Abstract
The formal oxidation state of carbon atoms in organic molecules depends on the covalent structure. In proteins, the average oxidation state of carbon (ZC) can be calculated as an elemental ratio from the chemical formula. To investigate oxidation–reduction (redox) patterns, groups of proteins from different subcellular locations and phylogenetic groups were selected for comparison. Extracellular proteins of yeast have a relatively high oxidation state of carbon, corresponding with oxidizing conditions outside of the cell. However, an inverse relationship between ZC and redox potential occurs between the endoplasmic reticulum and cytoplasm. This trend provides support for the hypothesis that protein transport and turnover are ultimately coupled to the maintenance of different glutathione redox potentials in subcellular compartments. There are broad changes in ZC in whole-genome protein compositions in microbes from different environments, and in Rubisco homologues, lower ZC tends to occur in organisms with higher optimal growth temperature. Energetic costs calculated from thermodynamic models are consistent with the notion that thermophilic organisms exhibit molecular adaptation to not only high temperature but also the reducing nature of many hydrothermal fluids. Further characterization of the material requirements of protein metabolism in terms of the chemical conditions of cells and environments may help to reveal other linkages among biochemical processes with implications for changes on evolutionary time scales.
Keywords: oxidation state, redox potential, subcellular location, protein metabolism, protein evolution
1. Introduction
Chemical reactions involving the transfer of electrons, known as oxidation–reduction or redox reactions, are ubiquitous in cellular and environmental systems [1,2]. In the cell, the oxidation of thiol groups in proteins to form disulfides has the potential to regulate (activate or inhibit) enzymatic function [3]. Because these reactions are reversible on short time scales, a regulatory network known as redox signalling is made possible by reactions of small-molecule metabolites, including glutathione (GSH) and reactive oxygen species [4]. On time scales of metabolism, complex oxidation–reduction reactions are required for the formation (anabolism) and degradation (catabolism) of proteins and other biomolecules. Although many individual steps in biomass synthesis are irreversible, much biomass is ultimately recycled through autophaghy and other degradation pathways [5] that support endogenous metabolism [6] and the dynamic turnover of biomass. On longer time scales, forces outside of individual cells and organisms sustain the redox disequilibria between inorganic and/or organic species that provide the energy sources for metabolisms suited to a multitude of environments [7]. In turn, the actions of organisms can alter the redox conditions on the Earth; the oxygenation of the atmosphere and oceans over geological time is linked to biological activity and changed the course of later evolution [8].
Through evolution, the sequences of genes, and their protein products, are progressively altered. The elemental stoichiometries (chemical formulae) and standard Gibbs energies of the molecules have a primary impact on the metabolic requirements for energy and elemental resources. The energetic cost for synthesis of biomass is a function not only of the composition of the biomass, but also of environmental parameters including temperature and the concentrations of inorganic or metabolic precursors. Temperature and oxidation–reduction potential (ORP) have profound effects on the relative energetic costs of formation of different amino acids [9] or proteins [10]. These energetic costs are sensitive to differences in the elemental compositions of biomolecules. To a first approximation, a shift to a more reducing environment alters the energetics of reactions in a direction that favours the formation of relatively reduced chemical compounds. In a field-based test of this notion, metagenomic sequences for the most highly reduced proteins were found in the hottest and most reducing zones of a hot spring [11].
The purpose of this study is to investigate a particular stoichiometric quantity, the average oxidation state of carbon (ZC, defined below), as a comparative tool for identifying compositional patterns at different levels of biological organization. By comparing a quantity derived from the elemental compositions of proteins, this study addresses one aspect of biochemical evolution. However, the questions raised here differ in important respects from conventionally defined biochemistry and evolutionary biology. Biochemical studies are most often concerned with the functions of molecules [12], including enzymatic catalysis and non-covalent interactions involved in the structural conformation of proteins and binding of ligands. Studies in molecular evolution often place emphasis on the historical relationships among sequences, but not their physical properties [12]. Combining these viewpoints, most current work assumes that structural stability of proteins is the primary criterion for molecular adaptation to high temperature [13]. By contrast, in this study, more attention is given to the differences in stoichiometric and energetic properties of chemical reactions that are implied by differences in the composition of the proteins. Because material replacement of proteins depends on metabolic outputs (the concept known as ‘metabolic closure’ [14]), the compositional differences among proteins have significant consequences for cellular organization and metabolism.
The following questions have been identified: (1) how does the relationship between ZC of amino acids and corresponding codons relate to the origin or form of the genetic code? (2) How do the differences in ZC between membrane proteins and others compare with properties of amino acids, e.g. hydrophobicity, known to favour localization to membranes? (3) How are the differences in ZC of proteins among eukaryotic subcellular compartments related to differences in redox potential? (4) How are the differences in ZC in different organisms, both in terms of bulk (genome-derived) protein composition and for homologues of a single family (ribulose bisphosphate carboxylase, Rubisco), related to environmental conditions, especially temperature and redox potential?
In the Results, each problem is briefly introduced, the empirical distribution of ZC is described and a discussion is developed to explore the biochemical and evolutionary implications. These short discussions, corresponding to questions (1)–(4), should be regarded as preliminary, and probably incomplete interpretations that await integration with a conceptual framework linking the biochemical reactions with the evolutionary processes that are implicit in all of the comparisons. The final subsection of the Results goes into more detail for the comparisons of Rubisco by examining the relative Gibbs energies of formation of proteins in environments of differing redox potential.
2. Material and methods
Throughout this study, ‘reducing’ and ‘oxidizing’ are used in reference to ORP, tied to a particular redox couple or to environmental conditions, often expressed as millivolts on the Eh scale. ‘Reduced’ and ‘oxidized’ are used to refer to variations in the oxidation state of carbon.
The formal oxidation state of a carbon atom in an organic molecule can be calculated by counting −1 for each carbon–hydrogen bond, 0 for each carbon–carbon bond and −1 for each bond of carbon to O, N or S [9,15]. In photosynthesizing organisms, and autotrophs in general, the carbon source is CO2, having the highest oxidation state of carbon (+4). The products of photosynthetic reactions include proteins and other biomolecules with a lower oxidation state of carbon. Even if the molecular structure is unknown, analytical elemental compositions can be used in calculations of the average oxidation state of carbon in biomass [16,17]. Because any gene or protein sequence corresponds to a definite canonical (non-ionized, unphosphorylated) chemical formula, the average oxidation state of carbon in these biomolecules is easily calculated.
In amino acids and proteins, the average oxidation state of carbon (ZC) can be calculated using
| 2.1 |
where Z is the charge on the molecule, and nC, nH, nN, nO and nS are the numbers of the subscripted elements in the chemical formula of the molecule. The coefficients on the terms in the numerator are derived from formal charges of atoms other than C, as follows: H (+1), N (−3), O (−2), S (−2). Negative formal charges reflect greater electronegativities of these elements compared with carbon. If two thiol groups react to form a disulfide bond, the oxidation states of the two sulfur atoms change from −2 to −1. Although H2 is produced in this reaction, the oxidation state of carbon in the protein remains constant. It follows that equation (2.1) is applicable only to chemical formulae of proteins in which the N, O and S are all fully reduced (bonded only to H and/or C).
The Z in equation (2.1) ensures that ionization by gain or loss of a proton, having an equal effect on Z and nH, does not change the ZC. Likewise, gain or loss of H2O, which affects equally the values 2nH and nO, does not alter the average oxidation state of carbon [15]. Accordingly, the ZC of a peptide formed by polymerization of amino acids (a dehydration reaction) is a weighted average of the ZC in the amino acids, where the weights are the numbers of carbon atoms in each amino acid. As an example, the ZC of hen egg white lysozyme, having a chemical formula of C613H959N193O185S10, is 0.016. This protein is oxidized compared to many other proteins, which commonly have negative values of ZC.
To aid in reproducibility, data files of protein sequences or amino acid composition and computer program files for the calculations are provided in the electronic supplementary material. The calculations and figures were generated using the R software environment [18] together with the CHNOSZ package [19].
3. Results
3.1. Comparison of ZC of amino acids with hydropathy and properties of codons
In contemplating the ancient origin of the genetic code, the chemical similarities of codons and the associated amino acids have been used to argue for coevolution (shared biosynthetic pathways) [20] or a tendency towards similar physico-chemical properties. Some advantages that have been identified for similar physico-chemical properties include enhancing the steric interactions between amino acids and codons [21] or increasing the similarity between different amino acids resulting from a single DNA base mutation in order to maintain protein structure [20].
