Abstract
Human mesenchymal stem cells (hMSCs) have three key properties that make them desirable for stem cell therapeutics: differentiation capacity, trophic activity, and ability to self-renew. However, current separation techniques are inefficient, time consuming, expensive, and, in some cases, alter hMSCs cellular function and viability. Dielectrophoresis (DEP) is a technique that uses alternating current electric fields to spatially separate biological cells based on the dielectric properties of their membrane and cytoplasm. This work implements the first steps toward the development of a continuous cell sorting microfluidic device by characterizing native hMSCs dielectric signatures and comparing them to hMSCs morphologically standardized with a polymer. A quadrapole Ti-Au electrode microdevice was used to observe hMSC DEP behaviors, and quantify frequency spectra and cross-over frequency of hMSCs from 0.010–35 MHz in dextrose buffer solutions (0.030 S/m and 0.10 S/m). This combined approach included a systematic parametric study to fit a core-shell model to the DEP spectra over the entire tested frequency range, adding robustness to the analysis technique. The membrane capacitance and permittivity were found to be 2.2 pF and 2.0 in 0.030 S/m and 4.5 pF and 4.1 in 0.10 S/m, respectively. Elastin-like polypeptide (ELP-) polyethyleneimine (PEI) copolymer was used to control hMSCs morphology to spheroidal cells and aggregates. Results demonstrated that ELP-PEI treatment controlled hMSCs morphology, increased experiment reproducibility, and concurrently increased hMSCs membrane permittivity to shift the cross-over frequency above 35 MHz. Therefore, ELP-PEI treatment may serve as a tool for the eventual determination of biosurface marker-dependent DEP signatures and hMSCs purification.
I. INTRODUCTION
Human mesenchymal stem cells (hMSCs) are an interesting cell source to researchers because of their regenerative1,2 and immunological properties.3,4 hMSCs are isolated from the bone marrow and other locations in the body5 due to their beneficial properties. They have a high differentiation capacity (adipocytes, chondrocytes, osteoblasts, etc.),6–10 self-renew,11,12 and secrete bioactive molecules (trophic activity).2,3,13,14 hMSCs trophic activity is substantial to their function because signals are sent to surrounding cells triggering tissue repair13 causing apoptosis inhibition, proliferation, and matrix production.13,14 hMSCs are being pursued as a therapeutic option for many chronic diseases such as lupus,15 diabetes mellitus,16 cardiomyopathy,17 liver cirrhosis,18 and Crohn's disease.19
For therapeutic treatments, obtaining hMSCs is a multistep process. hMSCs are a heterogeneous population;20,21 therefore, following bone marrow isolation, they are centrifuged via density gradient solution (step 1), adhered to plastic cell culture dish (step 2), and separated using trypsinization (step 3).17 This method is inefficient and time consuming,22 so other techniques are employed, fluorescent- and magnetic-activated cell sorting (FACS and MACS), which implement unique cell-surface antigens or other recognition elements to tag target cells. This ‘labeling’ of cells alters cellular function, which is not desirable.22–24 FACS and MACS require expensive raw materials and are labor intensive. There is not a unique biosurface marker that distinguishes hMSCs from other cell populations,21,25 the minimum requirements established by the International Society for Cellular Therapy are: (1) plastic adherence (generic cellular property6); (2) positive expression of CD105, CD73, and CD90; (3) negative expression for CD34, CD45, CD11a, CD19, CD14, and human leukocyte antigen-D related (HLA-DR) antigen; and (4) adipogenic, osteogenic, and chondrogenic differentiation potential.8,16 hMSCs have differing biosurface marker expressions on their membrane26 making it difficult to establish a unique biosurface marker for characterization.
Additionally, hMSCs morphology has variations, which can create subpopulations.27 Therefore a label-free, one-step cell purification technique that rapidly purifies hMSCs by accounting for their morphology without altering cellular function is needed.
Dielectrophoresis (DEP) is a separation technique that has potential to overcome the shortcomings of density gradient centrifugation, FACS, and MACS and could provide an electrical biomarker for hMSCs. DEP technologies enable a variety of particle polarizations with nonuniform AC electric fields on microchips28,29 to achieve particle manipulation. DEP has been used to study other cell systems such as red blood cells,30,31 cancer cells,32,33 white blood cells,34 and yeast cells.35,36 The red blood cell DEP studies are important because different ABO-Rh surface antigens were distinguishable from the DEP spectra. The long-term goals of characterizing hMSCs dielectric properties are to discern unique biosurface markers specific to hMSC subcultures.
Advances have been made in the study of stem cells using dielectrophoresis. Flanagan et al.,37 looked at the affects AC electric field expsosure times have on cell viabilty. They found that for human and mouse neural stem/progenitor cells, short exposure times (30 s-1 min) between 0.010–10 MHz did not affect cell viability and metabolic activity. However, longer exposure times, 5–30mins, at the same frequencies induced 20%–40% decrease in cell viability and metabolic activity. These results establish experimental parameters, and aligns with cell viability results found in this work after 90 s AC field exposure hMSC cell viability remained intact. Wu and Morrow38 conducted a one patient clinical study on stromal vascular fraction (SVF) cells separated via DEP. The SVF cells were obtained, separated at 15Vpp, 0.10–1.0 MHz and autologously transplanted to a patients hand to treat muscle atrophy. Positive results were achieved indicating that DEP treated cells were not harmful, and they accelerated the healing process over 3 months (as compared to control). The advantages to coupling DEP with microfluidics are microliter sample size, quick analysis (∼minutes to achieve results), little sample preparation, and minimal waste production. Disadvantages are that extended electric field exposure times (>5 min) negatively affect cells properties and viability.37
II. BACKGROUND
DEP utilizes nonuniform electric fields for cell movement based on the polarizability and dielectric properties (permittivity and conductivity) of their membrane, cytoplasm, and other structurally dominant organelles.39 Cells have distinct dielectric dispersions that can be used as an identification tool for cell purification. A cell's complex permittivity is frequency dependent and characterized by the α, β, and γ dielectric dispersion regions (ωα < ωβ < ωγ).40 At radio frequencies (β-region), 0.010–10 MHz, the dielectric dispersion of cells are affected by their membrane; high frequencies penetrate a cells surface and interogates the internal structure. Therefore, a plethora of information can be obtained about a cell population in the β-region; many researchers complete their experiments within this frequency range.
In the β-region, Maxwell-Wagner interfacial polarizations dominate the DEP response phenomena. Based on permittivity and conductivity, polarized cells will exhibit either positive DEP (pDEP), cells move to areas of high electric field gradient, or negative DEP (nDEP) force; cells move to areas of low electric field gradient.41,42 The DEP force is given by , and this cell motion in the electric field is defined by the Clausius-Mossotti factor, fCM, for core-shell spherical particles,39,41
| (1) |
| (2) |
| (3) |
where R is the radius of the cell cytoplasm and membrane (Rcyto and Rmem), is the complex permittivity, εi is the permittivity, and σi is the conductivity where i = cell, cytoplasm, membrane, or medium. The complex permittivity , is also dependent on the angular frequency ().41 fCM is dependent on the complex permittivity of the cell and the medium, Eq. (2). If a cell experiences pDEP, then fCM (Eq. (1)) is positive, indicating that the cell is more polarizable than the suspending medium, , and the cell moves toward areas of high electric field density.41 For nDEP, the fCM is negative and the cell is less polarizable than the suspending medium, , and the cell moves towards areas of low electric field density.41 When fCM is zero, known as the cross-over frequency fxo,39 cells experience no DEP force (FDEP = 0) as they transition from nDEP to pDEP or pDEP to nDEP. This fxo is an important component of a cell's DEP spectra because the dielectric properties of cells can be estimated along with other data points from the DEP spectra.
