Abstract
In human chromosomes, centromeric regions comprise megabase-size arrays of 171 bp alpha-satellite DNA monomers. The large distances spanned by these arrays preclude their replication from external sites and imply that the repetitive monomers contain replication origins. However, replication within these arrays has not previously been profiled and the role of alpha-satellite DNA in initiation of DNA replication has not yet been demonstrated. Here, replication of alpha-satellite DNA in endogenous human centromeric regions and in de novo formed Human Artificial Chromosome (HAC) was analyzed. We showed that alpha-satellite monomers could function as origins of DNA replication and that replication of alphoid arrays organized into centrochromatin occurred earlier than those organized into heterochromatin. The distribution of inter-origin distances within centromeric alphoid arrays was comparable to the distribution of inter-origin distances on randomly selected non-centromeric chromosomal regions. Depletion of CENP-B, a kinetochore protein that binds directly to a 17 bp CENP-B box motif common to alpha-satellite DNA, resulted in enrichment of alpha-satellite sequences for proteins of the ORC complex, suggesting that CENP-B may have a role in regulating the replication of centromeric regions. Mapping of replication initiation sites in the HAC revealed that replication preferentially initiated in transcriptionally active regions.
INTRODUCTION
Human centromere DNA comprises 171 bp AT-rich alpha-satellite monomers that are arranged into tandem arrays (or alphoid arrays) that typically span 3–5 Mb (1–6). Within each human centromeric region, alpha-satellites are organized as multimeric, higher-order repeats (HORs) arrays flanked by heterogeneous monomers lacking periodicity or hierarchy (monomeric alpha-satellite arrays) (5,7,8). Regions within the HORs directly interact with proteins involved in kinetochore assembly and HORs are competent for de novo centromere establishment (i.e. formation of human artificial chromosomes or HACs) (9,10). HOR units are enriched with a 17-bp binding motif for centromere protein B (CENP-B box) that is required for de novo kinetochore formation as well as for prevention of CENP-A nucleosome assembly at ectopic chromosomal sites (11,12).
Accurate and complete duplication of the eukaryotic genome relies on the initiation of DNA replication from specific chromosomal regions, termed replication origins, and follows a reproducible temporal program during S phase (13,14). Redundancy of centromeric repeats poses significant challenges to study their replication. Therefore, little is known about what determines the replication timing of centromeric domains and the spatial distribution of replication initiation within those domains has not yet been described.
In this work, we analyzed replication of alphoid DNA arrays in endogenous human centromeric regions and in the structurally characterized de novo formed alphoidtetO-HAC (15,16). The HAC was engineered from a synthetic alphoid DNA dimer with one monomer from a chromosome 17 HOR carrying CENP-B box linked to a consensus alpha-satellite monomer into which a tetracycline operator (tetO) sequence was incorporated. Replication initiation patterns were analyzed by different approaches, including fiber-FISH, chromatin immunoprecipitation and analysis of nascent-strand DNA (nsDNA).
Our data revealed that alpha-satellite monomers can function as replication origins, that alpha-satellite-containing centromeric repeats within heterochromatin and centrochromatin domains replicate during mid-late S phase, and that firing of replication origins throughout the centromeric repeats seems to be a stochastic process. In the context of the HAC, however, replication initiation is suppressed within alpha-satellite arrays, probably due to the proximity of transcriptionally active regions that preferentially initiate DNA replication. CENP-B depletion experiments support the role of this protein in regulating the replication of centromeric regions.
MATERIALS AND METHODS
Cell culture
For replication analysis, 11 human cells lines were used. HEK293 embryonic kidney cells, K562 erythroleukemia cells and MCF7 breast cancer cells were previously described (17). Ovarian cancer OV:ADR-RES cells and lung cancer LC:H266 cells are from a panel of the NCI-60 cell lines derived from nine tissue-of-origin types of cancer (18). Three HAC-containing cells, fibrosarcoma HT1080 cells–AB221821 (15), mesenchymal MSC cells–HAC-LoxP (19) and renal carcinoma 786-0 cells–HAC-VHL (20) were described in our previous publications. Three HT1080 clones, AB2-2-2, AB2-5-23 and AB2-5-30, with integration of the alphoidtetO-array into host chromosomes were isolated during experiments on de novo HAC formation (15). Conditions for culturing these cells are described in the corresponding publications.
Replication timing analysis
Timing analyses were made as previously described (21). S phase was divided into four fractions ranging from early-to-late S phase and designated S1 to S4.
Isolation of nascent-strand DNA
Nascent DNA was prepared using lambda exonuclease method as previously described (17,22).
Measuring of inter-origin distances
DNA molecular combing, detection of IdU and CldU and image scanning were carried out as previously described (23,24).
ChIP assay and real-time polymerase chain reaction
ChIP with antibodies against H3K4me2 (Upstate), H3K4me3 (Upstate), H3K27me3 (Upstate), Orc2 (Abcam), Cdt1 (Santa Cruz), Cdc6 (Santa Cruz), Treslin and CENP-B (Millipore) was carried out according to a previously described method (15,16). Anti-Treslin was a custom-made rabbit monoclonal antibody (epitomics) raised against a treslin peptide (aa 1566–1583: cASGLPKLRIKKIDPSSSL) recognizing a 220 kD protein in whole cell protein extracts and validated with truncated Treslin cDNA transfected cells. These antibodies were bound to the magnetic beads (Invitrogen) with constant rotation for 4 h at 4°C. DNA chromatin from nuclear lysate was prepared. HT1080 cells that were grown in DMEM supplemented with 10% FBS to 80% confluent were trypsinized and fixed with 1% Formaldehyde (Sigma) for 10 min at 37°C. The excess of Formaldehyde was quenched by 125 mM Glycine (Sigma) for 5 min at room temperature. Chromatins were harvested from approximately 1 × 106 cells by addition of 250 μl of lysis buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8, Proteinase inhibitor cocktail (Sigma) and incubated on ice for 10 min. DNA chromatin was shredded to 200–500 bp by sonication using Bioruptor (CosmoBio) with high setting pulse (30 s on, 30 s off) for 10 min at 4°C. Sonicated chromatin was diluted in 500 μl of washing buffer and kept on ice. Magnetic beads bound antibodies were washed and resuspended in sonicated chromatin. ChIP sample was rotated overnight at 4°C. After successive washing, antigen–DNA complexes were isolated from magnetic beads by elution buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, followed by addition of Proteinase K buffer containing 10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.35% SDS, 50 mg Proteinase K (Roche) and incubated at 42°C for 1 h. To reverse the cross-link of protein-DNA, sample was then incubated for 4 h at 65°C. DNA was subjected to phenol/chloroform extraction and pure DNA was precipitated with Ethanol in the presence of 20 μg/μl Glycogen (Roche). The recovery ratio of the immunoprecipitated DNA relative to input DNA was measured by real-time polymerase chain reaction (PCR) using iQ SYBR Green Supermix (Bio-Rad, USA). Data were analized using Bio-Rad iQ5 software (Bio-Rad) and Excell (Microsoft). A set of primers used for ChIP analyses and real-time PCR is listed in Supplementary Table S1. Antibodies used in this study are listed in Supplementary Table S2. All experiments were repeated three times with each sample prepared in triplicate.
