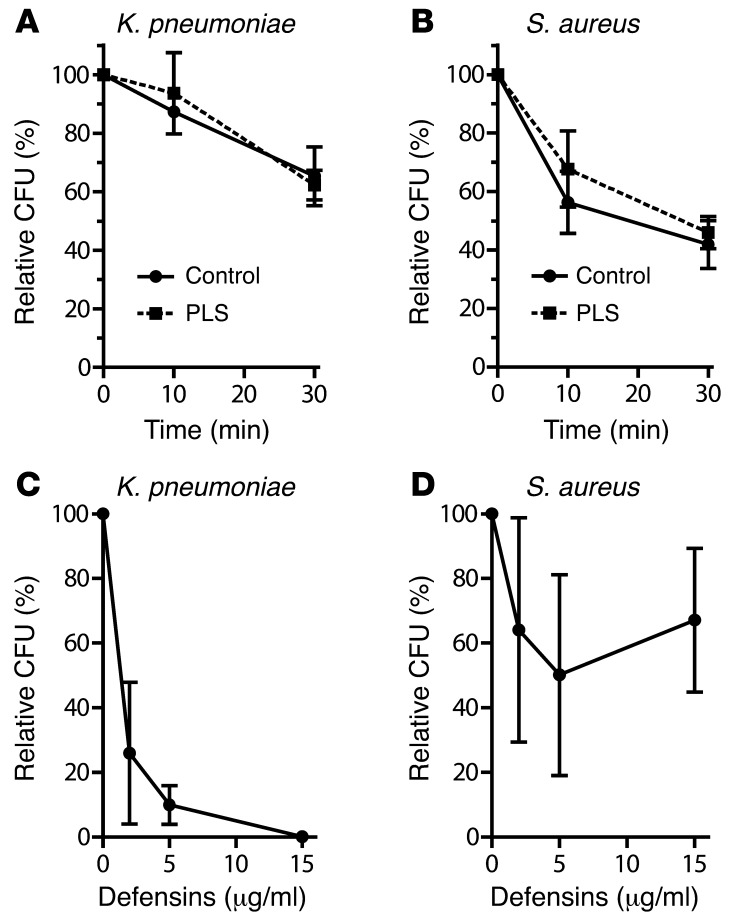
The ability of PLS neutrophils to kill Staphylococcus aureus and K. pneumoniae was tested in vitro. We found no defects in killing of either microorganism by PLS neutrophils (Figure 3). We tested the ability of isolated neutrophil α-defensins to kill S. aureus and K. pneumoniae. Defensins killed K. pneumoniae efficiently, but did not kill S. aureus to any major extent. As mice are devoid of neutrophil α-defensins, this may explain the susceptibility to infections by K. pneumonia of mice lacking serine proteases (17).
Figure 3. Bactericidal activity of neutrophils and defensins.
(A and B) Neutrophils isolated from blood as described above were incubated with S. aureus or K. pneumoniae at a MOI (bacteria: neutrophils) of 1:1 and 1:10, respectively, and incubated for 10 minutes, after which neutrophils were pelleted by centrifugation and resuspended in HBSS. Samples were taken immediately (time 0) and after 10 and 30 minutes. Neutrophils were lysed in water pH 11 to liberate intracellular bacteria. These were enumerated by CFU after overnight incubation. All experiments were run in triplicate. Results are expressed as CFU relative to time 0. (C and D) Defensins isolated from human neutrophil azurophil granules were incubated with S. aureus or K. pneumoniae for 1 hour at 37°C. The number of surviving bacteria was enumerated as CFU after overnight incubation. Results are expressed as CFU relative to control without defensin.
Proteome profiling of isolated subcellular fractions demonstrated that patient neutrophils lack or have significantly reduced amounts of NE, CTSG, PR3, and azurocidin (ref. 31, also known as CAP37, ref. 32; and HBP, ref. 33). Azurocidin is an inactive member of the neutrophil serine proteases, with strong chemotactic activity toward monocytes (34). A ranking was made of proteins based on the largest difference in signal intensities relative to the reference protein between PLS and control (Supplemental Table 1). NSP4 was not detected in patient or control neutrophils. Sequences of PR3 and azurocidin were detected in the PLS sample at a much reduced level (10% and 20% of control, respectively), but this may be due to degradation products still contained in azurophil granules, as intact PR3 was not detected by Western blotting (Figure 1).
