Abstract
The nephrotoxic food mutagen ochratoxin A (OTA) produces DNA adducts in rat kidneys, the major lesion being the C8-linked-2′-deoxyguanosine adduct (OTB-dG). Although research on other adducts stresses the importance of understanding the structure of the associated adducted DNA, site-specific incorporation of OTB-dG into DNA has yet to be attempted. The present work uses a robust computational approach to determine the conformational preferences of OTB-dG in three ionization states at three guanine positions in the NarI recognition sequence opposite cytosine. Representative adducted DNA helices were derived from over 2160 ns of simulation and ranked via free energies. For the first time, a close energetic separation between three distinct conformations is highlighted, which indicates OTA-adducted DNA likely adopts a mixture of conformations regardless of the sequence context. Nevertheless, the preferred conformation depends on the flanking bases and ionization state due to deviations in discrete local interactions at the lesion site. The structural characteristics of the lesion thus discerned have profound implications regarding its repair propensity and mutagenic outcomes, and support recent experiments suggesting the induction of double-strand breaks and deletion mutations upon OTA exposure. This combined structural and energetic characterization of the OTB-dG lesion in DNA will encourage future biochemical experiments on this potentially genotoxic lesion.
INTRODUCTION
Ochratoxin A (N−[[(3R)−5-chloro−8-hydroxy−3−methyl−1−oxo−7−isochromanyl] carbonyl]−3-phenyl−L−phenylalanine), abbreviated as OTA, is a mycotoxin produced by species of Aspergillus (1,2) and Penicillium (3), and a common agricultural contaminant. Humans are exposed to OTA through a wide variety of foodstuffs, such as cereals grains (4), legumes (5) and beverages, including coffee (5,6), wines and fruit juices (7). OTA is believed to be involved in a number of toxicological effects, such as nephrotoxicity (8), hepatotoxicity (9), teratogenicity (10), immunotoxicity (11), neurotoxicity (12), genotoxicity (13) and carcinogenicity (14). OTA exposure leads to a high occurrence of renal carcinogenesis in rats and has been implicated in urinary tract tumors (15) and testicular carcinogenesis (16) in humans. In fact, the nephrotoxic effects of OTA are so pronounced that it is the most potent renal carcinogen in rodents studied by the National Cancer Institute/National Toxicological Program to date (17). Due to sufficient evidence of OTA-mediated carcinogenicity in laboratory animals, OTA is classified as a possible (Group 2B) human carcinogen by the International Agency for Research on Cancer (18).
Several studies have been devoted to understanding the mechanism of action (MOA) of OTA-mediated toxicity and carcinogenicity. In addition to proposing several possible pathways for OTA bioactivation (19), these studies have generated considerable debate on the genotoxicity of OTA. Of particular interest are the contradictory results regarding whether OTA exerts carcinogenicity in rodents by an indirect mechanism or a direct interaction with DNA through the formation of adducts (addition products). Over the past decade, new studies have strengthened the argument that direct genotoxic effects contribute to OTA-induced tumor formation (13,20–28). Specifically, OTA-derived DNA adducts in OTA-exposed animal tissue have been detected in animal tissues (24,25). Most recently, an increase in mutant frequency, as well as induction of double-strand breaks and deletion (frameshift) mutations, at the red/gam gene at the carcinogenic target site of gpt delta transgenic rats strongly suggests the involvement of a genotoxic mechanism(s) in OTA-mediated carcinogenesis (20,26,27).
Despite the known carcinogenic effects of OTA, regular human exposure to this contaminant in foodstuffs varies throughout the world. Health Canada recommends a relatively stringent tolerable daily intake (TDI) of 28 ng/kg bw/week based on a nonthreshold model of risk assessment, which is generally applied to carcinogens that cause tumors through direct genotoxicity mechanisms (29). However, the European Food Safety Agency (EFSA) has established a relatively relaxed TDI for OTA of ∼120 ng/kg bw/week, which partially stems from a threshold-based approach of risk assessment that is normally implemented for nongenotoxic chemicals (30). The EFSA TDI assessment is mainly influenced by reports claiming an absence of the genotoxic MOA in OTA-mediated carcinogenicity (31,32). Therefore, structural and mechanistic studies have a critical role in influencing legislative attitudes related to the assessment of carcinogen exposure and consequent human health hazards.
Crucial information supporting the formation of DNA adducts in the genotoxic mechanism of OTA action has been provided by studies that have elucidated and characterized the chemical structure of covalent OTA adducts at specific DNA sites. In vitro assays on kidney microsomes prepared from male mice in the presence of four DNA nucleotides (dA, dC, dG and dT) and OTA suggest that OTA forms guanine-specific adducts (33,34). In particular, the photochemical reaction of OTA with 2′-deoxyguanosine (dG) indicates that OTA specifically reacts at the C8 site of dG to form the carbon-linked adduct (OTB-dG, Figure 1), which was characterized by mass spectrometry and 2D nuclear magnetic resonance (2D-NMR) spectroscopy (23). In addition to the major OTB-dG adduct, a minor oxygen-linked (OTA-dG) adduct has been characterized by mass spectroscopy (22,24). The formation of the OTB-dG C-linked adduct in animal tissues was established by 32P-postlabeling studies in the renal tissues of rats and pigs (24,25). Using mass spectrometry data of isolated OTB-dG as a standard, the OTA-DNA adducts were further characterized in vitro with calf thymus DNA (22).
Figure 1.
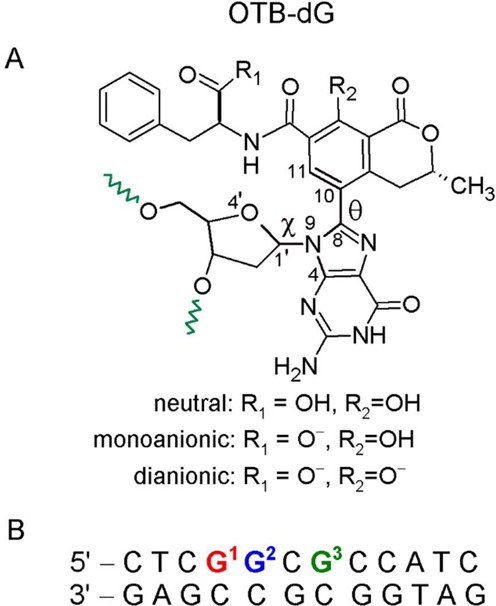
(A) Chemical structure of the OTB-dG adduct. Green wavy bonds represent the 5′ and 3′ sites where the adduct is linked to the DNA backbone. Torsion angles χ at the sugar–nucleobase linkage and θ at the nucleobase–substituent linkage are defined as follows: χ = ∠(O4′–C1′–N9–C4) and θ = ∠(N9–C8–C10–C11). The OTB-dG adduct can exist in neutral (non-ionized), monoanionic (carboxylic group ionized) and dianionic (carboxylic and phenolic groups ionized) forms. (B) The 12mer NarI recognition sequence used for MD simulations, where the OTB–dG lesion was incorporated at the G1, G2 or G3 position.
DNA damage through covalent adduct formation can alter the processing of genetic material by enzymes involved in DNA replication, which can lead to mutations (35). If the adducts persist in cells, such mutations may ultimately lead to cancer, especially when the lesion is located at the (proto) oncogene or tumor suppressor gene. Therefore, a number of studies have been devoted to understanding the effects of adduct formation on cellular DNA processing (36–38). However, in order to understand these effects, the conformational preferences of the damaged DNA duplex must be first understood (39). This is critical since dsDNA conformations correspond to the ‘mutagenic decision point’ that replication enzymes encounter before reaching the single-strand–double-strand junction at the replication fork, where the actual mutation takes place (40). In addition, a number of studies have shown that the conformational preference of a particular lesion also has a significant effect on its tendency to be recognized and removed by enzymes involved in the repair of bulky DNA adducts (35,39,41).
Although considerable progress has been made toward structurally characterizing the effects of a variety of potentially carcinogenic bulky DNA adducts, the site-specific incorporation of OTB-dG into DNA has yet to be attempted. However, previous NMR and computational studies on the well-studied carcinogenic N-linked aromatic amine DNA adducts suggest general DNA conformational themes that are prevalent among the broad class of C8-dG adducts (42). Specifically, a B-DNA type ‘major groove’ conformation places the C8-bulky moiety in the DNA major groove and maintains Watson–Crick hydrogen bonding with the opposing nucleotide (Figure 2, left). On the other hand, the ‘wedge’ conformation positions the C8-moiety in the minor groove and forms non-Watson–Crick pairing with the opposing base (Figure 2, middle). Finally, a ‘base-displaced intercalated’ (stacked) conformation has also been observed in which the C8-substituent is located between the neighboring base pairs in DNA, ruptures the Watson–Crick pairing with the opposing cytosine and thereby displaces C out of the helix (Figure 2, right) (42). Studies on a variety of C8-adducts have shown that the equilibrium between these DNA conformations is affected by a variety of local structural factors, including the strength of the Watson–Crick versus Hoogsteen binding arrangements, the stacking potential of the C8-moiety and the identity of the nearest and next-nearest nucleotides (35,39).
Figure 2.
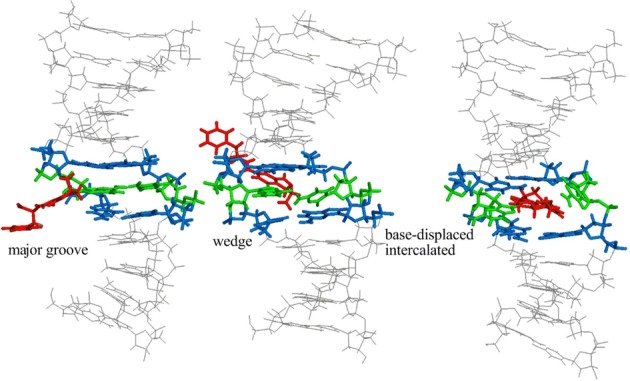
Representative possible conformations of adducted DNA with the OTB-dG lesion shown at the G3 position in the NarI recognition sequence. Although simulations are carried out using both the α and β-rotamers, the energetically more stable α-rotamer is shown here.
