Abstract
Heterochromatin protein 1 (HP1) is an evolutionarily conserved protein across different eukaryotic species and is crucial for heterochromatin establishment and maintenance. The silkworm, Bombyx mori, encodes two HP1 proteins, BmHP1a and BmHP1b. In order to investigate the role of BmHP1a in transcriptional regulation, we performed genome-wide analyses of the transcriptome, transcription start sites (TSSs), chromatin modification states and BmHP1a-binding sites of the silkworm ovary-derived BmN4 cell line. We identified a number of BmHP1a-binding loci throughout the silkworm genome and found that these loci included TSSs and frequently co-occurred with neighboring euchromatic histone modifications. In addition, we observed that genes with BmHP1a-associated TSSs were relatively highly expressed in BmN4 cells. RNA interference-mediated BmHP1a depletion resulted in the transcriptional repression of highly expressed genes with BmHP1a-associated TSSs, whereas genes not coupled with BmHP1a-binding regions were less affected by the treatment. These results demonstrate that BmHP1a binds near TSSs of highly expressed euchromatic genes and positively regulates their expression. Our study revealed a novel mode of transcriptional regulation mediated by HP1 proteins.
INTRODUCTION
Heterochromatin protein 1 (HP1) is a conserved non-histone chromosomal protein that is recruited to centric and telomeric heterochromatin (1). HP1 proteins are characterized by a common structure comprising two conserved domains, the N-terminal chromodomain (CD) and the C-terminal chromoshadow domain (CSD), which are separated by a poorly conserved and flexible hinge domain (2). HP1 proteins are recruited to chromatin via recognition of a specific histone modification, either the di- or trimethylation of Lys 9 on the histone H3 tail (H3K9me2/3). The HP1 protein CD mediates this interaction, resulting in the establishment of a heterochromatic domain (3,4). CSD is a structural motif required for dimerization and interaction with diverse HP1-associating proteins, such as the histone H3K9 methyltransferase Su(var)3–9. Su(var)3–9 methylates adjacent nucleosomes to establish HP1-binding sites; thus, initiating a spreading mechanism that causes chromatin condensation and inactivation (5–7).
Multiple HP1 isoforms exist in most eukaryotes, suggesting that these isoforms possibly exert stage and/or tissue-specific functions. In Drosophila, three of the five HP1 isoforms (HP1a, HP1b and HP1c) are ubiquitously expressed, while the other two (HP1d/Rhino and HP1e) are predominantly expressed in germline cells (8). The molecular mechanisms that determine the differential functional properties of these HP1 isoforms are largely unknown. Most of our knowledge about the modes of action of HP1 proteins has been acquired from studies involving Drosophila HP1a (DmHP1a), yeast Swi6 or mammalian HP1α. DmHP1a is generally known to bind H3K9me2/3-enriched heterochromatin regions and to recruit diverse factors, resulting in both heterochromatin assembly and gene silencing (5,9,10). However, recent studies have revealed a more complex feature of DmHP1a in the regulation of gene expression. DmHP1a was shown to be required for expression of the essential heterochromatic genes rolled and light (11). In addition, certain euchromatic genes require DmHP1a for their expression, suggesting a positive role for HP1 proteins in euchromatic gene expression (12,13). Moreover, DmHP1a is recruited to developmentally regulated genes and heat shock-induced puffs in an RNA-dependent manner (14,15). These results imply that HP1 proteins may play various context-dependent roles in the regulation of gene expression.
In contrast to Drosophila, Bombyx mori has only two HP1 orthologs, BmHP1a and BmHP1b (16–18), both of which are ubiquitously expressed and are relatively abundant in the gonads (Supplementary Figure S1). Biochemical experiments using bacterially expressed BmHP1 proteins showed that BmHP1a and BmHP1b could potentially form homo- and heterodimers and interact with silkworm Su(var)3–9 (16). Mitsunobu et al. (16) performed luciferase reporter assays in BmN4 cells using two vectors. The first vector expressed the BmHP1 proteins fused with GAL4 DNA-binding domain (GAL4DBD), whereas the other contained a luciferase gene with GAL4DBD-binding sequence and heat shock protein promoter. Co-transfection of BmN4 cells with these two vectors showed reduced luciferase activity, suggesting that both BmHP1 proteins may have transcriptional repressor activity in BmN4 cells (16). In addition, we previously performed chromatin immunoprecipitation followed by polymerase chain reaction (ChIP-PCR) and found BmHP1a enrichment in the SART1 transposon where H3K9me2 and H3K9me3 were enriched (17). These observations suggest that BmHP1 proteins likely play a role in the establishment and/or maintenance of heterochromatin. However, how BmHP1 proteins regulate gene expression via binding to the silkworm genome is unknown.
We recently constructed a comprehensive epigenome map of the silkworm ovary-derived BmN4 cell line (17) that possesses a complete PIWI-interacting RNA (piRNA) pathway (19). In this study, we used the BmN4 cell line and its epigenomic information to investigate a role of BmHP1a in transcriptional regulation and discovered the novel role for the HP1 protein. In other words, BmHP1a transcriptionally activates euchromatic genes, where the transcription start sites (TSSs) are located adjacent to BmHP1a-binding sites.
