Abstract
Sequence-dependent variation in structure and dynamics of a DNA duplex, collectively referred to as ‘DNA shape’, critically impacts interactions between DNA and proteins. Here, a method based on the technique of site-directed spin labeling was developed to experimentally map shapes of two DNA duplexes that contain response elements of the p53 tumor suppressor. An R5a nitroxide spin label, which was covalently attached at a specific phosphate group, was scanned consecutively through the DNA duplex. X-band continuous-wave electron paramagnetic resonance spectroscopy was used to monitor rotational motions of R5a, which report on DNA structure and dynamics at the labeling site. An approach based on Pearson's coefficient analysis was developed to collectively examine the degree of similarity among the ensemble of R5a spectra. The resulting Pearson's coefficients were used to generate maps representing variation of R5a mobility along the DNA duplex. The R5a mobility maps were found to correlate with maps of certain DNA helical parameters, and were capable of revealing similarity and deviation in the shape of the two closely related DNA duplexes. Collectively, the R5a probe and the Pearson's coefficient-based lineshape analysis scheme yielded a generalizable method for examining sequence-dependent DNA shapes.
INTRODUCTION
DNA shape refers to sequence-dependent structural and dynamic variations on a double-stranded duplex. At the global level, variations of DNA shape manifest as polymorphisms of the double helix (e.g. B-, A- and Z-DNA) and different propensity of bending; while at the local level (i.e. base-pair or base-pair step), shape may vary both in geometrical (structural) characteristics (e.g. narrowing of the minor groove in B-DNA; DNA kink) and in elastic properties (or deformability) that are characterized by the energetics of relative rotation and displacement of neighboring base pairs (1,2). The shape of a duplex, which is encoded by its sequence, critically impacts and influences interactions between DNA and other molecules, such as proteins, small ligands and metal ions (1,2). As such, information on DNA shape is essential for understanding and manipulating biological functions.
However, knowledge on sequence-dependent DNA shape, particularly in naked DNAs, is rather inadequate (1,2). Currently, our understanding of sequence-dependent shape in the DNA is derived largely from computational analyses. Most notably, Molecular Dynamics (reviewed in (3)) and Monte Carlo (4) simulations have been reported on DNAs. In addition, bio-informative approaches have been developed to analyze DNA and protein-DNA complex structures in the Protein Data Bank, and the resulting sequence-dependent helical parameters have been used to represent DNA shape (5). On the other hand, experimental probing of naked DNA shape, which is needed to validate and refine computational results, is challenging. Foot-printing experiments, such as those using hydroxyl radicals (6), have been used to probe DNA shape at the genomic scale, but their structural resolution is limited. X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy have provided high-resolution structures of DNAs; however, their number is small compared to available data for protein-DNA complexes (4). In addition, X-ray crystallography studies are hindered by crystal-packing biases, and NMR studies are constrained by the size of the DNA.
We have been exploring a biophysical technique, site-directed spin labeling (SDSL), to probe sequence-dependent shape of DNAs. In SDSL, a stable nitroxide radical is attached at a specific site of a bio-molecule, and electron paramagnetic resonance (EPR) spectroscopy is used to monitor the behavior of the nitroxide, from which structural and dynamic information at the labeling site can be derived (7). SDSL has matured as a technique for studying protein structure and dynamics (8,9). For SDSL studies of nucleic acids, information has been obtained primarily from nanometer distances measured between pairs of nitroxides, as well as mobility of nitroxides at 0.5–20 ns timescale, which is derived from X-band continuous-wave (cw-) EPR spectroscopy (10–15).
A powerful tool commonly used in protein SDSL studies is to scan a nitroxide probe through consecutive sites within a segment of the primary sequence. By collectively analyzing patterns of the measured cw-EPR spectra, one can obtain information, such as the underlying protein secondary structure as well as spatial organization of these secondary structure elements with respect to each other and to the environment (e.g. a lipid bilayer) (7,8). Conceptually, scanning a nitroxide consecutively through a DNA duplex could reveal structural and dynamic variations along the DNA, thus yielding information on DNA shape. However, to our knowledge, consecutive nitroxide scanning has not been reported in nucleic acids, although a number of studies have used nitroxide attached to multiple sites within the target molecule (e.g. (16–22)).
We have developed a family of nucleotide-independent nitroxide probes that can be covalently attached at the phosphate backbone of a DNA or RNA (23,24). We have shown that X-band cw-EPR spectra of this family of probes, particularly one designated as R5a (Figure 1), vary between different internal sites of DNA duplexes (21,25,26). Such spectral variations arise from differences in the modulation of nitroxide nanosecond rotational motions by the DNA, and therefore encode information on DNA local structural and dynamic features (21,25,26). Taking advantage that R5a can be attached to an arbitrary target DNA site, we present here work on using R5a ‘scanning’ to map DNA duplex shape. R5a was attached, consecutively and one nucleotide at a time, within two DNA duplexes that contain the p53 tumor suppressor response elements (REs) located respectively at the promoters of the BAX and p21 genes (Figure 1A) (27,28). X-band cw-EPR spectra were measured under conditions at which lineshape variations report on differences in the local DNA environment, and the degree of similarity among the ensemble of R5a spectra were characterized using a newly developed approach based on Pearson's coefficient analysis. The maps of the resulting Pearson's coefficients along these two sequences, which represent R5a mobility variations along the duplex, provided a measure of DNA shape at the level of individual base-pairs, and revealed sequence-dependent shape variations between the two closely related duplexes. Overall, the R5a probe and the Pearson's coefficient-based lineshape analysis tool yielded a generalizable method for examining sequence-dependent DNA shapes.
Figure 1.
