Abstract
Despite the myriad of soft lithography based micropatterning methods available to researchers, it is still challenging to define small features (10–100 μm) that are spaced far apart (1–10 mm). In this report, we describe a combined microfluidic-microstencil patterning method that can produce multifunctional substrates of small features, O(10 μm), with a large pitch, O(1 mm). In that, we fabricate microstencils using an UV curable polyurethane (Norland Optical Adhesive 81) with dense arrays of 10–100 μm holes. Overlaying arrays of microfluidic channels over these microstencils allow for the control of the spacing between features and the ability to pattern multiple substrates. We show that this method is capable of patterning soluble proteins, fibrillar insoluble collagen, liposomes, cells, and nanoparticles. We demonstrate the utility of the method by measuring platelet adhesion under flow to three adhesive proteins (insoluble fibrillar collagen, laminin, and reconstituted acid solubilized collagen fibers) in a single assay.
INTRODUCTION
Micropatterning biomolecules and cells is a useful tool for investigating how the spatial presentation of surface bound molecules and cells regulates cell adhesion, rolling, and spreading, as well as cell-cell communication, cancer metastasis, and outside-in signaling.1–3 Some patterning methods require specialized equipment such pin tools,4 dip-pen lithography,5 inkjet printer,6 and laser writers.7 Alternatively, soft-lithography based methods including microfluidic patterning,8 microcontact printing,2,9 and microstenciling10–13 are attractive to research laboratories because of their low cost and simple fabrication. Here, we report a soft-lithography method that combines microstencils and microfluidic patterning as an alternative to microcontact printing.
Microcontact printing is a versatile method for patterning proteins and lipids at the 100 μm to 100 nm scales but suffers from some important limitations. First, it is difficult to achieve consistent features,14 because the amount of material transferred depends on difficult to control parameters such as relative dryness and the applied pressure (rate and total).15 Second, the drying of material on stamps may cause inhomogeneity within patterns.13 Third, it is difficult to pattern fibrillar proteins such as native collagen fibers,16 which are the preferred substrate for some assays such as platelet adhesion.17 Finally, when using polydimethylsiloxane (PDMS) stamps, the size and spacing between features are limited due to mechanical instabilities such as roof collapse, buckling, and lateral collapse.9,18 To avoid these instabilities, patterned features need to be quite dense and feature size needs to be relatively uniform. This limits the use of microcontact stamping in flow-based adhesion assays because of cross-talk between adjacent features. For example, in the context of blood clot formation, the transport of thrombin from one feature to the next causes an increasing build-up of fibrin on each feature in the direction of flow.19 Alternatives to PDMS or supporting PDMS stamps on rigid materials can increase the spacing.20–22
Some of the limitations of microcontact printing have been addressed with microstencils, which are thin films of polymers (photoresists, PDMS, polyurethanes, and parylene) with holes in them that define the pattern.12,23–25 A solution of biomolecules or suspension of cells is incubated directly on the microstencil. The microstencil is then removed leaving behind a pattern defined by the holes. Since adhesion of the molecule is based on adsorption, the transfer process is easier to control than by microcontact stamping by changing the solute concentration and incubation time. Moreover, the molecules remain hydrated throughout the patterning process, thus preventing denaturation and potential loss in protein activity. Microstencils can be fabricated using soft lithography techniques. For example, a double molding process in which a PDMS master is used to define the holes in a polyurethane (Norland Optical Adhesive, NOA) membrane by capillary filling was recently reported.13 However, this method suffers similar limitations of PDMS microcontact stamping in that features need to be sufficiently close together to avoid mechanical instabilities in the PDMS master.9,14 In addition, microstencils do not easily lend themselves to pattern different materials (e.g., two different proteins) on the same substrate.
Microfluidic patterning uses microfluidic channels to define patterns and has the advantages of using small volumes of reagents, the ability to pattern several different molecules on a single substrate with high spatial precision, and is amenable to pattern fibrillar proteins and cells. One disadvantage of microfluidic patterning is that limited geometries are available for patterning, namely, rectangular strips. This is problematic in flow-based cell adhesion assays because the patterned molecules span the entire width of a channel. Since shear stress distribution across a rectangular channel varies with position, cells tend to accumulate in the low shear stress regions in the corner of channels. This non-uniform shear stress distribution can confound data analysis of cell adhesion assays.26 Previous reports have used a combination of microcontact printing and microfluidic patterning to yield multifunctional surfaces.27 In that, hydrophobic silanes were microcontact printed to define features with a polyethylene glycol (PEG) background, followed by microfluidic patterning of proteins that selectively adsorbed to the hydrophobic features. Here, we take a similar approach using microstencils and microfluidic patterning that provides the additional flexibility of patterning fibrillar proteins, cells, and particles without the need for modulating surface chemistry.