MATERIALS AND METHODS
Cell lines
BmN4 cells and NIAS-Bm-M1 cells (20) were cultured at 27°C in IPL-41 medium (Applichem GmBH, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS). BmVF cells (21) were cultured at 27°C in IPL-41 medium supplemented with 10% gamma-irradiated FBS. BmN4 cells that stably express Flag-BmHP1a were previously reported (17). B. mori larvae (p50T) were reared as previously described (18).
ChIP-PCR and ChIP followed by deep sequencing (ChIP-seq)
ChIP-PCR and ChIP-seq experiments were performed as previously described (17). Antibodies for histone modifications (anti-H3K4me, anti-H3K27ac, anti-H3K27me3 and anti-H3K36me3) were obtained from Millipore (Billerica, MA, USA). An anti-Flag M2 antibody (Sigma-Aldrich Corporation, St. Louis, MO, USA) was used in the ChIP experiments involving Flag-BmHP1a.
Mapping of ChIP-seq tags to transposons
ChIP-seq sequences were mapped to 121 well-annotated silkworm transposon sequences (22) using Bowtie (23) allowing no mismatches, and reads for each library were normalized to the total mapped reads for comparison.
Mapping of ChIP-seq and RNA-seq tags to the silkworm genome and genes
ChIP-seq and RNA-seq tags were mapped to the silkworm genome scaffold build2 and the silkworm gene models (14 623 genes) (24), respectively, using Bowtie (23) allowing one mismatch. The reads in each gene were defined as the reads per kilobase (kb) of exon model per million mapped reads (RPKM). We analyzed BmHP1a enrichment and histone modifications using the well-established MACS software (25) as previously described (17). A P-value cutoff (1.00e−05) was used to identify ChIP-seq data peaks that had been mapped to the genome. In addition, we calculated the distance from the end of each BmHP1a peak to the nearest gene model.
Identification of histone modifications and TSSs in a 2-kb-long region from the end of each BmHP1a peak
We determined the average coverage across each 50-base window and estimated the average level of each histone mark in a 2-kb-long region from the end of each BmHP1a peak. For each library, the average was normalized using the corresponding control (pIZ-BmN4 for BmHP1a-BmN4; rabbit immunoglobulin G (IgG-R) for H3K4me3, H3K4me2, H3K4me1, H3K27ac, H3K9ac, H3K9me2, H3K9me3, H3K27me3 and H3K36me3 and mouse IgG (IgG-M) for Pol2).
Mapping of TSSs to the silkworm genome
We mapped BmN4-derived TSS-tags (17) to the silkworm genome using Bowtie (23) while allowing one mismatch, and thus obtained the direction and location of each TSS. In order to determine the positional relationship between the BmHP1a-binding sites and TSSs, we calculated the distance from the end of each BmHP1a peak to the nearest TSS.
RNA interference (RNAi) in BmN4 cells
RNAi experiments in BmN4 cells were performed as previously described (18). Libraries for RNA sequencing were generated from BmN4 cells that had been transfected with double-stranded siRNAs against EGFP (control), BmHP1a or BmHP1b (FASMAC Co., Ltd., Kanagawa, Japan; Supplementary Table S1) using the TruSeq RNA Sample Preparation kit (Illumina Inc., San Diego, CA, USA) and were analyzed using the Illumina HiSeq 2500 platform with 100-bp paired-end reads according to the manufacturer's protocol (26). Expression of BmHP1a and BmHP1b in knocked down cells was 5.9% and 8.0%, respectively, compared with those in control cells.
RNAi in BmVF and NIAS-Bm-M1 cells
NIAS-Bm-M1 or BmVF cells (2.5 × 105 cells per 60-mm diameter dish) were transfected with siRNAs (250 pmol per dish) using X-tremeGENE HP (Roche) (18). Transfection was performed three times at 24 h intervals. Cells were harvested at 72 h after first transfection, and total RNA was isolated. Expression of BmHP1a in siRNA-transfected NIAS-Bm-M1 or BmVF cells was 15.2% and 15.9%, respectively, compared with those in control cells.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was prepared with the TRIzol reagent (Invitrogen/Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol and subjected to reverse transcription with avian myeloblastosis virus reverse transcriptase and oligo-dT primer (TaKaRa Bio Inc., Shiga, Japan). RT-qPCR was performed using the KAPA SYBR FAST qPCR kit (Kapa Biosystems Inc., Wilmington, MA, USA) and the specific primers listed in Supplementary Table S1.
Tissue distribution of BmHP1a-regulated genes
We selected BmHP1a-regulated genes whose RPKMs were more than 100 and 20% higher or lower in BmHP1a siRNA-transfected BmN4 cells compared with those in the EGFP siRNA-transfected cells. We estimated the patterns of tissue distribution via Basic Local Alignment Search Tool analyses that used 15 full-length cDNA libraries (Supplementary Table S2). cDNA libraries containing more than 10 000 clones were used in this analysis.