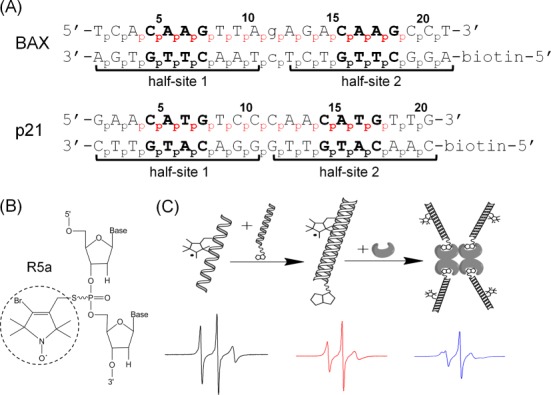
Experimental design. (A) Sequences of DNA duplexes studied. Both duplexes are REs of the p53 tumor suppressor, with two decameric half-sites (marked by brackets) each containing a ‘CWWG’ core (bold). Lower case letters in the BAX duplex mark the one-base-pair spacer between the two half-sites. No spacer is present in p21. For each duplex, the numbering scheme of the phosphates is shown, and the spin-labeled sites are indicated in red. (B) Chemical structure of the R5a spin label. (C) A schematic showing EPR sample assembly. One of the DNA strands was spin-labeled (left), paired with the complementary biotinylated strand (middle), then tethered to a streptavidin tetramer to form an ∼103 kD complex (right). Representative EPR spectra are shown below each step.
MATERIALS AND METHODS
Preparation of spin-labeled DNA
Figure 1A shows sequences of DNA studied. For each duplex, one of the strands was subjected to spin labeling, while the complementary strand had no modification at the nucleotides but a biotin attached at the 5′ terminus. All DNA oligonucleotides were synthesized by solid-phase chemical synthesis (Integrated DNA Technologies, Coralville, IA, USA).
Nitroxide spin labels, designated as R5a (Figure 1B), were site-specifically attached to DNAs using the phosphorothioate scheme as previously described (21,23,24). Briefly, during chemical synthesis, the phosphate group at the intended labeling site of a DNA strand is modified to a phosphorothioate. To attach the R5a label, crude DNA (∼0.6 mM) was incubated with 200 mM of 4-bromo-3-bromo-methyl-2,2,5,5-tetramethyl-1-oxylpyrroline (R5a precursor) in a solution of 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) (pH 5.8), and 40% (v/v) acetonitrile. The reaction mixture was incubated in dark at room temperature for 24 h under constant shaking. Labeled DNA was purified by anion-exchange high pressure liquid chromatography, followed by desalting using a reverse-phase column (21,24). Desalted oligonucleotides were then lyophilized, re-suspended in water and stored at −20°C. The final concentrations of DNA were determined by absorbance at 260 nm using extinction coefficients listed in Supplementary Table S1.
EPR sample preparation
Each EPR sample contained ∼50 μM of R5a-labeled DNA duplex, and was assembled in two steps (Figure 1C). First, a stock solution of the DNA duplex (200 μM) was prepared by annealing the R5a-labeled strand with the proper biotin-labeled complementary strand at a ratio of 1:1.1. After mixing appropriate amount of the respective strands, the mixture was heated at 95°C for 1 min, and then cooled down at room temperature for 1 min. The proper amount of salt was then added to the mixture to reach a final concentration of 50 mM Tris-HCl (Tris(hydroxymethyl)aminomethane hydrochloride) (pH 7.5) and 100 mM NaCl. The solution was left standing at room temperature for >1 h to allow duplex formation. In the second step, a spin-labeled DNA duplex was mixed with streptavidin (Amresco, Irvine, CA, USA) at a concentration ratio of 1:1.5 (duplex versus streptavidin monomer) in a solution containing 50 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (pH 7.5), 100 mM NaCl and 5 mM MgCl2. The mixture was incubated at room temperature for 2 h, and then used immediately for cw-EPR measurement. DNA tethering to streptavidin was confirmed by gel shift assays (Supplementary Figure S1).
Each labeled DNA sample was designated by its strand identity and labeling site. For example, ‘BAX_4’ represented the BAX duplex with an R5a attached at nucleotide 4, i.e. at the phosphorothioate between A3 and C4 (Figure 1A). Also note that Rp and Sp phosphorothioate diastereomers present at each attachment site were not separated (21).
X-band EPR spectroscopy
Each EPR spectrum was obtained using ∼5 μl sample loaded in a round glass capillary (0.6 mm i.d. × 0.8 mm o.d. Vitrocom, Inc., Mountain Lakes, NJ, USA) sealed at one end. X-band EPR spectra were acquired at room temperature on a Bruker EMX spectrometer using an ER4119HS cavity. The incident microwave power was 2 mW, and the field modulation was 1 G at a frequency of 100 kHz. Each spectrum was acquired with 512 points, corresponding to a spectral range of 100 G. All spectra reported had a signal-to-noise ratio (S/N), defined as the central line peak-to-peak height (i.e. signal) divided by the standard deviation of the first 25 points at the low field region (i.e. noise), over 350. Spectra were corrected for free spin-label if necessary, and normalized to the same number of spins using software kindly provided by the Hubbell group at UCLA. No baseline correction was carried out due to the high S/N of samples studied.
Computation of Pearson's coefficient
Pearson's coefficient (P), which measures the degree of linear correlation between data sets {Xi} and {Yi}, is defined as:
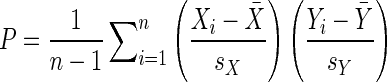 |
(1) |
where  and
and  are the mean of the respective data set, and sX and sY are the corresponding standard deviation. As defined, a Pearson's coefficient may vary between +1 and −1, with +1 indicating the two data sets positively correlate in a perfectly linear fashion, −1 indicating negative correlation in a perfectly linear fashion, and 0 indicating no linear correlation.
are the mean of the respective data set, and sX and sY are the corresponding standard deviation. As defined, a Pearson's coefficient may vary between +1 and −1, with +1 indicating the two data sets positively correlate in a perfectly linear fashion, −1 indicating negative correlation in a perfectly linear fashion, and 0 indicating no linear correlation.