Unlike the carboxylic and phenolic functional groups of OTB-dG, previously studied adducts, such as the N-linked aromatic amine lesions, do not contain an ionizable site. Therefore, to the best of our knowledge, the effect of the ionization states of a DNA adduct on the adducted DNA conformation has yet to be studied in the literature. In the case of OTB-dG, previous work indicates that the pKa of OTA is greatly influenced by environmental factors, such as macromolecular binding (43) and solvation (44). For example, binding of OTA to human serum albumin lowers the pKa of the OTA phenolic group to 3.9 (43). Furthermore, the OTA phenolic group is even more acidic than the carboxylic group in the presence of acetonitrile solvent (44). These results indicate that the surrounding environment of the lesion, and therefore likely DNA binding, will influence the pKa of the OTA moiety. Thus, understanding the conformational preferences of OTB-dG adducted DNA in different ionization states is critical for evaluating the effect of charge on adduct-induced conformational heterogeneity, which in turn will help elucidate the mutagenic potential of OTB-dG.
To explore the preferred conformations of OTB-dG adducted DNA, the present work undertakes an extensive molecular modeling study, including molecular dynamics (MD) simulations and post-processing free energy calculations, to determine the conformational preferences of OTB-dG adducted DNA in three protonation states at the G1, G2 or G3 sites in the NarI sequence (5′–CTCG1G2CG3CCATC–3′) opposite complementary cytosine. The center of this sequence (underlined) is the recognition site of the NarI Type II restriction endonuclease, which is the most vulnerable hotspot for deletion mutations induced by carcinogenic N-linked C8-dG adducts (45), and the site-specific incorporation of a variety of C8-linked adducts has been studied in this sequence (41,46–50). Since OTA exposure has been linked to an increase in the frequency of deletion mutations at carcinogenic target sites (20,26), we chose the NarI sequence to study the conformational preferences of the OTB-dG lesion. This choice also allows direct comparison of the conformational preferences of the OTB-dG adduct with widely studied C8-bonded N-linked carcinogenic aromatic amine lesions (41,49,51–54), and thereby the elucidation of the role of adduct structure in dictating the DNA conformational preferences. Our results indicate that OTB-dG adducted DNA has the tendency to adopt a mixture of major groove, wedge and base-displaced intercalated conformations when paired against complementary cytosine in the NarI sequence. The biological implications of these multiple conformations are discussed in detail and compared to the corresponding N-linked C8-dG adducts.
MATERIALS AND METHODS
OTB-dG models
OTA is a weak acid with a dissociation constant (pKa) of ∼4.4 for the carboxylic and 7.1 for the phenolic group (55). Thus, the OTB-dG adduct may exist in carboxylic ionized (monoanionic), and carboxylic and phenolic ionized (dianionic) forms at neutral or physiological pH. Thus, both the monoanionic and dianionic OTB-dG protonation states were considered in the present work. For comparison simulations were also conducted on the non-ionized (neutral) adduct, which may not be physiologically relevant.
MD simulations
MD simulations were carried out by incorporating neutral, monoanionic or dianonic OTB-dG at the G1, G2 or G3 position in the NarI helix opposite complementary cytosine. We investigated all conformational themes previously observed by NMR methods for other (N-linked) C8-dG modified duplexes (51), namely the major groove (anti), base-displaced intercalated (syn) and (minor groove) wedge (syn) structures. For each conformation, two rotamers along the axis passing through the fused six-member rings of the OTA isocoumarin moiety were considered, which are related by an ∼180° flip along the torsion angle θ (∠(N9C8C10C11), Figure 1). The rotamers are referred to as α (θ ∼ 0°) and β (θ ∼ 180°; Supplementary Figure S1). The unmodified control sequence was also studied for comparison.
For each structure, an MD simulation was carried out in explicit solvent (water and sodium counterions) for 40 ns using the AMBER program (56,57). The results of these simulations provided stable structures corresponding to each of the α and β-rotamers, where a unimodal distribution with respect to θ and χ was found for all conformers. Free energy calculations were carried out based on the ensemble of structures derived from the MD trajectories, using the Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) method (58), with the AMBER package, which yielded energy rankings for different conformations of adducted DNA. MM-PBSA free energy calculations have been successfully applied in the literature to study the conformational properties of natural and modified DNA (59–61). The ptraj and cpptraj modules (62) of AMBER were used for structural analyses. Additional details of the calculations, including force fields, MD protocol, free energy calculations and structural analyses are provided in the Supplementary Data.
RESULTS
Free energy ranking of different OTB-dG conformations paired against complementary cytosine
α-rotamers are energetically more stable than β-rotamers
Free energy calculations (Table 1) indicate that the α-rotamer (Supplementary Figure S1) is the energetically preferred conformation about the OTB-dG θ torsion angle (Figure 1), with the relative free energy difference being dependent on the conformation adopted (major groove, wedge or base-displaced intercalated) and the ionization state of the OTA-moiety (neutral, monoanionic or dianionic). For the wedge conformations, the α/β relative free energy difference is at least 9 kcal mol−1 in all cases, except the neutral adduct at the G3 position, where the β-rotamer is within 4 kJ mol−1 of the α-rotamer. Similarly, for the base-displaced intercalated structures, the relative free energy difference between the α and β-rotamers ranges from 4 to 22 kcal mol−1, being greatest for dianionic OTA. Thus, the β-rotamer can be substantially destabilized compared to the α-rotamer in both the wedge and base-displaced intercalated conformations. However, the free energies of the major groove conformations with the α and β-rotamers at the G1, G2 or G3 positions fall within 2 kcal mol−1 for at least one OTA ionization state, indicating that both rotamers are energetically accessible for this conformation. This can be rationalized according to reduced steric hindrance in the major groove conformations, which makes the β-rotamer accessible. Since the α-rotamer is more stable than the β-rotamer in all cases, results for the α-rotamer are presented in the main text, while the structural details of both the α and β-rotamers are provided in Supplementary Tables S1–S3 and Supplementary Figures S2–S4.
Table 1. Free energy rankings (kcal mol−1) of adducted DNA conformations with OTB-dG at the G1, G2 or G3 position in the NarI recognition sequencea.
| Lesion | Conformation | Rotamerb | Position | ||
|---|---|---|---|---|---|
| G1 | G2 | G3 | |||
| Major groove | α | 0.0 | 0.0 | 0.0 | |
| Neutral OTB-dGc | β | (8.3) | (0.9) | (2.3) | |
| Wedge | α | 1.6 | 5.1 | 1.7 | |
| β | (14.1) | (34.1) | (5.0) | ||
| Base-displaced (stacked) Intercalated | α | 1.8 | 2.1 | 1.7 | |
| β | (7.5) | (9.6) | (5.4) | ||
| Major groove | α | 0.0 | 0.0 | 7.0 | |
| Monoanionic OTB-dGc | β | (5.5) | (0.7) | (8.5) | |
| Wedge | α | 0.2 | 7.1 | 0.0 | |
| β | (9.5) | (16.5) | (16.3) | ||
| Base-displaced (stacked) | α | 6.3 | 0.7 | 2.9 | |
| Intercalated | β | (10.6) | (5.8) | (23.2) | |
| Major groove | α | 0.5 | 0.0 | 0.0 | |
| Dianionic OTB-dGc | β | (7.1) | (5.4) | (8.3) | |
| Wedge | α | 0.0 | 4.7 | 0.7 | |
| β | (12.7) | (18.3) | (30.2) | ||
| Base-displaced (stacked) | α | 7.5 | 4.4 | 8.3 | |
| Intercalated | β | (18.2) | (18.4) | (12.4) | |
aSee Supplementary Data for details of the free energy calculations.
bSee Supplementary Figure S1 for the definition of the α and β-rotamers.
cSee Figure 1 for the definition of OTA ionization states.
Relative stabilization of the different conformations depends on the sequence context
The α-rotamers of the major groove, wedge and base-displaced intercalated conformations lie within 9 kcal mol−1 when the OTB-dG adduct is at the G1, G2 or G3 position in the NarI recognition sequence (Table 1). The small free energy difference between helices suggests significant conformational heterogeneity for the OTB-dG lesion. Furthermore, this conformational heterogeneity occurs regardless of the bases flanking the OTB-dG (or the OTB-dG position) or the OTA ionization state. However, the free energy ranking of different conformations depends on the sequence context. Specifically, at the G1 position, the major groove and wedge conformations fall within 2 kcal mol−1, while the base-displaced intercalated structures are 2–8 kcal mol−1 higher in energy, for all OTA ionization states. Although the major groove structures remain the most stable regardless of the ionization state at G2, the stacked conformations are more stable than the wedge conformations (by up to 6.4 kcal mol−1). On the other hand, the conformational preference is dependent on the ionization state at the G3 position. Specifically, the wedge conformation is 7 kcal mol−1 more stable than the major groove conformation for the monoanionic adduct, while the major groove conformation is the most stable for the neutral and dianionic states (by 0.7 and 1.7 kcal mol−1 relative to the wedge conformation, respectively). However, the base-displaced intercalated structures lie ∼2–8 kcal mol−1 above the lowest energy conformation for all three OTA forms at G3. Regardless of the position or OTA ionization state considered, at least two of the major groove, wedge and base-displaced intercalated structures lie within 5 kcal mol−1, the energy cutoff used to determine the accessible conformations for an N-linked bulky adduct (63). Thus, a dynamic equilibrium between different conformations will be observed in OTB-dG damaged DNA helices with the lesion paired against cytosine.