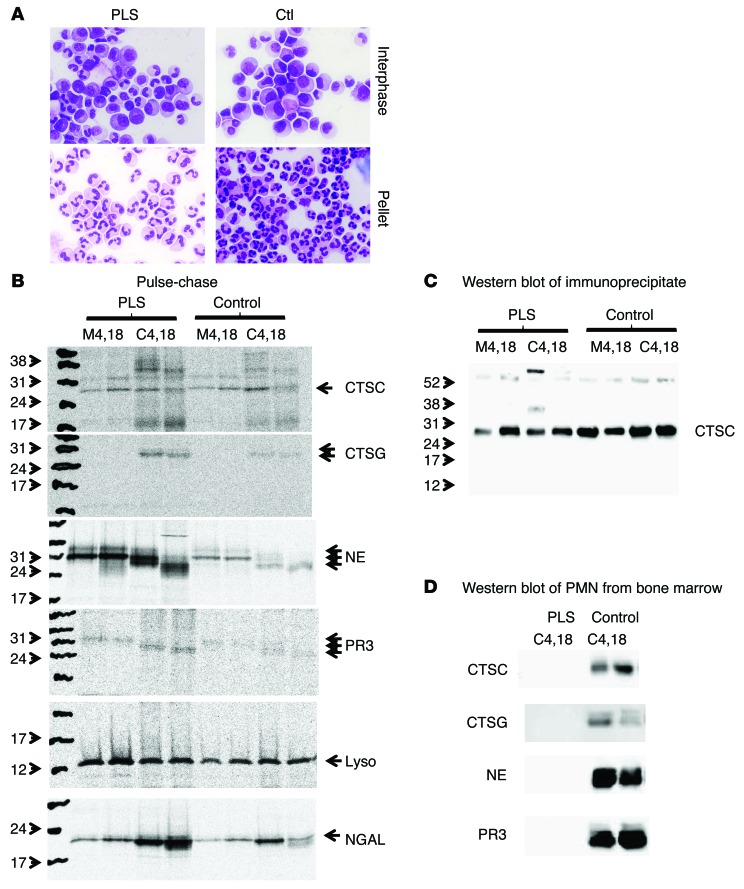
We next investigated the reason for the complete lack of serine proteases and CTSC from patient neutrophils. Our hypothesis was that in the absence of cleavage of the N-terminal 2 amino acids by CTSC, the serine proteases would escape sorting to granules and instead be secreted constitutively to the medium. This hypothesis was founded on previous publications demonstrating that unprocessed serine proteases are routed to medium during pulse-chase examination of synthesis of serine proteases in myeloid cell lines (10, 35–37). To test this hypothesis, myeloid cells were isolated from the patient’s bone marrow and separated on Lymphoprep. This procedure separates segmented, band cells, and most metamyelocytes that sediment to the pellet from more immature myeloid cells, in particular, blasts, promyelocytes, and myelocytes that do not enter the density medium and can be harvested from the interphase (38). Cytospins of the preparations demonstrated the characteristic morphology of the cells (Figure 4A). It was noted that the interphase from the Lymphoprep separation of PLS bone marrow cells was somewhat contaminated by band cells, indicating that the PLS neutrophils and their precursors are slightly lighter than corresponding cells from healthy controls. This corresponds to the slight skewing of the profile of azurophil granules toward lighter fractions seen in Supplemental Figure 1. This most likely is due to the absence of serine proteases from the granules.
Figure 4. Biosynthesis of serine proteases in immature neutrophils from bone marrow.
Bone marrow cells depleted of nonneutrophil lineage cells were separated into immature cells (less mature than band cells) and mature cells by density centrifugation on Lymphoprep. (A) Giemsa staining of cytospin of immature cells and mature cells from PLS and a normal control (Ctl). (B) Biosynthesis of CTSC, NE, CTSG, and PR3 in immature cells from bone marrow. Medium after 4 and 18 hours of chase (M4, 18). Cells after 4 and 18 hours of chase (C4, 18). 6.5 × 107 cells from PLS patient and 3.0 × 107 cells from control were used. (C) Western blotting of the immunoprecipitates from immature cells shown in B. (D) Western blotting of mature cells from bone marrow (pellet from Lymphoprep separation). These were treated, as were the immature cells for biosynthesis, and were chased for 4 and 18 hours, but were not subject to immunoprecipitation, and cell lysates were run directly for SDS-PAGE and Western blotting. The blots were stripped and reprobed with anti-lysozyme antibody as a loading control. This demonstrated equal loading, but is not shown.
Synthesis of azurophil granule serine proteases is known to take place at the promyelocyte stage (39–41). We confirm here that mRNA of these proteases is present in normal amounts in the immature cells isolated from the interphase, as determined by real-time PCR (Supplemental Figure 2).
Pulse-chase biosynthesis performed on the immature myeloid cells demonstrated that the serine proteases investigated were retained in cells after 4 and 18 hours of chase and were similarly processed to lower molecular weight forms than the initial 31 kD–sized proforms in PLS and control cells. If degradation of the proteases takes place in promyelocytes in which the proteases are synthesized or immediately after storage in granules, we would expect degradation products to be detected in these cells, which include promyelocytes and myelocytes (Figure 4A). Since the biosynthesis of CTSC showed several bands, the identity of CTSC was revealed by Western blotting of the immunoprecipitate used for fluorography (Figure 4C). Even chase for 18 hours did not reveal any major degradation in patient cells compared with control cells. This does not, however, rule out that degradation occurs in the more mature myelocytes, which are a few days more mature than promyelocytes (42). We therefore performed Western blotting of the bone marrow cells from the pellet after Lymphoprep separation. These were segmented and band cells (ref. 38 and Figure 4A). It is readily observed that serine proteases are completely absent from these mature bone marrow cells from the PLS patient (Figure 4D).
Discussion
The data demonstrate that a mutation in CTSC that results in exchange of tyrosine to cysteine at aa 168 of CTSC leads to a complete deficiency of CTSC in neutrophils and a complete deficiency of neutrophil serine proteases.
The fact that this patient, like most other PLS patients (15, 43), was remarkably free of major infections and largely only had clinical problems due to periodontitis, is a strong argument against the notion that azurophil granule serine proteases constitute a major antibacterial defense and that NADPH oxidase activity only works to unleash their antibacterial activity (44, 45). In fact, while periodontal disease is the key clinical feature of patients with PLS, it is not part of the clinical spectrum of chronic granulomatous disease (CGD), which is caused by NADPH oxidase deficiency (46). It therefore seems safe to abandon the hypothesis that the respiratory burst activity of neutrophils serves to activate the antibacterial activity of azurophil granule serine proteases (44, 45) and to accept that the NADPH oxidase is an important microbicidal armory by virtue of its capacity to generate ROS.