Metaphase FISH
Fluorescent in situ hybridization on metaphase spreads was performed as described in our previous publications (15,20). HACs were visualized with bio-tetO and dig-RCA vector probes. Images were captured using Zeiss axiophot with 1 × 40/oil ocular (Zeiss) and analyzed using IP lab software (Signal Analytic).
Immuno-FISH
Immunofluoresence FISH was performed as described previously (20).
Depletion of CENP-B by siRNA
Prevalidated siRNA used to deplete CENP-B (L003250-00-0005) as well as non-targeting control pool of siRNA (D-001810-10-05) were purchased from Dharmacon. HT1080 cells were grown on 6-well plates in DMEM supplemented by 10% FBS for overnight with 50% confluent. Cells were transfected with 20 nM of siRNA-CENP-B and siRNA pool (control) using Lipofectamin RNAi max (Invitrogen) reagent according to the protocol provided by manufacture. Transfected cells were incubated for 48 h. The level of CENP-B depletion in the cells was determined by western blot. The treated cells were used to evaluate a level of enrichment of alphoid DNA sequences in nsDNA using qualitative real-time PCR.
Nascent DNA mapping to alpha-satellite in endogenous centromeric regions
Available nascent DNA (nsDNA) datasets from K562 and MCF5 cell lines (17, SRA: SRR190808-23, including four genomic controls) were mapped to an extensive alpha-satellite DNA library from a single individual (HuRef, (25) and all annotated subsets included in the current human reference assembly (hg19/GRCh37, 26). Prior to mapping, alpha satellite containing reads genomic control datasets (SRR190812,13 and SRR190822,23) were identified by k-mer mapping (where k = 24, 36 and 50) to contain 2–2.4% alpha-satellite, concordant with previous genome-wide estimates (26,27). nsDNA datasets were largely consistent in alpha-satellite estimates of K562 and MCF7 replicates (K562, mean 0.27% SD 0.04, MCF7 mean 0.79%, SD 0.1, removing outlier SRR190818). Mapping of paired read datasets were performed initially using Burrows-Wheeler Aligner (BWA) (28), seeding 24 bp, selecting for best alignments with a threshold of two mismatches/35 bp, or 94.5%. Attention to concordant paired read mapping in hg19 and HuRef alpha-satellite reads identified a final dataset of representing an average of 86.3% reads with an observed published insert length (∼120 bp). nsDNA reads that map specifically to the DXZ1 array (27) were normalized to each respective dataset and studied with reference to the DXZ1 12-mer higher order repeat consensus.
Quantification of alphoid DNA transcripts in CENP-B-depleted cells
Real-time RT-PCR was carried out with RNA purified from CENP-B-depleted (L003250-00-0005) HT1080 cells, cells treated with si-Pool (D-001810-10-05) and cells without treatment. The level of CENP-B depletion in the cells was determined by western blot. The level of transcription was carried out using SYBR® Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. Total RNA was prepared with the RNeasy Mini Kit (Qiagen) with on-column DNase treatment using RNase-Free DNase Set (Qiagen). First-strand cDNA synthesis was performed using Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) (Invitrogen) according to the manufacturer's protocol. Reverse transcription and PCR were carried out with the primers for two high-order repeats, D5Z1 and D5Z2, in chromosome 5 listed in Supplementary Table S1.
Statistical analysis
Statistical analysis was made using Excel (Microsoft Inc., USA). An unpaired Student's t-test was used. P < 0.05 was considered significant.
RESULTS
New reagents for detection of DNA replication
Comparative analysis of replication initiation patterns within centromeric repeats requires positive standards comprising replication initiation sites (termed ‘replication origins’) that function reproducibly in a range of cell lines. However, we found that the activities of several previously well-characterized origins, which had been identified in several different cell lines, are not conserved among other cells. To identify a DNA sequence that we could use as a uniform replication origin standard in all our analyzes, we searched a recent database of nsDNA sequences (17) and identified two new origins of replication, one near the JUNB gene on chromosome 19 (29) and the other within the 5S rRNA gene cluster on chromosome 1 (30). Real-time PCR analysis of RNA-primed, short nascent DNA strands purified based on size fractionation and λ exonuclease mediated selection of RNA-primed DNA showed that the JUNB and 5S rDNA loci exhibited origin activity in HT1080 fibrosarcoma cells (Figure 1A-D). Moreover, JUNB seems to be a constitutive origin because it is functional in seven other randomly chosen human cell lines (Figure 1E). The JUNB locus initiates DNA replication with a variable initiation frequency within a relatively short genomic region (Figure 1D and E) similar to other previously described replication origins (13).
Figure 1.
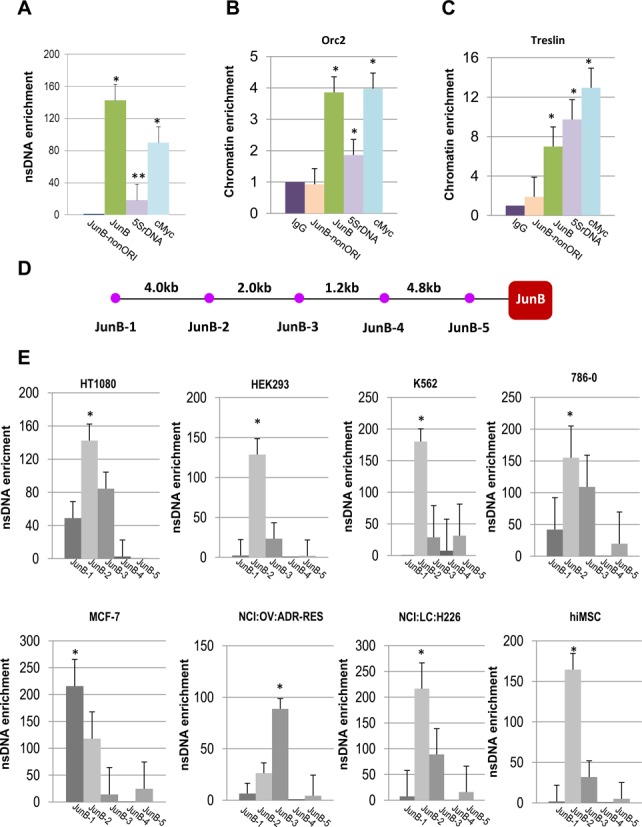
(A) Identification of new origins of replication, JunB and 5S rDNA in HT1080 cells. A real-time PCR-based nascent DNA enrichment assay (A) and ChIP assays with antibodies against components of pre-replication complex (B, C). cMyc origin was included as a positive control. Nascent DNA was normalized with JunB-nonORI (JunB-5). Precipitated chromatins for all antibodies were normalized with IgG. (D, E) Mapping of replication origin sites upstream the JunB gene in different human cell lines using nsDNA assay. Positions of primers used to map replication initiation sites within the JunB locus. A set of primers separated by 1.2 to 4.8 kb were chosen for mapping. Positions of primers sequences on the chromosome 19 in hg19 are shown in Supplementary Table S1. nsDNA enrichment in the JunB regions in different human cell lines. In each cell line, a profile of nascent DNA enrichment around JunB was characterized. In all analyzed cell lines, except for MCF-7 and NCI:OV:ADR-RES, the highest peak of nsDNA abundance corresponds to the JunB-2 sequence. In MCF-7 and NCI:OV:ADR-RES cells, origin of replication is mapped to the JunB-1 and JunB-3 sequences, respectively. Student's t-test was used: P < 0.01 (*) and P < 0.05 (**), ±SD.