The DEP behavior of cells is quantified experimentally by measuring the pDEP and nDEP at specific frequencies within a given buffer solution. The fxo can be extrapolated between the two nearest pDEP and nDEP frequencies. Cells typically display two fxo's, which are dominated by their membrane structure. The low fxo is determined by the cell's size, shape, and membrane with typical values between 0.010–0.10 MHz (β-region), but reported as high as ∼4 MHz.43 The high fxo is dominated by the cell's cytoplasm and is typically above 10 MHz in low conductivity media.44 For the experiments described in this work, the high fxo value falls outside of the range of our function generator (γ-region). Membrane capacitance, Cmem, is a function of fxo and given by44,45
| (4) |
where r is hMSCs radius (μm). Further, the membrane permittivity is proportional to Cmem,
| (5) |
where d is the membrane thickness and ε0 is the vacuum permittivity. Therefore, the fxo and Cmem can be used as indicators of treatment-induced hMSC changes and can be lumped into εmem, a key dielectric property.
To eliminate variation in cell morphology and increase DEP response reproducibility within our hMSC sample population, elastin-like polypeptide (ELP) polyethyleneimine (PEI) was employed. ELP-PEI is a positively charged copolymer that directs cells morphology to a spheroidal shape with a propensity to form aggregates. Similar to mammalian elastin, ELP is synthesized from amino acids including valine, proline, and glycine and has been utilized to induce spheroid formation and differentiation of H35 rat hepatocytes. ELP-PEI has also elucidated affects of free fatty acids and cytokines in 2D and 3D rat hepatoma cell cultures.46,47
In this paper, the first steps toward the development of a continuous cell sorting microdevice were completed by characterizing the dielectric signature of hMSCs and standardizing their morphology with ELP-PEI treatments. We demonstrate that DEP can quantify hMSCs εmem, σmem, and Cmem based on measured DEP spectra. We also characterize the DEP behavior of ELP-PEI treated hMSCs for comparison to the native hMSCs. Determing hMSCs dielectric properties and removing morphology variations is essential to increase reproducibility and magnify different membrane biosurface markers.
III. MATERIALS AND METHODS
A. Microdevice fabrication
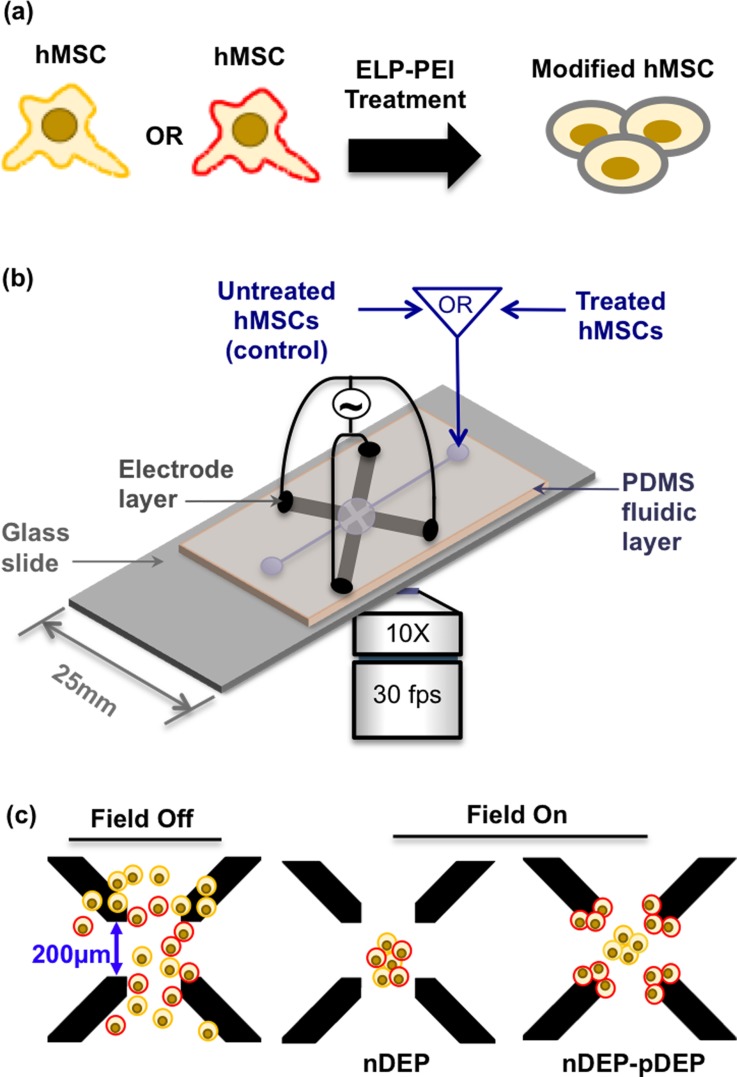
The microdevice in Figure 1(b) was fabricated using prior published techniques30 with 100μm wide and 200μm spaced electrodes. A 2000μm deep by 3000μm wide microwell made with polydimethylsiloxane (PDMS) was sealed on top of the quadrapole array. Figure 1(b) shows the quadrapole Ti-Au electrode microdevice setup.
FIG. 1.
(a) hMSCs cartooned to emphasize the differing cell membrane biosurface markers and thus properties. Cartooned spheroidal morphology after ELP-PEI treatment. (b) Quadrapole microdevice with 200 μm spaced Ti-Au electrodes used for DEP experiments. (c) hMSCs in device with field off, and the DEP response observed with field on. At a single frequency, both nDEP and pDEP were observed due to differing membrane biosurface markers within an hMSCs population.
B. hMSC cell culture
Nitrogen stored bone marrow derived hMSCs were recovered by thawing cells in 37 °C water bath. 500 ml alpha minimum essential medium (αMEM) with L-glutamine, without ribonucleosides or deoxyribonucleosides (Catalog # M20350, Atlanta Biologicals, Atlanta, GA) was completed with 50 ml fetal bovine serum (FBS, Catalog # S11550, Atlanta Biologicals), 3 ml L-glutamine (200 mM in 0.85% NaCl, Catalog # 25030081, Life Technologies, Carlsbad, CA), and 3 ml penicillin/streptomycin (Catalog # 15140122, Life Technologies). Thawed hMSCs were placed in 15 ml complete αMEM in sterile cell culture flask and incubated overnight at 37 °C, 5%CO2. After 24 h, phosphate buffer solution (PBS, Catalog # 6508-OP, EMD Millipore, Chicago, IL), 0.25% trypsin/1 mM EDTA (Catalog # 25200072, Life Technologies), and complete αMEM were warmed to 37 °C. Old complete αMEM was removed from the flask and attached hMSCs were washed with 10 ml PBS. 4 ml trypsin was added to the flask and hMSCs were incubated at 37 °C, 5% CO2 for 3–4 min for full cell detachment (monitored with microscope). After detachment, 5 ml of complete αMEM was added to flask to inactivate trypsin, and the hMSC suspension was placed in sterile centrifuge tubes. hMSCs were centrifuged for 10 min at 494 relative centrifugal force (RCF). The supernatant was removed and hMSCs were resuspended in 5 ml of fresh completed αMEM. 1 ml of hMSC suspension was placed in new cell culture flask with 15 ml complete αMEM for the next passage. hMSCs were incubated until 90% confluent (∼5 days) and passaged following the same trypsin detachment procedure. Multiple flasks of hMSCs were cultured for DEP tests, with an extra hMSC flask continually passaged every 4–5 days for subsequent DEP testing. DEP tests were completed when cells reached 106 cell/ml concentration (∼5–7 days).