It is not known why lack of neutrophil serine proteases results in severe periodontal disease. NETs were identified during the last 10 years as structures of potential importance for constraining bacteria by exposing them to a web of unfolded chromatin decorated with antibacterial proteins including histones and NE (26, 47). While NET formation may be elicited by different signal transduction pathways, NE and generation of a respiratory burst both seem to be required for neutrophils to make NETs (29, 48). Based on this, failure to generate NETs is an insufficient explanation for the severe periodontal disease, as CGD patients who, like this patient, also fail to generate NETs, do not suffer from periodontal disease, as discussed above. Also, the mild immunodeficiency of PLS patients indicates that NET formation is not a major antimicrobial defense mechanism.
Patients with severe congenital neutropenia often suffer major periodontal disease, even after normalization of their peripheral neutrophil count by granulocyte colony-stimulating factor (G-CSF). This has been ascribed to an unexplained deficiency of the neutrophil protein hCAP-18 (49–51), the antibacterial activity of which is unleashed when the bactericidal C-terminal peptide LL-37 is liberated from the propiece by proteolysis (52). We have shown that PR3 is responsible for this cleavage in neutrophils (30). In this respect, the documented lack of PR3 and the deficient processing of hCAP-18 in ionomycin-stimulated neutrophils may provide a clue to the particular propensity of periodontal disease in PLS patients (53). By the same token, the normal level of fully processed defensins in the azurophil granules of the patient neutrophils argues that prodefensins are not processed by any of the serine proteases during neutrophil development (54). More importantly, the fact that PLS neutrophils were capable of killing K. peumoniae to the same extent as normal neutrophils argues that defensins are important in protection against this pathogen, as mice that lack elastase and cathepsin G also lack neutrophil defensins and their neutrophils are deficient in killing of K. pneumoniae (17). In line with this, PLS patients have not been reported to be particularly susceptible to infections caused by K. pneumoniae.
Why does CTSC deficiency result in total lack of serine proteases of neutrophil granules? Our initial hypothesis was that lack of N-terminal processing would lead to constitutive secretion of newly formed protease protein. This hypothesis is clearly refuted by the study of biosynthesis of CTSC and neutrophil serine proteases. All are produced at the expected size. A fraction is routed unprocessed to the medium, as has been observed previously by the Lund group studying biosynthesis and processing of neutrophil serine proteases (10, 55). The fraction of newly synthesized protein routed to the medium was not larger in the PLS patient cells than in corresponding control cells. A remarkably similar trimming of newly synthesized protein was observed in PLS cells compared with control cells, even when the chase was extended to 18 hours, indicating that newly synthesized protein is not degraded immediately. In sharp contrast, CTSC and the serine proteases of azurophil granules were completely absent in mature PLS bone marrow cells, indicating that the inactive enzymes are degraded prior to this stage. Thus, while production and sorting of serine proteases occurs seemingly normally in PLS promyelocytes, degradation is halted until a more mature stage is reached, but clearly before the cells are released into circulation. This observation opens opportunities for novel hypotheses to be tested regarding processing and degradation of incorrectly processed proteins. It is possible that binding to the intracellular proteoglycan serglycin protects the proteases from degradation. Serglycin is present only in the early stages of myeloid cell development and disappears after maturation to neutrophils (56). This may leave the incorrectly processed proteases unprotected and allow degradation to occur at this stage of neutrophil development, but other mechanisms are possible and investigations to test these are planned.
Methods
Peripheral blood.
200 ml of peripheral blood was withdrawn and anticoagulated with acid citrate dextrose (ACD) (0.8% citric acid monohydrate, 2.2% Tri-Na citrate dihydrate, 2.7% glucose monohydrate). Dextran 500 (Sigma-Aldrich) was added to induce sedimentation of red cells. The leukocyte-rich supernatant was layered on Lymphoprep (Axix-Schield PoC AS) and centrifuged. The pellet was resuspended, and contaminating red cells lysed by hypotonic shock as described before (57). A sample of the isolated neutrophils was saved for functional studies and the rest were treated with diisopropyl fluorophosphate (DFP) (Calbiochem) and cavitated by nitrogen cavitation as described (57). The postnuclear supernatant was subjected to fractionation on a 3-layer Percoll density gradient. This gradient efficiently separates the organelles into fractions enriched in azurophil granules (α band, identified by MPO); specific granules (β1 band, identified by neutrophil gelatinase-associated lipocalin [NGAL]); gelatinase granules (β2 band, identified by gelatinase B [MMP9]), and fractions high in secretory vesicles (identified by albumin) and plasma membranes (identified by HLA) (58). Fractions of 1 ml each were collected by aspiration from the bottom, and fractions with peak MPO (fractions 1–8), peak NGAL (fractions 9–15) and peak gelatinase (fractions 16–20), peak albumin, and peak HLA (fractions 21–28), respectively, were pooled. Percoll was removed by ultracentrifugation and the biological material collected and resuspended in 200 μl PBS; all procedures were as described previously (57).
Bone marrow.