To determine whether pre-replication complexes are present at the JUNB and 5S rDNA putative origin loci, ChIP assays were performed using antibodies against Orc2 and Cdt1. We also used antibodies against Treslin recently developed in our lab (see Materials and Methods). Treslin is required for the transition from the pre-replication complex (pre-RC) to the pre-initiation complex (pre-IC) (31). ChIP assays clearly demonstrated that replication origins at the JUNB and 5S rDNA loci bind Orc2, Treslin and Cdt1 whereas adjacent sequences at the same loci that do not initiate replication do not (Figure 1B and C; Supplementary Figure S1A–C). As predicted, positions of replication initiation sites correlate with positions of open chromatin defined by ChIP analysis (Supplementary Figure S1D–F; Supplementary Figure S2). The newly identified JUNB replication origin and a known origin near the cMyc gene (32) along with antibodies against Orc2, Cdt1 and Treslin were used as standards to characterize the replication of centromeric regions.
Mapping origins of replication across a de novo formed centromere
The study of initiation of replication within natural alpha-satellite arrays is impeded by redundancy of nearly identical alphoid DNA units. In attempts to overcome this problem, we first mapped origins of replication on the alphoidtetO-HAC, which was engineered from an alphoid array that is distinguishable from the chromosomal arrays (15). This HAC was developed from a synthetic alphoid DNA dimer assembled into a 40-kb array and cloned into the 10-kb YAC/BAC vector (∼50 kb input DNA) (Figure 2A). Physical analysis of the HAC revealed that alphoidtetO-HAC formation was accompanied by an approximately 30-fold multimerization of the input DNA (16). A concatenated structure of the HAC DNA is clearly visualized by fiber-FISH (Figure 2B).
Figure 2.
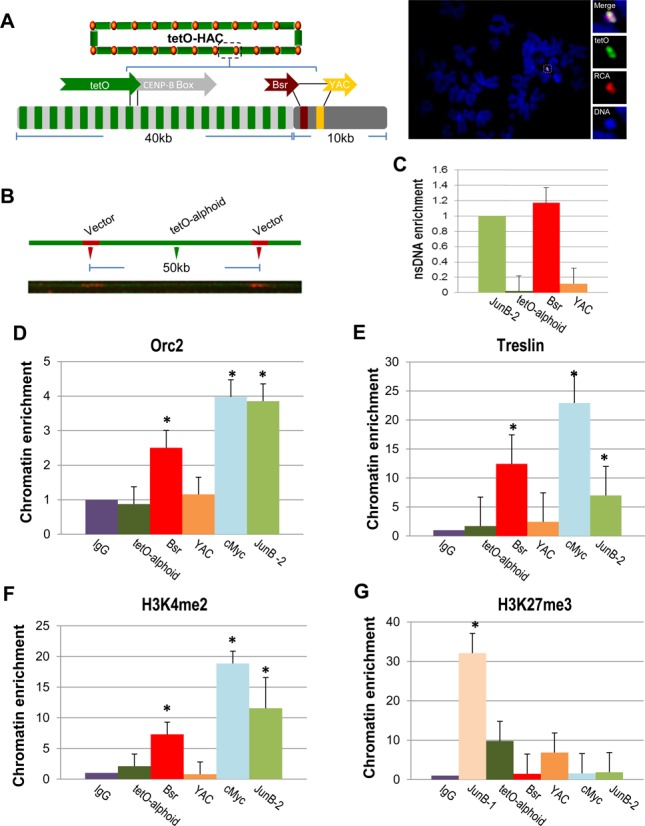
Mapping of replication initiation events within the alphoidtetO-HAC sequence. (A) Schematic of the HAC organization. The circular alphoidtetO-HAC was assembled from input DNA comprising ∼10 kb of RCA vector sequence (BAC/YAC) containing the Bsr gene and ∼40 kb of the synthetic tetO-alphoid array. The synthetic array is composed by tetO alphoid monomer (in green) arrange within every other monomer from chromosome 17 comprising CENP-B box (in gray). FISH demonstrates the presence of an autonomous form of alphoidtetO-HAC in HT1080 cells. The HAC was visualized using a vector probe (RCA) (in red) and tetO-alphoid probe (in green). (B) DNA fiber-FISH analysis using a YAC/BAC vector probe demonstrating a regular structure of the HAC, i.e. a 10 kb YAC/BAC vector signals (in red) are flanked with a 40 kb alphoidtetO array (in green). (C) Real-time PCR-based nascent DNA enrichment assay to map origins of replication along the HAC sequence. (D, E) ChIP-qPCR with antibodies against Orc2 and Treslin shows enrichment of ORC on the Bsr sequences. JUNB-2 and cMyc were used as positive controls. (F) Chromatin profiling of the alphoidtetO-HAC shows H3K4me2 accumulation on the Bsr sequences. JUNB-2 and cMyc were used as positive controls. (G) ChIP analysis of H3K27me3 chromatin in the HAC. Student's t-test was used: P < 0.01 (*), ±SD.
To investigate replication of the alphoidtetO-array in the HAC propagated in HT1080 cells, we used a real-time PCR-based nsDNA enrichment assay. To this end, we selected primer sets to amplify DNA regions corresponding to: (i) tetO-containing alpha-satellite DNA, (ii) the YAC backbone sequence in the YAC/BAC vector and (iii) the selectable marker sequence, blasticidin (Bsr), included into the vector to select the HAC in human cells. Reproducibly, the peaks of highest origin activity mapped to the Bsr gene sequence rather than tetO-alpha-satellite DNA (Figure 2C). The analysis of nsDNA enrichment also showed that the YAC sequence neighboring Bsr in the vector may also function as an origin of replication, but with a lower efficiency, thus forming a zone of replication similar to that observed for JUNB and other euchromatic origins (Figure 2C). This pattern of replication was not cell-type specific. Similar results were obtained when the HAC was transferred into mesenchymal human cells (MSC) and 786-0 renal carcinoma cells (Supplementary Figure S3A–D). In those cell lines, replication initiation zones also occurred within actively transcribed Bsr sequences. Mapping of replication origins to the Bsr sequences in the HAC was also validated by ChIP assays using antibodies against Treslin and Orc2 (Figure 2D and E).
ChIP analysis was also employed to characterize chromatin architecture across the HAC chromatin. The analysis revealed that H3K4me2 and H3K4me3, markers for active transcription, were predominantly associated with the Bsr sequence (Figure 2F; Supplementary Figure S3E). In contrast, H3K27me3, a marker for bivalent, repressible chromatin (33–35), was higher across the tetO-containing alphoid sequence than that at Bsr (Figure 2G).