C. DEP solution preparation
1M NaOH (Catalog #S318–500, Fisher Scientific, Waltham, MA) solution, 3M salt stock solution, 4% bovine serum albumin (BSA, Catalog # A7906–100 G, Sigma Aldrich, St. Louis, MO)-Epure H2O pH 10 solution, and 0.030 S/m and 0.10 S/m dextrose solutions were prepared to complete hMSCs DEP testing. The 3M salt stock was comprised of 1M KH2PO4 (Catalog # 7100, Mallinckrodt Chemicals, St. Louis, MO), 1M NaCl (Catalog # 7581, Macron Chemicals, Swedesboro, NJ), and 1M K2HPO4 (Catalog # BDH0266–500G, VWR, West Chester, PA) in 20 ml Epure H2O. A 4% BSA pH 10 solution was utilized to pretreat the microdevice surface. The 0.030 S/m dextrose solution was prepared to 0.30 M with 0.30 M dextrose (Catalog # D9434–500G, Sigma Aldrich) and 9.1 × 10−4M salt stock, then balanced to pH 7 with 1M NaOH (as necessary). 4% BSA was added and thoroughly vortexed. The 0.10 S/m dextrose solution was prepared to 0.28M with 0.27M dextrose and 7.5 × 10−3M salt stock, then balanced to pH 7 with 1 M NaOH (as necessary). 4% BSA was added and thoroughly vortexed. Final dextrose solution conductivities were verified using a conductivity meter (AB30, Fisher Scientific).
D. DEP experimental procedure
For DEP tests, centrifuged untreated hMSCs were resuspended to a concentration of 1.3 × 106cells/ml in either 0.030 S/m or 0.10 S/m dextrose solution. The microdevice was presoaked in 4% BSA-Epure H2O adjusted to pH 10 to prevent cell adhesion. The hMSC dextrose suspension was loaded into the quadrapole chamber using a micropipette. A function generator (Agilent 33250 A, Agilent, Santa Clara, CA) was connected to the microdevice via copper leads (Figure 1(b)). Static frequency measurements were completed using 10Vpp AC sine wave, 0.010 MHz to 35 MHz for 90 s. More than 300 untreated hMSCs (n > 300) were tested at each static frequency. Untreated hMSCs static DEP responses were compared to static measurements of ELP-PEI treated hMSCs. To achieve ELP-PEI hMSC treatment, ELP-PEI was synthesized as described in Refs. 46 and 47 and dissolved in Epure H2O (5 mg/ml) in a sterile centrifuge tube. 5 ml ELP-PEI Epure H2O solution was added to a vented cell culture flask and dried for 48 h at 37 °C. After 48 h, excess water was removed and the coated flask was decontaminated under UV light for 5min. 90% confluent hMSCs were trypsinized and placed in ELP-PEI coated flask for 24 h. hMSC images were taken before and after ELP-PEI treatment. After 24 h, ELP-PEI treated hMSCs were tested within the DEP microdevice using AC fields and static frequencies.
E. Data acquisition
30 fps video recordings using LabSmith SVM Synchronized Video Microscope 10× objective (LabSmith, Livermore, CA, USA) were collected. DEP behaviors of the untreated hMSCs and ELP-PEI treated hMSCs were quantified with ImageJ (NIH, Bethesda, MD) by recording cell position and number after 90 s in the AC electric field. Percent cell responses (Rc) were calculated by tabulating the number of cells, ni, exhibiting nDEP and pDEP behaviors (where i = nDEP, pDEP, pDEP-nDEP) divided by the total number of cells, nT, tested.
| (6) |
Rc was plotted as a stacked column chart to show hMSC nDEP, pDEP, and pDEP-nDEP behavior at individual static frequencies, Figures 3(a) and 3(b). Next, Rc was translated to a traditional DEP spectra plot (inspired by Ref. 48) by scaling the nDEP responses by fCM,min and pDEP responses by fCM,max calculated via Eqs. (1)–(3) for each conductivity. This scaling method was used because hMSCs measured DEP responses were binary displaying either nDEP or pDEP, or a combination of the two. The binary responses were weighted due to Rc, the percentage of cells displaying each phenomena. The transition from nDEP to pDEP, indicative of the low fxo, did not appear experimentally as a binary response but instead gradually transitioned. This scaling method was introduced to further enhance the nDEP to pDEP transition. Thus, scaling of the cells displaying nDEP by fCM,min enables the nDEP portion of the DEP curve to be reconstructed, while scaling with fCM,max of cells tallied to display pDEP enables the pDEP portion of the curve to be reconstructed. The scaled equations were
| (7) |
| (8) |
At 0.030 S/m, fCM,min = −0.45 and fCM,max = 0.72, and for 0.10 S/m, fCM,min = −0.49 and fCM,max = 0.36. If the magnitude of the scaled Rc,nDEP was larger than the scaled Rc,pDEP, then Rc,nDEP was selected, if not, then Rc,pDEP was selected. The DEP spectra transitions from nDEP to pDEP when Rc,nDEP Rc,pDEP = 0.50 (at the fxo) as illustrated in Figure 3(c).
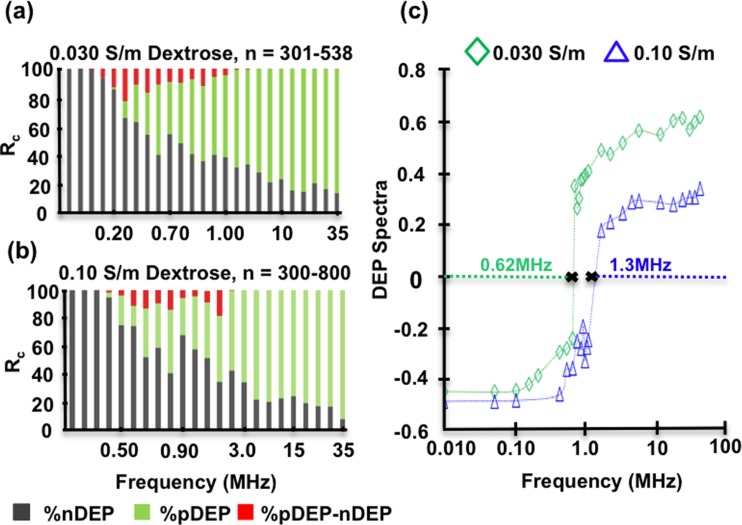
FIG. 3.
(a) Percent cell response Rc for untreated hMSCs in 0.030 S/m dextrose solution. (b) Rc for untreated hMSCs in 0.10 S/m dextrose solution. For both conductivities, nDEP dominates lower frequencies while pDEP dominates higher frequencies. (c) Data from (a) and (b) translated into DEP response spectra for untreated hMSCs at 0.030 S/m and 0.10 S/m. This format best illustrates 0.030 S/m fxo estimated as 0.62 MHz and 0.10 S/m fxo estimated as 1.3 MHz.
F. Comparison to model
First, Cmem was calculated from fxo to obtain parameters for εmem via Eqs. (4) and (5). Next, these initial dielectric parameter estimates were refined via a parametric analysis. The resulting DEP spectra for untreated and ELP-PEI treated hMSCs were separately fit to the core-shell spherical DEP polarization model. The starting literature values were εmem = 6.5,49–53 σmem = 10−6,49–52 εcyto = 60,49,50,52,53 σcyto = 0.63 S/m.49–53 These dielectric parameters (εmem, σmem, εcyto, and σcyto) were iteratively adjusted in Eqs. (1)–(3) starting from the literature values and values estimated from fxo, then varied to obtain the best curve fit to experimental data. R = 20 μm was used for each optimization.