In the Methods section, we describe how to fabricate large (25 mm × 25 mm) NOA microstencils combined with microfluidic patterning to provide control over both the size and spacing between patterned features. The NOA microstencils allow for the creation of discrete patterns down to 10 μm. The microfluidic channels allow for control of spacing between features and the ability to pattern multiple molecules on a single substrate. We demonstrate the utility of this approach by patterning proteins, lipids, nanoparticles, and cells, as well as by measuring platelet adhesion over three different adhesive proteins in a single assay.
MATERIALS AND METHODS
Materials
Anhydrous toluene, octadecyltrichlorosilane (OTS), fluorescein isothiocyanate (FITC), Texas Red®–2-sulfonamidoethyl methanethiosulfonate, dimethyl sulfoxide (DMSO), bovine serum albumin (BSA), collagen type I (acid soluble), laminin from human placenta, glutaraldehyde 50%, calcium chloride, magnesium chloride, sodium chloride, potassium chloride, disodium phosphate, dipotassium phosphate, 2-[4-(1-hydroxyethyl)piperazin-1-l]ethanesulfonic acid (HEPES), methanol, hydrochloride acid (37N), Pluronics F-127, gelatin (tissue culture grade), heparin sodium salt from porcine intestinal mucosa, trypsin, and endothelial cell growth supplement (ECGS), tetra-ethyl-ortho-silicate (TEOS), ammonium hydroxide ACS grade, and ethanol anhydrous (200 proof) were all obtained from Sigma Aldrich (St. Louis, MO, USA). Native collagen (type I) from equine tendons was purchased from Chrono-Log (Havertown, PA). SYLGARD® 184 Silicone Elastomer Kit was purchased form Dow Corning (Midland, MI, USA). NOA 81was purchased from Norland Products (NJ, USA). F-12K basal media and fetal bovine serum (FBS) were purchased from ATCC (Manassas, VA). L-α-phosphatidylcholine (PC) and L-a-phosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, USA) and Texas red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (DHPE) was purchased from Invitrogen (Carlsbad, CA, USA). KMPR 1050 was purchased from MicroChem Corporation (Westborough, MA). (Tridecafluoro-1,1,2,2-tetrahydooctyl) trichlorosilane was purchased from Gelest (Morrisville, PA). Anti-human CD41-Pacific Blue IgG was purchased from Southern Biotech (Birmingham, AL). Human recombinant tissue factor (TF) was purchased from Sekisui Diagnostics (Lexington, MA). HEPES Buffered Saline (HBS) was prepared by dissolving 150 mM NaCl and 25 mM HEPES in deionized (18 MΩ) water. 10× Phosphate Buffered Saline (PBS) was prepared by 1.37M NaCl, 27 mM KCl, 100 mM Na2HPO4, and 18 mM KH2PO4 in deionized water. 10× PBS was diluted to 1× using deionized water prior to use. Recalcification buffer was prepared by dissolving 75 mM CaCl2 and 35 mM MgCl2 in HBS. Wash buffer was prepared by dissolving 1 U/ml heparin, 7.5 mM CaCl2, and 3.5 mM MgCl2 in HBS. pH of the buffers was adjusted to 7.4 using 1M HCl and 1M NaOH. Buffers were filtered through a 0.22 μm pore size syringe filter (Millipore, Billerica, MA) prior to use.