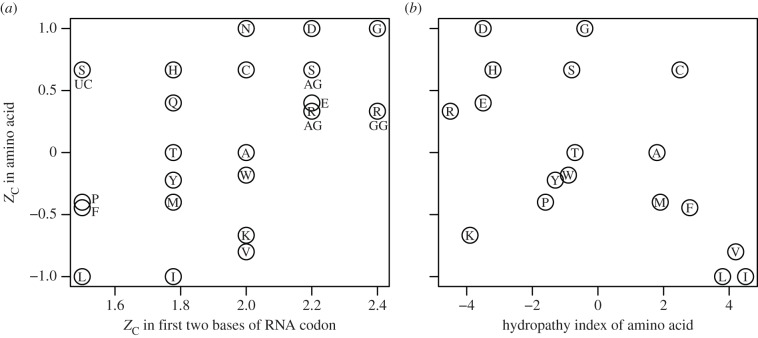
In the genetic code, the first two bases (a ‘doublet’) are more indicative of the amino acid than the third position of the codon [21]. The ZC of amino acids is compared with the values calculated for the corresponding RNA nucleobase doublets in figure 1a. Some of the doublets, e.g. UU (phenylalanine and leucine), CU (leucine), UC (serine) and CC (proline), have identical ZC (in this case, 1.5), leading to only five possible values of ZC for the doublets. The overall relationship suggested by figure 1a is weak correlation between the ZC of amino acids and of the RNA doublets. The most highly reduced amino acids, leucine (L) and isoleucine (I), are coded for by doublets having the two lowest ZC values.
Figure 1.
Average oxidation state of carbon (ZC) in amino acids compared with (a) ZC in the first two bases of the corresponding RNA codons and (b) hydropathy index of the amino acids taken from Kyte & Doolittle [22]. Standard one-letter abbreviations for the amino acids are used to identify the points. In (a), the different codon compositions for serine (S) and arginine (R) are indicated by letters below the symbols, and some amino acid labels are shifted for readability. In (b), labels for asparagine (N) and glutamine (Q) are omitted for clarity; they plot at the same positions as aspartic acid (D) and glutamic acid (E), respectively.
The relative Gibbs energies of formation reactions of amino acids differ considerably between hydrothermal (hot, reducing) and surface (cool, oxidizing) environments [9]. Accordingly, the increase in ZC going from leucine to alanine to glycine (figure 1) is reflected in the metastability fields of these amino acids, which occur in order of increasing oxidation potential, or oxygen fugacity [23]. (Metastable equilibrium refers to the equalization of the energies of reactions to form the amino acids; it is a partial equilibrium because the amino acids generally remain unstable with respect to inorganic species.) The parallel increase in ZC of the RNA doublets paired with these amino acids hints at the possibility for metastable equilibria among the amino acids and nucleobase sequences. Theoretical calculations of the energetics of synthesis reactions carried out using Gibbs energies available at high temperature [10,24] might be used to identify conditions favourable for abiotic or early biosynthesis of the amino acids and nucleobase sequences that are paired in the genetic code.
The hydropathy index, based on the relative hydrophobicity and hydrophilicity of amino acids [22], is commonly used for identifying probable membrane-spanning domains of proteins. In figure 1b, ZC is compared with the hydropathy values for individual amino acids. The three most hydropathic amino acids, isoleucine, leucine and valine, are also the three with the lowest ZC. Therefore, membrane proteins with hydrophobic domains are likely to be more reduced than other proteins. The following subsections examine the actual differences in human and yeast proteins.
3.2. Differences in ZC of membrane proteins
The lipid (fatty acid) components of membranes are reduced relative to many other biomolecules, including amino acids, nucleotides and saccharides (see fig. 1 of Amend et al. [25]). Proteins that are embedded in membranes tend to contain more hydrophobic amino acids, which enhance solubility of proteins in the membrane environment [22] and generally are relatively reduced (figure 1b).
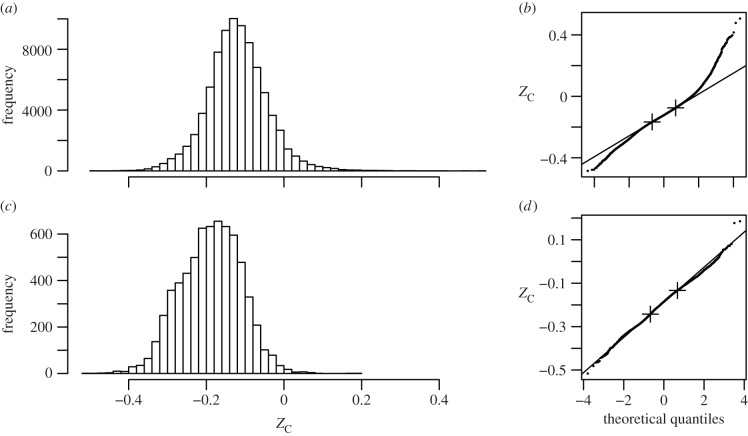
To compare membrane proteins with other human proteins, sequences for all human proteins were taken from the UniProt database [26], and sequences for predicted membrane proteins were taken from all FASTA sequence files provided in Additional File 2 of Almén et al. [27]. Only sequences at least 50 amino acids in length were considered. The distribution of ZC of all human proteins (figure 2a; n = 83 994) is centred on −0.123 (median), −0.120 ± 0.077 (mean ± s.d.). In the ZC of human membrane proteins, the distribution is shifted to lower values (figure 2c; n = 6627, −0.186 median, −0.189 ± 0.078 mean ± s.d.). The mean value is lower for membrane proteins than for all human proteins (Student's t-test: p < 2.2 × 10−16). Thus, the proteins located in the membranes are, on average, more reduced than other proteins in humans. A possible implication is that the coexistence of relatively reduced proteins with other relatively reduced biomolecules (lipids) reflects a compositional similarity that would contribute to energy optimization if metabolic pathways for proteins and lipids were operating under common redox potential conditions.
Figure 2.
Average oxidation state of carbon shown in histograms and normal probability plots for (a,b) all human proteins and (c,d) human membrane proteins. Only proteins of sequence length greater than or equal to 50 amino acids are considered. In the normal probability plots, the lines are drawn through the first and third quartiles, indicated by the crosses.
It should be noted that the mean values of ZC for groups of proteins given above and elsewhere in this study are calculated on a per-molecule basis, with no weighting for the different carbon numbers in the proteins. Hence, if the proteins were equally abundant, the per-molecule mean of ZC would be biased towards the smaller proteins compared with the carbon-weighted mean value, which is more representative. Specific calculations show that the per-molecule and carbon-weighted means have comparable differences between groups of proteins. For example, the carbon-weighted mean values of all human proteins and human membrane proteins are −0.117 and −0.165, respectively, with the membrane proteins again exhibiting a lower mean value of ZC. However, in the ideal case, calculations of the ZC of a group of proteins should also take account not only of the different carbon numbers of the proteins but also the protein abundances (i.e. expression levels). Actual protein abundances cover a large dynamic range, but a lack of abundance data for all the proteins that are considered here precludes the possibility of incorporating abundances in the present calculations. Therefore, the per-molecule weighted means of ZC for groups of proteins reported in this paper should be taken as a first-order description of these complex assemblages, which are subject to additional refinement as more data become available.
It follows from equation (2.1) that the values of ZC, as well as their differences, are formed by linear combinations of H/C, N/C, O/C and S/C ratios. The differences in the per-molecule mean values of these ratios for all human proteins and the human membrane proteins of at least 50 amino acids are, respectively, −0.0189, −0.0162, −0.0206 and 0.0012; that is, the membrane proteins have lower values of H/C, N/C and O/C, and higher S/C. The lower mean H/C ratio and higher S/C ratio in membrane proteins contribute to a more oxidized state of carbon (ΔZC = 0.019 and 0.002, respectively), which is offset by the lower N/C and O/C ratios (ΔZC = −0.049 and −0.041, respectively). The decrease in bonds to N and O accounts for the overall more reduced state of carbon in the membrane proteins.