IV. RESULTS AND DISCUSSION
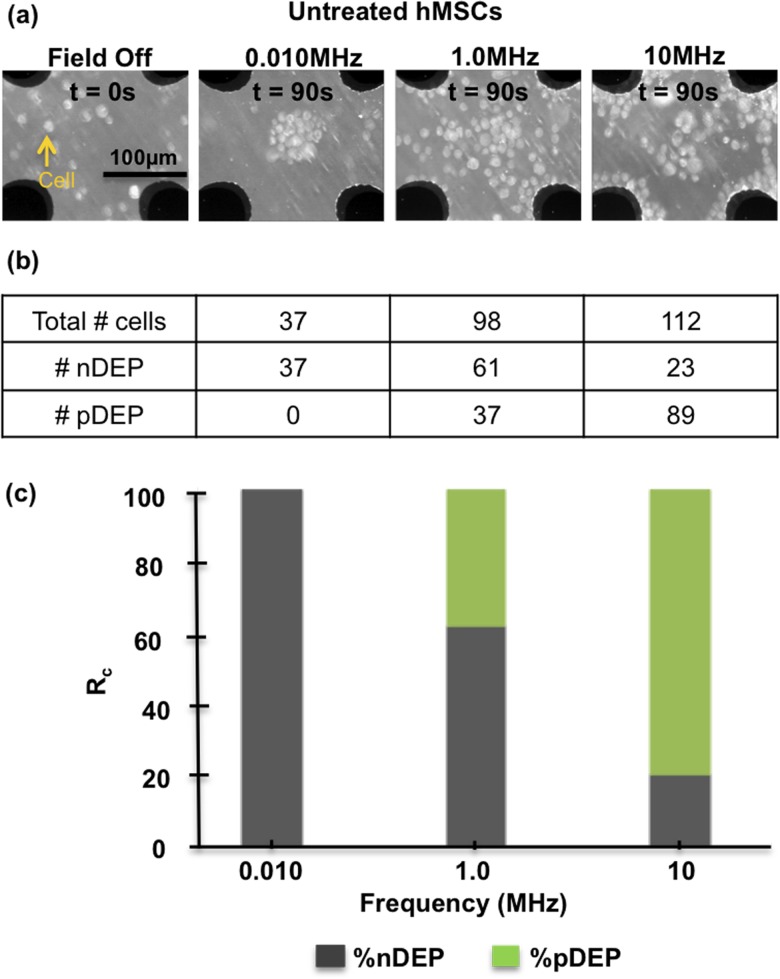
Static frequency measurements were completed from 0.010 MHz to 35 MHz to establish hMSCs DEP response spectra to then calculate untreated and ELP-PEI treated hMSCs dielectric properties. Static measurements were completed at 10 Vpp for 90 s at each individual frequency and Figure 2 illustrates the hMSC spatial cell counts and subsequent Rc at 0.010, 1.0, and 10 MHz for untreated hMSCs. The notation nDEP-pDEP indicates cells that initially exhibit pDEP and then transition to nDEP, a behavior that is pronounced near fxo (see Figure 3).
FIG. 2.
(a) Microscope images of untreated hMSCs DEP response with the field off and at 0.010, 1.0, and 10 MHz after 90 s in 0.10 S/m dextrose solution. At 0.010 MHz, hMSCs only display nDEP and display both nDEP and pDEP at 10 MHz. (b) Table of total cells for each image. (c) DEP responses tabulated as % cell response (Rc) into a stacked column chart. nDEP dominates at 0.010 MHz and at 10 MHz pDEP is 79% dominant.
To examine the medium conductivity dependence, hMSCs were tested in 0.030 S/m and 0.10 S/m at fixed 10Vpp for 90 s at each individual frequency. Experiments were rerun until n > 300 cells and were tabulated at each frequency. Figures 3(a) and 3(b) show the stacked column representation of hMSCs DEP response in 0.030 S/m and 0.10 S/m, respectively. In 0.030 S/m media, nDEP (gray) is the dominant behavior up to ∼0.65 MHz, at which point the DEP behavior transitions to pDEP (green) dominance. Individual cells experiencing both pDEP and nDEP (red) within one experimental run are seen between 0.10–2 MHz. In 0.10 S/m media, nDEP is dominant until ∼1 MHz and then transitions to pDEP behavior following a similar trend as the lower conductivity but with the frequency response shifted higher. Figure 3(c) illustrates the translation of the Rc responses to hMSCs DEP spectra at 0.030 S/m and 0.10 S/m, respectively. The DEP spectra curve shift is more apparent with this plot format. Thus, hMSCs display different fxo's dependent on the conductivity of the suspending solution, 0.030 S/m fxo = 0.62 MHz and 0.10 S/m fxo = 1.3 MHz. The frequency shift of 0.68MHz is consistent with other medium conductivity experiments in the literature.39 It was also observed that the DEP force weakened (slower hMSC movement) as the fxo was approached.
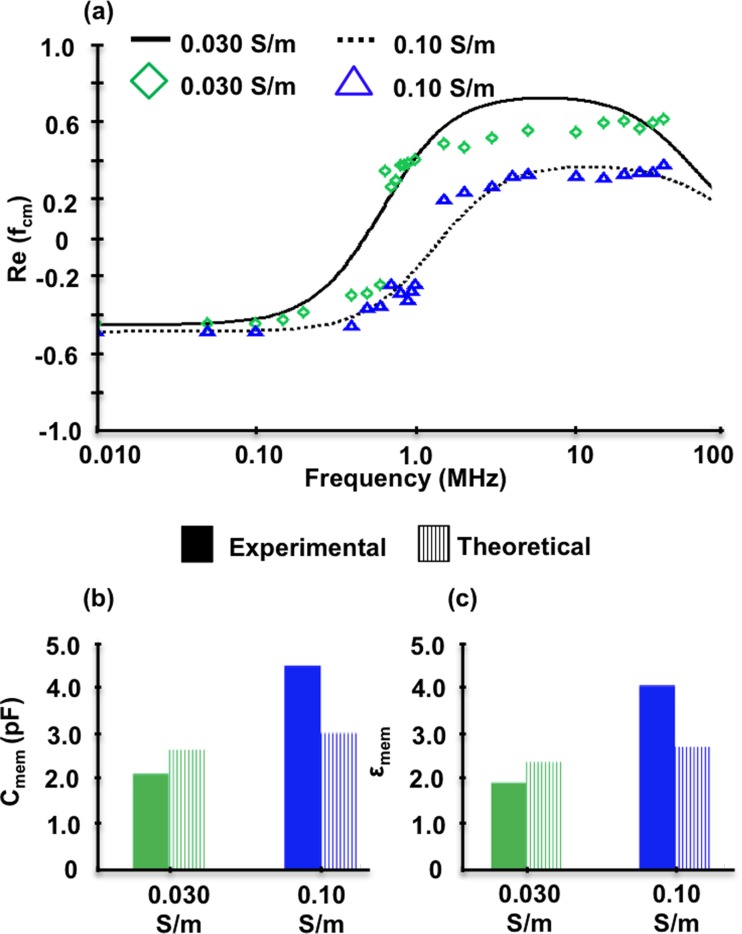
The experimental data in Figure 3 was compared to the core-shell spherical DEP polarization model (Eqs. (1)–(3)) as illustrated in Figure 4. The experimental fxo's were used in Eqs. (4) and (5) to calculate Cmem and εmem of 2.2 pF and 2.0, 4.5 pF and 4.1 for 0.030 S/m and 0.10 S/m, respectively. These initial dielectric parameter estimates were further optimized to fit the experimental data via a parametric analysis using the entire tested frequency range. The εmem's, starting model parameters specified in materials and methods were utilized along with the DEP response over the tested frequency range (0.01–35 MHz) to optimize εmem = 0.79, σmem = 10−6, εcyto = 60, and σcyto = 0.50 S/m for 0.030 S/m as shown in Figure 4(a) solid curve and Table I. The 0.030 S/m experimental data (open green diamonds) has good agreement, with the core-shell spherical polarization model with an overall 23% error. For 0.10 S/m, model parameters were separately optimized to εmem = 1.1, σmem = 10−6, εcyto = 60, and σcyto = 0.50 S/m (dashed curve in Figure 4(a) and Table I) and agreed well with the experimental data (open blue triangles), with an overall 17% error.