First, 15 ml bone marrow was aspirated from the posterior superior iliac crest under local anesthesia and collected in 5–ml sterile ACD. Then, 20 ml of 2% Dextran 500 in saline was added to induce sedimentation of red cells. The leukocyte-rich supernatant was aspirated and cells were pelleted by centrifugation and resuspended in 120 ml PBS. This suspension was divided in 4 equally sized aliquots that were each underlaid with 15 ml Lymphoprep and centrifuged. Contaminating erythrocytes in the pellet were lysed by hypotonic shock, and the leukocytes resuspended in 1 ml MACS buffer (Miltenyi Biotec). 20 μl anti CD49d was added per 108 cells, and the cells were incubated for 15 minutes, washed, and resuspended in 80 μl MACS buffer per 107 cells. Immunomagnetic anti-IgG beads were added, and CD49d–positive cells removed by magnetic absorption. Cells from the interphase were incubated under similar conditions, but with anti-CD2, anti-CD3, anti-CD5, anti-CD19, anti-CD56, anti-CD61, and anti-glycophorin A antibodies to remove nonmyeloid cells, as previously described (38). Samples of both preparations were saved for immunocytochemistry, Western blotting, and isolation of RNA. The remaining cells were resuspended at 2.0 × 107 cells/ml in methionine/cysteine free medium (DMEM; Gibco, Life Technologies) with 10% dialyzed fetal calf serum and incubated for 30 minutes at 37°C. The cells were then pelleted and resuspended at 3.0 × 107 cells/ml in identical medium to which 35S methionine/cysteine (EasyTag, PerkinElmer) was added to a final concentration of 200 μCi/ml. The cells were then incubated for 60 minutes at 37°C, washed twice in RPMI, and resuspended at 2 × 107 cells/ml in RPMI-1640 (Gibco-BRL, Life Technologies), containing 10% dialyzed fetal calf serum, penicillin, and streptomycin and divided into 2 equal aliquots that were incubated for 4 and 18 hours, respectively. Cells were then pelleted by centrifugation and resuspended at 107 cells/ml in RIPA buffer (150 mM NaCl, 1% [v/v] Triton X-100, 0.1% [w/v] SDS, 1% [w/v] sodium deoxycholate, 30 mM HEPES, pH 7.3). Protease inhibitor (complete mini; Roche), 1 tablet/10 ml RIPA buffer and 1 mM phenylmethylsulphonyl fluoride (Sigma-Aldrich) were added (39). The samples were incubated on ice for 2 hours and cleared by centrifugation at 20,000 g for 30 minutes. The supernatant was aspirated and subjected to immunoprecipitation. Also, the medium from the chase was subjected to immune precipitation after an equal volume of 2× RIPA buffer with proteinase inhibitors had been added. SDS-sample buffer was added and the samples run on 12% polyacrylamide (CTSC, CTSG, NGAL, NE) or 14% gels (PR3 and lysozyme). The gels were stained with Coomassie, destained, incubated with Amplifier (GE Healthcare), dried, and developed on Fuji BAS2500 PhosphoImager.
Protein analysis.
The resuspended samples from subcellular fractionation of peripheral blood neutrophils were diluted in SDS sample buffer with reduction as follows: NE and Def were diluted 10-fold, CTSG and PR3 were diluted 5-fold, and CTSC was diluted 3-fold; the mixture was then boiled. For Western blotting, 15 μl from each sample was applied in each lane and subjected to electrophoresis on a 1.5-mm–thick gradient gel, 4% to 12% polyacrylamide (Life Technologics, Novex). The blots were developed and digitalized by ChemiDoc (Bio-Rad). For proteome analysis, 20 μl of a 3-fold diluted sample was applied in each lane and subjected to electrophoresis on a 1.5-mm–thick gradient gel, 4% to 12% polyacrylaminde. Each lane was cut horizontally in 10 equally long sections that were each subjected to proteome analysis essentially as described before (59).
Antibodies used for Western blotting and immunoprecipitation.
Rabbit anti-lysozyme (DAKO A0099); rabbit anti-elastase (DAKO rabbit 1373, contract immunization); rabbit anti–cathepsin G (DAKO 588); rabbit anti-defensin (DAKO rabbit 5588, contract immunization); rabbit anti-prodefensin (DAKO rabbit # 8125, contract immunization) (60); goat anti-human CTSC (R&D AF1071); rabbit anti-proteinase 3 (Abcam 103632); rabbit preimmune IgG (DAKO X0903); rabbit anti-MPO (DAKO A0398); and mouse anti-NGAL clone 211.1 (in-house; ref. 61) were used. Horseradish peroxidase–coupled antibodies were as follows: goat anti-rabbit (DAKO P0448); rabbit anti-goat (DAKO P0449); and rabbit anti-mouse IgG (DAKO P0260). Fluorochrome-labeled antibodies were as follows: mouse IgG isotype control (BD 555749); mouse anti-CD62L (BD 555544); and mouse anti-CD11b (BD 555388).
FACS analysis on stimulated neutrophils.
Neutrophils were stimulated with 10–8 M fMLF as described previously (62). Unstimulated cells and equal amounts of stimulated cells were incubated for 30 minutes at 4°C, with isotype control diluted to 1:5, monoclonal anti-CD62L diluted to 1:5, or monoclonal anti-CD11b diluted to 1:100, washed once in PBS with 3% FCS, and centrifuged at 200 g for 6 minutes. The cells were resuspended in 400 μl PBS with 3% FCS and analyzed on FACSCalibur (BD) for cell-surface expression of CD62L and CD11b.
The ability to mount a respiratory burst was evaluated by dihydrorhodamine 123 (DHR) assay (63). Briefly, 2 × 105 neutrophils were suspended in 50 μl KRP+G (Krebs ringer phosphate, pH 7.4, with 5 mM glucose) and added to either DHR (5000 ng/ml) alone or DHR together with 10–8 M fMLF. The DHR sample was kept at 4°C, whereas the DHR+FMLF sample was incubated at 37°C for 15 minutes. Stimulation was stopped by addition of 132 μl ice-cold KRP-G followed by centrifugation at 200 g for 6 minutes. The samples were resuspended in 200 μl PBS with 3% FCS and rhodamine production analyzed by flow cytometry.