The observed distributions of ZC are each compared with a normal distribution in normal probability plots (theoretical quantile–quantile (Q–Q) plots) in figure 2b,d. The steeper trends in the low- and high-quantile range of figure 2b indicate that the distribution of ZC of human proteins has relatively long tails, especially at high ZC, compared with a normal distribution. Although an asymmetry is apparent in the uneven shape of the histogram in figure 2c and the wiggles in figure 2d, the overall distribution of ZC of the membrane proteins more closely resembles a normal distribution. According to the central limit theorem, a normal distribution can be generated through the addition of many small-scale, independent effects [28]. The shape of the distribution of ZC may have implications for the evolutionary processes that affect the chemical composition of organisms or their components. This theme, however, is not developed further here; instead, the overall differences in ZC of proteins in subcellular compartments are considered next.
3.3. Subcellular differences in ZC of proteins and comparison with subcellular redox potential
For some model organisms, including Saccharomyces cerevisiae (yeast), the identities of proteins associated with subcellular compartments are now available in databases. Here, calculations of ZC of proteins and a comparison with independent measurements of redox potential are used to investigate the oxidation–reduction features and dynamics of cellular structure.
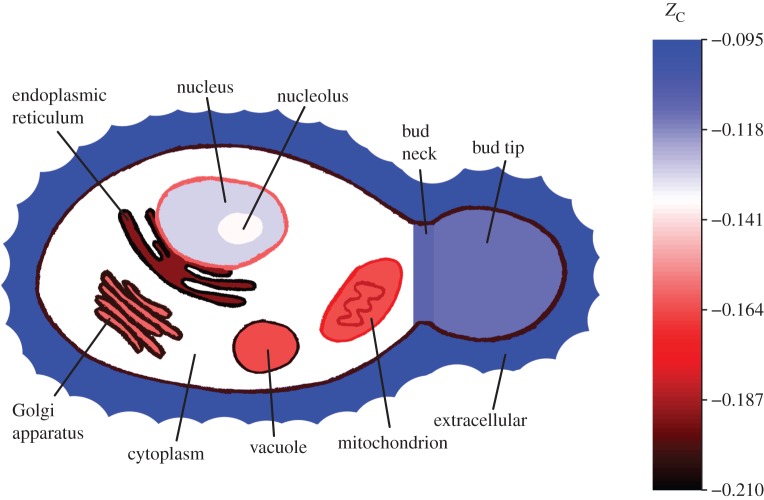
In a previous study, the limiting conditions for chemical transformations among proteins in subcellular compartments were quantified theoretically as a function of redox potential and hydration state [29]. In that study, the locations of proteins were taken from the ‘YeastGFP’ study of Huh et al. [30]. That dataset has the advantage that relative abundances of many of the proteins are available, but it is limited to 23 named locations in the cell. In order to consider more cellular components, including the membranes, a more extensive reference proteome is used in this study. This proteome is based on the Saccharomyces Genome Database (SGD) [31] annotations combined with the Gene Ontology (GO) [32] vocabulary for the ‘cellular component’ aspect, which describes many organelles and membranes within the cell. Major cellular components were selected for comparison, and the ZC was calculated for protein products of the genes, as summarized in table 1. The median values are portrayed in the drawing of a yeast cell in figure 3.
Table 1.
Summary of ZC of proteins in subcellular locations of yeast. Numbers of proteins (n) in SGD associated with the indicated GO terms are listed. The numerators of the fractions denote membrane-associated proteins that are also listed as ‘integral to membrane’ (GO:0016021); only these proteins were used in the calculations of ZC.
| cellular component | GO term | n | ZC (median) | ZC (mean ± s.d.) |
|---|---|---|---|---|
| cytoplasm | GO:0005737 | 2245 | −0.136 | −0.127 ± 0.064 |
| nucleus | GO:0005634 | 2073 | −0.129 | −0.121 ± 0.064 |
| mitochondrion | GO:0005739 | 1077 | −0.164 | −0.159 ± 0.060 |
| ER | GO:0005783 | 435 | −0.191 | −0.192 ± 0.070 |
| nucleolus | GO:0005730 | 263 | −0.137 | −0.128 ± 0.065 |
| Golgi apparatus | GO:0005794 | 215 | −0.160 | −0.167 ± 0.068 |
| cellular bud neck | GO:0005935 | 153 | −0.111 | −0.108 ± 0.063 |
| vacuole | GO:0005773 | 175 | −0.164 | −0.163 ± 0.079 |
| extracellular region | GO:0005576 | 95 | −0.096 | −0.098 ± 0.066 |
| cellular bud tip | GO:0005934 | 96 | −0.113 | −0.110 ± 0.071 |
| ER membrane | GO:0005789 | 283/338 | −0.209 | −0.211 ± 0.065 |
| plasma membrane | GO:0005886 | 224/427 | −0.200 | −0.188 ± 0.069 |
| mitochondrial inner membrane | GO:0005743 | 143/218 | −0.184 | −0.184 ± 0.061 |
| vacuolar membrane | GO:0005774 | 100/145 | −0.205 | −0.196 ± 0.072 |
| Golgi membrane | GO:0000139 | 76/121 | −0.203 | −0.200 ± 0.076 |
| nuclear membrane | GO:0031965 | 54/67 | −0.161 | −0.139 ± 0.084 |
| mitochondrial outer membrane | GO:0005741 | 51/92 | −0.176 | −0.177 ± 0.051 |
Figure 3.
A simplified drawing of a yeast cell depicting median values of the average oxidation state of carbon in proteins from the subcellular locations listed in table 1. All cellular components listed in the table are represented in the drawing. The colour scale is adjusted so that the cytoplasm has a neutral hue (white), and locations with relatively oxidized and reduced proteins are depicted by blue and red colours, respectively. Darker red colours are used for the more reduced groups of proteins in some of the membranes such as the ER, Golgi, vacuolar and plasma membranes. The nuclear and inner and outer mitochondrial membranes are shown with lighter reds because their proteins are relatively oxidized compared with those in the other membranes. Components not separated by membranes (or with membranes not shown) include the nucleolus, bud neck and bud tip.
It is apparent that the membrane proteins are highly reduced. However, not all membranes are equal; the proteins in the nuclear and inner and outer mitochondrial membranes are on average less reduced than those in the plasma membrane, and the endoplasmic reticulum (ER) membrane has very highly reduced proteins. Among the organellar proteins considered, the ER has the most highly reduced proteins, followed by Golgi, vacuoles and mitochondria; the nucleolar and cytoplasmic proteins are moderately reduced. The proteins in the nucleus, bud tip and bud neck are more oxidized than in the other compartments. The extracellular proteins are the most oxidized group in the system. The relative values of ZC of the proteins in the ER, mitochondrion, cytoplasm and nucleus are consistent with those of a previous study in which the calculations took account of the abundances of proteins [29].
For comparison with ZC of proteins, the values of subcellular redox potential (Eh, in mV) listed in table 2 were compiled from literature sources [1,33–40]. Measurements of reduced and oxidized glutathione (GSH and GSSG) in whole-cell extracts have been interpreted as reflecting cytoplasmic redox potential, but redox-sensitive green fluorescent protein (roGFP) probes [33] provide more specific data for subcellular locations. The data are not in all cases acquired from yeast, but it has been noted that cytoplasmic Eh values based on roGFP are similar for different model organisms [36].
Table 2.
Values of Eh compiled from literature sources, for yeast cells or culture except as noted. The ranges account for variation among different cell types, experimental techniques and published values, and are used to construct figure 4.
| location | range (mV) | references |
|---|---|---|
| mitochondrion | −360 to −255 | [33]: roGFP probe (−360 mV; human HeLa) [34]: rxYFP in matrix (−296 mV) and intermembrane space (−255 mV) |
| cytoplasm | −320 to −240 | [35]: GSH in proliferating mammalian cells (−240 mV) [36]: roGFP probe of GSH (−320 mV) |
| ER | −208 to −133 | [37]: NYTC peptide (−185 to −133 mV; murine hybridoma CRL-1606) [38]: roGFP (−208 mV; human HeLa) |
| vacuole | −160 to >−130 | see text |
| extracellular | −150 to >160 | [1]: aerobic (160 mV) and anaerobic (90 mV) cultures [39]: very high-gravity fermentation (−150 mV) [40]: H. sapiens plasma (−140 mV) |
The redox potentials in the vacuole and extracellular space are less well constrained than other locations. Under stress response, high amounts of GSSG, but not GSH, are sequestered in vacuoles [41]. A conservative lower range for the Eh of vacuoles (−160 to −130 mV) was calculated by taking a value of 80% GSSG and computing Eh from the GSH–GSSG equilibrium at concentrations of 1–10 mM GSH (see eqn (21) and fig. 4 in [35]). The redox potential would be higher if the GSSG/GSH ratio were in fact greater than 80/20. A high redox potential is also implicated by the presence of ferric iron species in vacuoles [42]. Extracellular redox state can vary greatly, but in aerobic organisms and laboratory culture it is likely to be generally oxidizing compared with subcellular compartments.