FIG. 4.
(a) Untreated hMSC DEP responses at 0.030 and 0.10 S/m compared to the core-shell spherical model. (b) hMSC membrane capacitance and (c) membrane permittivity based on experimentally determined cross-over frequency.
TABLE I.
Summary of literature and optimized parameters for core-shell spherical DEP polarization model for untreated and treated hMSCs in 0.030 S/m and 0.10 S/m.
| εmem | σmem (S/m) | εcyto | σcyto (S/m) | |
|---|---|---|---|---|
| Literature range | 6.5–1149–53 | 10−3–10−8 49–52 | 50–10049,50,52,53 | 0.30–0.8849–53 |
| (starting model values) | (0.50) | (10−6) | (60) | (0.63) |
| Untreated | 0.79 | 10−6 | 60 | 0.50 |
| 0.030 S/m | ||||
| Untreated | 1.1 | 10−6 | 60 | 0.50 |
| 0.10 S/m | ||||
| ELP-PEI treated | 0.050 | 10−6 | 60 | 6.3 × 10−3 |
| 0.10 S/m |
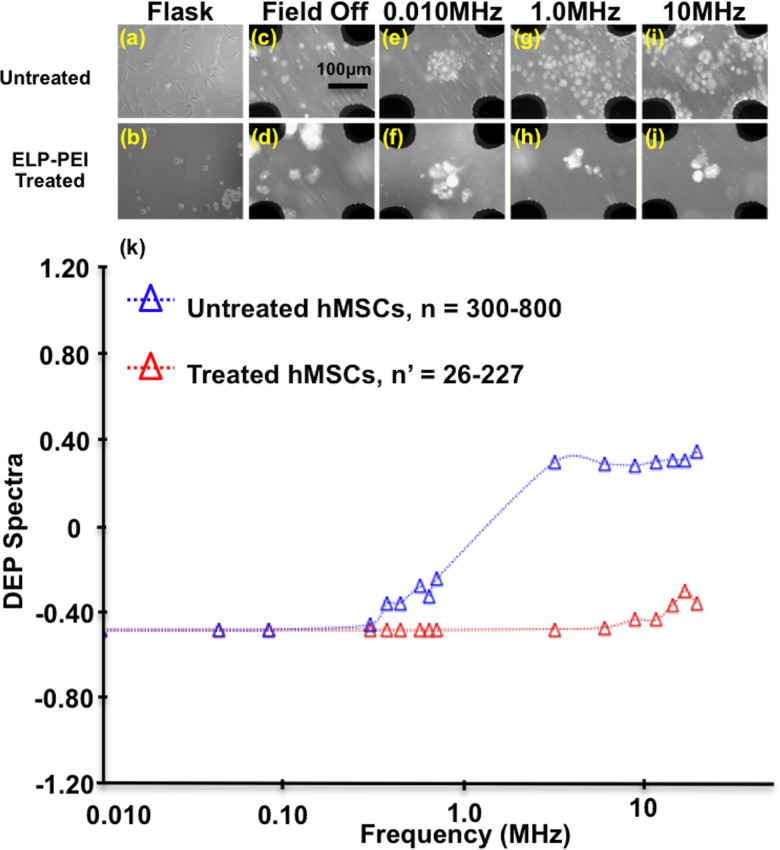
In order to elucidate the morphology contribution to the DEP response spectra, ELP-PEI treated hMSCs were examined following the same experimental procedures in 0.10 S/m. Figure 5 compares ELP-PEI treated to untreated hMSCs experimental images to DEP response spectra. Figures 5(a) and 5(b) illustrate the morphological changes between untreated hMSCs attached to the bottom of the cell culture flask and ELP-PEI treated hMSCs spheroidal cells and cell aggregates, which do not adhere to the flask bottom. These images indicate that treated hMSCs formed spheroidal aggregates consistent with liver cells, H35 rat hepatoma cells, and 3T3 mouse cells described in previous literature;46,47 therefore the ELP-PEI treatment on the hMSCs was successful in producing spheroidal morphology. Figures 5(c) and 5(d) compare untreated and ELP-PEI treated hMSCs within the DEP microdevice with the electric field off. With the field off, hMSC suspensions were randomly distributed within the microwell, although cell aggregates were apparent with the treated hMSCs. Figures 5(e) and 5(f) compare untreated and ELP-PEI treated hMSCs at 0.010 MHz after 90 s in the AC field. Both the untreated and treated hMSCs display nDEP behavior, but the treated hMSCs were more aggregated. At increased frequencies, 1.0MHz (Figure 5(g) and 5(h)) and 10 MHz (Figure 5(i) and 5(j)), untreated hMSCs exhibit both nDEP and pDEP behavior while ELP-PEI treated hMSCs only exhibit nDEP.
FIG. 5.
Untreated hMSCs (first row) compared to ELP-PEI treated hMSC spheroidal morphology (second row), both in 0.10 S/m dextrose solution. (a) Untreated hMSCs in cell culture flask, (b) treated hMSCs after 24 h in cell culture flask, (c) untreated and (d) treated hMSCs in microdevice with field off where black regions are the quadrapole electrodes. Untreated and treated hMSCs at 10 Vpp and 0.010 MHz ((e) and (f)), 1.0 MHz ((g) and (h)), and 10 MHz ((i) and (j)). (k) DEP response spectra of untreated and ELP-PEI treated hMSCs in 0.10 S/m. Untreated cells transition from nDEP to pDEP at ∼1.3 MHz, while treated cells predominantly exhibit nDEP behaviors and formed spheroidal aggregates.
Figure 5(k) shows the corresponding DEP spectra for untreated hMSCs (open red triangles) and treated hMSCs (open red triangles). At high frequencies (>10MHz), ELP-PEI treated hMSCs exhibit primarily nDEP with minor pDEP behaviors. However, untreated hMSCs reveal nDEP behavior at lower frequencies, and pDEP at higher frequencies with the fxo ∼ 1.3 MHz. There was not a fxo value within the tested frequency range for treated hMSCs, so Cmem and εmem were estimated to be >0.13 pF and >0.12 with fxo > 35 MHz. For untreated hMSCs, cell counts were at least 300 for each static frequency tested. Treated hMSCs were aggregated spheroids, therefore the cell counts are denoted n’. Lower n’ numbers were tolerated at lower frequencies where nDEP clearly dominated the responses.