Fabrication of NOA 81 microstencils
The fabrication of NOA 81 microstencils was achieved by a double molding process as depicted in Fig. 1. Features were defined by photolithography on silicon wafers using negative tone photoresist KMPR 1050 (height ∼42 μm). Wafers were incubated with (tridecafluoro-1,1,2,2-tetrahydooctyl) trichlorosilane for at least 4 h under ∼10 psi vacuum. Next, PDMS was molded off of the wafer with 1:10 catalyst:base and cured at 80 °C for 4 h. PDMS substrates consisted of an array of pillars 90 μm in diameter and a pitch of 225 μm or staggered array (600 μm between two spots in one column and 300 μm between columns). Following removal from the silicon, PDMS substrates were cleaned in 1M HCl, acetone, and ethanol for 5 min each and dried in an oven at 80 °C for at least 2 h before use. Glass slides were cleaned in 1:1 HCl:methanol (37N HCl) for 2 h, rinsed thoroughly in deionized water (18 MΩ), and dried at 80 °C before use. NOA 81 (10–60 μl) was pipetted onto the cleaned glass slide. The PDMS master was placed on the NOA 81 and gently pressed down by hand (Fig. 1(a)). The assembly was then placed under water with a 200 g weight on the slide to promote contact between the PDMS substrate and the glass slide to ensure through holes (Fig. 1(b)). NOA 81 was polymerized by UV exposure (365 nm) for 3–10 min under water at a distance of 1–5 cm. The resultant NOA stencil was then rinsed thoroughly with ethanol to remove any unpolymerized NOA. After using microstencils for patterning, they were sonicated in 1M HCl and ethanol for 5 min each to remove proteins that may be present on the membrane and stored in a humid environment at 4 °C for reuse. When reused, the microstencils were sonicated in ethanol for 7 min prior to use to remove any debris that may have adhered during storage.
FIG. 1.
Fabrication and patterning using the microfluidic-microstencil method. (a) A clean glass slide with liquid NOA81 pipetted on the surface is brought in contact with a PDMS master with desired features. (b) Uniform pressure is applied to the assembly by placing a 200 g weight on the PDMS master and placed under water. The assembly is then exposed to UV radiation from the bottom of the assembly to cure the NOA. (c) The PDMS master is removed leaving a microstencil through the holes. (d) The microstencil is then placed on the substrate to be patterned. (e) A microfluidic device is placed on the stencil, reversibly vacuum bonded, and the channels are filled with the desired solution or suspension. The different colors represent different solutions/suspensions. Vacuum bonding is achieved by connecting a vacuum pump to the device via a tubing. (f) Upon removal of the stencil and the microfluidic device, the patterned surface is revealed on the substrate.
Cell lines and cell culture
Human Umbilical Vein Endothelial Cells (HUVECs) were purchased from ATCC (CRL-1730) and were grown as recommended by ATCC in F-12 K complete medium under humidified conditions at 37 °C and 5% CO2. The base medium was supplemented with 0.1 mg/ml heparin, 0.03–0.05 mg/ml ECGS, and 10% FBS to make the complete medium.
Microfluidic-microstencil patterning
NOA microstencils were wetted with HBS for 5 min to fill the wells. A PDMS microfluidic patterning device with three channels (500 μm wide, 50 μm high, and 50 mm long; channels separated by 5 mm) was then held in place on the stencil using vacuum-assisted bonding (Fig. 1(d)). A vacuum chamber surrounds the channels. This chamber is a recess in the microfluidic device and is connected to a vacuum pump by tubing. When the pump is turned on, the reduced pressure bonds the device to the stencil. Following the patterning, the vacuum is released and the device is removed.26 Solutions of molecules to be patterned were manually drawn into the channels with a 1 ml syringe and allowed to incubate for 1–2 h (Fig. 1(e)). Following incubation, unbound biomolecules were washed with HBS. The device and the microstencil are then removed to reveal patterned surfaces (Fig. 1(f)).
Scanning electron microscopy
Microstencils were placed on a silicon wafer and sputter-coated with ∼10 nm of gold. Images were captured at an accelerating voltage of 1.5 kV and 6 mm working distance (JEOL 7000 Field Emission Scanning Electron Microscopy (FESEM), Hitachi, Tokyo, Japan).
Patterning of proteins
OTS modified glass slides as previously described1 or clean unmodified glass slides were used to pattern proteins. Protein solutions (10–150 μl) were either pipetted directly on the membrane or perfused through microfluidic patterned devices, and left in a sealed humidified dark box at 4 °C overnight or at room temperature for 1–2 h. After incubation, excess protein from the surface of the microstencil was removed and the microstencil was gently peeled off the glass surface. The slide was then washed with HBS to remove excess and unbound protein. The pH of type I acid solubilized collagen was raised to 7.4 using 0.1M NaOH and diluted with PBS to the desired concentrations (1 mg/ml) immediately prior to patterning.1 Patterned proteins were visualized using either an Olympus BX60 microscope equipped with 5× (NA 0.15), 10× (NA 0.30), 20×(NA 0.40), and 50× (NA 0.50) objectives, a dark field filter, and a digital camera (Infinity 2, Lumenera, Ontario, Canada) or an Olympus IX 81 equipped with equipped with a motorized stage 2× (NA 0.06), 10× (NA 0.3), 20× (NA 0.45), and 40× (NA 0.6) objectives and a 16 bit CCD camera (Hamamatsu, San Jose, CA). BSA was conjugated to FITC for visualization as previously described.1
Patterning silica beads
Silica beads were synthesized as previously described.28 Briefly, 3 ml of TEOS was added drop wise to a mixture of ethanol (30.8 ml) and ammonium hydroxide (13 ml) at room temperature with constant stirring. After 2 h of stirring, the contents were centrifuged and the pellet was resuspended in ethanol and centrifuged again. The pellet was washed in HBS twice to remove any residual reactants. This process yields silica beads 800 nm in diameter as determined by dynamic light scattering (Zetasizer Nano, Brookhaven Instruments). Beads suspended in HBS (∼10 μl) were perfused through the microfluidic patterning device and allowed to incubate in a dark humidified box for 2 h at room temperature. Following incubation, the channel was rinsed with HBS to remove unbound beads. The device and the microstencil were gently peeled from the glass slide.