A set of programs written in Matlab (R2011b 7.13.0.564, The MathWorks, Inc.) were developed in-house to automatically compute Pearson's coefficients between any pair of EPR spectra. To compute a Pearson's coefficient, two spectra were first aligned at the center, which was defined as the point at which the amplitude change signs within the center manifold. Then spectra corresponding to 40 G at either side of the center point were selected and digitized into 412 data points. Following spectral alignment and range selection, the pair-wise Pearson's coefficient was calculated using the ‘corr(x,y)’ function built-in within Matlab.
Sequence-dependent DNA shape parameters prediction
Sequence-dependent DNA helical parameters were obtained using the webserver of ‘High-throughput DNA shape prediction’ (http://rohslab.cmb.usc.edu/DNAshape/) (4), which currently provides predictions for Roll, Helical Twist, Minor Groove Width and Propeller Twist.
RESULTS
Nitroxide scanning of DNA duplexes using an optimized streptavidin tethering scheme
Studies reported here used two DNA duplexes, designated as BAX and p21 (Figure 1A). Both duplexes are REs of the p53 tumor suppressor, and conform to a consensus sequence of two decameric ‘half-sites’ (RRRCWWGYYY; R = A,G; W = A,T; Y = C,T) that are either consecutive or separated by a spacer (27,28). Recently, using EPR measured inter-nitroxide distances, we demonstrated these two REs both deform at the central region between the two half-sites in response to binding by the p53 core domain, but their modes of deformation differ between each other (29,30).
The BAX and p21 duplex each has a molecular weight of approximately 13 kD (Figure 1A). In aqueous solution at room temperature, the rotational correlation time of each duplex is estimated to be ∼7 ns (31). This global tumbling of the duplex averages the anisotropic magnetic tensors of nitroxide in a site-independent manner, and at X-band (∼9.5 GHz), diminishes the observable site-dependent spectral variations. Interference due to site-independent global tumbling is not a problem unique to the current work. One of the commonly used approaches to address this issue is to increase solution viscosity by including a co-solvent (e.g. sucrose (16,18,21,32)). However, the co-solvent may affect not only global tumbling, but also local motions at the target molecules and/or the spin labels (33), thus potentially introducing artifacts.
In this study, instead of adding co-solvent to minimize spectral interference due to global tumbling of BAX and p21, a previously reported tethering scheme (34) was further optimized and implemented (see Materials and Methods, Figures 1C and Supplementary Figure S1). In this scheme, the spin-labeled DNA duplex was tethered to streptavidin to form a complex, thus increasing the overall molecular weight by ∼8-fold. Tethering efficiently reduced the overall rotational motion of the duplex without interfering with local motions at the labeling sites, rendering the observed X-band EPR spectrum more sensitive to local DNA environment.
Using this tethering scheme, spectra were obtained for 16 and 15 consecutive internal sites along one strand of the BAX and p21 duplex, respectively (Figure 2). Spectral variations, such as hyperfine splitting, central line width and features of the low- and high-field manifolds, can be observed among these spectra. Prior studies have shown that R5a minimally perturbs the native local environment of a DNA duplex (21,35). Furthermore, observed R5a spectra were not affected by changing the relative location of the labeling site with respect to the tethering point (Supplementary Figure S2), indicating that R5a did not directly contact streptavidin. In addition, while four DNA duplexes were present on one streptavidin tetramer (Figure 1B and Supplementary Figure S1), under the experimental conditions the observed spectra were not biased by spin–spin interactions between the duplexes (Supplementary Figure S3). Overall, the observed lineshape variations report on differences at the local DNA environment.
Figure 2.
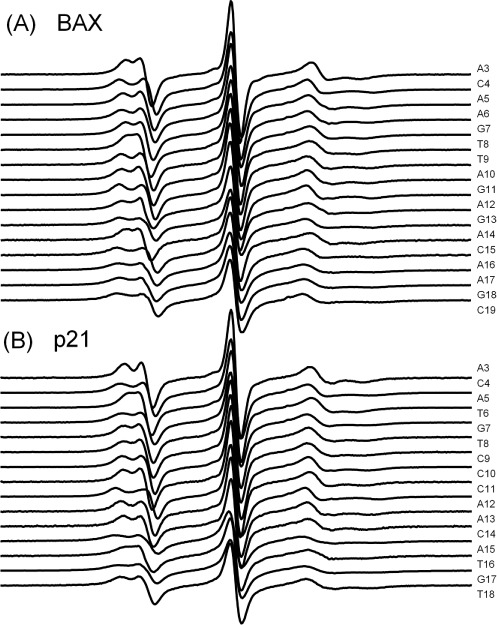
X-band cw-EPR spectra of R5a-labeled streptavidin-tethered BAX (panel A) and p21 (panel B) duplexes. All spectra were corrected for free spin-label (<5%) if necessary, normalized and aligned at the central line. The S/N ratio ranged between 350 and 2300.
Pair-wise Pearson's coefficients for assessment of EPR lineshape variation
To retrieve information on DNA shape from the ensemble of R5a spectra, a numerical value encoding relevant spectral characteristics is needed. In protein nitroxide scanning, this can be accomplished by using parameters, such as central line width (ΔHpp) or effective hyperfine splitting (2Aeff), to empirically represent nitroxide mobility (7,8). However, 3 of the 31 R5a spectra measured did not show discernable 2Aeff (BAX_9, p21_6, p21_16, Figure 2). The ΔHpp values varied within a narrow range (1.5–2.3 G), as difference along a DNA duplex is likely smaller than that in a folded protein. As such, the traditional lineshape parameters are not as effective for analyzing R5a spectra in DNA mapping.