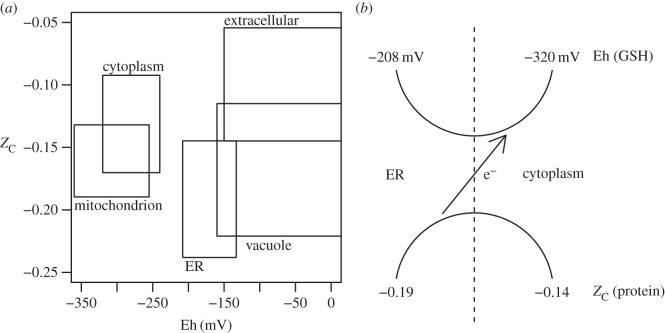
The values in table 2 are not comprehensive and should be taken as a rough guide, but even with the uncertainties, a comparison with the interquartile range of ZC of the proteins reveals some trends (figure 4a). The difference in both ZC and Eh is positive going from any subcellular compartment shown, except for vacuoles, to the extracellular space. This pattern has an intuitive explanation: by evolutionary adjustment to optimize proteins for their environment, the inside of the cell, which is more reducing, would be expected to have more reduced proteins compared with the outside.
Figure 4.
The plot (a) compares the average oxidation state of carbon in proteins from different subcellular locations of yeast with Eh values taken from glutathione (GSH–GSSH) or other redox indicators (see table 2). The heights of the boxes indicate the interquartile ranges of ZC values, and the widths represent the ranges of Eh listed in table 2. The scheme (b) invokes electron transfer to account for the differences between ER and cytoplasm of redox potential and ZC of proteins (see text).
The more surprising trend in figure 4a is an inverse relationship between ZC and Eh of the cytoplasm and ER. Does this contrast have any biochemical significance? The ER is a component of the secretory pathway, which transports proteins to membranes and to outside the cell [38]. Let us hypothesize that the populations of proteins in the ER and cytoplasm are connected through common metabolic intermediates. This is a plausible scenario if the formation and degradation of proteins are part of the recycling of biomass and endogenous metabolism (e.g. [5,6]). It follows that the formation of proteins of a higher ZC in the cytoplasm entails the loss of electrons from proteins into metabolic pathways. Perhaps through these pathways electrons are ultimately transferred to reactions that drive the formation of GSH in the cytoplasm.
This scheme is depicted in figure 4b. The vertical dashed line represents a physical, but not impermeable, boundary between ER and cytoplasm; the curved lines represent reactions and transport within glutathione and protein systems that cross the compartments, and the arrow represents a linkage between glutathione and protein systems, which is effectively a coupled oxidation–reduction reaction. The scheme represents a redox mass-balance interpretation of the overall stoichiometric relationships, not a mechanism applied to individual GSH or protein molecules; the complete picture of metabolic connectivity in the cell is certainly more complex. Note that this scheme refers to the relative oxidation states of carbon of the proteins, not the oxidation of protein thiol groups to form disulfides. Disulfide bond formation takes place during folding and secretion of proteins and may also contribute to glutathione metabolism [4]. Although there is growing awareness of the pathways of glutathione metabolism in the cell, including compartmentalization between ER and cytoplasm, it is not known how they connect with non-thiol systems (e.g. [40]). Integrating the oxidation–reduction requirements of protein metabolism into existing metabolic models may help to complete the balance sheet of redox interactions in the cell.
Experiments on the connections between redox conditions and protein metabolism at the subcellular level can help elucidate the possible effects of coupling of protein metabolism to the glutathione redox system. If the net transfer of high-ZC proteins to the cytoplasm is stopped, then a decrease in GSH/GSSG redox potential in the ER relative to the cytoplasm would be expected based on the scheme shown in figure 4. This prediction is consistent with the outcome of experiments showing that puromycin-induced halting of protein synthesis causes a decrease in the redox potential monitored by roGFP in the ER [43]. A further untested implication of this hypothesis is that in ER-stress experiments, the ZC of the protein population in the ER would increase. Reactions involved in protein turnover might also intersect with redox pathways other than the oxidation and reduction of glutathione. A linkage of this type has been documented in plants, where degradation of aromatic and branched-chain amino acids was identified as a source of electrons for the mitochondrial electron transport chain [44].
3.4. Phylogenetic variation in ZC of proteins and comparison with optimal growth temperature
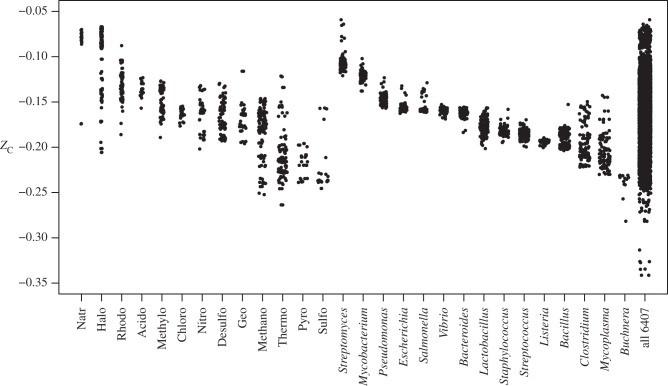
A comparison of ZC of the combined proteins from selected microbial genomes in the NCBI Reference Sequence (RefSeq) database [45] is shown in figure 5. The sets of proteins shown on the left-hand side of figure 5 correspond to those organisms whose scientific names contain the indicated substring. In many cases, the names of the organisms reflect their environments and/or metabolic strategies. Examples of the matching genus names are Natronobacterium, Haloferax, Rhodobacter, Acidovorax, Methylobacterium, Chlorobium, Nitrosomonas, Desulfovibrio, Geobacter, Methanococcus, Thermococcus, Pyrobaculum and Sulfolobus. Most terms, however, match more than one genus (e.g. Pyrobaculum and Pyrococcus). On the right-hand side of figure 5 are shown genera containing many groups with clinical and technological relevance; by the numbers of points, it is apparent that their representation in RefSeq is greater than that of the environmental microbes.
Figure 5.
Average oxidation state of carbon in total combined proteins from sequenced microbial genomes. Sequences of microbial proteins were taken from NCBI RefSeq release 61. Only organisms with total sequenced protein length greater than 50 000 amino acids were used, leaving 6407 organisms. The group on the left-hand side is identified by substring matches in the scientific name of the organism; the terms were chosen to emphasize environmental variation. The group on the right-hand side consists of the indicated genera, emphasizing organisms of clinical and biotechnological relevance. The final category represents total proteins from all microbial genomes meeting the minimal size requirement, many of which are not shown in the other categories.
A general trend towards lower ZC of proteins in organisms from hot environments (e.g. represented by Thermo and Pyro in the names) is apparent. Organisms with the highest ZC inhabit saline evaporative waters (Natr and Halo), whereas other aquatic organisms (e.g. Rhodo) have less highly oxidized proteins. The variation within groups is notable; for example, among the halophilic organisms, the highest values of ZC are associated with the archaeal Halosimplex carlsbadense [46] and aerobic species of Halorubrum [47], whereas Halothermothrix orenii and Halobacteroides halobius, both anaerobic bacteria [48,49], have the lowest values of ZC. Within a given genus (right-hand side of plot), the clusters of ZC tend to be tighter, reflecting conserved compositional trends. Streptomyces, common in soils, has the highest ZC of the genera shown here. Buchnera is notable because its proteins are highly reduced. Buchnera aphidicola BCc is the primary endosymbiont of the cedar aphid (Cinara cedri) and, at the time of sequencing, had one of the smallest known bacterial genomes [50]. Being relatively closely related [51], some species of Mycoplasma and Clostridium also have relatively low ZC. Mycoplasma are known for their small genomes and dependence on metabolic products of the host. ‘Candidatus Zinderia insecticola’ and ‘Candidatus Carsonella ruddii’ are endosymbionts with even smaller genomes [52,53], and they are the organisms with the most reduced proteins in the dataset (ZC < −0.321; lower right corner of figure 5). The low ZC of proteins in these organisms may be indicative of constraints imposed by growth in reducing intracellular or intraorganismal environments.