Patterning lipid bilayers
Multilamellar lipid vesicles made from PC, PS, and DHPE (PC:PS:DHPE molar ratios 80:19:1) with and without recombinant human TF were prepared as previously described.29 Suspensions of vesicles (∼10 μl) were perfused through the microfluidic patterning device and allowed to incubate in a dark humidified box for 2 h at room temperature. Following incubation, excess lipids were removed and the channel was washed with HBS to remove unbound lipids. The device and the microstencil were gently peeled from the glass slide. The surface was again rinsed with HBS and kept hydrated for imaging.
Patterning cells
The NOA microstencil was placed over a clean glass slide coated with gelatin by applying a layer of 0.2% tissue culture grade gelatin and then placing a coverslip to ensure a uniform thickness. Confluent HUVEC cells were gently detached from the tissue culture flask using trypsin-EDTA, washed, and resuspended at 250 000/ml in complete media. The channels of the microfluidic device were primed with complete media to allow entry of cells into the channel. After 45 min of incubation, the device and microstencil were removed. The slide with patterned cells was washed gently with HBS to remove any unbound cells and placed in complete media at culture condition. The slides were then allowed to incubate for an additional 4 h to allow the cells to attach and spread over the gelatin coated glass slide.
Blood collection and preparation
Blood was collected from healthy donors by venipuncture into vacutainer tubes containing 3.2% sodium citrate. Phlebotomy was conducted in accordance with the Declaration of Helsinki and in accordance with the institutional review board of the University of Colorado, Boulder. Donors had not consumed alcohol within 48 h prior to the draw, nor had they taken any prescription or over-the-counter drugs within the previous 10 days. The first tube of blood collected was treated as waste to eliminate any activated platelets due to venipuncture.
Platelet adhesion flow assays
Clean glass was patterned with fibrillar collagen, laminin, and acid soluble collagen using the microfluidic-microstenciling technique described above for the flow assay. Texas Red (1–5 μg/ml) was added to the protein solutions to aid in visualizing the patterned features. To prevent denaturation of proteins, the surfaces were not allowed to dry rather were kept hydrated with a small amount of buffer at all times. Following patterning, slides were incubated with 5% BSA for 30 min at room temperature. Texas Red was removed below detectable levels during the BSA incubation (pacification step to prevent non-specific adhesion) and the subsequent wash (to remove excess BSA). Prior to the flow assay, the whole blood and a fluorescently labeled anti-human CD41 antibody (1:100 blood:antibody) were incubated together in a 37 °C water bath for 10 min to resensitize platelets.30 Immediately before the assay, divalent cations were reintroduced into the whole blood by adding 9:1 whole blood:recalcification buffer (7.5 mM CaCl2 and 3.5 mM MgCl2 final concentration). The flow rate of the whole blood was controlled with a syringe pump (PHD2000, Harvard Apparatus, Holliston, MA) to achieve the desired wall shear rates. Wall shear rates were related to volumetric flow rate by
where Q is the volumetric flow rate, γwall is the wall shear rate, and ε is the channel aspect ratio (width/height). The microfluidic channel used for the flow assay was 250 μm wide and 50 μm high and was placed perpendicular to the long arrays of protein spots defined by the microfluidic patterning steps. Whole blood was perfused through the channel at a wall shear rate of 100 s−1 for 7 min. This wall shear rate is the characteristic of venous blood flow and is recommended by Biorheology Subcommittee of the International Society of Thrombosis and Hemostasis.31 Following the flow assay, blood was removed from channels by perfusing a washing buffer and the clots were fixed by perfusing 2% glutaraldehyde in HBS for 5 min. Both washing and fixing steps were performed at the same wall shear rates as the platelet adhesion assay.