We also investigated whether centromere assembly on the alphoidtetO-array affects the position of replication origins. For this purpose, we analyzed three independent integrations of the alphoidtetO-array into chromosome arms in HT1080 cells. None of these sites exhibited evidence of kinetochore assembly (15). Origins of replication were associated with the Bsr sequences in all three cell lines (Supplementary Figure S4).
We conclude that neither host cells nor the presence of kinetochore proteins affect the selection of Bsr sequences as origins of DNA replication within the megabase-size alphoidtetO -array.
Timing of replication of centromeric regions
We next examined the replication of endogenous natural centromeric regions. First, we determined the timing of replication of centromeric DNA. For this purpose, an asynchronous population of human fibrosarcoma HT1080 cells was pulse labeled with BrdU. An asynchronous population of cells was used to avoid artifacts introduced by drug-induced synchronization procedures. BrdU-treated cells were sorted by FACS to six fractions: G1, S1 (early S-phase), S2 (early-middle S-phase), S3 (late-middle S-phase), S4 (late S-phase) and G2/Mitosis. In each fraction, replicating sequences were quantified by qPCR using primers designed from an alpha-satellite consensus sequence (2). Notably that sequencing of the PCR products confirmed that these primers recognized centromeric repeats of different chromosomes (Supplementary Table S3). As internal controls, the replication timing of three unique genomic regions, cMyc, JUNB and 5S rDNA, was determined. These origins correspond to transcribed loci and, therefore, as expected, replicate in early S phase (Figure 3A–D). These experiments demonstrated two features of centromere replication. First, centromeres replicate asynchronously. Second, most of the alpha-satellite sequences replicate in the middle to late S phase (Figure 3E). These results are consistent with previous reports indicating that DNA replication and centromere assembly do not overlap during the cell cycle (36–40).
Figure 3.
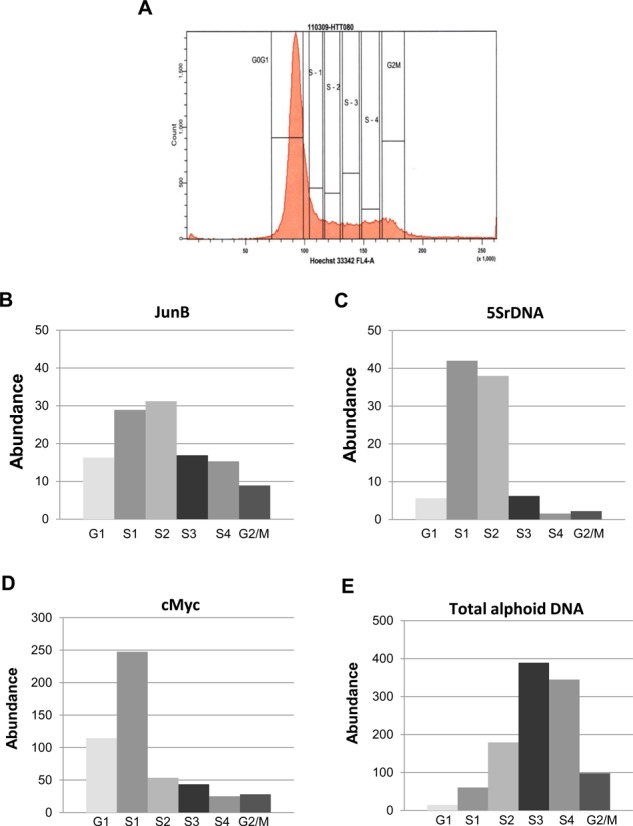
Replication timing of alphoid arrays. (A) Replication timing was measured by sorting BrdU-labeled cells into different S-phase fractions followed by BrdU-IPs. BrdU-labeled cells were stained with Hoechst 33342 and sorted to six fractions by FACS: G1, S1, S2, S3, S4 and G2/M. (B–D) Replication timing of JunB, 5S rDNA and cMyc sites. They replicate early in S phase. (E) Enrichment of BrdU-labeled alpha-satellite DNA in fractions identified by PCR using primers for the alpha-satellite consensus sequence. This experiment suggests late replication of alphoid DNA.
It is known that alpha-satellite sequences forming kinetochore are organized into higher-order repeats (HORs). In most human chromosomes, alpha-satellite DNA forms a single HOR. Therefore, both heterochromatin and centrochromatin are formed on the same DNA sequences. This feature had previously prevented analysis of timing of replication of these two different chromatin domains separately using the standard tools. Therefore, we compared the timing of replication of heterochromatin and centrochromatin domains forming a functional kinetochore by taking advantage of human chromosome 5 that comprises two HORs, D5Z1 and D5Z2, corresponding to different types of chromatin (33). Earlier chromatin immunoprecipitation analysis revealed that the kinetochore forming CENP-A domain (centrochromatin) is predominantly localized to D5Z2 while largely excluded from D5Z1 (heterochromatin) in a range of human cell lines (41). These two arrays can be distinguished by PCR using specific pairs of primers (41). To determine if the timing of replication of these HORs is different or the same, an asynchronous population of human fibrosarcoma HT1080 cells was pulse labeled with BrdU, then sorted by FACS to six fractions as described above (G1, S1 (early), S2 (early-middle), S3 (late-middle), S4 (late) and G2/Mitosis). In each fraction replicating sequences were quantified by qPCR using a set of primers specific to each HOR array. As internal controls, two known chromosomal origins, β-globin origin that replicates in late S phase, and collagen origin that replicates in early S phase, were used (17). As seen, the majority of alpha-satellite sequences in D5Z2 replicate earlier (S2) than those in heterochromatic D5Z1 region (S3) (Figure 4). These data indicate that chromatin status of alpha-satellite repeats may affect their timing of replication.
Figure 4.
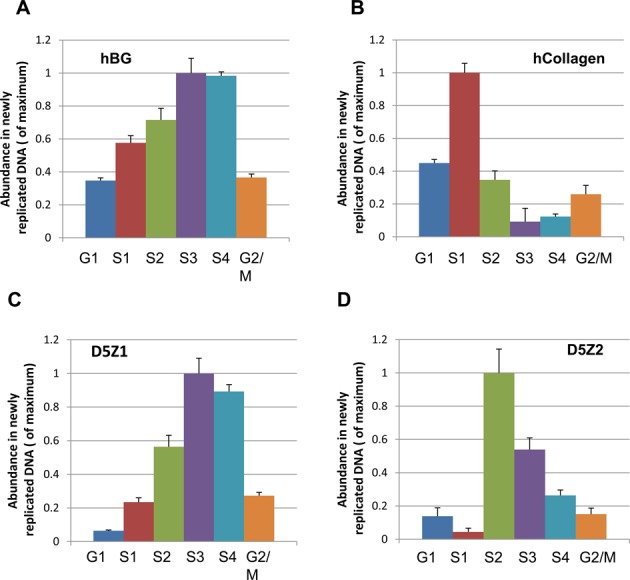
Replication timing of two different HORs, D5Z1 and D5Z2, on chromosome 5. Replication timing was measured by sorting BrdU-labeled cells into different S-phase fractions followed by BrdU-IPs. BrdU-labeled cells were stained with Hoechst 33342 and sorted to six fractions by FACS: G1, S1, S2, S3, S4 and G2/M. (A, B) Replication timing of β-globin and collagen origins. They replicate in early and late S phase, correspondingly. (C, D) Enrichment of BrdU-labeled alpha-satellite DNA in fractions identified by PCR using specific primers for D5Z1 and D5Z2 HORs. Statistically significant difference was observed between S2 and S3 phases for D5Z1 and D5Z2 in three experiments (t-test, N = 3, P < 0.05).