We developed a new spectral analysis tool based on the Pearson's coefficient to retrieve information collectively embedded in the R5a spectra (see Materials and Methods). The Pearson's coefficient, which ranges from +1 to −1, assesses the degree of linear correlation between two sets of variables. A Pearson's coefficient of +1 indicates two perfectly positively correlated data sets, and in this work, EPR spectra with identical lineshape. Indeed, when BAX_19 spectra (see Materials and Methods for nomenclatures) obtained from three different sample preparations were analyzed, the pair-wise Pearson's coefficients (P) were averaged to 0.999 with a standard deviation of 0.001 (Figure 3A). Furthermore, control studies showed that the P-values remain invariant to the third significant figure upon: (i) decreasing the S/N ratio down to 300 (Supplementary Figure S4); (ii) spectral normalization (Supplementary Figure S5) and (iii) extending the spectral range from 80 G to 100 G. Overall, for the reported R5a spectra, the Pearson's coefficients were deemed accurate to three significant figures.
Figure 3.

Characteristics of pair-wise Pearson's coefficients. (A) Overlay of BAX_19 spectra obtained from three independent sample preparations. The pair-wise P-values were 0.99892, 0.99968 and 0.99822, respectively. (B) Examples of pair-wise spectral comparisons with their corresponding P-values shown. (C) Examples showing that a pair of spectra (BAX_5 and p21_12, center) with a high P-value compared similarly to a third spectrum (top: p21_5; bottom: BAX_13).
Figure 3B shows examples of pair-wise spectral comparison, which demonstrates that decreasing P-values report on increasing spectral differences. The P-value was 0.995 between BAX_4 and p21_4, both of which were obtained at an ApC di-nucleotide step, and direct comparison revealed highly similar lineshapes. When BAX_4 was compared to BAX_5, larger spectral difference in splitting at the low- and high-field manifolds can be observed, and a lower P-value of 0.938 was obtained. Among the 465 pairs of spectra investigated, the lowest P-value was 0.885, and was obtained between BAX_4 and p21_15, which indeed showed the most lineshape variations, with the largest difference in the splitting at both low- and high-field manifolds and clear deviation in central line width. Note that a P-value of 0.885 indicated that the two data sets (i.e. spectra) remain positively correlated, which reflects the fact that BAX_4 and p21_15 remain within the same motional regime (Figure 3B).
P-values not only provide a numeric measure of similarity/dissimilarity for one pair of spectra, but also allow assessment between different pairs. For example, P(BAX_4/p21_15) < P(BAX_4/BAX_5), and direct spectral superimposition showed that the difference is larger between BAX_4/p21_15 as compared to that between BAX_4/BAX_5 (Figure 3B). Also note that if two spectra are highly similar (i.e. P > 0.980), each of them would compare similarly with respect to a third spectrum, and therefore give similar P-values. This is illustrated in Figure 3C, in which two highly similar spectra, BAX_5 and p21_12 (P = 0.987), were compared with either p21_5 (Figure 3C, top) or BAX_13 (Figure 3C, bottom).
Pearson's coefficients referenced to TEMPOL spectrum report nitroxide mobility
In addition to analyzing R5a spectra by pair-wise Pearson's coefficients, we developed another parameter, PTEMPOL, defined as the Pearson's coefficient between an EPR spectrum and that of a free radical compound, TEMPOL (4-hydroxy-2,2,6,6-tetramethylpiperidinyloxy) (Figure 4). The TEMPOL spectrum, measured at room temperature in aqueous buffer without added molecular crowders, shows three sharp lines with equal amplitude (Figure 4A, bottom) and represents a nitroxide undergoing isotropic rotation at the fast motion limit. As such, a high PTEMPOL value indicates similarity between the sample spectrum and that of TEMPOL, and likely reports high nitroxide mobility. A low PTEMPOL indicates deviation of the sample spectrum from that of TEMPOL, and in the context of R5a spectra analyzed here, low nitroxide mobility.
Figure 4.
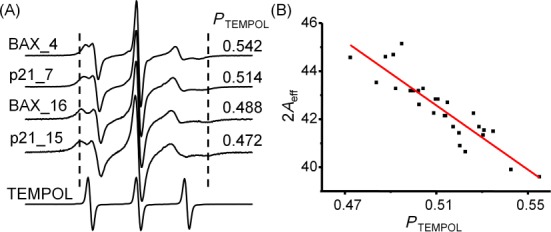
Characteristics of PTEMOPL. (A) Examples of R5a spectra showing the correlation between PTEMOPL and lineshape variations. The reference TEMPOL spectrum was shown at the bottom. The R5a spectra were scaled to the same height at the center line, while the TEMPOL reference was scaled to 1/2 of that of R5a. Dashed lines are shown to aid visualization of 2Aeff differences. (B) Correlation between PTEMPOL and 2Aeff. Data points (black dots) were obtained from spectra shown in Figure 2, but excluded BAX_9, p21_6 and p21_16, in which no clear 2Aeff could be measured. Linear fit (red line) yielded a coefficient of determination R2 of 0.81.
To validate the connection between PTEMPOL and R5a mobility, we examined the correlation between PTEMPOL and 2Aeff, a well-established lineshape parameter that generally increases as nitroxide mobility decreases (11,32). Figure 4A shows examples of R5a spectra, where the computed PTEMPOL value decreases as the corresponding 2Aeff increases. Furthermore, for the 28 spectra with measurable 2Aeff, PTEMPOL negatively correlates with the corresponding 2Aeff in a linear fashion (Figure 4B). Overall, the analyses demonstrated that PTEMPOL indeed reports R5a mobility, with a larger PTEMPOL value indicating higher mobility.
R5a mobility maps reveal DNA duplex shape at the level of individual base-pairs
For each DNA duplex studied, we obtained a PTEMPOL map by plotting the measured PTEMPOL value according to the corresponding residue number (Figure 5A). PTEMPOL acts as a measurement of R5a mobility, which previously has been shown to report structural and dynamic features at the labeling site (21,25,26). As such, the PTEMPOL map provided a map of the DNA local environment as sensed by R5a.
Figure 5.