Image analysis
To quantify the fluorescence intensity within each patterned feature, the mean gray values was calculated using ImageJ.32 The image processing consists of the following steps: (1) The background was subtracted using a rolling ball filter in the Subtract Background function. (2) The background-subtracted image was converted to a binary image by manually setting the threshold value to include all features in the Threshold function. Note that the same threshold value was used for all analysis on a given substrate. (3) The Set Measurements features were set to redirect measurements of mean gray value to the original grayscale image. Then, the Analyze Particle function was applied on the binary image. This procedure allows for calculation of the mean gray value within each feature in the original grayscale image as defined by the binary image. Intensity histograms were obtained by measuring the line intensity across the diameter of the spot.
Statistical analysis
Statistical analysis was performed using R version 3.0.2 (http://www.R-project.org/). Where appropriate students' t-test was used at a significance level of 0.05. Values reported are mean ± standard deviation unless noted otherwise.
RESULTS AND DISCUSSION
Characterization of microstencils and fidelity of patterning
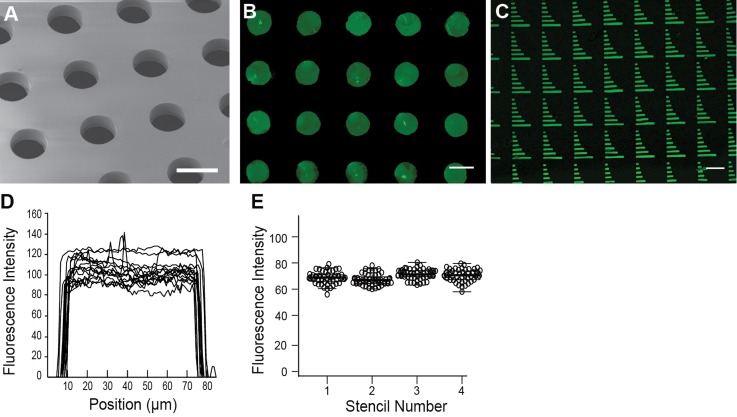
Fig. 1 shows the workflow for the fabrication of microstencils and subsequent patterning via microfluidic channels. Microstencils were fabricated by placing a PDMS master over liquid, uncured NOA81, and then applying a 200 g weight atop the master. The NOA was cross-linked with UV light while the entire assembly was immersed in water. The water immersion ensures that the NOA stencil does not bond to the glass slide. Fig. 2(a) shows a scanning electron micrograph of the NOA membrane through holes. Fig. 2(b) shows a pattern of BSA-FITC generated from a microstencil with 90 μm holes spaced 225 μm apart (center-to-center). The thickness of the microstencils was 35 ± 2 μm. An array of different sized rectangles shows that holes as small as 10 μm can be successfully fabricated by this technique (Fig. 2(c)). Line intensity profiles across 25 spots on each microstencil were used as a measure to assess homogeneity of protein within spots (Fig. 2(d)). The fluorescence intensity was relatively constant across each spot. The patterned spots are 71.2 ± 5 μm in diameter, thus there is approximately an 18 μm reduction in the diameter of spots from the original PDMS master pillars. This reduction in size is likely a result of by a thin ring of NOA at the bottom of the stencil holes that is visible in electron microscopy images (Fig. 2(a)). To test the repeatability of patterning across different microstencils, we measured the average fluorescence intensity of 90 spots from four separate stencils (Fig. 2(e)). The results show repeatable patterning within a stencil and between stencils.
FIG. 2.
Characterizing the microstencil. (a) Electron micrograph of the NOA membrane through the holes. Scale bar = 100 μm. (b) BSA-FITC was patterned using the NOA microstencil into spots (scale bar = 100 μm) and (c) bars ranging from 150 μm to 10 μm in length (scale bar = 100 μm). (d) Line fluorescence intensity profiles measured across 25 spots. (e) Average fluorescence intensity of spots on four stencils (n = 90 for each stencil).
Patterning of proteins, lipids, and cells using microfluidic-microstencil method
Adsorption using microstencils alone can provide dense arrays of biomolecules and cells (Fig. S1),36 similar to previous reports using microstencils.13 Overlaying microfluidic channels on the microstencils during the adsorption step allow for greater control over the spatial presentation of patterned features. Here, microfluidic devices were reversibly vacuum-bonded to the microstencils followed by the introduction of protein solutions or suspensions of nanoparticles or cells. Direct adsorption onto stencils without microfluidic channels required ∼150 μl of solution/suspension. Patterning with microfluidic channels only required ∼10 μl of fluid/suspension. The flexibility of this approach was demonstrated by patterning a variety of biomolecules and cells (Fig. 3).