Inter-origin distances on centromeric and non-centromeric regions are identical
In separate experiments, we measured the inter-origin distance within centromeric regions. For this purpose, a single-molecule approach based on molecular combing was used (16,23,24). An asynchronous population of cells was sequentially pulse labeled with the nucleotide analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) for an equal time of 20 min each. Cells were immediately harvested and embedded in agarose plugs to prepare high-molecular-weight genomic DNA. DNA fibers were then prepared by molecular combing with a constant and sequence-independent stretching factor. Centromeric regions were visualized by a biotinylated probe developed from the 171-bp alpha-satellite DNA consensus (2). This probe recognizes all centromeric regions (Figure 5A). Newly synthesized DNA labeled with IdU and CldU was detected by fluorescent antibodies (in blue and in red, respectively). Images were acquired with an epifluorescence microscope (Figure 5B). Using this approach, more than 100 inter-origin distances corresponding to alphoid arrays and random chromosomal regions were measured and plotted. Our analysis did not reveal any significant difference between the periodicity of inter-origin distances on centromeric regions and on random chromosomal fragments (Figure 5C and D). Both exhibited a peak at about 140 kb, suggesting that inter-origin distances are roughly similar between euchromatin and heterochromatin.
Figure 5.
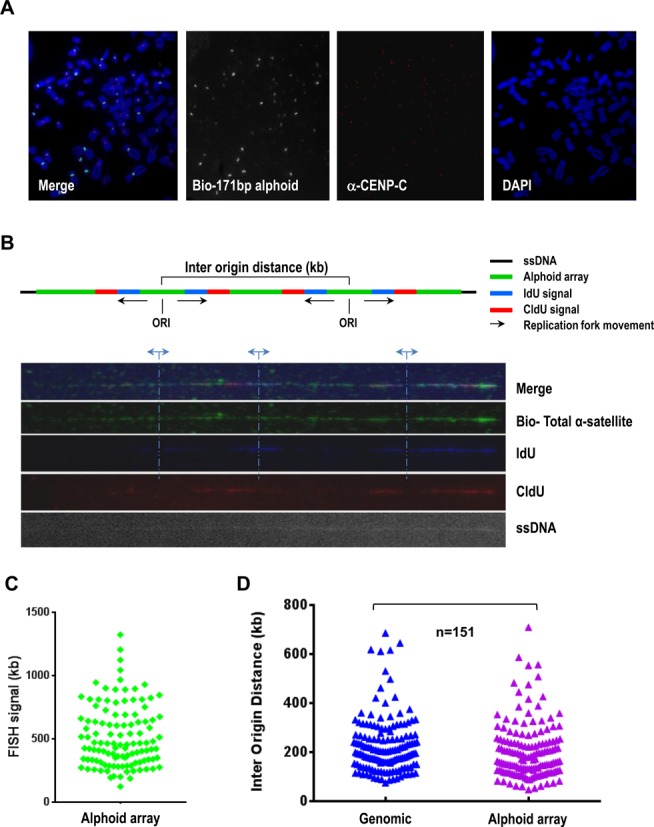
Measuring of replication fork speeds and inter-origin distances within alphoid DNA arrays. (A) Immuno-metaphase FISH assay. A biotinylated alpha-satellite consensus probe was used to visualize human centromeres. Abs against CENP-C were used to stain functional kinetochores (in red). (B) Adjacent replication bubbles were visualized on a combed DNA molecule. Cells were sequentially labeld with IdU (in blue) and CldU (in red) for 20 min. Replication origins were determined by the patterns of IdU-blue and CldU-red signals as described previously (23,24). The distance between adjacent replication origins represents the inter-origin distance. Alphoid DNA arrays were probed with a biotinylated alpha-satellite consensus probe (in green). DNA fibers were visualized by anti-single-strand DNA antibodies (ssDNA). (C) Histogram of the lengths of alphoid DNA arrays identified by an alpha-satellite consensus probe (median = 550). (D) Histogram of inter-origin distances within the alphoid DNA arrays (median = 140). Each triangle represents an inter-origin distance. The shortest measured distance is ∼48 kb and the largest is ∼710 kb. Inter-origin distances on centromeric repeats and on random chromosomal fragments were not distinguishable. P = 0.0134, Mann–Whitney test, n = 151.
Replication of centromeric regions initiates from alpha-satellite repeats
The results of the molecular combing analyses could not exclude that initiation of replication might start from non-alpha-satellite sequence elements such as LINEs and other mobile elements that might potentially be present within alphoid arrays (42–44). To clarify whether the pre-replication complex forms on alphoid DNA and not on adjacent sequences, we performed ChIP assays and nascent DNA strand abundance analyses using primers and probes specific for alphoid DNA. Cross-linked chromatin was purified and tested using antibodies against components of the pre-replication complex. DNA was recovered from antibody–chromatin complexes and tested by PCR using a pair of primers designed from the alpha-satellite consensus sequence (2). The consensus primers recognize ∼50,000 monomers corresponding to both HORs and monomeric arrays. Because HORs represent >90% of all alpha-satellite DNA, these primers should predominantly amplify this fraction of alphoid DNA. This analysis revealed that alphoid arrays could be pulled down by Orc2 and Treslin antibodies (Figure 6A and B in pink). Thus, we conclude that the alpha-satellite repeats investigated above indeed participate in the formation of pre-replication complexes. At the same time, ChIP analysis using the primers ChrX-CEN, corresponding to another fraction of mega-base HOR array (DXZ1–derived from chromosome X that represents approximately a 100-kb sequence (15)) did not detect any enrichment for Orc2 and Treslin (Figure 6A and B in yellow). These data indicate that not all blocks of alpha-satellite repeats are recognized equally by the replication machinery and participate in the formation of pre-replication complexes.
Figure 6.