Salient structural characteristics of the major groove OTB-dG adducted DNA conformations
Major groove conformations are structurally similar to the unmodified control
The major groove conformations of OTB-dG (Figure 3) retain strong Watson–Crick hydrogen bonding with more than 96% occupancy of each of the three hydrogen bonds (Supplementary Table S4). The corresponding base pairing interaction energy is at least 22 kcal mol−1 (Supplementary Table S1), which is ∼2 kcal mol−1 smaller for neutral OTB-dG compared to the unmodified control (Supplementary Table S1). Interestingly, the deviation from natural DNA increases with OTA ionization, possibly because of enhanced repulsive interactions due to the close proximity of charged OTA and the DNA phosphate groups. Nevertheless, the slight decreases in the hydrogen-bond strength are compensated for by enhanced stacking interactions (by up to 4 kcal mol−1 compared to the natural helix), which are less affected by adduct ionization. Furthermore, the stacking interactions display no significant sequence dependence (Figure 4) in any OTA ionization state. In addition to discrete interactions, general structural features of the adducted helices at the lesion site, such as shift, slide, rise, tilt, roll, twist and minor groove dimensions, are similar to the natural helix for the major groove structures (Supplementary Table S1). The Watson–Crick hydrogen bonding of the flanking base pairs also remains similar to the unmodified control for all adducted helices (Supplementary Table S4). Nevertheless, contrary to the natural NarI helix, which acquires a mixture of C2′-endo and C1′-exo puckers at G1 and G3 and a dynamic range of puckers at G2 (Supplementary Figure S17), OTB-dG predominantly acquires an O4′-endo sugar pucker in the major groove adducted conformation at all three positions, possibly to minimize steric interactions between the bulky moiety and the sugar ring (Supplementary Figures S14).
Figure 3.
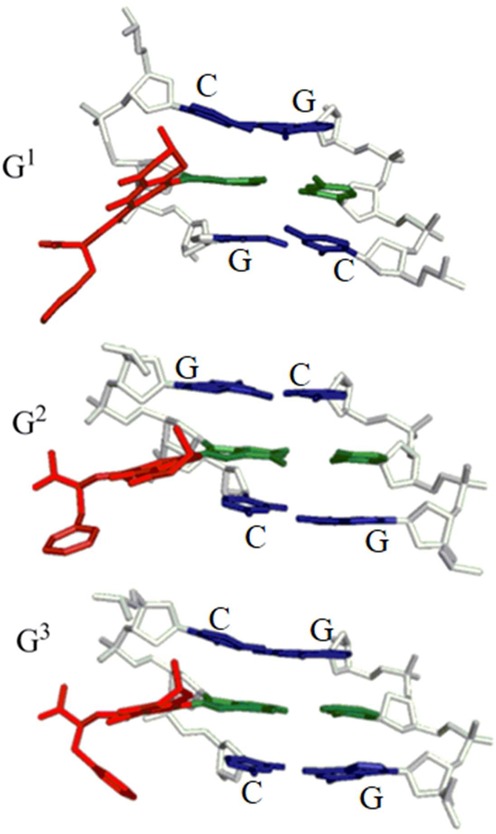
Representative structures of the major groove conformation of adducted DNA with the neutral OTB-dG adduct (α-rotamer) incorporated at the G1, G2 or G3 position in the NarI recognition sequence. Central trimers are shown that include the lesion–base pair (green, with bulky moiety in red) and the flanking base pairs (blue) viewed from the major groove side. The sugar-phosphate backbone is in white and hydrogen atoms are removed for clarity.
Figure 4.
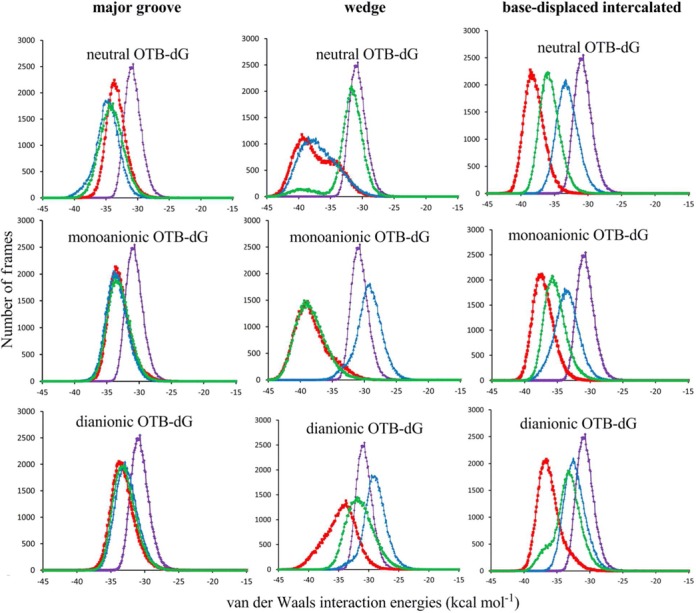
Comparison of the stacking interaction energies involving the OTB-dG adduct in different ionization states at G1 (red), G2 (blue) or G3 (green) in the NarI recognition sequence. The average of the G1, G2 and G3 stacking interactions in the natural sequence is shown in purple.
Salient structural characteristics of the wedge OTB-dG adducted DNA conformations
Strength of the Hoogsteen hydrogen bonding in the wedge conformations depends on the sequence context and adduct ionization state
The wedge conformations of OTB-dG adducted DNA can possess two Hoogsteen hydrogen bonds involving the N4 amino group of the opposing cytosine at G1 and G3, and a single Hoogsteen hydrogen bond at G2, where the occupancies of these hydrogen bonds increase with the charge of the lesion (Figure 5 and Supplementary Table S5). The change in Hoogsteen hydrogen bonding is related to the identity of the 5′ nucleotide with respect to the adduct. Specifically, at the G2 position, the purine (guanine) nucleotide on the 5′-side of the adduct sterically repels the C8-moiety, and thereby pushes the adducted nucleotide slightly out of the helix toward the minor groove. This results in the loss of a Hoogsteen hydrogen bond with the opposing cytosine. However, such steric interactions are greatly reduced when a pyrimidine (cytosine) flanks the 5′-side of the adduct at the G1 and G3 positions.
Figure 5.
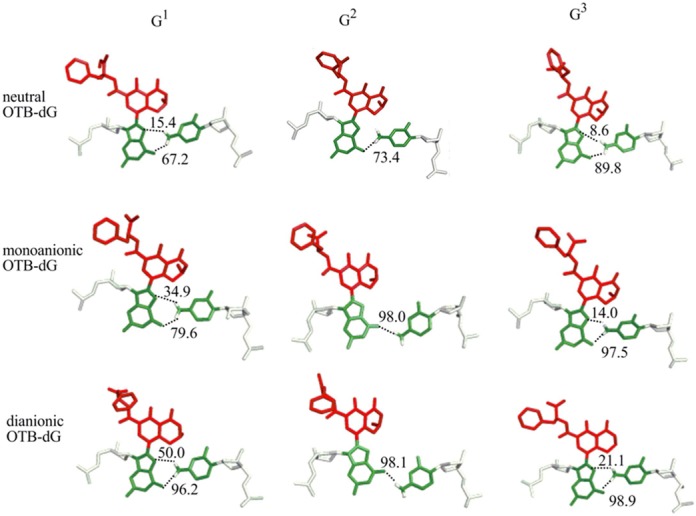
Hydrogen-bonding interactions between OTB-dG in different ionization states at the G1, G2 or G3 positions and the opposing cytosine in adducted helices adopting the wedge conformation. The hydrogen bonds are indicated by dashed lines, and the corresponding occupancies are provided.
In addition to sequence context, the adduct ionization state affects the Hoogsteen hydrogen-bonding interactions between the lesion and the opposing cytosine. Although adduct ionization affects the occupancies of Hoogsteen hydrogen bonds at all three positions, there is also a structural effect at G2. Specifically, the neutral lesion at G2 leads to greater penetration of the C8-moiety into the helix, which is accompanied by a change in the Hoogsteen hydrogen-bonding pattern at the lesion site compared to the ionized OTB-dG lesions (Figure 5).
With the exception of the increase in rise parameter (by at least 0.7 Å) and minor groove width (by at least 2 Å) in the case of the anionic OTB-dG lesions at G2 due to the change in the hydrogen-bonding pattern (Figure 6), no significant structural perturbations are observed at the lesion site compared to the unmodified control helix (Supplementary Table S2). In addition, the base pairs flanking the lesion, as well as those at the terminal ends of the helix, remain intact during the simulations (Supplementary Tables S5 and S7). Nevertheless, the wedge conformations at all three studied positions predominantly acquire a C1′-exo sugar pucker (Supplementary Figure S16).
Figure 6.
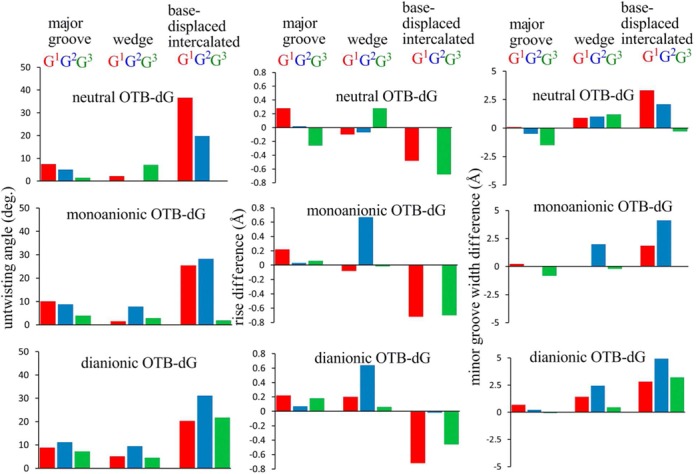
The difference between the twist (represented as the untwisting angle), rise and minor groove width for different conformations of the OTB-dG modified NarI DNA duplex relative to the unmodified control.