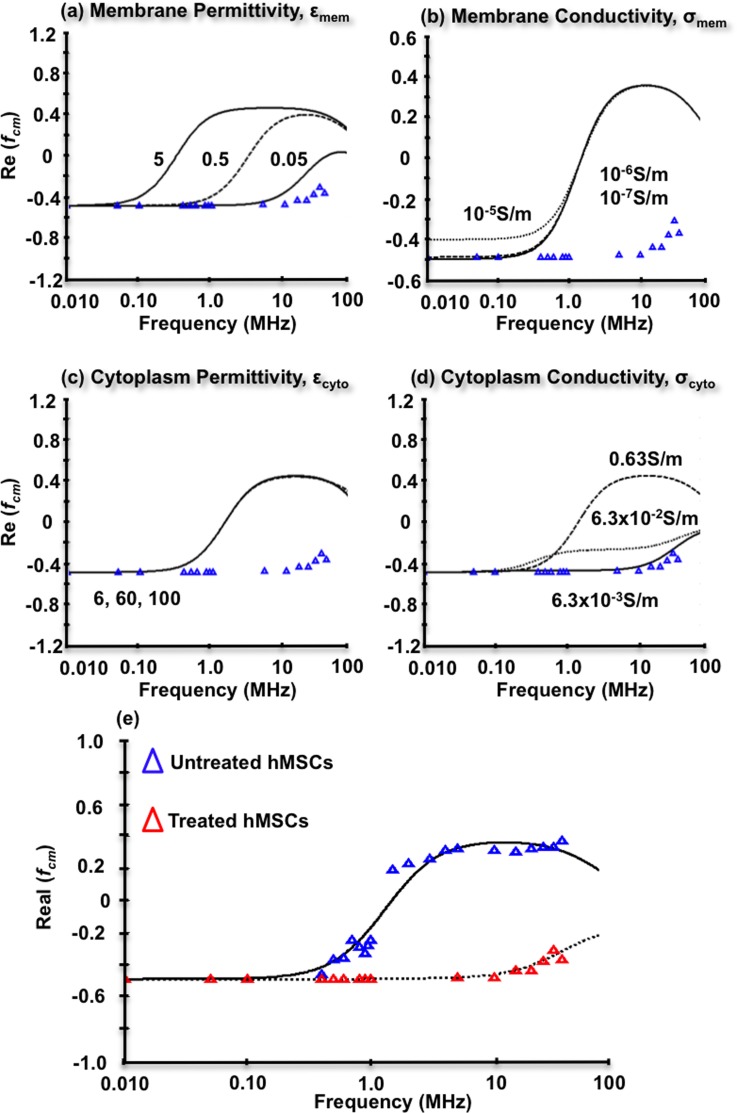
The ELP-PEI treated hMSCs DEP experimental data at 0.10 S/m were also fit to the core-shell spherical DEP polarization model as shown in Figure 6. Because fxo could not be directly measured, model parameters εmem, σmem, εcyto, and σcyto were systematically adjusted from theoretical values to arrive at best fit parameters in Figures 6(a) through 6(d), respectively. For each individual parameter optimization, the other three literature values were held constant as illustrated in Table I. Membrane permittivity adjustments in Figure 6(a) revealed that decreasing εmem from 549–53 to 0.050 better matched the experimental data. Membrane conductivity adjustments in Figure 6(b) from σmem 10−5 down to 10−7 (Refs. 49–52) had very little impact on achieving model/data agreement. Cytoplasm permittivity adjustments in Figure 6(c) from εmem 6 up to 100 (Refs. 49, 50, 52, and 53) also had very little impact on achieving model/data agreement. Cytoplasm conductivity adjustments in Figure 6(d) from σcyto 0.63 S/m (Refs. 49–53) down to 6.3 × 10−3 S/m, achieved good agreement with experimental data, and may account for the nDEP behavior displayed by the ELP-PEI treated hMSCs. Figure 6(e) compares the optimized model parameters for untreated and treated hMSC. These values are simultaneously summarized in Table I. All final model parameters lie within the range of previously reported literature values, except the membrane permittivity and cytoplasm conductivity, which were slightly lower than the lowest reported value of 6.5 (Refs. 49–53) and 0.63 S/m,49–53 respectively.
FIG. 6.
Key parameter optimizations for the core-shell spherical DEP polarization model for ELP-PEI treated hMSCs at 0.10 S/m. (a) Decreases from literature values in membrane permittivity better capture ELP-PEI treated hMSCs nDEP behaviors. (b) Increases or decreases in membrane conductivity do not contribute to the experimentally observed nDEP. (c) Decreases in cytoplasm permittivity also do not contribute. (d) Decreases in cytoplasm conductivity may contribute to the nDEP behavior with σcyto = 6.3 × 10−3 S/m fitting well. (e) Model prediction for both untreated and treated hMSCs at 0.10 S/m using all optimal parameters for each. Agreement between the model and the data is quite good with an overall 2.5% error.
In review, there was an observable conductivity dependence on the DEP behavior of untreated hMSCs in 0.030 S/m and 0.10 S/m. This dependency was most discernable near the hMSCs frequency transition region from nDEP to pDEP and the fxo value, both were encompassed in the range of 0.61 and 1.4 MHz. Maxwell-Wagner interfacial polarization mechanisms dominate in the frequency range from 0.010 MHz to 10 MHz in the β-dispersion region,40 such that an observed fxo is influenced by the ionic interactions of the hMSCs membrane with the surrounding medium. As the frequency increases in the MHz range, this interfacial polarization transitions to being dominated by the membrane permittivity, which was more consistent with the optimized model parameters in Table I. Thus, at 0.10 S/m, the hMSCs membrane was electrically more permissive to the ions driven by the AC field evidenced by the shift in the DEP spectra to higher frequencies. These experimental findings were consistent with the core-shell DEP polarization model optimization whereby the cytoplasm properities did not vary, but the membrane parameters did (Table I, summary). These experimental observations were corroborated with increases in the calculated membrane capacitance, 2.2 pF and 4.5 pF at 0.030 S/m and 0.10 S/m, respectively. Small deviations between the experimental data and the core-shell spherical DEP polarization model likely occurred because the model does not account for all of the hMSCs morphological and biosurface marker complexities.
To reduce the hMSCs morphological complexity, the cells were treated with ELP-PEI to yield a uniform spheroidal cell shape. The ELP-PEI treatment concurrently caused some hMSCs to form spheroidal aggregates consistent with previous work.46,47 For low frequencies, 0.010–0.10 MHz, untreated and ELP-PEI treated hMSCs similarly exhibited nDEP, suggesting comparable membrane resistance at those frequencies. Above 0.10 MHz, ELP-PEI treated hMSCs only exhibited nDEP behavior, which deviated from untreated hMSCs that transitioned to pDEP behavior. There are a few possible scenarios to explain this behavior: (1) the ELP-PEI may be incorporating itself into the hMSCs membrane, (2) small concentrations of ELP-PEI could be present in the dextrose solution surrounding the cells, and/or (3) cells aggregating into spheroids could shield membrane polarization effects.
Within hypothesis (1) context, that ELP-PEI may have intercolated into the hMSCs membrane, the core-shell spherical DEP polarization model key parameters in Figure 6 and Table I illustrate that decreases in hMSCs membrane permittivity were necessary to match experimental data (εmem is 0.050). A decrease in cytoplasm conductivity from 0.63 S/m to 6.3 × 10−3 S/m also supports this claim (Figure 6(d)), but since the DEP tests were completed in the β-dispersion region, it is unlikely that the DEP responses recorded were representative of the cytoplasm structure inside of hMSCs.