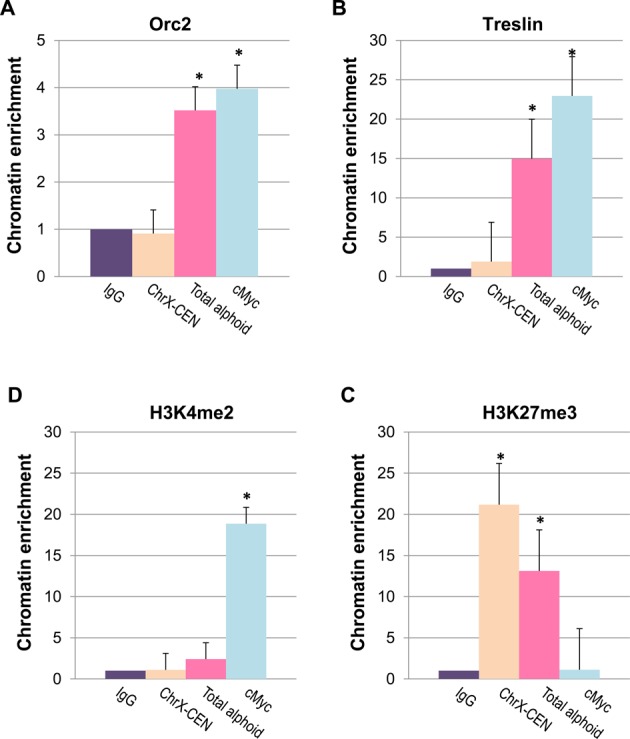
Origin replication complex is assembled on alphoid DNA monomers. (A, B) ChIP-qPCR with antibodies against Orc2 and Treslin shows enrichment of ORC and Treslin on alpha-satellite monomers. The enrichment was observed with the primers developed from the alpha-satellite consensus sequence. The primers recognize a major fraction of monomers in the human genome. No enrichment was detected with other primers, ChrX-CEN, that recognize a 100 kb centromeric region on the chromosome X. (C, D) ChIP for markers of transcriptionally competent or neutral chromatin and bivalent, repressible chromatin, H3K4me2 and H3K27me3, correspondingly. Precipitated chromatins for all antibodies are normalized with IgG. Student's t-test was used: P < 0.01 (*), ±SD.
To determine if some specific repeat monomers may preferentially initiate DNA replication, we compared the published nsDNA sequences isolated from two human female cell lines, K562 and MCF7 (17), with a recently published complete sequence representation of alpha-satellite monomers across a centromeric region of one of human chromosomes (27). Supplementary Figure S5 demonstrates alignments of the 2-kb chromosome X-assigned higher-order repeat units relative to nsDNA. As seen, there is a correlation between sites of nsDNA enrichments and monomers that contain CENP-B boxes. Significance of this observation may be clarified when a complete sequence representation of other chromosome-specific HORs is available.
ChIP with antibodies against various post-translational modifications of histone H3 revealed that centromeric sequences recognized by the consensus primers are associated with H3K27me3 (Figure 6C), a marker for a bivalent chromatin that has previously been linked with centromeres (33). They are also associated to a lesser extent with H3K4me2, a marker for a transcriptionally competent or neutral chromatin (Figure 6D). This result may be interpreted to suggest that replication of alphoid DNA sequences may be initiated both in heterochromatin and centrochromatin corresponding to transcriptionally competent chromatin (45,46).
Depletion of CENP-B activates initiation of replication on alphoid DNA
CENP-B binding to CENP-B boxes influences nucleosome positioning in centromeric regions (47), which can be a factor for origin selection in yeast (48). Also, one of the CENP-B homologs in fission yeast was initially identified as an ARS-binding protein (49). Therefore, we analyzed if the CENP-B protein plays a role in replication of human centromeric repeats. For this purpose, we depleted CENP-B from HT1080 cells. We observed a significant decrease in CENP-B levels in the cells treated by siRNA (Figure 7A). We used ChIP analysis to compare the assembly of pre-replication complexes on alphoid sequences from CENP-B depleted and control cells. Three pairs of primers were chosen: two primer pairs specific for two regions of centromeric HORs on the chromosome X, ChrX-CEN (see above) and ChrX-HOR (these primers recognize most of higher-order repeats), as well as consensus primers recognizing total alpha-satellite DNA. CENP-B depletion was accompanied by CENP-B protein loss from alpha-satellite DNA and increase of the level of H3K4me2, a marker for a transcriptionally competent or neutral chromatin assembled on all three analyzed regions (Figure 7B). At the same time, ChIP assays with antibodies directed against protein members of the pre-replication complex, including Treslin and Orc2, revealed an increased enrichment of alpha-satellite sequences in CENP-B-depleted cells than in control cells (Figure 7C). These data correlate with a 4-fold increase in the abundance of alpha-satellite sequences in nsDNA isolated from CENP-B-depleted cells while no detectable change in enrichment of the cMyc origin sequence was observed (Figure 7D). Importantly, ChIP with Orc2 and Treslin antibodies revealed the same level of enrichment of JunB and cMyc sequences in control and CENP-B-depleted cells (Supplementary Figure S6), suggesting that the observed increase in replication origins is specific for centromeric arrays. These results suggest that the loss of CENP-B protein from alpha-satellite chromatin causes a change of that chromatin, permitting a general increase in replication origin assembly and activity. However, DNA molecular combing analysis did not reveal a significant difference in the inter-origin distances on alphoid DNA arrays between CENP-B-depleted cells and control cells (Supplementary Figure S7A). The results taken together imply that there are more initiation events within the alphoid array, but that those events are not necessarily close to each other. Therefore, we cannot detect a decrease in inter-origin distances using the combing technique.
Figure 7.
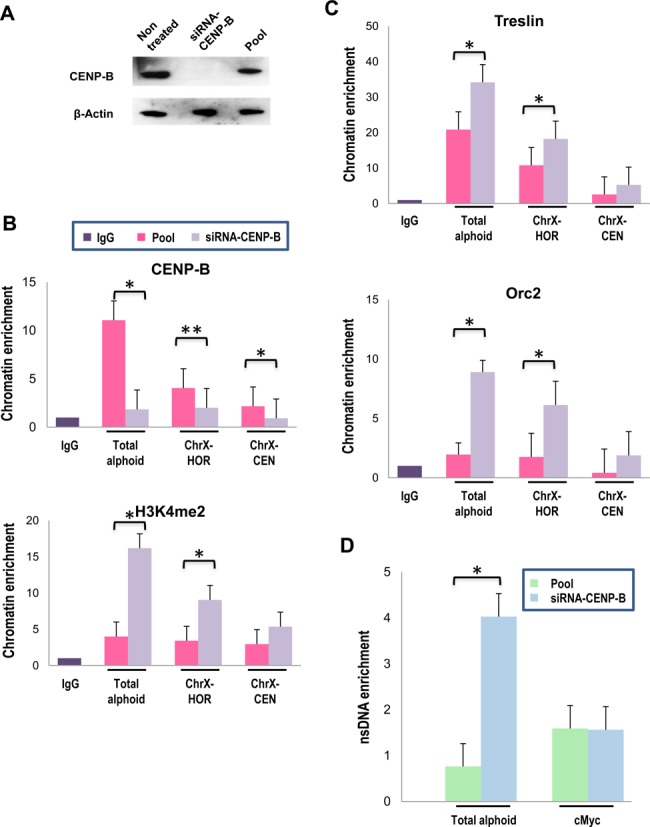
Depletion of the centromeric protein CENP-B activates initiation of replication from alpha-satellite DNA monomers. (A) Western blot analysis demonstrates the level of the CENP-B protein in the cells treated by siRNA-CENP-B and control cells treated by a pool of non-target-siRNA. β-actin was used as a loading control. (B) ChIP-qPCR analysis of CENP-B-depleted cells with antibodies against CENP-B and H3K4me2. A level of alpha-satellite DNA sequences was quantified using the primers for the alpha-satellite consensus sequence and for HORs array from the chromosome X. (C) Abundance of alpha-satellite sequences in the fraction of nsDNA isolated from CENP-B depleted and control cells. ChIP-qPCR with antibodies against Orc2 and Treslin. Precipitated chromatins for all antibodies were normalized with IgG. (D) nsDNA enrichment analysis of alphoid regions after CENP-B depletion. Levels of enrichment are presented relatively to the enrichment for the JunB origin of replication. Student's t-test was used: P < 0.01 (*) and P < 0.05 (**), ±SD.