Hydrogen-bonding pattern in the wedge conformation influences the sequence dependence of the free energy rankings
As discussed, the wedge conformation of OTB-dG adducted DNA involves two hydrogen bonds at G1 and G3, and a single hydrogen bond at G2 due to the identity of the 5′-flanking base (vide supra). This decrease in hydrogen-bonding strength explains why the wedge conformation is energetically less accessible at G2 compared to G1 and G3 (Table 1). Such a sequence dependence of the conformational preferences has been observed for C8-linked aromatic amine adducts, where the identity of the nearest and next-nearest nucleotides dictates the equilibrium between conformations (35,39,64).
Stacking interactions determine the adduct ionization-dependent stability of the wedge conformation at G2 and G3
Although all three OTA ionization states possess a single hydrogen bond at G2, the hydrogen bonding of the neutral form differs in the cytosine amino hydrogen that participates in hydrogen bonding compared to the ionic forms due to the greater penetration of the opposing C into the helix (Figure 5). This geometric difference leads to deviations in the base pairing energy and stacking strengths of the neutral versus charged lesions at G2. Specifically, the neutral stacking and hydrogen-bonding energies are larger (by up to 7 and 14 kcal mol−1, respectively) than those of the charged forms. Interestingly, this hydrogen-bonding mediated enhancement in stacking in the neutral form at G2 also explains why the stacking interactions at G2 are comparable to those at G1 and G3 for neutral OTB-dG, but weaker for ionic OTB-dG.
Table 1 indicates that the major groove conformations are generally more stable compared to the wedge conformation at all three positions, with the notable exception of the more stable wedge conformation (by 7 kcal mol−1 relative to the major groove conformer) when monoanionic OTB-dG is at G3. Figure 4 indicates that the stacking interactions of the wedge conformation at G3 are significantly enhanced in the monoanionic form compared to the corresponding neutral and dianionic states (by on average 6–7 kJ mol−1, Supplementary Table S2). Furthermore, the average stacking energy of the monoanionic wedge conformation is (5.3 kcal mol−1) greater than for the corresponding major groove conformation. On the other hand, the wedge conformation of the neutral and dianionic forms at G3 have weaker stacking compared to the corresponding major groove conformations (Supplementary Tables S1 and S2). The enhancement in stacking for the monoanionic lesion is partly driven by the formation of an out-of-plane hydrogen bond between N4 of the opposing cytosine and O2 of the cytosine on the 3′ side of the adduct (Figure 7).
Figure 7.
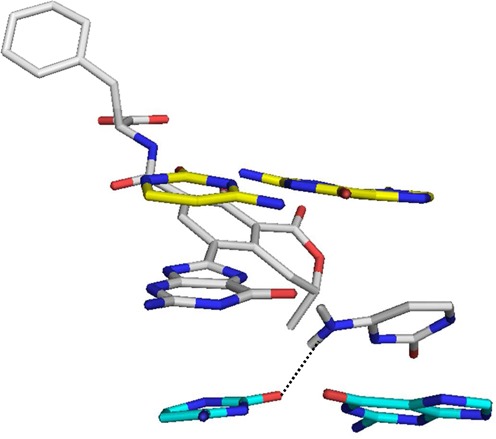
Hydrogen bond between N4 of cytosine opposing the monoanionic OTB-dG adduct at the G3 position and O2 of the 3′ cytosine with respect to the adduct in the NarI adducted wedge conformation.
Salient structural characteristics of the base-displaced intercalated OTB-dG adducted DNA conformations
Base-displaced intercalated structures exhibit sequence and adduct ionization state-dependent increases in duplex untwisting, rise and minor groove dimensions
Intercalation of OTA into the helix to adopt the (base-displaced intercalated) stacked structures results in significant backbone untwisting (by up to 37°) and minor groove enlargement (by up to 5 Å) in the region specific to the adduct, compared to the unmodified control (Figure 6 and Supplementary Table S3). The extent of duplex untwisting is greater with adduction at G1 and G2, with little distortion occurring upon lesion incorporation at G3, especially for neutral and monoanionic OTB-dG. Of the three OTB-dG forms, the dianionic state inflicts the greatest perturbation to the duplex twist and minor groove dimensions (Figure 6 and Supplementary Table S3), possibly due to increased electrostatic repulsion when the highly negatively charged OTB-moiety intercalates into the helix. In addition to untwisting and minor groove enlargement, sequence-dependent differences are found in the rise parameter calculated between the base pairs flanking the lesion (Figure 6 and Supplementary Table S3). Specifically, the rise increases by up to 0.7 Å compared to the unmodified control with the lesion at G1 and G3 irrespective of the ionization state, while no perturbation to the natural parameters occurs upon damage at G2. In addition, the base-displaced intercalated conformations of OTB-dG adducted DNA acquire either an O4′-endo or twisted C1′-exo O4′-endo pucker of 2′-deoxyribose (Supplementary Figure S15).
Opposing cytosine stabilization determines the free energy rankings of the base-displaced intercalated structures
Figure 4 indicates a strong sequence dependence of the stacking interactions in the base-displaced intercalated structures. Specifically, the adduct at G2 exhibits the weakest stacking, followed by G3, while G1 damage leads to the strongest stacking interactions. Although this suggests that the base-displaced intercalated structures will be the least stable when the adduct is at G2, the free energy rankings (Table 1) indicate that the base-displaced intercalated structures are of comparable (or even greater) stability at G2, than G1 or G3. Thus, the stacking interactions do not explain the free energy trend observed for the base-displaced intercalated structures.
Analysis of representative structures (Figure 8) indicates that additional interactions involving the opposing cytosine stabilize the base-displaced intercalated structure at G2. Specifically, when the adduct is at G2, an interaction exists between N4 of the opposing cytosine and the phosphate group of the 5′-guanosine nucleotide (48, 61 and 76% occupancy for the neutral, monoanionic and dianionic lesions, respectively). This base-phosphate interaction is only possible when the cytosine opposing the adduct is flanked on the 5′-side by a purine (guanine) followed by a pyrimidine (cytosine). Such a sequence context brings the phosphate group in close proximity to the extrahelical cytosine. In addition to this base-phosphate interaction, a second minor interaction occurs between O2 of the opposing cytosine and N4 of the 3′-cytosine for the neutral lesion (27% occupancy). However, this interaction only occurs for 7% of the total simulation time for the monoanionic lesion and is not observed for the dianionic lesion. Due to differences in sequence context, no such interactions occur when the adduct is at G1 or G3 (Figure 8).
Figure 8.
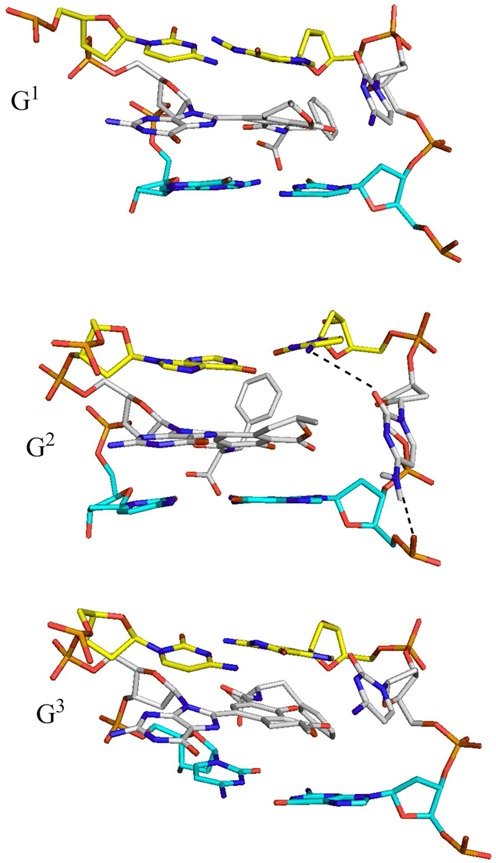
Base-phosphate and nucleobase–nucleobase interactions between the cytosine opposing the dianionic OTB-dG adduct at G2 and its flanking nucleotides in the base-displaced intercalated duplex, as well as the corresponding structures with the adduct at G1 and G3 that lack such interactions.
DISCUSSION
Factors affecting the conformational equilibrium of the OTB-dG lesion
Our extensive MD and free energy analysis suggests that the conformational preferences of OTB-dG adducted DNA are determined by a combination of two factors: (1) the sequence context and (2) the OTA ionization state.
Effect of the sequence context
To understand the sequence dependence of the conformational preferences, we focus on the monoanionic OTB-dG lesion, which is likely to be one of the prevalent OTA ionization states at physiological pH. The monoanionic OTB-dG lesion displays a conformational mixture of the major groove and wedge conformations (within 0.2 kcal mol−1) at G1, major groove and base-displaced intercalated structures (within 0.7 kcal mol−1) at G2 and wedge and base-displaced intercalated conformations (within 2.9 kcal mol−1) at G3 in the NarI sequence (Table 1). This sequence-dependent conformational heterogeneity is driven by discrete hydrogen-bonding interactions within the adducted base pair, and between the adducted pair and the neighboring residues. Specifically, although the major groove conformation is of comparable stability at the G1 and G2 positions, the wedge conformation is destabilized at G2 because of the loss of one Hoogsteen hydrogen bond due to steric hindrance posed by the 5′-guanosine with respect to the adduct. On the other hand, additional base-phosphate interactions involving the opposing extrahelical cytosine stabilize the base-displaced intercalated conformation at G2. The required close proximity between the cytosine and phosphate in the 5′-nucleotide is only observed at G2 when the sequence 5′ with respect to the extrahelical cytosine contains a pyrimidine (cytosine) as the next-nearest neighbor. Similarly, greater stabilization of the wedge conformation at G3 occurs due to an additional hydrogen bond between the opposing cytosine and the 3′-cytosine with respect to the adduct (Figure 7). In addition, enhanced stacking interactions between the OTA moiety at G3 and the 3′ and 5′-guanosines with respect to the opposing cytosine in the wedge and base-displaced conformation may explain why the major groove conformation is energetically less favored upon adduct formation at this position. Thus, there exists an intricate connection between the nature of interbase interactions and the overall stability of adducted DNA.