FIG. 3.
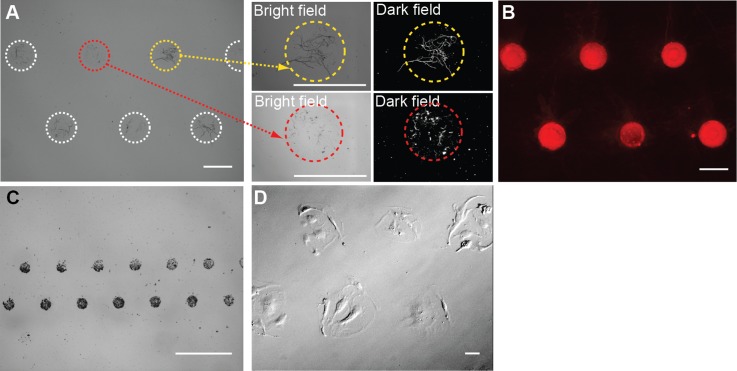
Patterning insoluble proteins, particles, and cells using the microfluidic-microstencil method. (a) Insoluble fibrillar collagen fibers, (b) Texas–Red conjugated lipids, (c) 800 nm silica beads, and (d) HUVECs were successfully patterned. After seeding, the cells were allowed to attach and spread resulting in an irregular and larger patterned area. Scale bars = 100 μm.
We patterned insoluble fibrillar collagen, which, in our hands, is difficult to pattern using microcontact stamping (Fig. 3(a)).1 There was some variation in the number of large collagen fibers per spot as indicated by the high magnification dark field microscopy images. However, this is likely a function of the size distribution of fibers in the solution rather than any inherent defect in the patterning.
We patterned particles and cells in suspension using the microfluidic-microstencil method. Suspensions of liposomes (100 nm) were selectively patterned into spots (Fig. 3(b)). Dense arrays of silica beads (800 nm) were also successfully patterned into spots with no observable beads between spots (Fig. 3(c)). This is an improvement over a microblotting method we have previously used to pattern silica nanoparticle that tends to leave some particles between features.28 Note the areas of dense features surrounded by areas with no features (Fig. 3(c)). The ability to combine dense and sparse feature is an advantage of this patterning approach. Endothelial cells selectively adhered to gelatin surfaces within the microstencil wells (Fig. 3(d)). Between 3 and 7 endothelial cells were patterned within each spot. A tighter tolerance on the cell density may be achieved by optimizing the cell concentration and/or seeding conditions (continuous or periodic perfusion). Alternatively, matrix proteins could first be patterned into the wells of the microstencils, followed by the introduction of cells. Taken together, these results show that the microfluidic-microstencil approach can produce patterns of insoluble fibrillar proteins and particles and cells that would be challenging, if not impossible, to achieve by microcontact stamping. Further, we were able to define discrete spots of particles and cells, rather than long strips, which are characteristic of microfluidic patterning.
Platelet adhesion on multifunctional substrates
A second feature of the microfluidic-microstencil method is the ability to generate multifunctional substrates. To demonstrate this feature, we patterned type I fibrillar collagen, laminin, and reconstituted fibers of collagen from an acid soluble collagen solution on a single substrate. Following patterning, a second microfluidic flow device was reversibly vacuum-bonded to the substrate with channels running perpendicular to the array of patterned proteins (Fig. 4(a)). Whole blood was perfused through the channels at a wall shear rate of 100 s−1 for 7 min, and platelet adhesion was monitored by fluorescence. Platelet adhesion was observed at locations corresponding to the patterned features (Fig. 4(b)). There was no evidence of non-specific adhesion outside the spots. Platelet accumulation was greatest on fibrillar collagen, as expected owing to its strong activation through the glycoprotein VI receptor.17,33 Platelets adhered but did not aggregate on laminin, as previously reported.34,35 Reconstituted fibers from acid solubilized collagen showed the lowest adhesion. Importantly, there was no evidence of cross-talk between adjacent spots of different compositions owing to the relatively large spacing (5 mm) between features as controlled by the microfluidic channels.
FIG. 4.