Because the ELP-PEI treatment caused hMSCs to form spheroidal aggregates, size effects were examined. Using the treated hMSCs starting model parameters (Table I) in the core-shell spherical DEP polarization model, the size would have to reduce to 0.75 μm to exhibit nDEP behavior similar to the observed experimental data. This was not feasible for the treated cells and their aggregates, which had average sizes of 17 ± 4 μm and 43 ± 16 μm, respectively. The aggregates increased hMSCs overall size, and it might be inferred this would increase the overall DEP force on the aggregates. Recent work in our group has shown this is not the case due to the electric field shielding effects, groups of cells experience a slightly smaller DEP force than a single cell.54 However, there was no evidence that this size increase would prevent interrogation of biosurface markers present on hMSCs membranes. Data did show strong media/membrane polarization behaviors, which are prerequisites for surface biosurface marker-induced stem cell separations in nonlinear AC electric fields. We conclude that the ELP-PEI treatment did achieve standardization of the hMSCs morphology and simultaneously altered DEP behavior.
V. CONCLUSIONS
hMSCs are an important cell system due to their differentiation capacity, trophic activity, and their ability to self-renew. Their therapeutic potential is currently being explored through clinical trials to treat lupus, Crohn's disease, and diabetes mellitus. For successful and effective stem cell therapies, hMSCs purification after bone marrow isolation could be considerably improved to increase selective recovery of the most highly viable cells. Separation techniques like FACS and MACS are expensive and alter cellular function and viability due to antigen tagging. User-friendly DEP microdevices have previously demonstrated rapid cell separation and no loss in cell viability with low voltage AC electric fields.37 hMSCs populations are heterogeneous with varied morphology, so the development of a continuous cell sorting microdevice needs to concurrently control for and accurately measure (1) hMSCs dielectric signatures and (2) cell morphology variations. The latter was accomplished in this work by morphologically standardizing the cell population with ELP-PEI. Additionally, hMSC population heterogeneity correlates with biosurface marker expression predisposing cells for adipocytes, chondrocytes, and osteoblasts. Thus, after morphological standardization, precise measurements of DEP dielectric signatures may facilitate biosurface marker-dependent cell separations.
hMSCs have a distinct dielectric dispersion defined by internal structures, cell membrane and cytoplasm. hMSCs membrane mostly affects the DEP response at radio frequencies in the β-dispersion region; the radio frequencies polarize the hMSCs surface, facilitating a DEP force along the field gradient, and thus allowing interrogation of the cell membrane dielectric properties. The DEP results in this work reveal that the conductivity of hMSCs suspending solution is critical to the membrane polarization. Higher conductivity solutions increase the membrane permittivity shifting the DEP spectra to higher frequencies for 0.10 S/m than 0.030 S/m. This translates to a membrane capacitance increase from 2.2 pF for 0.030 S/m to 4.5 pF for 0.10 S/m. Similarly, the membrane permittivity increases from 2.0 for 0.030 S/m to 4.1 for 0.10 S/m. These dielectric properties were extracted using a robust technique that combined the fxo approach with a systematic parameteric study to fit a core-shell model to hMSCs DEP spectra over the tested frequency range. The solution conductivity dependence was also apparent at the fxo for hMSCs; 0.030 S/m yields a lower fxo at 0.62 MHz and 0.10 S/m yields a lower fxo at 1.3 MHz. Frequency ranges tested did not enable measurement of the higher fxo. This dielectric signature is unique for hMSCs because typical low fxo's for other cell systems fall in the range of 0.010–0.10 MHz.44 These differing DEP spectra could be harnessed for hMSC cell separations from these other cell systems. Complicating any separation endeavor is that the untreated hMSC DEP responses at static frequencies varied within a single culture, thus reproducible separations need to consider and control for biosurface marker expression.
The ELP-PEI treatment successfully standardized hMSCs population morphology, although spheroidal cell aggregates were concurrently observed. The ELP-PEI treatment concurrently increased the DEP response reproducibility. This standardization occurred because the polymer interacted with the cell membrane and shifted hMSCs polarization behaviors. Only nDEP responses were observed from 0.010–10 MHz, which was substantially different from untreated hMSCs. The spheroidal cell aggregates present after ELP-PEI treatment should increase the DEP force, which is proportional to cell radius cubed. However, the strong media/membrane polarization behaviors observed suggest that this hMSC cell system is a good candidate for future surface biosurface marker-induced stem cell separations. With optimization of the ELP-PEI treatment, detection of molecular level differences in hMSCs may be realizable to aid understanding of biological functions and cell population purification for stem cell therapies.
ACKNOWLEDGMENTS
The research completed in this paper was supported by Michigan Tech's Finishing Fellowship, Kings-Chavez-Parks Future Faculty Fellowship, and partially supported by the National Science Foundation (Award No. 1033525) to A. V. J. The authors would like to thank Dr. Caryn Heldt for the use of her cell culture hood, and both Lijun Zhang and Qi Xing for assistance with hMSC samples.
References
- 1.Kolios G. and Moodley Y., Respiration 85, 3 (2013). 10.1159/000345615 [DOI] [PubMed] [Google Scholar]
- 2.Caplan A., J. Cell. Physiol. 213, 341 (2007). 10.1002/jcp.21200 [DOI] [PubMed] [Google Scholar]
- 3.Caplan A. and Correa D., Cell Stem Cell 9, 11 (2011). 10.1016/j.stem.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F., JSM Biotechnol. Biomed. Eng. 1, 1012 (2013). [PMC free article] [PubMed] [Google Scholar]
- 5.Bobis S., Jarocha D., and Majka M., Folia Histochem. Cytobiol. 44, 215 (2006). [PubMed] [Google Scholar]
- 6.Stewart M. C. and Stewart A. A., Vet. Clin. N. Am-Equine. 27, 243 (2011) 10.1016/j.cveq.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 7.Wu H. W., Lin C. C., and Lee G. B., Biomicrofluidics 5, 013401 (2011). 10.1063/1.3528299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salem H. K. and Thiemermann C., Stem Cells 28, 585 (2010) 10.1002/stem.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao F., Veldhuis J. J., Duan Y., Yang Y., Christoforou N., Ma T., and Leong K. W., Mol. Ther. 18, 1010 (2010). 10.1038/mt.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao F., Grayson W. L., Ma T., and Irsigler A., J. Cell. Physiol. 219, 421 (2009). 10.1002/jcp.21688 [DOI] [PubMed] [Google Scholar]