To further confirm that CENP-B depletion results in a specific change of alpha-satellite chromatin, we applied qRT-PCR analysis to measure the level of transcription of well-characterized HORs, D5Z1 and D5Z2, on chromosome 5. As shown in Supplementary Figures S7B and S7C, we did not observe any significant changes in the level of alpha-satellite transcripts from the D5Z1 region lacking CENP-B boxes in CENP-B-depleted cells. At the same time, a significant increase in the transcription level was detected for D5Z2 array after depletion of CENP-B. To summarize, our data indicate a potential role of the CENP-B protein in replication of centromeric regions.
DISCUSSION
The study of centromeric DNA replication in humans has been impeded by the massive redundancy of centromeric repeats. Indeed, each human centromere contains 10,000–20,000 alpha-satellite monomers. Most (>90%) of these comprise highly homogeneous HORs. This organization prevents physical contig assembly of centromeric regions, which still remain as the largest gaps in the human genome (50).
Here, for the first time we present data showing that alpha-satellite monomers initiate DNA replication in endogeneous human centromeric regions. Moreover, DNA fiber-FISH analysis revealed no significant difference between the ∼140-kb inter-origin distance on centromeric repeats and on randomly selected chromosomal regions. For tandem alpha-satellite repeats, this distance corresponds to ∼800 monomers. Therefore, approximately one in 800 monomers might initiate replication. Due to a high homology between monomers, the initiation of replication within alphoid DNA arrays is likely to be a stochastic process. Thus, each monomer in the arrays could potentially have a chance to initiate replication. However, our findings do not exclude the possibility that some monomers are recognized by the pre-replication complex better than others. For example, chromosome X monomers containing the CENP-B box are enriched in the nsDNA fraction (see Supplementary Figure S5).
It is commonly accepted that transcriptionally active regions replicate early during S phase while regions organized into heterochromatin typically replicate later during S phase (51,52). Thus, a relatively broad distribution of replication timing observed across alphoid DNA might indicate that heterochromatin and centrochromatin domains of kinetochores replicate in different time. Indeed, a comparison of two HOR arrays on chromosome 5, that are organized into compact constitutive heterochromatin (D5Z1) and relatively open centrochromatin (D5Z2), revealed that the centrochromatin region replicates earlier than D5Z1.
Roughly one third of alpha-satellite monomers contain a recognition site for the kinetochore CENP-B protein. In this work, siRNA-induced depletion of CENP-B was exploited to investigate a possible role of CENP-B boxes in replication of alphoid arrays. ChIP assay revealed a significant enrichment for proteins of the pre-replication complex, Orc2 and Treslin, on alpha-satellite sequences in CENP-B-depleted cells compared to control cells. This suggests that CENP-B is involved in regulation of assembly of pre-replicative complexes within long alphoid arrays. Two possible mechanisms may be considered. The first one is based on the known data that binding of CENP-B can promote heterochromatin formation (11). In that case, histone H3 heterochromatic modifications of the alphoid arrays may limit the number of sites that participate in pre-replication complex assembly. A second possible mechanism is based on the known phenomenon of nucleosome positioning that is induced by CENP-B (47) and observation that CENP-B boxes locate on the linker DNA separating two neighboring CENP-A nucleosomes (53,54). In yeast, nucleosome positioning is a major factor for origin selection (48). Therefore, we can speculate that binding of CENP-B to linker DNA may suppress assembly of multiple origins on arrays. Both mechanisms suggest a change of chromatin structure on alpha-satellite repeats after removal of the CENP-B protein. It is worth noting that the observed enrichment for proteins of the pre-replication complex in CENP-B-depleted cells seems to not affect the kinetochore function because no detectable HAC loss was observed after CENP-B depletion (data not shown), supporting plasticity of the replication program for centromeric repeats. Further experiments are required to elucidate if enrichment for proteins of the pre-replication complex in CENP-B-depleted cells results in activation of additional functional origins of replication (the combing technique does not allow to detect a predicted decrease in inter-origin distances if initiation events are not close to each other) and effects the timing of replication. The consequences of the local enrichment for proteins of the pre-replication complex or increase of activation of origin of replication in the centromeric regions on cell adaptation and transformation deserves a further investigation. For these studies, cells with a complete deletion of CENP-B (55–57) may be suitable.
Interestingly, mapping of origins of replication in the structurally characterized alphoidtetO-HAC (15,16) revealed that a megabase-size array of alpha-satellite monomers in this HAC, which generally possesses the same chromatin architecture as kinetochores of natural chromosomes, does not participate in initiation of DNA replication. Instead, origins are localized in multiple copies of the Bsr sequence that are organized into transcriptionally active chromatin and interrupt the alphoid repeats approximately every ∼50 kb. This may be explained by the fact that the alphoid DNA array in the HAC is assembled from a synthetic dimer that is not recognized efficiently by the pre-replication complex or, alternatively, by origin dominance. Indeed, being organized into open and transcribed chromatin, the Bsr sequences would be expected to replicate in early S phase (51,52). Therefore, initiation of DNA replication from multiple Bsr origins may preclude activation of origins of replication on alpha-satellite monomers that would be timed to replicate in the middle-to-late S phase. Future experiments with other HACs generated de novo from natural alphoid DNA arrays and carrying different selectable markers (9,10,26,58–60) may help to reveal how origins of replication are selected on HACs.
To summarize, this study represents the first systematic analysis of replication of human centromeric regions. A further extension of this work may shed light on mechanism controlling maintenance of kinetochore integrity during cell divisions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank Barbara Taylor for helping in FACS sorting, Dr Tonia Carter and Dr Jinki Lozano for help in statistical analysis and Dr Urbain Weyemi for advice in siRNA experiments.