We now turn to a comparison of homologues of a specific protein. Rubisco is an essential enzyme for carbon fixation. Sequence comparison of homologues (related sequences that appear in different organisms) has provided the basis for many phylogenetic studies [54]. Major divergent forms of the enzyme are Forms I and II, found in aerobic organisms, and Form III and ‘Rubisco-like proteins’, found in anaerobic organisms [55]. The organisms listed in table 3 have in common the occurrence of Rubisco in their genome. Forms I, II or III were included in this comparison, but Rubisco-like proteins were excluded; however, some of these were found to have considerably lower ZC. The selection of organisms was made in order to represent a variety of optimal growth temperatures (Topt) as reported in the studies cited in the table [56–82].
Table 3.
Names of species, optimal growth temperatures (Topt), UniProt [26] accession numbers (IDs) and calculated ZC for the large subunit of Rubisco. Literature references for Topt are indicated in brackets. A, Archaea; B, Bacteria; E, Eukaryota. The numbers are used to identify the points in figure 6a (duplicated numbers occur in different temperature ranges).
| no. | domain | species | Topt (°C) | reference | ID | ZC |
|---|---|---|---|---|---|---|
| 1 | E | Phaeocystis antarctica | 0–6 | [56] | G9FID8 | −0.137 |
| 2 | B | Octadecabacter antarcticus 307 | 4–10 | [57] | M9R7V1 | −0.114 |
| 3 | A | Methanolobus psychrophilus | 18 | [58] | K4MAK9 | −0.152 |
| 4 | A | Methanococcoides burtonii | 23 | [59] | Q12TQ0 | −0.147 |
| 5 | B | Bradyrhizobium japonicum | 25–30 | [60] | Q9ZI34 | −0.125 |
| 6 | B | Thiobacillus ferrooxidans | 28–30 | [61] | P0C916 | −0.110 |
| 7 | E | Zea mays | 30 | [62] | P00874 | −0.117 |
| 8 | B | Mariprofundus ferrooxydans | 30 | [63] | Q0EX22 | −0.114 |
| 9 | B | Desulfovibrio hydrothermalis | 35 | [64] | L0RHZ1 | −0.097 |
| 1 | A | Methanosarcina acetivorans | 35–40 | [65] | Q8THG2 | −0.145 |
| 2 | B | Acidithiobacillus caldus | 45 | [66] | F9ZLP0 | −0.105 |
| 3 | E | Cyanidium caldarium | 45 | [67] | P37393 | −0.160 |
| 4 | B | Sulfobacillus acidophilus | 45–50 | [68] | P72383 | −0.126 |
| 5 | B | Pseudomonas hydrogenothermophila | 52 | [69] | Q51856 | −0.138 |
| 6 | B | Synechococcus sp. (strain JA-2-3B'a(2-13)) | 50–55 | [70] | Q2JIP3 | −0.117 |
| 7 | A | Methanosaeta thermophila | 55–60 | [71] | A0B9K9 | −0.112 |
| 8 | B | Thermosynechococcus elongatus | 57 | [72] | Q8DIS5 | −0.124 |
| 9 | B | Clostridium clariflavum | 60 | [73] | G8LZL2 | −0.181 |
| 1 | B | Bacillus acidocaldarius | 65 | [74] | F8IID7 | −0.143 |
| 2 | B | Thermotoga lettingae | 65 | [75] | A8F7V4 | −0.187 |
| 3 | B | Thermomicrobium roseum | 70 | [76] | B9KXE5 | −0.132 |
| 4 | A | Archaeoglobus fulgidus | 76 | [77] | O28635 | −0.161 |
| 5 | A | Methanocaldococcus jannaschii | 85 | [78] | Q58632 | −0.188 |
| 6 | A | Thermofilum pendens | 85–90 | [79] | A1RZJ5 | −0.175 |
| 7 | A | Staphylothermus marinus | 85–92 | [80] | A3DND9 | −0.189 |
| 8 | A | Pyrococcus horikoshii | 98 | [81] | O58677 | −0.175 |
| 9 | A | Pyrococcus furiosus | 100 | [82] | Q8U1P9 | −0.190 |
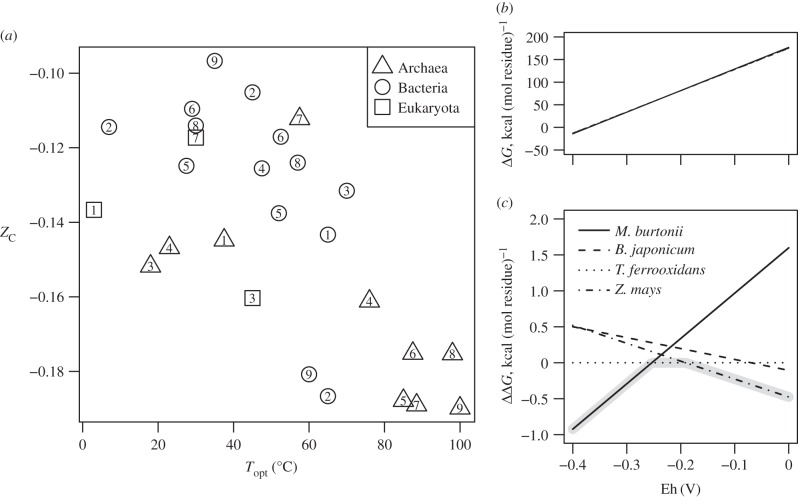
A comparison between Topt and ZC of Rubisco is presented in figure 6. The ZC of Rubisco are somewhat higher than the bulk protein content of the organisms; compare for example the values for Pyrococcus horikoshii (−0.175) and Pyrococcus furiosus (−0.190) (the highest temperature points labelled 8 and 9 in figure 6a) with the range of values for ‘Pyro’ in figure 5 (−0.239 to −0.196). At lower temperatures (0–50°C), the differences between the phylogenetic domains are most apparent; Rubiscos of the Bacteria in this sample set are more oxidized than those of Archaea and Eukaryota. There is a tendency for the Rubiscos of the Archaea to have lower ZC; this appears to be characteristic of anaerobic methanogenesis and Form III Rubiscos. An interesting exception is the high-ZC (−0.112) Rubisco of Methanosaeta thermophila; this organism grows on acetate to produce both CH4 and CO2 [71]. The major pattern that emerges is that higher temperatures are associated with a lower average oxidation state of carbon in proteins. As outlined below, a decrease in oxidation state of carbon of the proteins is associated with energetic savings for overall protein synthesis in reducing environments.
Figure 6.
(a) Average oxidation state of carbon in Rubisco compared with optimal growth temperature (Topt) of organisms. Numbers are used to identify the organisms (see table 3). (b,c) Gibbs energies of formation reactions, per residue, of Rubisco from selected organisms in the 23–30°C range of optimal growth temperature (points labelled 4–7). (b) Total Gibbs energies of individual reactions as a function of Eh and (c) difference between the reaction for T. ferrooxidans and the others are shown. The grey highlight indicates the protein with the lowest ΔG along the range of Eh values.
3.5. Beyond ZC: energetics of protein formation as a function of environmental redox potential
To a first approximation, energetic considerations predict that more reducing conditions tend to favour the formation of proteins with relatively lower ZC, and vice versa. To assess the directionality and magnitude of chemical forces on the evolutionary transformations of proteins, the energetics of reactions can be calculated using thermodynamic models. Because the time scales of evolution are much longer than transformations of biomolecules during metabolism, a discussion of the assumptions underlying the application of thermodynamic theory to biochemical evolution is warranted.
In energetic terms, adaptation can be defined as a problem of optimal efficiency, with trade-offs between energy utilization and power [83, p. 140]. Overall minimization of biomass synthesis costs can be expected from the energy utilization standpoint. The links between energetics and evolutionary outcome implicitly depend on the proposal that greater fitness is associated with mutations that lower the synthesis costs of proteins for a given function (e.g. [84]).