Platelet adhesion assay on multifunctional surfaces of adhesive proteins. (a) A microfluidic flow device is placed over patterned features of insoluble fibrillar collagen, laminin, and acid soluble collagen. The flow device is placed perpendicular to the pattern on the glass slide and reversibly bonded using vacuum. (b) Platelet adhesion in the whole blood on patterned spots during a 7 min flow assay at a wall shear of 100 s−1. Platelets were labeled with Pacific Blue conjugated anti-human CD41. Scale bars = 100 μm.
CONCLUSIONS
In this report, we have described a combined microfluidic-micropatterning method for patterning biomolecules, lipids, nanoparticles, and cells. This method allows for the definition of multifunctional substrates where the size and geometry of patterned features are controlled by the microstencil and the spacing of features is controlled independently using microfluidic channels. As such, we can define features at multiple length scales, the individual features size, as small as 10 μm, and feature spacing as much as several millimeters. Moreover, this approach allows for direct patterning of particles and cells in suspension, which would be impossible using microcontact printing. We showed one application of this method by measuring platelet adhesion under flow on three different adhesive proteins within a single assay. Since the method uses soft lithography techniques, it is readily implemented in research laboratories without the need for expensive equipment.
ACKNOWLEDGMENTS
This work was supported by a National Science Foundation CAREER Award, Grant-in-Aid award from the American Heart Association, Bayer Hemophilia Awards Program, and the National Institutes of Health (R21NS082933 and R01HL120728). The authors gratefully acknowledge assistance from Abimbola Onasoga-Jarvis for her help with making silica beads and lipids.
References
- 1.Hansen R. R., Tipnis A. A., White-Adams T. C., Paola J. A. Di, and Neeves K. B., Langmuir 27, 13648 (2011). 10.1021/la2023727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D. and King M. R., Biotechnol. Prog. 24, 1052 (2008). 10.1002/btpr.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rana K., Liesveld J. L., and King M. R., Biotechnol. Bioeng. 102, 1692 (2009). 10.1002/bit.22204 [DOI] [PubMed] [Google Scholar]
- 4.Piner R. D., Science 283, 661 (1999). 10.1126/science.283.5402.661 [DOI] [PubMed] [Google Scholar]
- 5.Salaita K., Wang Y., and Mirkin C. A., Nat. Nanotechnol. 2, 145 (2007). 10.1038/nnano.2007.39 [DOI] [PubMed] [Google Scholar]
- 6.Doraiswamy A., Dunaway T. M., Wilker J. J., and Narayan R. J., J. Biomed. Mater. Res., Part B 89, 28 (2009). 10.1002/jbm.b.31183 [DOI] [PubMed] [Google Scholar]
- 7.Scott M. A., Wissner-Gross Z. D., and Yanik M. F., Lab Chip 12, 2265 (2012). 10.1039/c2lc21105j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sylman J. L., Lantvit S. M., Reynolds M. M., and Neeves K. B., Cell. Mol. Bioeng. 7, 421 (2014). 10.1007/s12195-014-0331-1 [DOI] [Google Scholar]
- 9.Ruiz S. Alom and Chen C. S., Soft Matter 3, 168 (2007). 10.1039/b613349e [DOI] [PubMed] [Google Scholar]
- 10.Bartolo D., Degré G., Nghe P., and Studer V., Lab Chip 8, 274 (2008). 10.1039/b712368j [DOI] [PubMed] [Google Scholar]
- 11.Morel M., Bartolo D., Galas J.-C., Dahan M., and Studer V., Lab Chip 9, 1011 (2009). 10.1039/b819090a [DOI] [PubMed] [Google Scholar]
- 12.Wright D., Rajalingam B., Karp J. M., Selvarasah S., Ling Y., Yeh J., Langer R., Dokmeci M. R., and Khademhosseini A., J. Biomed. Mater. Res., Part A 85, 530 (2008). 10.1002/jbm.a.31281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masters T., Engl W., Weng Z. L., Arasi B., Gauthier N., and Viasnoff V., PLoS One 7, e44261 (2012). 10.1371/journal.pone.0044261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perl A., Reinhoudt D. N., and Huskens J., Adv. Mater. 21, 2257 (2009). 10.1002/adma.200801864 [DOI] [Google Scholar]