Effect of the OTA ionization state
The conformational preferences of the OTB-dG adduct are affected by the charge of the OTA moiety, albeit to a lesser extent than the sequence context. In general, ionization of the OTA phenolic group destabilizes the base-displaced intercalated structure at all three NarI positions. This mainly occurs because of enhanced (unfavorable) electrostatics with increasing charge upon intercalation of OTA into the DNA helix. In the case of the wedge conformations, increased OTA charge at G2 leads to relatively smaller penetration of the C8-moiety into the helix, which in turn changes the hydrogen-bonding pattern and relative free energies. On the other hand, since the OTA moiety remains extrahelical in the major groove conformations, the relative free energies of these conformations are less affected by the charge of the OTA moiety.
Conformational heterogeneity of the OTB-dG lesion
Despite the sequence and OTA ionization state dependence of the relative stabilization of the three adducted DNA conformations, the small energetic separation indicates that OTB-dG comparably stabilizes different conformations, including those with the carcinogenic moiety in the major groove, the minor groove and the base-displaced intercalated position. This suggests a possible conformational equilibrium in cells, which is affected by the sequence context. Such tendency to adopt a mixture of conformations has been reported for other adducts including the polycyclic aromatic hydrocarbons (40) and aromatic amine adducts (35,51,53), and is believed to lead to varied mutagenic outcomes (40,65). Thus, it is plausible to hypothesize that the OTB-dG adduct may exist in a mixture of three conformations, each of which may have a different mutagenic outcome.
Comparison of OTB-dG and aromatic amine modified double helices at G1, G2 or G3 in the NarI sequence
Previous studies on the model aromatic amine carcinogen N-acetyl-2-aminofluorene (AAF) bonded to the C8-position of dG have shown that the NarI sequence is a hotspot for deletion mutations in Escherichia coli (39,41,66,67). Therefore, a number of studies have structurally characterized AAF-dG (41) (Figure 9), as well as other aromatic amine adducts bonded to C8-dG, such as those formed from aminofluorene (AF-dG) (52,53) and 2-amino-3-methylimidazo[4,5-f]quinolone (IQ-dG) (Figure 9) (49,54), in the NarI sequence using NMR and molecular modeling techniques. Existing studies on these aromatic amine adducts indicate that subtle changes in the chemical structure of the adduct may strongly influence the conformational preference(s) (41,42,68). However, the OTB-dG adduct is inherently distinct from other C8-DNA (aromatic amine) adducts in a number of ways. First, the OTB-dG belongs to one of the relatively less studied class of C-linked C8-dG adducts in which the bulky carcinogenic moiety is bonded to C8 of guanine through a direct C–C linkage. On other hand, all aromatic amine C8-dG lesions (e.g. AF-dG, AAF-dG, IQ-dG) are N-linked adducts in which the bulky moiety is bonded at C8 through a C–N–C linkage. In other words, the C8-linkage is more flexible in case of aromatic amines. Second, the C8-moiety in OTB-dG contains a fused two ring system (one aromatic carbocyclic ring and one saturated heterocyclic ring), which are connected via an amide linkage to a phenylalaninic moiety. In contrast, the aromatic amine adducts generally contain carbocyclic or heterocyclic ring systems consisting of a single or multiple (fused or biphenyl–type) rings. Third, the identity of functional groups is unique in OTB-dG (phenolic, carboxylic groups and carbonyl groups) compared to most aromatic amines (i.e. acetyl group in AAF-dG), while other aromatic amine adducts do not contain specific functional groups. Most importantly, whereas the OTB-dG adduct can exist in three possible ionization states (neutral, monoanionic and dianionic), no examples of different ionization states of the aromatic amine adducts exist in literature. Therefore, comparison of the conformational preferences and structural details of the OTB-dG adduct to other bulky DNA lesions is of interest to understand how the chemical structure of the bulky moiety governs the conformation of adducted DNA, which may affect the associated mutagenic outcomes.
Figure 9.
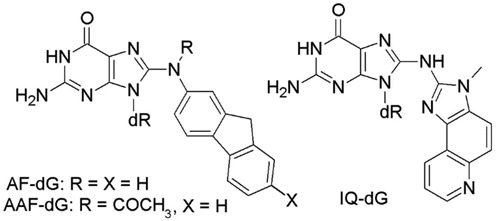
Structures of the AF-dG, AAF-dG and IQ-dG adducts previously studied in the NarI recognition sequence.
Although AAF-dG (41) and OTB-dG adducted DNA (in three ionization states) can acquire three (major groove, wedge and base-displaced intercalated) major helical conformations, the AF-dG (41,52,53) and IQ-dG (54) adducts only acquire two of the three conformations in the NarI sequence. Specifically, the AF-dG adduct only leads to the major groove and base-displaced intercalated conformations when paired against C (41,66,69). In contrast, the IQ-dG lesion acquires a minor groove conformation at G1 and G2, but a base-displaced intercalated structure at G3, and preferentially excludes the major groove conformation observed for other two aromatic amine adducts when paired against C (49,54). Thus, the greater conformational heterogeneity observed for AAF-dG and OTB-dG appears to be due to the presence of additional and flexible groups (the acetyl group in AAF-dG and the amide-linked phenylalaninic moiety in OTB-dG) compared to AF-dG and IQ-dG (no flexible groups in the bulky moiety).
Previous studies on the AF-dG and AAF-dG adducts indicate that the major groove conformations are structurally similar to the unmodified helix, while the base-displaced conformations lead to duplex untwisting and minor groove enlargement (41). Similar results were found in the present study for OTB-dG, indicating that regardless of charge on the C8-moiety, these are general structural characteristics of bulky lesion-modified duplexes. On the other hand, the OTB-dG and AF-dG (41) base-displaced intercalated structures lack the base-phosphate interactions between the adducted guanine and the phosphate atom of the 5′ nucleotide with respect to the adduct that are present upon incorporation of AAF-dG (41) and IQ-dG (49). However, a sequence context exists for (both neutral and charged) OTB-dG such that the base-displaced intercalated conformation can acquire additional stabilization through base-phosphate interactions between the opposing base and its flanking nucleotide (Figure 8). Similar stabilizing hydrogen-bonding interactions have yet to be reported for (neutral) aromatic amine adducts.
Although the OTB-dG and AAF-dG wedge conformations generally have similar Hoogsteen hydrogen bonding at the three guanine positions, adduct ionization-dependent variations in the hydrogen-bonding interactions are seen for OTB-dG. For example, an interstrand and interstep hydrogen bond occurs between the amino group of cytosine opposing the lesion and O2 of the cytosine on the 3′-side of the adduct in the predominant monoanionic form of the OTB-dG lesion (Figure 7), which compensates for the reduced accessibility of the hydrogen bond between amino group of the opposing cytosine and N7 of the adducted guanine (Figure 5). This conformation mainly originates due to steric crowding at the C8-linkage of adducted guanine, which makes N7 less accessible for hydrogen bonding. Furthermore, this steric crowding only occurs in the case of C-linked adducts that, unlike N-linked aromatic amine adducts, lack a flexible (C–N–C) linkage at the C8-site in dG. This reiterates the important finding that adducts with a C–C linkage at C8 (e.g. OTB-dG) display different structural characteristics compared to the relatively well-studied aromatic amine adducts with a C–N–C linkage at C8.
In addition to hydrogen bonding, the trends in the stacking interactions between the lesion-containing base pair and the neighboring pairs vary in the wedge conformation. Specifically, whereas the wedge structure of AAF-dG adducted DNA possess stacking interactions comparable to the unmodified control at all three guanine positions, the stacking interactions are significantly greater at G1 position for all three ionization states of OTB-dG. However, the effect of adduct ionization is more pronounced at G2 position, where the stacking interactions are significantly enhanced for the neutral lesion, and weaker in the case of the anionic forms, relative to the unmodified control. On the contrary, the stacking interactions are comparable to the unmodified control at the G3 position for the neutral form, and significantly enhanced for the anionic forms. Thus, the availability of different ionization of OTB-dG leads to a more complex sequence dependence in the stacking interactions at the lesion site compared to AAF-dG, a representative aromatic amine adduct.
In summary, the differences in size, linkage, composition and charge of the bulky moiety affect discrete interactions in the helix, which in turn affect the sequence dependence of the conformational preferences. Nevertheless, significant conformational heterogeneity is observed for many C8-dG lesions.
Relevance to OTA mutagenesis
Nucleotide excision repair efficiency of the OTB-dG adduct
The mutational outcome of a particular DNA adduct depends on its persistence in the genome, which in turn depends on its ability to be recognized by repair enzymes. It is widely believed that lesions that thermodynamically destabilize and distort DNA are better nucleotide excision repair (NER) substrates (35,39,41), a common pathway for repair of bulky DNA adducts (70). A previous crystal structure of the yeast ortholog of XPC-RAD23B (a recognition factor involved in the initial steps of NER recognition in eukaryotes) bound to DNA reveals that insertion of the β-hairpin through the major groove side of DNA combined with flipping of the opposing base(s) out of the helix are important factors in initial steps of NER recognition (71). Furthermore, modeling studies on polycyclic aromatic hydrocarbon lesions indicate that intercalation of the adducted moiety from the major groove side can possibly obstruct β-hairpin insertion and flipping of the opposing base(s) (72). In contrast, minor groove intercalation of the adducted moiety facilitates β-hairpin insertion and flipping of the opposing base into the minor groove (73).