It has been argued previously that the propensity for some evolutionary changes can be modelled using the related concepts of equilibration, energy minimization and maximum entropy. In an evolutionary context, these concepts have often been defined by analogy to their definitions in thermodynamics [83,85]. The current discussion instead considers the possibility of application of chemical thermodynamic models based on Gibbs energy calculations to formulate a quantitative description of patterns of protein composition. The major assumption in the following discussion is that the energetic demands of protein formation primarily depend on the amino acid composition of the protein and the environmental conditions.
An example calculation is carried out here for selected Rubiscos from organisms with optimal temperatures in the range of 23–30°C (table 3, numbers 4–7). The basis species, representing inorganic starting materials, and their chemical activities used for this example are CO2 (10−3), H2O (1), NH3 (10−4), H2S (10−7) and H+ (10−7, i.e. pH = 7). The activity of the electron, representing the effects of the redox variable (Eh) through the Nernst equation, is left to vary. The overall Gibbs energies of the reactions to form the proteins from the basis species at 25°C (which is in the range of Topt of the selected organisms) were calculated as described previously [10,19] and are plotted in figure 6b. From this figure, it can be seen that the Gibbs energies of the reactions to form the proteins (ΔG), normalized by protein length, are positive over nearly the entire range, and steadily increase (become less favourable) with increasing Eh. As a consequence of the large differences in ΔG as a function of Eh, the differences between the proteins cannot be discerned easily in figure 6b. In figure 6c, the values of ΔG are shown relative to the reaction for Thiobacillus ferrooxidans, giving a difference in Gibbs energy of formation (ΔΔG) that can be used to assess the relative energetics of the reactions. Lower energies indicate the most stable protein (in the sense of chemical formation, not structural conformation), and these relative energies depend on the chemical conditions of the environment, measured in part by the ORP.
By using Gibbs energy calculations such as those shown in figure 6c, one can make an assessment of which protein in a given redox environment demands lower energy for overall synthesis than others. Where two lines cross in figure 6c, the energies to form the two proteins are equal. The three least costly proteins in this example, going from low to high Eh, are those from Methanococcoides burtonii, T. ferrooxidans and Zea mays. Methanogenic archaea such as M. burtonii [59] inhabit reducing environments, where Eh values of −400 mV or lower have been documented [2]. By contrast, the metal-leaching activity of T. ferrooxidans is associated with an increase in ORP (which carries the same units as Eh) [86]. Compared to the Rubisco from M. burtonii (ZC = −0.147), that from T. ferrooxidans is more oxidized (ZC = −0.110) and is also more stable at higher Eh. The correspondence between redox conditions of habitats and the Eh range where ΔG of protein formation for an organism is minimized relative to homologues from other organisms provides evidence for an environmental signal in the adaptation of the proteins.
The protein from T. ferrooxidans, which has the highest ZC of the four considered, does not have the lowest value of ΔΔG in the most oxidizing (highest Eh) conditions. Instead, the Rubiscos from Bradyrhizobium japonicum and Z. mays, even though they have lower ZC (−0.125 and −0.117, respectively), are calculated to be relatively more stable at the highest Eh values considered. This result shows that comparing values of ZC provides only an approximation of the dependence of the relative energetics of protein formation on redox changes; chemical thermodynamic models integrate more information about reaction stoichiometry and the effects of multiple environmental variables.
Previously, comparisons of Rubisco in Cyanobacteria found at different temperatures revealed an increase in the frequency of hydrophobic amino acids at high temperature that was linked to greater conformational stability of the proteins [87]. It may also be possible to interpret changes of amino acid composition attributed to thermophilic adaptation in terms of the relative energetic costs of synthesis of amino acids [88]. ZC is defined by the amino acid composition of proteins, so changes in amino acid composition that correlate with adaptation to high temperature also imply a correlation with ZC. However, trends in ZC carry implications for the overall energetic demands of forming the proteins as a function of redox conditions. Higher temperatures are often associated with more reducing environments. For example, compared with seawater, activities of dissolved hydrogen in hydrothermal fluids are higher, and mixed hydrothermal-seawater fluids have a reducing potential that favours formation of relatively reduced amino acids [9]. Redox potential is a major variable affecting the energetics of protein formation at different temperatures; therefore, adaptations that minimize biosynthetic costs in high-temperature environments are likely to have both redox- and temperature-dependent characteristics.
4. Conclusion
Comparisons of the average oxidation state of carbon in proteins can be used to visualize the compositional diversity of proteins in relation to redox chemistry in subcellular compartments and external environments. Proteins in different subcellular locations of yeast have a wide compositional variation, spanning the range from relatively reduced membrane proteins to relatively oxidized extracellular proteins. An opposing trend between redox potential and ZC of proteins in the ER and cytoplasm may be an indication of links between protein metabolism and oxidation–reduction reactions involving glutathione and other metabolites, though further work is needed in this area. Compositional divergences among proteins are also apparent in phylogenetic comparisons, showing that more reduced proteins are found in organisms inhabiting anaerobic extracellular or intracellular environments. The possibility that evolutionary adaptation confers energetic savings is supported by calculations that show a correspondence between relative minima of Gibbs energies of overall formation reactions of Rubiscos from different organisms and their environmental redox conditions. These findings are independent from, but complementary to, models for thermophilic adaptation that are based on considerations of conformational stability of proteins.
Owing to the broad distributions of ZC within natural populations of proteins, the interpretation offered here does not presume the existence of definite correlations between ZC and environmental or subcellular conditions. Instead, it is hoped that the comparisons provide a broad context that aids discussion and generation of hypotheses about chemical processes that may play a role in the evolution of the proteins.
Supplementary Material
Acknowledgements
Thanks to Svenja Tulipani and Katy Evans for their comments on an earlier version of the manuscript.
References
- 1.Hewitt LF. 1950. Oxidation–reduction potentials in bacteriology and biochemistry, 6th edn Edinburgh, UK: E. and S. Livingstone Ltd. [Google Scholar]
- 2.Baas Becking LGM, Kaplan IR, Moore D. 1960. Limits of the natural environment in terms of pH and oxidation–reduction potentials. J. Geol. 68, 243–284. ( 10.1086/626659) [DOI] [Google Scholar]
- 3.Ziegler DM. 1985. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu. Rev. Biochem. 54, 305–329. ( 10.1146/annurev.bi.54.070185.001513) [DOI] [PubMed] [Google Scholar]
- 4.Bindoli A, Rigobello MP. 2013. Principles in redox signaling: from chemistry to functional significance. Antioxid. Redox Signal. 18, 1557–1593. ( 10.1089/ars.2012.4655) [DOI] [PubMed] [Google Scholar]
- 5.Knecht E, Aguado C, Cárcel J, Esteban I, Esteve JM, Ghislat G, Moruno JF, Vidal JM, Sáez R. 2009. Intracellular protein degradation in mammalian cells: recent developments. Cell. Mol. Life Sci. 66, 2427–2443. ( 10.1007/s00018-009-0030-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawes EA, Ribbons DW. 1962. The endogenous metabolism of microorganisms. Annu. Rev. Microbiol. 16, 241–264. ( 10.1146/annurev.mi.16.100162.001325) [DOI] [PubMed] [Google Scholar]