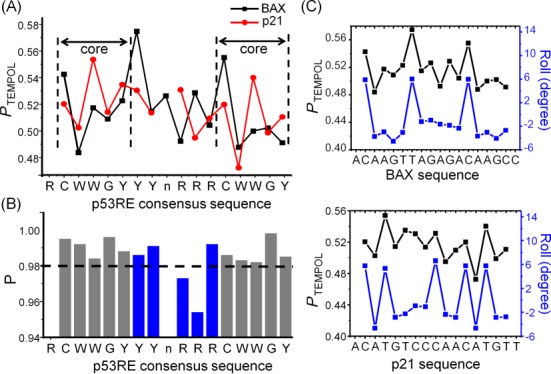
R5a mobility maps reveal DNA duplex shape. (A) Plots of PTEMPOL along BAX (black) and p21 (red) sequence. (B) Pair-wise P-values between spectra acquired at corresponding sites along the two DNAs. The CWWG core regions were shown in gray, and the central region in blue. A dashed line was drawn at P = 0.980 to aid visualization. (C) Comparisons between the PTEMPOL map (black) and that of Roll (blue) for BAX (top panel) and p21 (bottom panel). PTEMPOL value was aligned to the Roll value of the base-pair step 3′ of the spin-labeled phosphate. Pearson's coefficients between the two maps were 0.848 and 0.685, respectively, for BAX and p21.
The PTEMPOL maps were used to compare shapes between BAX and p21. As targets of the p53 tumor suppressor, these two DNAs follow the same consensus pattern but differ in their exact sequences (Figure 1A). At each of the corresponding CWWG core regions (marked by dashed lines, Figure 5A), the PTEMPOL maps showed very similar patterns, indicating similarity in local shapes between the CWWG cores. This is further supported by the observation that the pair-wise P-values for spectra obtained at the corresponding sites were all >0.980 (Figure 5B), which indicate high degrees of spectral similarity. For example, BAX_5 and p21_5, obtained at CpW of the first core, give a P of 0.992 (Figure 3C, top left). However, at the central region between the CWWG cores, variations of R5a mobility were observed between the two DNAs. Specifically, at the ‘RRR’ segment located 5′ of the second half-site, opposite trends of PTEMPOL alteration can be observed (Figure 5A), and the two corresponding pair-wise P-values were below 0.980 (Figure 5B). Examination of the corresponding spectra indeed confirmed a high degree of spectral variation (e.g. P(BAX_13/p21_12) = 0.954, Figure 3C, bottom right). This indicates that local shapes at the central regions of these two DNA are more divergent. Furthermore, maps of ΔHpp and 2Aeff along BAX and p21 supported the conclusions drawn from the PTEMPOL maps, although the 2Aeff map had gaps and the ΔHpp map had a narrow dynamic range (Supplementary Figure S6).
To better understand how PTEMPOL maps report on DNA local shape, we compared PTEMPOL maps to that of average DNA helical parameters, which represent local geometric variations of the duplex. Using an extensively validated high-throughput server (4), we obtained predicted maps of Roll, Helical Twist (HelT), Minor Groove Width (MGW) and Propeller Twist (ProT), for BAX and p21. In both duplexes, a high degree of positive linear correlations can be observed between the PTEMPOL map and that of Roll (Figure 5C), indicating higher R5a mobility at sites with larger Roll values. On the other hand, correlations between PTEMPOL/MGW (Supplementary Figure S7) and PTEMPOL/ProT (Supplementary Figure S8) were below 0.5 in both BAX and p21, indicating that R5a mobility is likely not correlated with variations of MGW and ProT. For HelT, correlation with PTEMPOL was low in BAX but high in p21 (Supplementary Figure S9), so how R5a mobility reflects HelT variation is not clear.
Another aspect of local DNA shape variation is sequence-dependent deformability (1,5). Using parameters reported by Olson et al. (5), we predicted base-pair step elastic force constant maps of Roll–Roll and HelT–HelT for BAX and p21, which serve as representations of deformability variation. However, correlations between these maps and that of PTEMPOL were all below 0.6, with no consistent pattern in the two duplexes (Supplementary Figure S10). As such, the current analysis did not reveal a clear correlation between PTEMPOL and DNA deformability.
Overall, analyses demonstrated that PTEMPOL maps report on certain physical attributes of DNA duplex shape, and reveal that between BAX and p21 DNA shapes are very similar at the CWWG cores but deviate at the central region between the two cores.
P matrix for global lineshape analyses
To globally analyze lineshape features of 31 R5a spectra obtained, we constructed a 2D matrix (the P-matrix), in which each cell was assigned the P-value between the two spectra placed at the respective row and column (Figure 6). For this purpose, we placed the 31 spectra into row and column following the same order of decreasing PTEMPOL (i.e. decreasing nitroxide mobility) (Figure 6, Supplementary Table S2). To aid analyses, we color-coded the cells so that those with the highest P (i.e. 1 at the diagonal cells) were pure black, the one with the lowest P (i.e. 0.885) was pure white, and those in between were represented by a linear percentage of gray. As expected, this matrix was symmetric; the diagonal cells (representing self-comparison) all had P = 1.000; and P-values generally decreased from the diagonal toward the outer edges, as the outer edge cells compared spectra with different mobility and hence increasing lineshape deviations.
Figure 6.