FUNDING
Intramural Research Program of the NIH; National Cancer Institute; Center for Cancer Research, USA (to V.L. and M.I.A.); Wellcome Trust Principal Research Fellowship [073915] (to W.C.E.); Grand-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology of Japan [23247030, 23114008 to H.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Willard H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 2.Choo K.H, Visel B., Naggi A., Earle E., Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vissel B., Choo K.H. Human alpha satellite DNA-consensus sequence and conserve region. Nucleic Acids Res. 1991;15:6751–6752. doi: 10.1093/nar/15.16.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waye J.S., Willard H.F. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987;15:7549–7569. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willard H.F., Waye J.S. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- 6.Wu J.C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J. Mol. Biol. 1980;142:363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]
- 7.Alexandrov I, Kazakov A, Tumeneva I, Shepelev V., Yurov Y. Alpha satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- 8.Hayden K.E. Human centromere genomics: now it's personal. Chromosome Res. 2012;20:621–633. doi: 10.1007/s10577-012-9295-y. [DOI] [PubMed] [Google Scholar]
- 9.Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 10.Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N.I., Cooke H., Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 11.Okada T., Ohzeki J.I., Nakano M., Yoda K., Brinkley W.R., Larionov V., Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Tachiwana H., Miya Y., Shono N., Ohzeki J., Osakabe A., Otake K., Larionov V., Earnshaw W.C., Kimura H., Masumoto H., et al. Nap1 regulates proper CENP-B binding to nucleosomes. Nucleic Acids Res. 2013;41:2869–2880. doi: 10.1093/nar/gks1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner A.M., Aladjem M.I. The chromatin backdrop of DNA replication: lesson from genetics and genome scale analyses. Biochim. Biophys. Acta. 2012;7:794–801. doi: 10.1016/j.bbagrm.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aparicio O.M. Location, location, location: it's all in the timing for replication origins. Genes Dev. 2013;27:117–128. doi: 10.1101/gad.209999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano M., Cardinale S., Noskov V.N., Gassman R., Vagnarelli P., Kandels-Lewis S., Earnshaw W.C., Larionov V., Masumoto H. Inactivation of a human kinetochore by a specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouprina N., Samoshkin A., Erliandri I., Nakano M., Lee H.S., Fu H., Aladjem M.I., Masumoto H., Earnshaw W.C., Larionov V. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth. Biol. 2012;1:590–601. doi: 10.1021/sb3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M.M., Ryan M., Kim R.G., Zakas A.L., Fu H., Lin C.M., Reinhold W.C., Davis S.R., Bilke S., Liu H., et al. Genome wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 19.Iida Y., Kim J.H., Kazuki Y., Hoshiya H., Takiguchi M., Hayashi M., Erliandri I., Lee H.S., Samoshkin A., Masumoto M., et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.H., Kononenko A, Erliandri I., Kim T.A., Nakano N., Iida Y., Oshimura M., Masumoto H., Earnshaw W.C., Larionov V., et al. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu H., Wang L., Lin C.M., Singhania S., Bouhassira E.E., Aladjem M.I. Preventing gene silencing with human replicators. Nat. Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 22.Gerbi S.A., Bielinsky A.K. Replication initiation point mapping. Methods. 1997;13:271–280. doi: 10.1006/meth.1997.0526. [DOI] [PubMed] [Google Scholar]
- 23.Bensimon A., Simon A., Chiffaudel A., Croquette V., Heslot F., Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 24.Conti C., Saccà B., Herrick J., Lalou C., Pommier Y., Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G. The diploid genome sequence of an individual human. PLoS Biology. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayden K.E., Strome E.D., Merrett S.L., Lee H.R., Rudd M.K., Willard HF. Sequences associated with centromere competency in the human genome. Mol. Cell. Biol. 2013;33:763–772. doi: 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., Kent W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piechaczyk M., Farràs R. Regulation and function of JunB in cell proliferation. Biochem. Soc. Trans. 2008;36:864–867. doi: 10.1042/BST0360864. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen P.D., Frederiksen S. Characterization of human 5S rRNA genes. Nucleic Acids Res. 1991;19:4147–4151. doi: 10.1093/nar/19.15.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waltz S.E., Trivedi A.A., Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;24:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mravinac B., Sullivan L.L., Reeves J.W., Yan C.M., Kopf K.S., Farr C.J., Schueler M.G., Sullivan B.A. Histone modifications within the human X centromere region. PLoS One. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeld J.A., Wang Z., Schones D.E., Zhao K., DeSalle R., Zhang M.Q. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sengoku T., Yokoyama S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 2011;25:2266–2277. doi: 10.1101/gad.172296.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ten Hagen K.G., Gilbert D.M., Willard H.F., Cohen S.N. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell. Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific a-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haaf T., Ward D.C. Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 1994;3:697–709. doi: 10.1093/hmg/3.5.697. [DOI] [PubMed] [Google Scholar]
- 39.Shelby R.D., Monier K., Sullivan K.F. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell. Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slee R.B., Steiner C.M., Herbert B.S., Vance G.H., Hickey R.J., Schwarz T., Christan S., Radovich M., Schneider B.P., Schindelhauer D., et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;231:3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 42.Erickson R.P., Glover T.W., Hall B.K., Witt M. Polymerase chain reactions with alphoid-repeat primers in combination with Alu or LINEs primers, generate chromosome-specific DNA fragments. Ann. Hum. Genet. 1991;55:199–211. doi: 10.1111/j.1469-1809.1991.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 43.Cooper K.F., Fisher R.B., Tyler-Smith C. Structure of the pericentric long arm region of the human Y chromosome. J. Mol. Biol. 1992;228:421–432. doi: 10.1016/0022-2836(92)90831-4. [DOI] [PubMed] [Google Scholar]
- 44.Laurent A.M., Marçais B., Muleris M., Roizès G. A rapid and simple method to isolate and characterize highly polymorphic markers from the centromeric regions of the human chromosomes. Nucleic Acids Res. 1994;22:194–199. doi: 10.1093/nar/22.2.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V., et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan F.L., Wong L.H. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012;40:11178–11188. doi: 10.1093/nar/gks921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka Y., Tachiwana H., Yoda K., Masumoto H., Okazaki T., Kurumizaka H., Yokoyama S. Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. J. Biol. Chem. 2005;280:41609–41618. doi: 10.1074/jbc.M509666200. [DOI] [PubMed] [Google Scholar]
- 48.MacAlphine D.M., Almouzni G. Chromatin and DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami Y., Huberman J.A., Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. U.S.A. 1996;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eichler E.E., Clark R.A., She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 51.Schwaiger M., Schubeler D. A question of timing: emerging links between transcription and replication. Curr. Opin. Genet. Dev. 2006;16:177–183. doi: 10.1016/j.gde.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Zink D. The temporal program of DNA replication: new insights into old questions. Chromosoma. 2006;115:273–287. doi: 10.1007/s00412-006-0062-8. [DOI] [PubMed] [Google Scholar]
- 53.Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A., Warburton P.E., Black B.E. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson D.F., Fowler K.J., Earle E., Saffery R., Kalitsis P., Trowell H., Hill J., Wreford N.G., de Kretser D.M., Cancilla M.R., et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell. Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapoor M., Montes de Oca Luna R., Liu G., Lozano G., Cummings C., Mancini M., Ouspenski I., Brinkley B.R., May G.S. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- 57.Fowler K.J., Hudson D.F., Salamonsen L.A., Edmondson S.R., Earle E., Sibson M.C., Choo K.H. Uterine dysfunction and genetic modifiers in centromere protein B-deficient mice. Genome Res. 2000;10:30–41. [PMC free article] [PubMed] [Google Scholar]
- 58.Ebersole T.A., Ross A., Clark E., McGill N., Schindelhauer D., Cooke H., Grimes B. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 59.Kouprina N., Ebersole T., Koriabine M., Pak E., Rogozin I.B., Katoh M., Oshimura M., Ogi K., Peredelchuk M., Solomon G., et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucl. Acids Res. 2003;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moralli D., Simpson K.M., Wade-Martins R., Monaco Z.L. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep. 2006;7:911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