- 15.Xia Y. and Whitesides G. M., Langmuir 13, 2059 (1997). 10.1021/la960936e [DOI] [Google Scholar]
- 16.Hansen R. R., Wufsus A. R., Barton S. T., Onasoga A. A., Johnson-Paben R. M., and Neeves K. B., Ann. Biomed. Eng. 41, 250 (2013). 10.1007/s10439-012-0658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heemskerk J. W. M., Sakariassen K. S., Zwaginga J. J., Brass L. F., Jackson S. P., Farndale R. W., and Biorheology Subcommittee of the SSC of the ISTH, J. Thromb. Haemostasis 9, 856 (2011). 10.1111/j.1538-7836.2011.04230.x [DOI] [PubMed] [Google Scholar]
- 18.Hui C. Y., Jagota A., Lin Y. Y., and Kramer E. J., Langmuir 18, 1394 (2002). 10.1021/la0113567 [DOI] [Google Scholar]
- 19.Okorie U. M., Denney W. S., Chatterjee M. S., Neeves K. B., and Diamond S. L., Blood 111, 3507 (2008). 10.1182/blood-2007-08-106229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byun I., Coleman A. W., and Kim B., RSC Adv. 3, 24872 (2013). 10.1039/c3ra44072a [DOI] [Google Scholar]
- 21.Lee J., Yun Y.-K., Kim Y., and Jo K., Bull. Korean Chem. Soc. 30, 1793 (2009). 10.5012/bkcs.2009.30.8.1793 [DOI] [Google Scholar]
- 22.Wägli P., Homsy A., and Rooij N. F. de, Sens. Actuators, B 156, 994 (2011). 10.1016/j.snb.2011.02.005 [DOI] [Google Scholar]
- 23.Ostuni E., Kane R., Chen C. S., Ingber D. E., and Whitesides G. M., Langmuir 16, 7811 (2000). 10.1021/la000382m [DOI] [Google Scholar]
- 24.Zheng Y., Dai W., Ryan D., and Wu H., Biomicrofluidics 4, 036504 (2010). 10.1063/1.3491474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang J. H., Um E., and Park J.-K., J. Micromech. Microeng. 19, 045027 (2009). 10.1088/0960-1317/19/4/045027 [DOI] [Google Scholar]
- 26.Neeves K. B., Maloney S. F., Fong K. P., Schmaier A. A., Khan M. L., Brass L. F., and Diamond S. L., J. Thromb. Haemostasis 6, 2193 (2008). 10.1111/j.1538-7836.2008.03188.x [DOI] [PubMed] [Google Scholar]
- 27.Ghosh M., Alves C., Tong Z., Tettey K., Konstantopoulos K., and Stebe K. J., Langmuir 24, 8134 (2008). 10.1021/la8006525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onasoga-Jarvis A. A., Puls T. J., O'Brien S. K., Kuang L., Liang H. J., and Neeves K. B., J. Thromb. Haemostasis 12, 373 (2014). 10.1111/jth.12491 [DOI] [PubMed] [Google Scholar]
- 29.Onasoga-Jarvis A. A., Leiderman K., Fogelson A. L., Wang M., Manco-Johnson M. J., Paola J. A. Di, and Neeves K. B., PLoS One 8, e78732 (2013). 10.1371/journal.pone.0078732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Kruchten R., Cosemans J. M. E. M., and Heemskerk J. W. M., Platelets 23, 229 (2012). 10.3109/09537104.2011.630848 [DOI] [PubMed] [Google Scholar]
- 31.Zwaginga J. J., Sakariassen K. S., Nash G., King M. R., Heemskerk J. W., Frojmovic M., and Hoylaerts M. F., J. Thromb. Haemostasis 4, 2716 (2006). 10.1111/j.1538-7836.2006.02178.x [DOI] [PubMed] [Google Scholar]
- 32.Schneider C. A., Rasband W. S., and Eliceiri K. W., Nat. Methods 9, 671 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heemskerk J. W. M., Mattheij N. J. A., and Cosemans J. M. E. M., J. Thromb. Haemostasis 11, 2 (2013). 10.1111/jth.12045 [DOI] [PubMed] [Google Scholar]
- 34.White-Adams T. C., Berny M. A., Patel I. A., Tucker E. I., Gailani D., Gruber A., and McCarty O. J. T., J. Thromb. Haemostasis 8, 1295 (2010). 10.1111/j.1538-7836.2010.03850.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue O., Suzuki-Inoue K., and Ozaki Y., J. Biol. Chem. 283, 16279 (2008). 10.1074/jbc.C700241200 [DOI] [PubMed] [Google Scholar]
- 36.See supplementary material at http://dx.doi.org/10.1063/1.4896231E-BIOMGB-8-013405 for Figure S1-patterning by microstencils without microfluidic channels.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4896231E-BIOMGB-8-013405 for Figure S1-patterning by microstencils without microfluidic channels.