- 7.Amend JP, Shock EL. 2001. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25, 2175–2243. ( 10.1016/S0168-6445(00)00062-0) [DOI] [PubMed] [Google Scholar]
- 8.Sleep NH, Bird DK. 2008. Evolutionary ecology during the rise of dioxygen in the Earth's atmosphere. Phil. Trans. R. Soc. B 363, 2651–2664. ( 10.1098/rstb.2008.0018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amend JP, Shock EL. 1998. Energetics of amino acid synthesis in hydrothermal ecosystems. Science 281, 1659–1662. ( 10.1126/science.281.5383.1659) [DOI] [PubMed] [Google Scholar]
- 10.Dick JM, LaRowe DE, Helgeson HC. 2006. Temperature, pressure, and electrochemical constraints on protein speciation: group additivity calculation of the standard molal thermodynamic properties of ionized unfolded proteins. Biogeosciences 3, 311–336. ( 10.5194/bg-3-311-2006) [DOI] [Google Scholar]
- 11.Dick JM, Shock EL. 2011. Calculation of the relative chemical stabilities of proteins as a function of temperature and redox chemistry in a hot spring. PLoS ONE 8, e22782 ( 10.1371/journal.pone.0022782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms MJ, Thornton JW. 2013. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat. Rev. Genet. 14, 559–571. ( 10.1038/nrg3540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterner R, Liebl W. 2001. Thermophilic adaptation of proteins. Crit. Rev. Biochem. Mol. Biol. 36, 39–106. ( 10.1080/20014091074174) [DOI] [PubMed] [Google Scholar]
- 14.Cornish-Bowden A, Cárdenas ML, Letelier J-C, Soto-Andrade J. 2007. Beyond reductionism: metabolic circularity as a guiding vision for a real biology of systems. Proteomics 7, 839–845. ( 10.1002/pmic.200600431) [DOI] [PubMed] [Google Scholar]
- 15.Helgeson HC. 1991. Organic/inorganic reactions in metamorphic processes. Can. Mineral. 29, 707–739. (http://canmin.geoscienceworld.org/content/29/4.toc) [Google Scholar]
- 16.Masiello CA, Gallagher ME, Randerson JT, Deco RM, Chadwick OA. 2008. Evaluating two experimental approaches for measuring ecosystem carbon oxidation state and oxidative ratio. J. Geophys. Res. Biogeosci. 113, G03010 ( 10.1029/2007JG000534) [DOI] [Google Scholar]
- 17.Kroll JH, et al. 2011. Carbon oxidation state as a metric for describing the chemistry of atmospheric organic aerosol. Nat. Chem. 3, 133–139. ( 10.1038/NCHEM.948) [DOI] [PubMed] [Google Scholar]
- 18.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org) [Google Scholar]
- 19.Dick JM. 2008. Calculation of the relative metastabilities of proteins using the CHNOSZ software package. Geochem. Trans. 9, 10 ( 10.1186/1467-4866-9-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JT-F. 1981. Coevolution of genetic code and amino acid biosynthesis. Trends Biochem. Sci. 6, 33–36. ( 10.1016/0968-0004(81)90013-X) [DOI] [Google Scholar]
- 21.Woese CR, Dugre DH, Saxinger WC, Dugre SA. 1966. The molecular basis for the genetic code. Proc. Natl Acad. Sci. USA 55, 966–974. ( 10.1073/pnas.55.4.966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. ( 10.1016/0022-2836(82)90515-0) [DOI] [PubMed] [Google Scholar]
- 23.Shock EL. 1990. Do amino acids equilibrate in hydrothermal fluids? Geochim. Cosmochim. Acta 54, 1185–1189. ( 10.1016/0016-7037(90)90450-Y) [DOI] [Google Scholar]
- 24.LaRowe DE, Helgeson HC. 2006. Biomolecules in hydrothermal systems: calculation of the standard molal thermodynamic properties of nucleic-acid bases, nucleosides, and nucleotides at elevated temperatures and pressures. Geochim. Cosmochim. Acta 70, 4680–4724. ( 10.1016/j.gca.2006.04.010) [DOI] [Google Scholar]
- 25.Amend JP, LaRowe DE, McCollom TM, Shock EL. 2013. The energetics of organic synthesis inside and outside the cell. Phil. Trans. R. Soc. B 368, 20120255 ( 10.1098/rstb.2012.0255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt Consortium. 2012. Reorganizing the protein space at the universal protein resource (UniProt). Nucleic Acids Res. 40, D71–D75. ( 10.1093/nar/gkr981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almén M, Nordström KJV, Fredriksson R, Schiöth HB. 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 7, 50 ( 10.1186/1741-7007-7-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank SA. 2009. The common patterns of nature. J. Evol. Biol. 22, 1563–1585. ( 10.1111/j.1420-9101.2009.01775.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dick JM. 2009. Calculation of the relative metastabilities of proteins in subcellular compartments of Saccharomyces cerevisiae. BMC Syst. Biol. 3, 75 ( 10.1186/1752-0509-3-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425, 686–691. ( 10.1038/nature02026) [DOI] [PubMed] [Google Scholar]
- 31.Cherry JM, et al. 2012. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705. ( 10.1093/nar/gkr1029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. ( 10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13 044–13 053. ( 10.1074/jbc.M312846200) [DOI] [PubMed] [Google Scholar]
- 34.Hu JJ, Dong LX, Outten CE. 2008. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 283, 29 126–29 134. ( 10.1074/jbc.M803028200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer FQ, Buettner GR. 2001. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212. ( 10.1016/S0891-5849(01)00480-4) [DOI] [PubMed] [Google Scholar]
- 36.Morgan B, Ezeriņa D, Amoako TNE, Riemer J, Seedorf M, Dick TP. 2013. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 9, 119–125. ( 10.1038/nchembio.1142) [DOI] [PubMed] [Google Scholar]
- 37.Hwang C, Sinskey AJ, Lodish HF. 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502. ( 10.1126/science.1523409) [DOI] [PubMed] [Google Scholar]
- 38.Birk J, Meyer M, Aller I, Hansen HG, Odermatt A, Dick TP, Meyer AJ, Appenzeller-Herzog C. 2013. Endoplasmic reticulum: reduced and oxidized glutathione revisited. J. Cell Sci. 126, 1604–1617. ( 10.1242/jcs.117218) [DOI] [PubMed] [Google Scholar]
- 39.Lin Y-H, Chien W-S, Duan K-J. 2010. Correlations between reduction-oxidation potential profiles and growth patterns of Saccharomyces cerevisiae during very-high-gravity fermentation. Process Biochem. 45, 765–770. ( 10.1016/j.procbio.2010.01.018) [DOI] [Google Scholar]
- 40.Go Y-M, Jones DP. 2008. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780, 1271–1290. ( 10.1016/j.bbagen.2008.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Queval G, Jaillard D, Zechmann B, Noctor G. 2011. Increased intracellular H2O2 availability preferentially drives glutathione accumulation in vacuoles and chloroplasts. Plant Cell Environ. 34, 21–32. ( 10.1111/j.1365-3040.2010.02222.x) [DOI] [PubMed] [Google Scholar]
- 42.Singh A, Kaur N, Kosman DJ. 2007. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 282, 28 619–28 626. ( 10.1074/jbc.M703398200) [DOI] [PubMed] [Google Scholar]
- 43.van Lith M, Tiwari S, Pediani J, Milligan G, Bulleid NJ. 2011. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J. Cell Sci. 124, 2349–2356. ( 10.1242/jcs.085530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. 2011. Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498. ( 10.1016/j.tplants.2011.05.008) [DOI] [PubMed] [Google Scholar]
- 45.Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135. ( 10.1093/nar/gkr1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vreeland RH, Straight S, Krammes J, Dougherty K, Rosenzweig WD, Kamekura M. 2002. Halosimplex carlsbadense gen. nov., sp. nov., a unique halophilic archaeon, with three 16S rRNA genes, that grows only in defined medium with glycerol and acetate or pyruvate. Extremophiles 6, 445–452. ( 10.1007/s00792-002-0278-3) [DOI] [PubMed] [Google Scholar]
- 47.Tindall BJ, Trüper HG. 1986. Ecophysiology of the aerobic halophilic archaebacteria. Syst. Appl. Microbiol. 7, 202–212. ( 10.1016/S0723-2020(86)80007-8) [DOI] [Google Scholar]
- 48.Cayol J-L, Ollivier B, Patel BKC, Prensier G, Guezennec J, Garcia J-L. 1994. Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. Int. J. Syst. Bacteriol. 44, 534–540. ( 10.1099/00207713-44-3-534) [DOI] [PubMed] [Google Scholar]