Processing of hCAP-18.
Isolated neutrophils from control and PLS patients were each resuspended in 2 vials at 3 × 107 cells in 1 ml KRP+G and incubated at 37°C for 5 minutes. Ionomycin was added to a final concentration of 1 μM to one vial while the other served as control, and both were incubated for an additional 15 minutes. Cells were then pelleted by centrifugation, the supernatant was aspirated, and the cell pellet resuspended to the original volume in ice-cold KRP+G. Samples were taken for SDS-PAGE (14% polyacrylamide) and Western blotting with a rabbit antibody against hCAP-18 (64).
NETs.
Coverslips were washed once with PBS, followed by incubation in 12-well plates with 0.01% poly-l-lysine in sterile PBS overnight at 37°C. The coated coverslips were washed once in PBS, and 400 μl of 5 × 105 PMNs/ml was added to each well. PMNs were allowed to adhere for 15 minutes at room temperature, followed by 15 minutes at 37°C in 5% CO2. Unstimulated neutrophils in RPMI with 0.2% (w/v) albumin were used as control. NETs were induced by either 20 nM PMA or 100 mU/ml glucose oxidase for 150 minutes at 37°C. After stimulation of neutrophils, the medium was removed and the coverslips were washed once in PBS. Cells were fixed with 4% paraformaldehyde for 20 minutes at 37°C and 5% CO2, followed by washing 3 times for 5 minutes each time with PBS. Cells were permeabilized by addition of 0.5% Triton X-100 in PBS for 1 minute. Cells were washed 3 × 1 minutes in PBST (PBS + 0.05% Tween-20), followed by incubation in blocking buffer (5% goat serum in PBST) for 30 minutes at 37°C. Cells were incubated in primary antibody against human MPO at a dilution of 1:500 (diluted in blocking buffer) for 1 hour at 37°C and washed 3 × 5 minutes in PBS. Alexa Fluor 594 F(ab′)2 fragment of goat anti-rabbit (Molecular Probes) diluted to 1:1000 in blocking buffer was added for 1 hour at 37°C, followed by washing in PBS for 3 × 5 minutes. Coverslips were mounted using PROLONG Gold Anti-Fade reagent with DAPI (Life Technologies). Slides were allowed to dry in the dark at room temperature overnight before examination of PMNs and NETs using fluorescence microscopy. Images were acquired using a Nikon Eclipse TE200 equipped with a Hamamatsu C4742-95 CCD camera using Plan Apochromat ×20 and ×100 objectives. NIS-elements 3.1 (Nikon) software was used for image acquisition and processing. Quantification of NETosis was performed as described (65) with minor modifications. Twenty random ×20 images with a minimum of 175 cells in total were used for quantification for each condition. The DAPI channel was used to identify nuclei. MPO staining was used to quantitate the MPO positive area in unstimulated cells with normal polymorphonuclear morphology. An increase in the MPO-positive area of 33% was used as a cutoff for NET formation. Image analysis was performed with the public domain software (Fiji).
Bacterial cultures for neutrophil bactericidal activity.
In vitro bactericidal assays were performed using the 2 wild-type strains: K. pneumoniae strain KP4 and S. aureus strain NCTC 8325. A single colony from each of the 2 strains was incubated overnight in LB-medium (State Serum Institute) at 37°C in a shaking incubator at 150 rpm. After 18 hours of incubation, the bacteria were washed 2 times in PBS and resuspended in HBSS (Gibco; Life Technologies). Cultures were diluted in HBSS until they reached the desired OD (S. aureus, OD 500 0.60; K. pneumoniae, OD 500 0.45) corresponding to 1 × 108 CFU and were further diluted until desired concentrations were obtained (S. aureus, 1 × 107 CFU/ml; K. pneumoniae, 1 × 106 CFU/ml). The bacteria were pelleted by centrifugation and resuspended in HBSS containing 10% ABpos human serum and tumbled for 20 minutes at 37°C for opsonization.
PMN bactericidal assay.
PMNs from the PLS patient and a healthy control were isolated as described above and suspended at 1 × 107 cells/ml in HBSS containing 10% AB serum. 1500 μl cells were immediately mixed with bacteria at a final ratio of bacteria to neutrophils of 1:1 for S. aureus and 1:10 for K. pnemoniae and incubated for 10 minutes at 37°C while tumbling. The cells were pelleted by centrifugation at 500 g, resuspended in HBSS with 10% serum at 37°C, and further incubated while tumbling. 100 μl samples were taken at the times indicated (0 minutes, 10 minutes, and 30 minutes). A 20-fold excess of ice-cold H2O, freshly adjusted to pH 11.0, was added to the samples to lyse neutrophils and liberate bacteria. 80 μl DNAse mix containing 250 mM Tris-HCl, pH 7.4, 25 mM CaCl2, 12.5 mM MgCl2, and 2000 U/ml DNAse (Roche) was added, and the samples were incubated for 10 minutes at 37°C to degrade nuclear material. Finally, the samples were diluted in H2O, pH 11, and plated on blue agar (K. pneumoniae) or 5% blood agar (S. aureus), 4 plates per sample, and incubated overnight at 37°C.
Bacterial cultures for defensin bactericidal activity.