Our work indicates that the greatest structural distortions to DNA upon OTB-dG incorporation occur in the base-displaced intercalated conformation. Furthermore, in this conformation, the OTA moiety intercalates from the minor groove side, which may assist insertion of the β-hairpin of the NER recognition factor through the major groove side of the adducted DNA. In addition, the β-hairpin insertion will likely be facilitated due to a reduced energetic penalty required for flipping the already extrahelical opposing base out of the helix, which in turn will enhance the adduct repair propensity. Nevertheless, significant thermodynamic stabilization provided by lesion–base stacking interactions to the adducted helix may prevent damage recognition (72), which could in turn prevent recruitment of subsequent NER factors and may provide resistance to NER (73). Regardless, the greater structural distortions and extrahelical position of opposing cytosine likely combine to make the base-displaced intercalated structure prone to repair. However, since the OTA moiety resides in the minor groove in the wedge conformation, it is possible that NER recognition is most favourable in this conformation due to facilitation of β-hairpin insertion. Nevertheless, due to negligible distortion to the DNA helix, the nearly energetically equivalent major groove and wedge conformations may elude repair.
Based on the anticipated greater repair propensity of the base-displaced stacked structure, the neutral and monoanionic OTB-dG will likely be more prone to repair because of a greater preference for this conformation. However, since the dianionic form favors the structurally less distorting major groove and wedge conformations, this charged lesion may evade repair. Furthermore, in addition to the ionization state, the NER recognition of the adduct may depend on the sequence context. For example, due to greater accessibility of the base-displaced intercalated conformation at G2, the monoanionic form of OTB-dG may more likely undergo repair at G2 than G1 and G3. Thus, the differential repair of different conformations of the OTB-dG adduct will lead to its complicated mutagenic profile in cells.
OTA-mediated deletion mutations
As discussed in the Introduction, several studies (13,22,23) strongly suggest the involvement of direct genotoxic effects in OTA-induced mutagenesis, which includes OTA-DNA adduct formation. One OTA-induced mutation that has been experimentally observed is the formation of deletion mutations (28). Since the major groove and wedge conformations are less likely to be repaired, it is possible that adducted DNA adopts these conformations in dsDNA and at the start of replication. However, the small calculated energy difference between the three unique conformations suggests that the major groove or wedge conformation may interconvert to the intercalated conformation at the replication fork (74). This may cause the polymerase to stall and introduce deletion mutations through a base-slippage mechanism (75). Indeed, deletion mutations may be stabilized by the base-displaced intercalated conformation as reported for select aromatic amine adducts (76,77). Thus, the greater accessibility of the base-displaced intercalated conformation observed in the neutral and monoanionic forms of OTB-dG at all three guanine positions indicates that these forms are more likely induce deletion mutations compared to the dianionic form, where the base-displaced intercalated structures are the least accessible. However, in addition to greater accessibility of base-displaced intercalated structure, the neutral lesion exhibits the greatest conformational flexibility at all three guanine positions among the three OTB-dG charged states, being the most flexible at G3. Since increased conformational flexibility at the lesion site has been linked to base misincorporation (78), neutral OTB-dG is likely to induce base-substitution rather than frameshift mutations. Thus, the monoanionic OTA ionization state is the most likely candidate to induce deletion mutations.
Interestingly, a study of gpt delta rats given a carcinogenic dose of OTA using in vivo mutation assays indicates that OTA-induced genotoxic effects only occur in specific regions (outer stripe of the outer medulla) of the kidney rather than the whole kidney (20). Such a differential expression of OTA-induced carcinogenesis in renal tissues can be ascribed to a number of factors, including the difference in metabolic activation of OTA between cells of different tissues, the level of OTA exposure of a particular tissue and the nature of the OTA adduct formed at a specific DNA site (the major (OTB-dG) carbon-linked or the minor (OTA-dG) oxygen-linked adduct) and the existence of alternate adduct conformations. Indeed, such factors are believed to influence the mutagenesis of a variety carcinogenic DNA adducts (76,79–82). Our work verifies for the first time that multiple OTA-dG conformations are energetically accessible. It is possible that some of these (potentially mutagenic) conformations may be more dominant in particular cellular environments compared to other (potentially nonmutagenic) conformations, which thereby explains the observed selective mutagenicity depending on the tissue considered.
OTA-mediated induction of double-strand breaks
Previous studies have shown that the genes associated with homologous recombination repair are activated upon treatment of cells with OTA, indicating the induction of double-strand breaks in DNA (28). The induction of double-strand breaks indicates that OTB-dG is not easily bypassed during replication by alternate enzymes (e.g. during translesion synthesis (83)). Although the present study does not predict the structure of OTB-dG during replication, the active site of replicative enzymes may alter the relatively small energy differences between different conformations, thereby changing the preferred conformation. Double-strand breaks can occur when a DNA lesion blocks replication by inhibiting the replication fork (84). Based on studies on other adducts, OTA replication blockage is likely caused by either a base-displaced intercalated structure that has evaded repair (48) or an intercalated structure formed at the template-primer junction (85). However, our calculated structures highlight the larger and more flexible amide-linked phenylalaninic moiety of OTB-dG in DNA compared to C8-substituents in other C8-dG adducts. As a result, the protrusion of the OTA moiety into either the major or minor DNA groove (i.e. adoption of the major groove or wedge conformation) may also block replicative enzymes. Thus, a combination of the conformational flexibility and large size of this adduct is likely responsible for the observed varied mutagenic outcomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
Computational resources provided by Westgrid and Compute/Calcul Canada are greatly appreciated.
FUNDING
Natural Sciences and Engineering Research Council of Canada (249598–07); Canada Research Chain Program (950–228175); Canadian Foundation of Innovation (22770). Funding for open access charge: Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bayman P., Baker J.L., Doster M.A., Michailides T.J., Mahoney N.E. Ochratoxin production by the aspergillus ochraceus group and aspergillus alliaceus. Appl. Environ. Microbiol. 2002;68:2326–2329. doi: 10.1128/AEM.68.5.2326-2329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abarca M.L., Bragulat M.R., Castellá G., Cabañes F.J. Ochratoxin A production by strains of aspergillus niger var. Niger. Appl. Environ. Microbiol. 1994;60:2650–2652. doi: 10.1128/aem.60.7.2650-2652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabañes F.J., Bragulat M.R., Castellá G. Ochratoxin A producing species in the genus penicillium. Toxins. 2010;2:1111–1120. doi: 10.3390/toxins2051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelhardt G., Barthel J., Sparrer D. Fusarium mycotoxins and ochratoxin A in cereals and cereal products. Mol. Nutr. Food Res. 2006;50:401–405. doi: 10.1002/mnfr.200500191. [DOI] [PubMed] [Google Scholar]
- 5.Jörgensen K. Survey of pork, poultry, coffee, beer and pulses for ochratoxin A. Food Addit. Contam. 1998;15:550–554. doi: 10.1080/02652039809374680. [DOI] [PubMed] [Google Scholar]
- 6.Batista L.R., Chalfoun S.M., Silva C.F., Cirillo M., Varga E.A., Schwan R.F. Ochratoxin A in coffee beans (coffea arabica l.) processed by dry and wet methods. Food Contr. 2009;20:784–790. [Google Scholar]
- 7.Bellí N., Marín S., Sanchis V., Ramos A.J. Review: Ochratoxin A (OTA) in wines, musts and grape juices: occurrence, regulations and methods of analysis. Food Sci. Technol. Int. 2002;8:325–335. [Google Scholar]
- 8.Mally A., Dekant W. Mycotoxins and the kidney: modes of action for renal tumor formation by ochratoxin A in rodents. Mol. Nutr. Food Res. 2009;53:467–478. doi: 10.1002/mnfr.200800149. [DOI] [PubMed] [Google Scholar]
- 9.Qi X., Yang X., Chen S., He X., Dweep H., Guo M., Cheng W.-H., Xu W., Luo Y., Gretz N. Ochratoxin A induced early hepatotoxicity: new mechanistic insights from microRNA, mRNA and proteomic profiling studies. Sci. Rep. 2014;4:5163–5163. [Google Scholar]