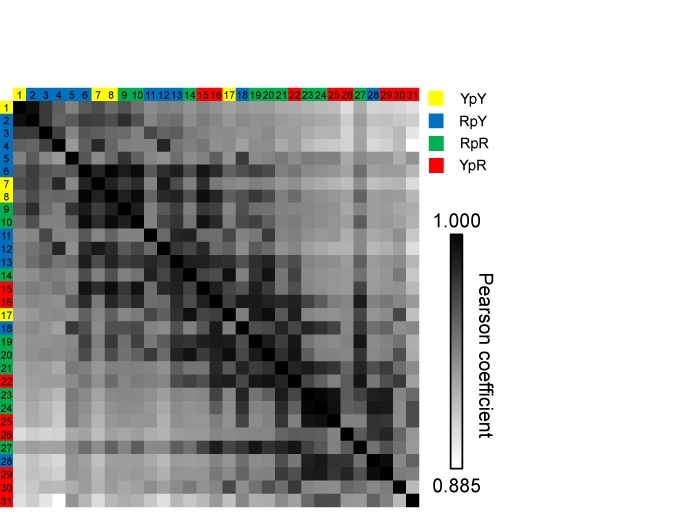
A matrix of the pair-wise Pearson's coefficients obtained with 31 R5a spectra. For clarity each spectrum was assigned a number, with the key given in Supplementary Table S2. Each cell was color-coded in gray-scale based on the P-value between spectra identified by the respective row and column labels. The row/column labels are color-coded based on the di-nucleotide step as shown in the figure legend.
With the P-matrix constructed, direct visual inspection revealed a number of interesting features. First, off-diagonal cells in the last column on the right, which contained pair-wise P-values computed between p21_15 (spectrum #31, Supplementary Table S2) and others, all showed relatively low P-values (Figure 6). This indicated that the p21_15 spectrum, obtained from an R5a attached at a CpA step, differed from any other spectra within the ensemble. The same is true for BAX_5 (spectrum #30, Supplementary Table S2), another CpA step, which occupied the second last column on the right (Figure 6). The other three spectra obtained at ‘CpA’ steps (p21_5, p21_12 and BAX_16, spectra #22, #26 and #29, respectively, Supplementary Table S2) also showed relatively low R5a mobility, although they did have high P-values with a few spectra obtained at non-CpA sites (Figure 6). Interestingly, pyrimidine-purine steps, including CpA, are the most easily deformed among all di-nucleotide steps, and usually contribute the largest flexibility in DNA/protein complexes (5). It is possible that in the naked DNA, the high deformability/flexibility of the CpA step manifests as sampling, at rates slower than nanosecond, of micro-conformations that vary depending on sequence context. This would result in broad spectra (apparent low R5a mobility) that are divergent among CpA sites (Figure 6).
For the remaining spectra, a number of off-diagonal cells had high P-values, and clusters of dark gray cells were observed (Figure 6). For example, BAX_18, BAX_17 and p21_17 (spectra #23, #24 and #25, respectively, Supplementary Table S2) cluster at the lower right corner of the matrix, and direct comparisons revealed that these spectra were indeed highly similar (Supplementary Figure S11). Spectra within each cluster hold a relatively high degree of similarity in lineshape features, possibly indicating similarity in DNA shape between these sites. Between clusters, low P-values are observed, indicating divergence of spectral features and possible DNA shape differences.
DISCUSSION
Site-specifically attached nitroxides and their EPR lineshapes have been used widely to investigate local environments in DNA and RNA (10–11,13). However, contrary to protein studies, to the best of our knowledge, nitroxide scanning of consecutive sites has not been reported in nucleic acid SDSL. In this work, we scanned consecutive sites within two DNA duplexes using a nucleotide-independent nitroxide probe, R5a. EPR spectral variations, which report on the local environment at the respective labeling site, were analyzed using a newly developed method based on Pearson's correlation coefficients. The resulting Pearson's coefficients, when plotted along the DNA sequence, provided maps that allow examination and comparison of local DNA shape in the two duplexes. The studies established an objective method to analyze EPR spectral variation, and together with the R5a probe, yielded an experimental approach for examining sequence-dependent DNA shape.
Pearson's coefficients for EPR lineshape analysis and comparison
The Pearson's coefficient-based protocol was developed to address issues arisen when scanning DNA using R5a, namely, sites lacking discernible hyperfine splitting (2Aeff) and the narrow spread of central line width (ΔHpp). The Pearson's coefficient accounts for differences in all three manifolds of the 14N nitroxide spectrum, and therefore globally reflects, in one numerical value, differences characterized by other spectral parameters, including ΔHpp and 2Aeff. Two types of Pearson's coefficients were demonstrated in mapping DNA shape: the pair-wise P-values, which can be used for comparing different DNA sites (Figures 3C and 5B); and PTEMPOL, which reports R5a mobility variations along the DNA strand (Figures 4 and 5A). Pearson's coefficients were robust with respect to a number of experimental factors, including a certain degree of deterioration in S/N (Supplementary Figure S4). Furthermore, they are invariant with respect to linear scaling of either spectrum, and are not impacted by, and indeed do not require, spectral normalization (Supplementary Figure S5). This is in contrast to root mean square deviation (RMSD) between two spectra, which is based on differences in absolute value (Supplementary Figure S5). Additionally, RMSD is dominated by the high amplitude region, which is usually the central line in a nitroxide spectrum, rendering it suboptimal for comparing R5a spectra, most of which have sharp central lines.
Similar to 2Aeff and ΔHpp, Pearson's coefficients are semiquantitative parameters derived empirically from the cw-EPR spectrum. They are not expected to provide a complete description of nitroxide motion, and their effectiveness on reporting variations of nitroxide mobility likely depends on the motional regime. For example, for R5a data obtained in the two DNA duplexes studied here, PTEMPOL was shown to correlate with 2Aeff in a linear fashion (Figure 4), and sufficiently represent differences in R5a mobility to allow DNA mapping (Figure 5). In principle, one can construct a Pearson's scale using an arbitrary data set as the reference spectrum, although its effectiveness on representing the particular nitroxide mobility for mapping DNA shape would need to be examined. Nonetheless, the Pearson's coefficients are obtained in an automated fashion without the highly subjective and rather qualitative assessment by a user, and should be applicable to other labels and systems. This expands the repertoire of parameters for extracting information from an ensemble of EPR spectra.