- 11.Fuchs E. and Chen T., EMBO J. 14, 39 (2013) 10.1038/embor.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He S., Nakada D., and Morrison S. J., Annu. Rev. Cell Dev. Biol. 25, 377 (2009). 10.1146/annurev.cellbio.042308.113248 [DOI] [PubMed] [Google Scholar]
- 13.Caplan A. and Dennis J. E., J. Cell. Biochem. 98, 1076 (2006). 10.1002/jcb.20886 [DOI] [PubMed] [Google Scholar]
- 14.Wu L., Prins H. J., Helder M. N., van Blitterswijk C. A., and Karperien M., Tissue Eng., Part A 18, 1542 (2012). 10.1089/ten.tea.2011.0715 [DOI] [PubMed] [Google Scholar]
- 15.Parekkadan B. and Milwid J. M., Annu. Rev. Biomed. Eng. 12, 87 (2010). 10.1146/annurev-bioeng-070909-105309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G. P., Ge M. H., Shu Q., Rojas M., and Xu J., World J. Pediatr. 9, 197 (2013). 10.1007/s12519-013-0425-1 [DOI] [PubMed] [Google Scholar]
- 17.Williams A. R. and Hare J. M., Circ. Res. 109, 923 (2011). 10.1161/CIRCRESAHA.111.243147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamnig A. and Lepperdinger G., Can. J. Physiol. Pharmacol. 90, 295 (2012). 10.1139/y11-109 [DOI] [PubMed] [Google Scholar]
- 19.Lanzoni G., Alviano F., Marchionni C., Bonsi L., Costa R., Foroni L., Roda G., Belluzzi A., Caponi A., Ricci F., Tazzari P. L., Pagliaro P., Rizzo R., Lanza F., Baricordi O. R., Pasquinelli G., Roda E., and Bagnara G. P., Cytotherapy 11, 1020 (2009). 10.3109/14653240903253840 [DOI] [PubMed] [Google Scholar]
- 20.Kolf C. M., Cho E., and Tuan R. S., Arthritis Res. Ther. 9, 204 (2007). 10.1186/ar2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pevsner-Fischer M., Levin S., and Zipori D., Stem Cell Rev. Rep. 7, 560 (2011). 10.1007/s12015-011-9229-7 [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Gonzalez M., Vazquez-Villegas P., Garcia-Salinas C., and Rito-Palomares M., J. Chem. Technol. Biotechnol. 87, 2 (2012). 10.1002/jctb.2723 [DOI] [Google Scholar]
- 23.Flanagan L. A., Lu J., Wang L., Marchenko S. A., Jeon N. L., Lee A. P., and Monuki E., Stem Cells 26, 656 (2008). 10.1634/stemcells.2007-0810 [DOI] [PubMed] [Google Scholar]
- 24.Vykoukal J., Vykoukal D. M., Freyberg S., Alt E. U., and Gascoyne P. R. C., Lab Chip 8, 1386 (2008). 10.1039/b717043b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosna F., Sensebe L., and Krampera M., Stem Cells Dev. 19, 1449 (2010). 10.1089/scd.2010.0140 [DOI] [PubMed] [Google Scholar]
- 26.Mafi P., Hindocha S., Mafi R., Griffin M., and Khan W. S., Open Orthop. J. 5, 253 (2011). 10.2174/1874325001105010253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haasters F., Prall W. C., Anz D., Bourquin C., Pautke C., Endres S., Mutchler W., Docheva D., and Schieker M., J. Anat. 214, 759 (2009). 10.1111/j.1469-7580.2009.01065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pethig R., Menachery A., Pells S., and Sousa P. De, J. Biomed. Biotechnol. 2010, 182581 (2010) 10.1155/2010/182581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C., Khoshmanesh K., Mitchell A., and Kalantar-zadeh K., Anal. Bioanal. Chem. 396, 401 (2010). 10.1007/s00216-009-2922-6 [DOI] [PubMed] [Google Scholar]
- 30.Leonard K. M. and Minerick A. R., Electrophoresis 32, 2512 (2011). 10.1002/elps.201100077 [DOI] [PubMed] [Google Scholar]
- 31.Srivastava S. K., Daggolu P. R., Burgess S. C., and Minerick A. R., Electrophoresis 29, 5033 (2008). 10.1002/elps.200800166 [DOI] [PubMed] [Google Scholar]
- 32.Salmanzadeh A., Kittur H., Sano M. B., Roberts P. C., Schmelz E. M., and Davalos R. V., Biomicrofluidics 6, 024104 (2012). 10.1063/1.3699973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmanzadeh A., Romero L., Shafiee H., Gallo-Villanueva R. C., Stremler M. A., Cramer S. D., and Davalos R. V., Lab Chip 12, 182 (2012). 10.1039/c1lc20701f [DOI] [PubMed] [Google Scholar]
- 34.Khoshmanesh K., Nahavandi S., Baratchi S., Mitchell A., and Kalantar-zadeh K., Biosens. Bioelectron. 26, 1800 (2011). 10.1016/j.bios.2010.09.022 [DOI] [PubMed] [Google Scholar]
- 35.Patel S., Showers D., Vedantam P., Tzeng T. R., Qian S. Z., and Xuan X. C., Biomicrofluidics 6, 034102 (2012). 10.1063/1.4732800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razak M. A. A., Hoettges K. F., Fatoyinbo H. O., Labeed F. H., and Hughes M. P., Biomicrofluidics 7, 064110 (2013). 10.1063/1.4842395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J., Barrios C. A., Dickson A. R., Nourse J. L., Lee A. P., and Flanagan L. A., Integr. Biol. 4, 1223 (2012). 10.1039/c2ib20171b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu A. Y. and Morrow D. M., J. Transl. Med. 10, 99 (2012). 10.4252/wjsc.v3.i4.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pethig R., Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams T. N. G., Leonard K. M., and Minerick A. R., Biomicrofluidics 7, 064114 (2013). 10.1063/1.4833095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gagnon Z. R., Electrophoresis 32, 2466 (2011). 10.1002/elps.201100060 [DOI] [PubMed] [Google Scholar]
- 42.Hawkins B. G., Huang C., Arasanipalai S., and Kirby B. J., Anal. Chem. 83, 3507 (2011). 10.1021/ac2002017 [DOI] [PubMed] [Google Scholar]
- 43.Alshareef M., Metrakos N., Perez E. J., Azer F., Yang F., Yang X., and Wang G., Biomicrofluidics 7, 011803 (2013). 10.1063/1.4774312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salmanzadeh A., Elvington E. S., Roberts P. C., Schmelz E. M., and Davalos R. V., Integr. Biol. 5, 843 (2013). 10.1039/c3ib00008g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gascoyne P. R. C., Shim S., Noshari J., Becker F. F., and Stemke-Hale K., Electrophoresis 34, 1042 (2013). 10.1002/elps.201200496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner P. A., Harris L. M., Purser C. A., Baker R. C., and Janorkar A. V., Biotechnol. Bioeng. 111, 174 (2014). 10.1002/bit.25099 [DOI] [PubMed] [Google Scholar]
- 47.Janorkar A. V., Harris L. M., Murphey B. S., and Sowell B. L., Biotechnol. Bioeng. 108, 1171 (2011). 10.1002/bit.23025 [DOI] [PubMed] [Google Scholar]
- 48.Rozitsky L., Fine A., Dado D., Nussbaum-Ben-Shaul S., Levenberg S., and Yossifon G., Biomed. Microdevices 15, 859 (2013). 10.1007/s10544-013-9773-9 [DOI] [PubMed] [Google Scholar]
- 49.Cho S. and Thielecke H., Microelectron. Eng. 85, 1272 (2008). 10.1016/j.mee.2008.01.004 [DOI] [Google Scholar]
- 50.Biasio A. Di and Cametti C., Colloid Surf. B 84, 433 (2011). 10.1016/j.colsurfb.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 51.Gagnon Z., Gordon J., Sengupta S., and Chang H. C., Electrophoresis 29, 2272 (2008). 10.1002/elps.200700604 [DOI] [PubMed] [Google Scholar]
- 52.Wu L., Yung L. Y. L., and Lim K. M., Biomicrofluidics 6, 014113 (2012). 10.1063/1.3690470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao K., Bai W., and Mi H., Bioelectrochem. 69, 49 (2006). 10.1016/j.bioelechem.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 54.Moncada-Hernandez H., Nagler E., and Minerick A. R., Electrophoresis 35, 1803 (2014). 10.1002/elps.201300636 [DOI] [PubMed] [Google Scholar]