- 49.Oren A, Weisburg WG, Kessel M, Woese CR. 1984. Halobacteroides halobius gen. nov., sp. nov., a moderately halophilic anaerobic bacterium from the bottom sediments of the Dead Sea. Syst. Appl. Microbiol. 5, 58–70. ( 10.1016/S0723-2020(84)80051-X) [DOI] [Google Scholar]
- 50.Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314, 312–313. ( 10.1126/science.1130441) [DOI] [PubMed] [Google Scholar]
- 51.Rogers MJ, et al. 1985. Construction of the mycoplasma evolutionary tree from 5S rRNA sequence data. Proc. Natl Acad. Sci. USA 82, 1160–1164. ( 10.1073/pnas.82.4.1160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267 ( 10.1126/science.1134196) [DOI] [PubMed] [Google Scholar]
- 53.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26. ( 10.1038/nrmicro2670) [DOI] [PubMed] [Google Scholar]
- 54.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. 2008. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524. ( 10.1093/jxb/erm361) [DOI] [PubMed] [Google Scholar]
- 55.Nisbet EG, Grassineau NV, Howe CJ, Abell PI, Regelous M, Nisbet RER. 2007. The age of Rubisco: the evolution of oxygenic photosynthesis. Geobiology 5, 311–335. ( 10.1111/j.1472-4669.2007.00127.x) [DOI] [Google Scholar]
- 56.Wang XD, Tang KW, Wang Y, Smith WO., Jr 2010. Temperature effects on growth, colony development and carbon partitioning in three Phaeocystis species. Aquat. Biol. 9, 239–249. ( 10.3354/ab00256) [DOI] [Google Scholar]
- 57.Gosink JJ, Herwig RP, Staley JT. 1997. Octadecabacter arcticus gen. nov., sp. nov., and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from polar sea ice and water. Syst. Appl. Microbiol. 20, 356–365. ( 10.1016/S0723-2020(97)80003-3) [DOI] [Google Scholar]
- 58.Zhang GS, Jiang N, Liu XL, Dong XZ. 2008. Methanogenesis from methanol at low temperatures by a novel psychrophilic methanogen, ‘Methanolobus psychrophilus’ sp. nov., prevalent in Zoige wetland of the Tibetan plateau. Appl. Environ. Microbiol. 74, 6114–6120. ( 10.1128/AEM.01146-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franzmann PD, Springer N, Ludwig W, Demacario EC, Rohde M. 1992. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst. Appl. Microbiol. 15, 573–581. ( 10.1016/S0723-2020(11)80117-7) [DOI] [Google Scholar]
- 60.Jordan DC. 1982. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32, 136–139. ( 10.1099/00207713-32-1-136) [DOI] [Google Scholar]
- 61.Hallmann R, Friedrich A, Koops H-P, Pommerening-Röser A, Rohde K, Zenneck C, Sand W. 1992. Physiological characteristics of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans and physicochemical factors influence microbial metal leaching. Geomicrobiol. J. 10, 193–206. ( 10.1080/01490459209377920) [DOI] [Google Scholar]
- 62.Miedema P. 1982. The effects of low temperature on Zea mays. Adv. Agron. 35, 93–128. ( 10.1016/S0065-2113(08)60322-3) [DOI] [Google Scholar]
- 63.Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, Moyer CL. 2007. A novel lineage of Proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2, e667 ( 10.1371/journal.pone.0000667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alazard D, Dukan S, Urios A, Verhé F, Bouabida N, Morel F, Thomas P, Garcia J-L, Ollivier B. 2003. Desulfovibrio hydrothermalis sp. nov., a novel sulfate-reducing bacterium isolated from hydrothermal vents. Int. J. Syst. Evol. Microbiol. 53, 173–178. ( 10.1099/ijs.0.02323-0) [DOI] [PubMed] [Google Scholar]
- 65.Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47, 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hallberg KB, Lindström EB. 1994. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140, 3451–3456. ( 10.1099/13500872-140-12-3451) [DOI] [PubMed] [Google Scholar]
- 67.Doemel WN, Brock TD. 1970. The upper temperature limit of Cyanidium caldarium. Arch. Mikrobiol. 72, 326–332. ( 10.1007/BF00409031) [DOI] [PubMed] [Google Scholar]
- 68.Norris PR, Clark DA, Owen JP, Waterhouse S. 1996. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology 142, 775–783. ( 10.1099/00221287-142-4-775) [DOI] [PubMed] [Google Scholar]
- 69.Goto E, Kodama T, Minoda Y. 1977. Studies on hydrogen utilizing microorganisms. 5. Isolation and culture conditions of thermophilic hydrogen bacteria. Agric. Biol. Chem. 41, 685–690. ( 10.1271/bbb1961.41.685) [DOI] [Google Scholar]
- 70.Allewalt JP, Bateson MM, Revsbech NP, Slack K, Ward DM. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus Spring microbial mat community of Yellowstone National Park. Appl. Environ. Microbiol. 72, 544–550. ( 10.1128/AEM.72.1.544-550.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamagata Y, Kawasaki H, Oyaizu H, Nakamura K, Mikami E, Endo G, Koga Y, Yamasato K. 1992. Characterization of three thermophilic strains of Methanothrix (‘Methanosaeta’) thermophila sp. nov. and rejection of Methanothrix (‘Methanosaeta’) thermoacetophila. Int. J. Syst. Bacteriol. 42, 463–468. ( 10.1099/00207713-42-3-463) [DOI] [PubMed] [Google Scholar]
- 72.Yamaoka T, Satoh K, Katoh S. 1978. Photosynthetic activities of a thermophilic blue-green alga. Plant Cell Physiol. 19, 943–954. [Google Scholar]
- 73.Shiratori H, Sasaya K, Ohiwa H, Ikeno H, Ayame S, Kataoka N, Miya A, Beppu T, Ueda K. 2009. Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int. J. Syst. Evol. Microbiol. 59, 1764–1770. ( 10.1099/ijs.0.003483-0) [DOI] [PubMed] [Google Scholar]
- 74.Darland G, Brock TD. 1971. Bacillus acidocaldarius sp.nov., an acidophilic thermophilic spore-forming bacterium. J. Gen. Microbiol. 67, 9–15. ( 10.1099/00221287-67-1-9) [DOI] [Google Scholar]
- 75.Balk M, Weijma J, Stams AJM. 2002. Thermotoga lettingae sp nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 52, 1361–1368. ( 10.1099/ijs.0.02165-0) [DOI] [PubMed] [Google Scholar]
- 76.Wu DY, et al. 2009. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS ONE 4, e4207 ( 10.1371/journal.pone.0004207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beeder J, Nilsen RK, Rosnes JT, Torsvik T, Lien T. 1994. Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl. Environ. Microbiol. 60, 1227–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136, 254–261. ( 10.1007/BF00425213) [DOI] [Google Scholar]
- 79.Zillig W, Gierl A, Schreiber G, Wunderl S, Janekovic D, Stetter KO, Klenk HP. 1983. The archaebacterium Thermofilum pendens represents a novel genus of the thermophilic, anaerobic sulfur respiring Thermoproteales. Syst. Appl. Microbiol. 4, 79–87. ( 10.1016/S0723-2020(83)80035-6) [DOI] [PubMed] [Google Scholar]
- 80.Fiala G, Stetter KO, Jannasch HW, Langworthy TA, Madon J. 1986. Staphylothermus marinus sp. nov. represents a novel genus of extremely thermophilic submarine heterotrophic archaebacteria growing up to 98°C. Syst. Appl. Microbiol. 8, 106–113. ( 10.1016/S0723-2020(86)80157-6) [DOI] [Google Scholar]
- 81.González JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. 1998. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough . Extremophiles 2, 123–130. ( 10.1007/s007920050051) [DOI] [PubMed] [Google Scholar]
- 82.Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C . Arch. Microbiol. 145, 56–61. ( 10.1007/BF00413027) [DOI] [Google Scholar]
- 83.Wicken JS. 1987. Evolution, thermodynamics, and information. New York, NY: Oxford University Press. [Google Scholar]
- 84.Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA 99, 3695–3700. ( 10.1073/pnas.062526999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sella G, Hirsh AE. 2005. The application of statistical physics to evolutionary biology. Proc. Natl Acad. Sci. USA 102, 9541–9546. ( 10.1073/pnas.0501865102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lombardi AT, Garcia O., Jr 2002. Biological leaching of Mn, Al, Zn, Cu and Ti in an anaerobic sewage sludge effectuated by Thiobacillus ferrooxidans and its effect on metal partitioning. Water Res. 36, 3193–3202. ( 10.1016/S0043-1354(02)00008-8) [DOI] [PubMed] [Google Scholar]
- 87.Miller SR. 2003. Evidence for the adaptive evolution of the carbon fixation gene rbcL during diversification in temperature tolerance of a clade of hot spring cyanobacteria. Mol. Ecol. 12, 1237–1246. ( 10.1046/j.1365-294X.2003.01831.x) [DOI] [PubMed] [Google Scholar]
- 88.McDonald JH. 2010. Temperature adaptation at homologous sites in proteins from nine thermophile–mesophile species pairs. Genome Biol. Evol. 2, 267–276. ( 10.1093/gbe/evq017) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.