Mapping DNA shape using the nucleotide-independent nitroxide probe
We have previously shown that EPR lineshapes of the family of nucleotide-independent nitroxide probes (e.g. R5a) vary among different sites of DNA duplexes, and such variations report on variation in the local DNA environment (21,25,26). Using semi-empirical lineshape parameters that report R5a mobility, including the Pearson's coefficients developed here (Figure 5) and the more traditional ΔHpp and 2Aeff values (Supplementary Figure S6), we generated ‘maps’ to globally and systematically examine structural and dynamic variation along a DNA duplex. This allowed us to assess similarities and differences in local DNA shape between duplex segments. Data obtained for BAX and p21, both of which are p53 REs, revealed that DNA shape is similar between the corresponding CWWG cores, but varies at the central region bridging the two cores (Figure 5). Previously, we have reported that the central regions of these two REs deform in different fashions upon interacting with the p53 core DNA-binding domain: BAX undergoing further unwinding, while p21 incurring a pronouncing shift of the helix axis (30). Variability of DNA shape at the central region in the naked DNA is consistent with those observations.
The strategy of attaching labels at multiple sites to gain a comprehensive understanding of the target DNA/RNA has been implemented in many prior SDSL studies (e.g. (16–22)). However, work presented here, to the best of our knowledge, is the first report of scanning a nitroxide consecutively through strands of nucleic acids. This is enabled by two key elements: (i) the nucleotide-independent R5a probe, which allows efficient labeling at each target site; and (ii) the method of analyzing R5a spectral variation using the Pearson's coefficient, which complements the traditional ΔHpp and 2Aeff parameters. With the R5a maps including each individual nucleotide within the target DNA strand, a high resolution is achieved. This is advantageous for comparing DNA shape, as demonstrated by the ability to pick up the highly localized differences between the central regions of BAX and p21 (Figure 5).
Previous work has established that within a DNA duplex R5a motions can be described as Brownian diffusion under a restricted potential, and the observed EPR spectrum reports a combined effect due to the rate and the ordering of R5a motion (25,26). The local DNA environment modulates the sterically allowed rotameric space at the labeling site, thereby affecting R5a motional ordering and the resulting EPR spectrum. The rotameric space likely has a direct correlation to local geometric variation of the duplex, giving rise to the positive correlations observed between maps of PTEMPOL and Roll (Figure 5C).
We also reported that the R5a spectrum can be modulated by DNA backbone motions (i.e. the BI/BII transition) at the 5′ nearest neighbor and by specific contacts with the 3′ nearest neighbor (26). However, current analyses did not reveal clear correlations between PTEMPOL and predicted elastic force constants representing variations of DNA deformability (Supplementary Figure S10). It is possible that DNA deformability and backbone motions are not directly correlated. In addition, the elastic force constant maps were predicted using a dimer model (5). This is likely not sufficient as R5a lineshape is certainly impacted by sequences beyond a dimer (e.g. the variability of the CpA spectra, Figure 6).
Work reported here provided the first indication that EPR spectral parameters can be connected to actual DNA helical parameters. However, an R5a spectrum likely reflects only certain aspect of DNA local shape, and may be affected by 5′ and 3′ neighboring nucleotides (21,26). A lot more work is required for a better understanding between the EPR observables and the actual physical shape of a DNA duplex. Toward this goal, we have proposed to establish an empirical library in which R5a spectra are obtained and categorized according to DNA sequence (34), thus allowing systematic analyses of nitroxide/DNA coupling (26). A key requirement for such a library is to identify clusters of sites with similar lineshape features. This now becomes possible with the P-matrix analysis, which allows global assessments on variations among the 465 pairs of R5a spectra (Figure 6). The P-matrix did reveal clustering in the observed spectra (Figure 6), supporting the feasibility of establishing an R5a library.
R5a is attached at the phosphate, and the di-nucleotide step sandwiching the probe likely exerts the most effect on its EPR spectra. As a first step toward identifying converging physical attributes that result in certain lineshape features, we examined the distribution of di-nucleotide step among the different clusters (Figure 6). We found that the YpY (Y: pyrimidine; C or T) steps are located almost exclusively at the upper left region of the matrix, indicating that R5a attached to YpY shows a similar lineshape with characteristics of a higher degree of mobility. This likely reflects the weaker stacking between pyrimidine bases due to their smaller surface areas (36,37), and is consistent with prior studies of R5a (21). Furthermore, RpY (R: purine; A or G) steps are mostly located at the left half on the matrix, while YpR di-nucleotides favor strongly the right half. On the other hand, the RpR di-nucleotide steps spread throughout the matrix, which may suggest that these sites are subjected to influences beyond the nearest di-nucleotides. Note that while 31 spectra were analyzed here, the ensemble does not allow sufficient coverage of all the di-nucleotide steps, let alone further analysis on effects beyond the nearest neighbors. Future work should expand the library size.
In summary, this work established a method for experimentally examining sequence-dependent DNA shape. Further studies, particularly those combine experimental and computational approaches (26), should advance our understanding on how DNA modulates R5a spectrum, thus enhancing our ability to map DNA shape using SDSL.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Elizabeth M. Backus for help in manuscript preparation.