Newly plated bacteria were cultured overnight in LB medium. 200 μl of the overnight culture was subcultured in 10 ml of fresh LB-medium for 3 hours. Bacteria were washed once in Tris-glucose (10 mM Tris, 5 mM glucose, pH 7.5). OD620 were adjusted to 0.2 in Tris-glucose. Bacteria were diluted 1:100, and 2.5 μl was used for CFU assay.
Defensins.
Defensins were isolated from azurophil granules of human neutrophils essentially as described (66), with the modification that defensins were eluted from Mono-S by 20 mM NaOH and that 2 rounds of purification by ion exchange chromatography on Mono-S were used instead of one. Defensins were diluted to a 100 μg/ml solution in Tris-glucose buffer and used as follows: control (2.5 μl bacteria plus 47.5 μl Tris-glucose); 15 μg/ml defensins (2.5 μl bacteria plus 7.5 μl defensins + 40 μl Tris-glucose); 5 μg/ml defensins (2.5 μl bacteria plus 2.5 μl defensins plus 45 μl Tris-glucose); 2 μg/ml defensins (2.5 μl bacteria plus 1 μl defensins plus 46.5 μl Tris-glucose). Samples were incubated for 1 hour at 37°C and plated out in dilutions of 2-, 20-, and 100-fold.
Real-time PCR.
RNA was extracted by TRIzol Reagent (Invitrogen) for RNA isolation. Real-time PCR was performed in triplicate using 5 μl cDNA as described (62): MPO (FAM) (HS00165162_m1), LCN2 (FAM)(HS00194353_m1) ELANE (FAM)( Hs00236952_m1), PTRN3 (FAM) (Hs01597752_m1), CTSG (FAM)(Hs00175195_m1), and CTSC (FAM)(Hs00175188_m1). ACTB (HEX)(4326315E) was used as an internal control for normalization. All probes were provided by Applied Biosystems. Reaction conditions included incubation at 95°C for 10 minutes followed by 40 cycles at 95°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute. The amount of fluorescent FAM or HEX was measured at the end of each cycle. Results were expressed as fluorescence relative to number of PCR cycles.
Statistics.
Statistical calculations were performed with GraphPad 5.0 (GraphPad Software Inc.). Tests were 2-tailed, and the significance level was set at P < 0.05. Bacterial killing assays were set up in triplicate and assessed using a nonparametric test, the Mann-Whitney test.
Study approval.
Written informed consent was obtained from the patient to carry out the studies reported and to report the results. The study was approved by the ethics committee of the Capital Region of Denmark (H-1-2011-165).
Supplementary Material
Acknowledgments
The work was supported by grants to N. Borregaard from the Danish Medical Research Council, the Augustinus Foundation, and The Vilhelm Petersen and Hustru Foundation after recommendation by the Novo Nordisk Foundation. The expert technical assistance of Charlotte Horn is greatly appreciated.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(10):4539–4548. doi:10.1172/JCI76009.
See the related Commentary beginning on page 4237.
References
- 1.Toomes C, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23(4):421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt C, et al. The role of cathepsin C in Papillon-Lefevre syndrome, prepubertal periodontitis, and aggressive periodontitis. Hum Mutat. 2004;23(3):222–228. doi: 10.1002/humu.10314. [DOI] [PubMed] [Google Scholar]
- 3.Noack B, et al. Functional Cathepsin C mutations cause different Papillon-Lefevre syndrome phenotypes. J Clin Periodontol. 2008;35(4):311–316. doi: 10.1111/j.1600-051X.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 4.Rao NV, Rao GV, Hoidal JR. Human dipeptidyl-peptidase I. Gene characterization, localization, and expression. J Biol Chem. 1997;272(15):10260–10265. doi: 10.1074/jbc.272.15.10260. [DOI] [PubMed] [Google Scholar]
- 5.McGuire MJ, Lipsky PE, Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. J Biol Chem. 1993;268(4):2458–2467. [PubMed] [Google Scholar]
- 6.Perera NC, et al. NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity. J Immunol. 2013;191(5):2700–2707. doi: 10.4049/jimmunol.1301293. [DOI] [PubMed] [Google Scholar]
- 7.Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci U S A. 1999;96(15):8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ, Caughey GH. Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J Biol Chem. 2001;276(21):18551–18556. doi: 10.1074/jbc.M100223200. [DOI] [PubMed] [Google Scholar]
- 9.Salvesen G, Enghild JJ. An unusual specificity in the activation of neutrophil serin proteinase zymogens. Biochemistry. 1990;29(22):5304–5308. doi: 10.1021/bi00474a013. [DOI] [PubMed] [Google Scholar]
- 10.Lindmark A, Persson A-M, Olsson I. Biosynthesis and processing of cathepsin G and neutrophil elastase in the leukemic myeloid cell line U-937. Blood. 1990;76(11):2374–2380. [PubMed] [Google Scholar]
- 11.Dalgic B, Bukulmez A, Sari S. Eponym: Papillon-Lefevre syndrome. Eur J Pediatr. 2011;170(6):689–691. doi: 10.1007/s00431-010-1367-4. [DOI] [PubMed] [Google Scholar]
- 12.Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefevre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol. 2004;173(12):7277–7281. doi: 10.4049/jimmunol.173.12.7277. [DOI] [PubMed] [Google Scholar]
- 13.Vethanayagam RR, et al. Role of NADPH oxidase versus neutrophil proteases in antimicrobial host defense. PLoS One. 2011;6(12):e28149. doi: 10.1371/journal.pone.0028149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallen-St CJ, Pham CT, Villalta SA, Caughey GH, Wolters PJ. Mast cell dipeptidyl peptidase I mediates survival from sepsis. J Clin Invest. 2004;113(4):628–634. doi: 10.1172/JCI19062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haneke E. The Papillon-Lefevre syndrome: keratosis palmoplantaris with periodontopathy. Report of a case and review of the cases in the literature. Hum Genet. 1979;51(1):1–35. doi: 10.1007/BF00278288. [DOI] [PubMed] [Google Scholar]