- 10.Wangikar P.B., Dwivedi P., Sinha N., Sharma A.K., Telang A.G. Teratogenic effects in rabbits of simultaneous exposure to ochratoxin A and aflatoxin B1 with special reference to microscopic effects. Toxicology. 2005;215:37–47. doi: 10.1016/j.tox.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Al-Anati L., Petzinger E. Immunotoxic activity of ochratoxin A. J. Vet. Pharmacol. Ther. 2006;29:79–90. doi: 10.1111/j.1365-2885.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 12.Sava V., Reunova O., Velasquez A., Harbison R., Sánchez-Ramos J. Acute neurotoxic effects of the fungal metabolite ochratoxin-A. NeuroToxicology. 2006;27:82–92. doi: 10.1016/j.neuro.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Pfohl-Leszkowicz A., Manderville R.A. An update on direct genotoxicity as a molecular mechanism of ochratoxin A carcinogenicity. Chem. Res. Toxicol. 2012;25:252–262. doi: 10.1021/tx200430f. [DOI] [PubMed] [Google Scholar]
- 14.Pfohl-Leszkowicz A., Manderville R.A. Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007;51:1192–1192. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- 15.Petkova-Bocharova T., Castegnaro M. Ochratoxin A in human blood in relation to balkan endemic nephropathy and urinary tract tumours in bulgaria. IARC Sci. Publ. 1991;115:135–137. [PubMed] [Google Scholar]
- 16.Schwartz G.G. Hypothesis: does ochratoxin A cause testicular cancer. Cancer Causes Control. 2002;13:91–100. doi: 10.1023/a:1013973715289. [DOI] [PubMed] [Google Scholar]
- 17.Boorman G. NIH Publication No. 89–2813, U.S. Department of Health and Human Services. Research Triangle Park, NC: National Institutes of Health; 1989. NTP technical report on the toxicology and carcinogenesis studies of ochratoxin A (CAS no. 303–47–9) in F344/N rats (gavage studies) [Google Scholar]
- 18.Kujawa M. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 56. 38:351–351. [Google Scholar]
- 19.Manderville R.A. A case for the genotoxicity of ochratoxin A by bioactivation and covalent DNA adduction. Chem. Res. Toxicol. 2005;18:1091–1097. doi: 10.1021/tx050070p. [DOI] [PubMed] [Google Scholar]
- 20.Hibi D., Suzuki Y., Ishii Y., Jin M., Watanabe M., Sugita-Konishi Y., Yanai T., Nohmi T., Nishikawa A., Umemura T. Site-specific in vivo mutagenicity in the kidney of gpt delta rats given a carcinogenic dose of ochratoxin A. Toxicol. Sci. 2011;122:406–414. doi: 10.1093/toxsci/kfr139. [DOI] [PubMed] [Google Scholar]
- 21.Klarić M., Daraboš D., Rozgaj R., Kašuba V., Pepeljnjak S. Beauvericin and ochratoxin A genotoxicity evaluated using the alkaline comet assay: single and combined genotoxic action. Arch. Toxicol. 2010;84:641–650. doi: 10.1007/s00204-010-0535-7. [DOI] [PubMed] [Google Scholar]
- 22.Mantle P.G., Faucet-Marquis V., Manderville R.A., Squillaci B., Pfohl-Leszkowicz A. Structures of covalent adducts between DNA and ochratoxin A: a new factor in debate about genotoxicity and human risk assessment. Chem. Res. Toxicol. 2009;23:89–98. doi: 10.1021/tx900295a. [DOI] [PubMed] [Google Scholar]
- 23.Dai J., Wright M.W., Manderville R.A. Ochratoxin A forms a carbon-bonded C8-deoxyguanosine nucleoside adduct: implications for C8 reactivity by a phenolic radical. J. Am. Chem. Soc. 2003;125:3716–3717. doi: 10.1021/ja034221r. [DOI] [PubMed] [Google Scholar]
- 24.Faucet V., Pfohl-Leszkowicz A., Dai J., Castegnaro M., Manderville R.A. Evidence for covalent DNA adduction by ochratoxin A following chronic exposure to rat and subacute exposure to pig. Chem. Res. Toxicol. 2004;17:1289–1296. doi: 10.1021/tx049877s. [DOI] [PubMed] [Google Scholar]
- 25.Dai J., Park G., Perry J.L., Il'ichev Y.V., Bow D.A.J., Pritchard J.B., Faucet V., Pfohl-Leszkowicz A., Manderville R.A., Simon J.D. Molecular aspects of the transport and toxicity of ochratoxin A. Acc. Chem. Res. 2004;37:874–881. doi: 10.1021/ar0302134. [DOI] [PubMed] [Google Scholar]
- 26.Hibi D., Kijima A., Suzuki Y., Ishii Y., Jin M., Sugita-Konishi Y., Yanai T., Nishikawa A., Umemura T. Effects of p53 knockout on ochratoxin A-induced genotoxicity in p53-deficient gpt delta mice. Toxicology. 2013;304:92–99. doi: 10.1016/j.tox.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Hibi D., Kijima A., Kuroda K., Suzuki Y., Ishii Y., Jin M., Nakajima M., Sugita-Konishi Y., Yanai T., Nohmi T., et al. Molecular mechanisms underlying ochratoxin A-induced genotoxicity: global gene expression analysis suggests induction of DNA double-strand breaks and cell cycle progression. J. Toxicol. Sci. 2013;38:57–69. doi: 10.2131/jts.38.57. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda K., Hibi D., Ishii Y., Takasu S., Kijima A., Matsushita K., Masumura K.-i., Watanabe M., Sugita-Konishi Y., Sakai H., et al. Ochratoxin A induces DNA double-strand breaks and large deletion mutations in the carcinogenic target site of gpt delta rats. Mutagenesis. 2014;29:27–36. doi: 10.1093/mutage/get054. [DOI] [PubMed] [Google Scholar]
- 29.Kuiper-Goodman T., Hilts C., Billiard S.M., Kiparissis Y., Richard I.D.K., Hayward S. Health risk assessment of ochratoxin A for all age-sex strata in a market economy. Food Addit Contam. A. 2009;27:212–240. doi: 10.1080/02652030903013278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.EFSA. Opinion of the scientific panel on contaminants in the food chain on a request from the commission related to ochratoxin A in food, efsa-q-2005–154. EFSA J. 2006;365:1–56. [Google Scholar]
- 31.Mally A., Zepnik H., Wanek P., Eder E., Dingley K., Ihmels H., Völkel W., Dekant W. Ochratoxin A: lack of formation of covalent DNA adducts. Chem. Res. Toxicol. 2004;17:234–242. doi: 10.1021/tx034188m. [DOI] [PubMed] [Google Scholar]
- 32.Mally A., Pepe G., Ravoori S., Fiore M., Gupta R.C., Dekant W., Mosesso P. Ochratoxin A causes DNA damage and cytogenetic effects but no DNA adducts in rats. Chem. Res. Toxicol. 2005;18:1253–1261. doi: 10.1021/tx049650x. [DOI] [PubMed] [Google Scholar]
- 33.Obrecht-Pflumio S., Dirheimer G. In vitro DNA and dGMP adducts formation caused by ochratoxin A. Chem.-Biol. Inter. 2000;127:29–44. doi: 10.1016/s0009-2797(00)00169-1. [DOI] [PubMed] [Google Scholar]
- 34.Obrecht-Pflumio S., Dirheimer G. Horseradish peroxidase mediates DNA and deoxyguanosine 3′-monophosphate adduct formation in the presence of ochratoxin A. Arch. Toxicol. 2001;75:583–590. doi: 10.1007/s00204-001-0289-3. [DOI] [PubMed] [Google Scholar]
- 35.Cho B.P. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J. Environ. Sci. Heal. C. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 36.Donny-Clark K., Broyde S. Influence of local sequence context on damaged base conformation in human DNA polymerase ι: molecular dynamics studies of nucleotide incorporation opposite a benzo[a]pyrene-derived adenine lesion. Nucleic Acids Res. 2009;37:7095–7109. doi: 10.1093/nar/gkp745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rechkoblit O., Kolbanovskiy A., Malinina L., Geacintov N.E., Broyde S., Patel D.J. Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by y-family polymerase Dpo4. Nat. Struct. Mol. Biol. 2010;17:379–388. doi: 10.1038/nsmb.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitri A., Jia L., Shafirovich V., Geacintov N.E., Broyde S., Scicchitano D.A. Transcription of DNA containing the 5-guanidino-4-nitroimidazole lesion by human RNA polymerase ii and bacteriophage T7 RNA polymerase. DNA Repair. 2008;7:1276–1288. doi: 10.1016/j.dnarep.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel D.J., Mao B., Gu Z., Hingerty B.E., Gorin A., Basu A.K., Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine−DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 40.Kozack R., Seo K.-Y., Jelinsky S.A., Loechler E.L. Toward an understanding of the role of DNA adduct conformation in defining mutagenic mechanism based on studies of the major adduct (formed at N2-dG) of the potent environmental carcinogen, benzo[a]pyrene. Mutat. Res.-Fund. Mol. M. 2000;450:41–59. doi: 10.1016/s0027-5107(00)00015-4. [DOI] [PubMed] [Google Scholar]
- 41.Mu H., Kropachev K., Wang L., Zhang L., Kolbanovskiy A., Kolbanovskiy M., Geacintov N. E., Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;40:9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millen A.L., Sharma P., Wetmore S.D. C8-linked bulky guanosine DNA adducts: experimental and computational insights into adduct conformational preferences and resulting mutagenicity. Future Med. Chem. 2012;4:1981–2007. doi: 10.4155/fmc.12.138. [DOI] [PubMed] [Google Scholar]
- 43.Il'ichev Y.V., Perry J.L., Simon J.D. Interaction of Ochratoxin A with Human Serum Albumin. Preferential binding of the dianion and pH effects. J. Phys. Chem. B. 2002;106:452–459. [Google Scholar]
- 44.Calcutt M. W., Gillman I. G., Noftle R. E., Manderville R. A. Electrochemical oxidation of Ochratoxin A: correlation with 4-Chlorophenol. Chem. Res. Toxicol. 2001;14:1266–1272. doi: 10.1021/tx015516q. [DOI] [PubMed] [Google Scholar]
- 45.Fuchs R.P.P., Schwartz N., Daune M.P. Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature. 1981;294:657–659. doi: 10.1038/294657a0. [DOI] [PubMed] [Google Scholar]
- 46.Kuska M.S., Witham A.A., Sproviero M., Manderville R.A., Majdi Yazdi M., Sharma P., Wetmore S.D. Structural influence of C8-phenoxy-guanine in the NarI recognition DNA sequence. Chem. Res. Toxicol. 2013;26:1397–1408. doi: 10.1021/tx400252g. [DOI] [PubMed] [Google Scholar]
- 47.Sharma P., Manderville R.A., Wetmore S.D. Modeling the conformational preference of the carbon-bonded covalent adduct formed upon exposure of 2′-deoxyguanosine to ochratoxin A. Chem. Res. Toxicol. 2013;26:803–816. doi: 10.1021/tx4000864. [DOI] [PubMed] [Google Scholar]