FUNDING
NSF [CHE-1213673, P.Z.Q.]; NIH [GM069557, P.Z.Q.]. Funding for open access charge: NSF [CHE-1213673, P.Z.Q.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Ann. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egli M., Pallan P.S. The many twists and turns of DNA: template, telomere, tool, and target. Curr. Opin. Struct. Biol. 2010;20:262–275. doi: 10.1016/j.sbi.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez A., Luque F.J., Orozco M. Frontiers in molecular dynamics simulations of DNA. Account. Chem. Res. 2012;45:196–205. doi: 10.1021/ar2001217. [DOI] [PubMed] [Google Scholar]
- 4.Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–W62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker S.C., Hansen L., Abaan H.O., Tullius T.D., Margulies E.H. Local DNA topography correlates with functional noncoding regions of the human genome. Science (N.Y.) 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbell W.L., Altenbach C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol. 1994;4:566–573. [Google Scholar]
- 8.Fanucci G.E., Cafiso D.S. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Hubbell W.L., López C.J., Altenbach C., Yang Z. Technological advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 2013;23:725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowa G.Z., Qin P.Z. Site-directed spin labeling studies on nucleic acid structure and dynamics. Prog. Nucleic Acids Res. Mol. Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Cekan P., Sigurdsson S.T., Qin P.Z. Studying RNA using site-directed spin-labeling and continuous-wave electron paramagnetic resonance spectroscopy. Methods Enzymol. 2009;469:303–328. doi: 10.1016/S0076-6879(09)69015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reginsson G.W., Schiemann O. Studying biomolecular complexes with pulsed electron-electron double resonance spectroscopy. Biochem. Soc. Trans. 2011;39:128–139. doi: 10.1042/BST0390128. [DOI] [PubMed] [Google Scholar]
- 13.Krstic I., Endeward B., Margraf D., Marko A., Prisner T.F. Structure and dynamics of nucleic acids. Topics Curr. Chem. 2012;321:159–198. doi: 10.1007/128_2011_300. [DOI] [PubMed] [Google Scholar]
- 14.Shelke S.A., Sigurdsson S.T. Site-directed spin labelling of nucleic acids. Eur. J. Org. Chem. 2012;2012:2291–2301. [Google Scholar]
- 15.Fedorova O.S., Tsvetkov Y.D. Pulsed electron double resonance in structural studies of spin-labeled nucleic acids. Acta Naturae. 2013;5:9–32. [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards T.E., Okonogi T.M., Robinson B.H., Sigurdsson S.T. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 2001;123:1527–1528. doi: 10.1021/ja005649i. [DOI] [PubMed] [Google Scholar]
- 17.Edwards T.E., Okonogi T.M., Sigurdsson S.T. Investigation of RNA-protein and RNA-metal ion interactions by electron paramagnetic resonance spectroscopy. The HIV TAR-Tat motif. Chem. Biol. 2002;9:699–706. doi: 10.1016/s1074-5521(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 18.Qin P.Z., Hideg K., Feigon J., Hubbell W.L. Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemistry. 2003;42:6772–6783. doi: 10.1021/bi027222p. [DOI] [PubMed] [Google Scholar]
- 19.Edwards T.E., Robinson B.H., Sigurdsson S.T. Identification of amino acids that promote specific and rigid TAR RNA-tat protein complex formation. Chem. Biol. 2005;12:329–337. doi: 10.1016/j.chembiol.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Kim N.K., Murali A., DeRose V.J. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J. Am. Chem. Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popova A.M., Kalai T., Hideg K., Qin P.Z. Site-specific DNA structural and dynamic features revealed by nucleotide-independent nitroxide probes. Biochemistry. 2009;48:8540–8550. doi: 10.1021/bi900860w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cekan P., Jonsson E.O., Sigurdsson S.T. Folding of the cocaine aptamer studied by EPR and fluorescence spectroscopies using the bifunctional spectroscopic probe C. Nucleic Acids Res. 2009;37:3990–3995. doi: 10.1093/nar/gkp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin P.Z., Butcher S.E., Feigon J., Hubbell W.L. Quantitative analysis of the GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 24.Qin P.Z., Haworth I.S., Cai Q., Kusnetzow A.K., Grant G.P.G., Price E.A., Sowa G.Z., Popova A., Herreros B., He H. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nat. Protocols. 2007;2:2354–2365. doi: 10.1038/nprot.2007.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popova A.M., Qin P.Z. A nucleotide-independent nitroxide probe reports on site-specific stereomeric environment in DNA. Biophys. J. 2010;99:2180–2189. doi: 10.1016/j.bpj.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popova A.M., Hatmal M.M., Frushicheva M.P., Price E.A., Qin P.Z., Haworth I.S. Nitroxide sensing of a DNA microenvironment: mechanistic insights from EPR spectroscopy and molecular dynamics simulations. J. Phys. Chem. B. 2012;116:6387–6396. doi: 10.1021/jp303303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley T., Sontag E., Chen P., Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell. Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 28.Menendez D., Inga A., Resnick M.A. The expanding universe of p53 targets. Nat. Rev. Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y., Zhang X., Dantas Machado A.C., Ding Y., Chen Z., Qin P.Z., Rohs R., Chen L. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 2013;41:8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Dantas Machado A.C., Ding Y., Chen Y., Lu Y., Duan Y., Tham K.W., Chen L., Rohs R., Qin P.Z. Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling. Nucleic Acids Res. 2014;42:2789–2797. doi: 10.1093/nar/gkt1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tirado M.M., Garciadelatorre J. Rotational-dynamics of rigid, symmetric top macromolecules—application to circular-cylinders. J. Chem. Phys. 1980;73:1986–1993. [Google Scholar]
- 32.McHaourab H.S., Lietzow M.A., Hideg K., Hubbell W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 33.Galiano L., Blackburn M.E., Veloro A.M., Bonora M., Fanucci G.E. Solute effects on spin labels at an aqueous-exposed site in the flap region of HIV-1 protease. J. Phys. Chem. B. 2009;113:1673–1680. doi: 10.1021/jp8057788. [DOI] [PubMed] [Google Scholar]
- 34.Qin P.Z., Iseri J., Oki A. A model system for investigating lineshape/structure correlations in RNA site-directed spin labeling. Biochem. Biophys. Res. Commun. 2006;343:117–124. doi: 10.1016/j.bbrc.2006.02.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Q., Kusnetzow A.K., Hubbell W.L., Haworth I.S., Gacho G.P.C., Van Eps N., Hideg K., Chambers E.J., Qin P.Z. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006;34:4722–4734. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kool E.T. Hydrogen bonding, base stacking, and steric effects in dna replication. Ann. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Bommarito S., Peyret N., SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