- 16.Belaaouaj A, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4(5):615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 17.Tkalcevic J, Novelli M, Phylactides M, Iredale JP, Segal AW, Roes J. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity. 2000;12(2):201–210. doi: 10.1016/S1074-7613(00)80173-9. [DOI] [PubMed] [Google Scholar]
- 18.Rice WG, Ganz T, Kinkade JM, Selsted ME, Lehrer RI, Parmley RT. Defensin-rich dense granules of human neutrophils. Blood. 1987;70(3):757–765. [PubMed] [Google Scholar]
- 19.Adkison AM, Raptis SZ, Kelley DG, Pham CT. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest. 2002;109(3):363–371. doi: 10.1172/JCI13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cigic B, Dahl SW, Pain RH. The residual pro-part of cathepsin C fulfills the criteria required for an intramolecular chaperone in folding and stabilizing the human proenzyme. Biochemistry. 2000;39(40):12382–12390. doi: 10.1021/bi0008837. [DOI] [PubMed] [Google Scholar]
- 21.Turk D, et al. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20(23):6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molgaard A, Arnau J, Lauritzen C, Larsen S, Petersen G, Pedersen J. The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2. Biochem J. 2007;401(3):645–650. doi: 10.1042/BJ20061389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glenthoj A, Cowland JB, Heegaard NH, Larsen MT, Borregaard N. Serglycin participates in retention of alpha-defensin in granules during myelopoiesis. Blood. 2011;118(16):4440–4448. doi: 10.1182/blood-2011-06-362947. [DOI] [PubMed] [Google Scholar]
- 24.Lollike K, Kjeldsen L, Sengeløv H, Borregaard N. Purification of lysozyme from human neutrophils and development of an ELISA for quantification in cells and plasma. Leukemia. 1995;9(1):206–209. [PubMed] [Google Scholar]
- 25.Horiuchi K. A brief history of tumor necrosis factor α–converting enzyme: an overview of ectodomain shedding. Keio J Med. 2013;62(1):29–36. doi: 10.2302/kjm.2012-0003-RE. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yipp BG, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. 2012;18(9):1386–1393. doi: 10.1038/nm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122(16):2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 29.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191(3):677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen OE, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97(12):3951–3959. doi: 10.1182/blood.V97.12.3951. [DOI] [PubMed] [Google Scholar]
- 31.Campanelli D, Detmers PA, Nathan CF, Gabay JE. Azurocidin and a homologous serine protease from neutrophils. Differential antimicrobial and proteolytic properties. J Clin Invest. 1990;85(3):904–915. doi: 10.1172/JCI114518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohl J, Pereira HA, Martin NM, Spitznagel JK. Amino acid sequence of CAP37, a human neutrophil granule-derived antibacterial and monocyte-specific chemotactic glycoprotein structurally similar to neutrophil elastase. FEBS Lett. 1990;272(1):200–204. doi: 10.1016/0014-5793(90)80484-z. [DOI] [PubMed] [Google Scholar]
- 33.Ostergaard E, Flodgaard H. A neutrophil-derived proteolytic inactive elastase homologue (hHBP) mediates reversible contraction of fibroblasts and endothelial cell monolayers and stimulates monocyte survival and thrombospondin secretion. J Leuk Biol. 1992;51(4):316–323. doi: 10.1002/jlb.51.4.316. [DOI] [PubMed] [Google Scholar]
- 34.Pereira HA, Erdem I, Pohl J, Spitznagel JK. Synthetic bactericidal peptide based on CAP37: A 37-kDa human neutrophil granule-associated cationic antimicrobial protein chemotactic for monocytes. Proc Natl Acad Sci U S A. 1993;90(10):4733–4737. doi: 10.1073/pnas.90.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garwicz D, Lindmark A, Persson AM, Guilberg U. On the role of the proform-conformation for processing and intracellular sorting of human cathepsin G. Blood. 1998;92(4):1415–1422. [PubMed] [Google Scholar]
- 36.Lindmark A, Gullberg U, Olsson I. Processing and intracellular transport of cathepsin g and neutrophil elastase in the leukemic myeloid cell line U- 937- modulation by brefeldin a, ammonium chloride, and monensin. J Leuk Biol. 1994;55(1):50–57. doi: 10.1002/jlb.55.1.50. [DOI] [PubMed] [Google Scholar]
- 37.Lindmark A, Garwicz D, Rasmussen PB, Flodgaard H, Gullberg U. Characterization of the biosynthesis, processing, and sorting of human HBP/CAP37/azurocidin. J Leukoc Biol. 1999;66(4):634–643. doi: 10.1002/jlb.66.4.634. [DOI] [PubMed] [Google Scholar]
- 38.Mora-Jensen H, Jendholm J, Fossum A, Porse B, Borregaard N, Theilgaard-Monch K. Technical advance: immunophenotypical characterization of human neutrophil differentiation. J Leukoc Biol. 2011;90(3):629–634. doi: 10.1189/jlb.0311123. [DOI] [PubMed] [Google Scholar]
- 39.Borregaard N, Sehested M, Nielsen BS, Sengeløv H, Kjeldsen L. Biosynthesis of granule proteins in normal human bone marrow cells. Gelatinase is a marker of terminal neutrophil differentiation. Blood. 1995;85(3):812–817. [PubMed] [Google Scholar]