- 48.Stavros K.M., Hawkins E.K., Rizzo C.J., Stone M.P. Base-displaced intercalation of the 2-amino-3-methylimidazo[4,5-f]quinolone N2-dG adduct in the NarI DNA recognition sequence. Nucleic Acids Res. 2014;42:3450–3463. doi: 10.1093/nar/gkt1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F., DeMuro N.E., Elmquist C.E., Stover J.S., Rizzo C.J., Stone M.P. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for −2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rankin K.M., Sproviero M., Rankin K., Sharma P., Wetmore S.D., Manderville R.A. C8-heteroaryl-2′-deoxyguanosine adducts as conformational fluorescent probes in the NarI recognition sequence. J. Org. Chem. 2012;77:10498–10508. doi: 10.1021/jo302164c. [DOI] [PubMed] [Google Scholar]
- 51.Patnaik S., Cho B.P. Structures of 2-acetylaminofluorene modified DNA revisited: insight into conformational heterogeneity. Chem. Res. Toxicol. 2010;23:1650–1652. doi: 10.1021/tx100341u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao B., Hingerty B.E., Broyde S., Patel D.J. Solution structure of the aminofluorene [AF]-intercalated conformer of the syn-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998a;37:81–94. doi: 10.1021/bi972257o. [DOI] [PubMed] [Google Scholar]
- 53.Mao B., Hingerty B.E., Broyde S., Patel D.J. Solution structure of the aminofluorene [AF]-external conformer of the anti-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998b;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 54.Wang F., Elmquist C.E., Stover J.S., Rizzo C.J., Stone M.P. DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme. Biochemistry. 2007;46:8498–8516. doi: 10.1021/bi700361u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiodini A.M., Scherpenisse P., Bergwerff A.A. Ochratoxin A contents in wine: comparison of organically and conventionally produced products. J. Agric. Food Chem. 2006;54:7399–7404. doi: 10.1021/jf0613482. [DOI] [PubMed] [Google Scholar]
- 56.Case D.A., Darden T.A., Cheatham T.E.I., Simmerling C.L., Wang J., Duke R.E., Luo R., Crowley M., Walker R.C., Zhang W., et al. Amber 11. San Francisco, CA: University of California; 2010. [Google Scholar]
- 57.Case D.A., Darden T.A., Cheatham T.E.I., Simmerling C.L., Wang J., Duke R.E., Luo R., Walker R.C., Zhang W., Merz K.M., et al. Amber 12. San Francisco, CA: University of California; 2012. [Google Scholar]
- 58.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.Py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 59.Suresh G., Priyakumar U.D. Structures, dynamics, and stabilities of fully modified locked nucleic acid (β-D-LNA and α-L-LNA) duplexes in comparison to pure DNA and rna duplexes. J. Phys. Chem. B. 2013;117:5556–5564. doi: 10.1021/jp4016068. [DOI] [PubMed] [Google Scholar]
- 60.Yan S., Wu M., Patel D.J., Geacintov N.E., Broyde S. Simulating structural and thermodynamic properties of carcinogen-damaged DNA. Biophys. J. 2003;84:2137–2148. doi: 10.1016/S0006-3495(03)75020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding S., Shapiro R., Cai Y., Geacintov N.E., Broyde S. Conformational properties of equilenin−DNA adducts: stereoisomer and base effects. Chem. Res. Toxicol. 2008;21:1064–1073. doi: 10.1021/tx800010u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Case D.A., Darden T.A., Cheatham T.E., III, Simmerling C.L., Wang J., Duke R.E., Luo R., Crowley M., Walker R.C., Zhang W., et al. Amber Tools. San Francisco, CA: University of California; 2008. [Google Scholar]
- 63.Nolan S.J., Vyas R.R., Hingerty B.E., Ellis S., Broyde S., Shapiro R., Basu A.K. Solution properties and computational analysis of an oligodeoxynucleotide containing N-(deoxyguanosin-8-yl)-1-aminopyrene. Carcinogenesis. 1996;17:133–144. doi: 10.1093/carcin/17.1.133. [DOI] [PubMed] [Google Scholar]
- 64.Meneni S.R., D'Mello R., Norigian G., Baker G., Gao L., Chiarelli M.P., Cho B.P. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kozack R.E., Shukla R., Loechler E.L. A hypothesis for what conformation of the major adduct of (+)-anti-B[a]PDE (N2-dG) causes G→T versus G→A mutations based upon a correlation between mutagenesis and molecular modeling results. Carcinogenesis. 1999;20:95–102. doi: 10.1093/carcin/20.1.95. [DOI] [PubMed] [Google Scholar]
- 66.Jain V., Hilton B., Patnaik S., Zou Y., Chiarelli M.P., Cho B.P. Conformational and thermodynamic properties modulate the nucleotide excision repair of 2-aminofluorene and 2-acetylaminofluorene dG adducts in the NarI sequence. Nucleic Acids Res. 2012;40:3939–3951. doi: 10.1093/nar/gkr1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burnouf D., Koehl P., Fuchs R.P. Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc. Natl Acad. Sci. U.S.A. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho B. Structure–function characteristics of aromatic amine-DNA adducts. In: Geactinov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. pp. 217–238. [Google Scholar]
- 69.Abuaf P., Hingerty B.E., Broyde S., Grunberger D. Solution conformation of the N-(deoxyguanosin-8-yl)aminofluorene adduct opposite deoxyinosine and deoxyguanosine in DNA by NMR and computational characterization. Chem. Res. Toxicol. 1995;8:369–378. doi: 10.1021/tx00045a008. [DOI] [PubMed] [Google Scholar]
- 70.Costa R.M.A., Chiganças V., da Silva Galhardo R., Carvalho H., Menck C.F.M. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 71.Min J.-H., Pavletich N.P. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 72.Cai Y., Zheng H., Ding S., Kropachev K., Schwaid A.G., Tang Y., Mu H., Wang S., Geacintov N.E., Zhang Y., et al. Free energy profiles of base flipping in intercalative polycyclic aromatic hydrocarbon-damaged DNA duplexes: energetic and structural relationships to nucleotide excision repair susceptibility. Chem. Res. Toxicol. 2013;26:1115–1125. doi: 10.1021/tx400156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reeves D.A., Mu H., Kropachev K., Cai Y., Ding S., Kolbanovskiy A., Kolbanovskiy M., Chen Y., Krzeminski J., Amin S., et al. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion–base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rechkoblit O., Zhang Y., Guo D., Wang Z., Amin S., Krzeminsky J., Louneva N., Geactinov N.E. Trans-lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 75.Hoffmann G.R., Fuchs R.P.P. Mechanisms of frameshift mutations: insight from aromatic amines. Chem. Res. Toxicol. 1997;10:347–359. doi: 10.1021/tx960128n. [DOI] [PubMed] [Google Scholar]
- 76.Mao B., Hingerty B.E., Broyde S., Patel D.J. Solution conformation of [AF]dG opposite a -2 deletion site in a DNA duplex: intercalation of the covalently attached aminofluorene ring into the helix with base displacement of the C8-modified syn guanine and adjacent unpaired 3'-adenine into the major groove. Biochemstry. 1997;34:16641–16653. doi: 10.1021/bi00051a012. [DOI] [PubMed] [Google Scholar]
- 77.Mao B., Cosman M., Hingerty B.E., Broyde S., Patel D.J. Solution conformation of [AF]dG opposite a -1 deletion site in a DNA duplex: intercalation of the covalently attached aminofluorene ring into the helix with base displacement of the C8-modified syn guanine into the major groove. Biochemistry. 1995;34:6226–6238. doi: 10.1021/bi00018a027. [DOI] [PubMed] [Google Scholar]
- 78.Perlow-Poehnelt R.A., Likhterov I., Wang L., Scicchitano D.A., Geactinov N.E., Broyde S. Increased flexibility enhancesmisincorporation. Temperature effects on nucleotide incorporation opposite abulky carcinogen-DNA adduct by a Y-familyDNA polymerase. J. Biol. Chem. 2007;82:1397–1408. doi: 10.1074/jbc.M606769200. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez H., Loechler E.L. Mutational specificity of the (+)-anti-diol epoxide of benzo[a]pyrene in a supf gene of an Escherichia coli plasmid: DNA sequence context influences hotspots, mutagenic specificity and the extent of SOS enhancement of mutagenesis. Carcinogenesis. 1993;14:373–383. doi: 10.1093/carcin/14.3.373. [DOI] [PubMed] [Google Scholar]
- 80.Fernandes A., Liu T., Amin S., Geacintov N.E., Grollman A.P., Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2–2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 81.Makino H., Ishizaka Y., Tsujimoto A., Nakamura T., Onda M., Sugimura T., Nagao M. Rat p53 gene mutations in primary zymbal gland tumors induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Proc. Natl Acad. Sci. U. S. A. 1992;89:4850–4854. doi: 10.1073/pnas.89.11.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watanabe M., Ohta T. Analysis of mutational specificity induced by heterocyclic amines in the lacZ gene of Escherichia coli. Carcinogenesis. 1993;14:1149–1153. doi: 10.1093/carcin/14.6.1149. [DOI] [PubMed] [Google Scholar]
- 83.Stallons L.J., McGregor W.G. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J. Nucleic Acids. 2010;2010:643857. doi: 10.4061/2010/643857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bi X., Slater D.M., Ohmori H., Vaziri C. DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced s-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 85.Mao B., Gu Z., Gorin A., Hingerty B.E., Broyde S., Patel D.J. Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction. Biochemistry. 1997;36:14491–14501. doi: 10.1021/bi972206r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