- 40.Le Cabec V, Cowland JB, Calafat J, Borregaard N. Targeting of proteins to granule subsets determined by timing not by sorting: the specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc Natl Acad Sci U S A. 1996;93(4):6454–6457. doi: 10.1073/pnas.93.13.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theilgaard-Monch K, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105(4):1785–1796. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 42.Bainton DF, Ullyot JL, Farquhar M. The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J Exp Med. 1971;143(4):907–934. doi: 10.1084/jem.134.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haneke E, Hornstein OP, Lex C. Increased susceptibility to infections in the Papillon-Lefevre syndrome. Dermatologica. 1975;150(5):283–286. doi: 10.1159/000251443. [DOI] [PubMed] [Google Scholar]
- 44.Reeves EP, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416(6878):291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 45.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nussbaum G, Shapira L. How has neutrophil research improved our understanding of periodontal pathogenesis? J Clin Periodontol. 2011;38(suppl 11):49–59. doi: 10.1111/j.1600-051X.2010.01678.x. [DOI] [PubMed] [Google Scholar]
- 47.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 48.Bianchi M, et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 2009;114(13):2619–2622. doi: 10.1182/blood-2009-05-221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002;360(9340):1144–1149. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- 50.Andersson M, Karlsson J, Carlsson G, Putsep K. Expression of granule-associated proteins in neutrophils from patients with severe congenital neutropenia. Blood. 2007;110(7):2772–2773. doi: 10.1182/blood-2007-03-083063. [DOI] [PubMed] [Google Scholar]
- 51.Ye Y, et al. Mutations in the ELANE gene are associated with development of periodontitis in patients with severe congenital neutropenia. J Clin Immunol. 2011;31(6):936–945. doi: 10.1007/s10875-011-9572-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B, Salcedo R. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur J Biochem. 1996;238(2):325–332. doi: 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- 53.Puklo M, Guentsch A, Hiemstra PS, Eick S, Potempa J. Analysis of neutrophil-derived antimicrobial peptides in gingival crevicular fluid suggests importance of cathelicidin LL-37 in the innate immune response against periodontogenic bacteria. Oral Microbiol Immunol. 2008;23(4):328–335. doi: 10.1111/j.1399-302X.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tongaonkar P, Golji AE, Tran P, Ouellette AJ, Selsted ME. High fidelity processing and activation of the human alpha-defensin HNP1 precursor by neutrophil elastase and proteinase 3. PLoS One. 2012;7(3):e32469. doi: 10.1371/journal.pone.0032469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindmark A, Garwicz D, Rasmussen PB, Flodgaard H, Gullberg U. Characterization of the biosynthesis, processing, and sorting of human HBP/CAP37/azurocidin. J Leukoc Biol. 1999;66(4):634–643. doi: 10.1002/jlb.66.4.634. [DOI] [PubMed] [Google Scholar]
- 56.Niemann CU, Cowland JB, Klausen P, Askaa J, Calafat J, Borregaard N. Localization of serglycin in human neutrophil granulocytes and their precursors. J Leukoc Biol. 2004;76(2):406–415. doi: 10.1189/jlb.1003502. [DOI] [PubMed] [Google Scholar]
- 57.Borregaard N, Heiple JM, Simons ER, Clark RA. Subcellular localization of the b-cytochrome component of the human neutrophil microbicidal oxidase: translocation during activation. J Cell Biol. 1983;97(1):52–61. doi: 10.1083/jcb.97.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kjeldsen L, Sengeløv H, Lollike K, Nielsen MH, Borregaard N. Isolation and characterization of gelatinase granules from human neutrophils. Blood. 1994;83(6):1640–1649. [PubMed] [Google Scholar]
- 59.Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013;94(4):711–721. doi: 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- 60.Faurschou M, et al. Prodefensins are matrix proteins of specific granules in human neutrophils. J Leukoc Biol. 2005;78(3):785–793. doi: 10.1189/jlb.1104688. [DOI] [PubMed] [Google Scholar]
- 61.Kjeldsen L, Koch C, Arnljots K, Borregaard N. Characterization of two ELISAs for NGAL, a newly described lipocalin in human neutrophils. J Immunol Methods. 1996;198(2):155–164. doi: 10.1016/S0022-1759(96)00153-6. [DOI] [PubMed] [Google Scholar]
- 62.Rorvig S, et al. Ficolin-1 is present in a highly mobilizable subset of human neutrophil granules and associates with the cell surface after stimulation with fMLP. J Leukoc Biol. 2009;86(6):1439–1449. doi: 10.1189/jlb.1008606. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y, Junger WG. Measurement of oxidative burst in neutrophils. Methods Mol Biol. 2012;844:115–124. doi: 10.1007/978-1-61779-527-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorensen O, Cowland JB, Askaa J, Borregaard N. An ELISA for hCAP-18, the cathelicidin present in human neutrophils and plasma. J Immunol Methods. 1997;206(1–2):53–59. doi: 10.1016/s0022-1759(97)00084-7. [DOI] [PubMed] [Google Scholar]
- 65.Brinkmann V, Goosmann C, Kuhn LI, Zychlinsky A. Automatic quantification of in vitro NET formation. Front Immunol. 2012;3:413. doi: 10.3389/fimmu.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faurschou M, Sorensen OE, Johnsen AH, Askaa J, Borregaard N. Defensin-rich granules of human neutrophils: characterization of secretory properties. Biochim Biophys Acta. 2002;1591(1):29–35. doi: 10.1016/s0167-4889(02)00243-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.