RESULTS
BmHP1a binding to the telomeric regions of the silkworm genome
Our previous ChIP-PCR experiments demonstrated that BmHP1a is enriched for the SART1 transposon, which is known to massively accumulate in the silkworm heterochromatic telomere regions (17,27). This suggests that BmHP1a presumably plays a canonical role, such as heterochromatin establishment and/or maintenance (28,29). In order to identify the BmHP1a-binding loci in the silkworm, we performed ChIP-seq experiments with BmN4 cells that expressed Flag-tagged BmHP1a (BmHP1a-BmN4 cells). We first mapped the ChIP-seq tags from BmHP1a-BmN4 and control (pIZ-BmN4) cells to 121 well-annotated silkworm transposons and compared the mapped reads from the two libraries. We found that BmHP1a was extensively enriched for telomere-specific transposons (Figure 1 and Supplementary Figure S2), which suggested the involvement of BmHP1a in the establishment and/or maintenance of the silkworm telomeric regions.
Figure 1.
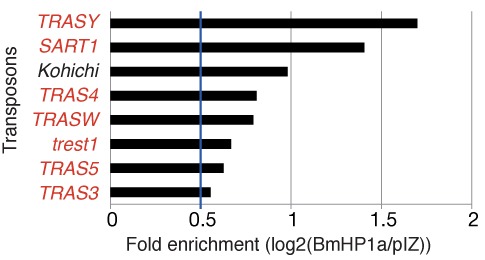
BmHP1a enrichment in silkworm transposons. We mapped the ChIP-seq data from BmHP1a-BmN4 and pIZ-BmN4 (control) cells to 121 well-annotated silkworm transposons. We selected those transposons for which enrichment in the BmHP1a-BmN4 cells was more than 1.5-fold of that in the control cells. Transposons shown in red letters are known to belong to the telomere-specific transposon family.
Genome-wide identification of BmHP1a-binding loci in the silkworm genome
We recently constructed an epigenomic map of BmN4 cells (17). In the present study, we upgraded this map by adding ChIP-seq data for four histone marks, H3K4me (enhancer mark), H3K27ac (euchromatic mark) and H3K27me3 and H3K36me3 (heterochromatic marks; see Supplementary Figures S3 and S4). This enabled us to obtain more precise and detailed information regarding histone modifications at the genomic loci of interest in the silkworm genome. In order to investigate the genome-wide properties of the BmHP1a-binding loci, we performed further analyses with this epigenome map and ChIP-seq reads that were uniquely mapped to the silkworm genome scaffolds.
First, we attempted to visually identify the BmHP1a-enriched loci with a genome browser and detected several distinct peaks in the data from BmHP1a-BmN4 cells relative to those from the control cells. One of these examples is shown in Figure 2A. At this locus, BmHP1a was enriched in the region immediately upstream of the predicted gene BGIBMGA011975 and BGIBMGA011922. This enrichment was statistically verified by ChIP-qPCR (Figure 2B). Although this BmHP1a-enriched locus did not co-occur with all of the histone modifications, this position was closely adjacent to the peaks for the euchromatic modifications H3K4me3, H3K4me2 (Figure 2A) and H3K27ac (Supplementary Figure S3); however, no distinct peaks for heterochromatin marks (H3K9me2/3, H3K27me3 and H3K36me3) were observed in adjacent regions (Figure 2A and Supplementary Figure S3). The ChIP-qPCR results validated this positional relationship among the BmHP1a enrichment and histone modifications; in other words, the BmHP1a-binding locus was situated closely adjacent to three euchromatic modifications (Figure 2B–D). A similar pattern was often observed throughout the genome (see Supplementary Figure S4. Another example was observed on chromosome 23; at this locus, BmHP1a binds to the upstream regions of two head-to-head located genes, BGIBMGA011491 and BGIBMGA011523). These results suggested that BmHP1a likely binds the upstream regions of euchromatic protein-coding gene loci.
Figure 2.
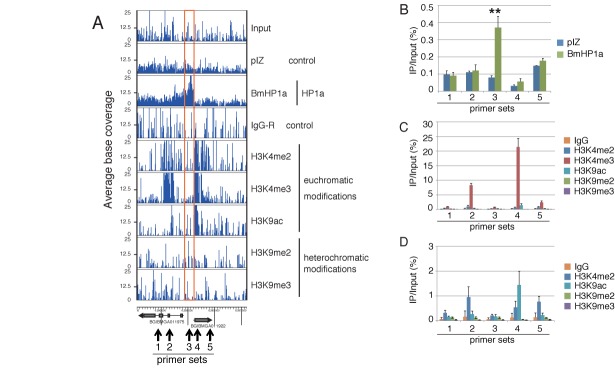
A representative picture of histone modifications in a BmHP1a-binding region. (A) The BmHP1a-binding locus on chromosome 11: 6 287 750–6 298 000. The mapped reads were visualized using the Genome studio (Illumina). The BmHP1a-binding region is indicated by an orange box. (B–D) ChIP-qPCR experiments. The ChIP-seq data in Figure 2A were validated by ChIP-qPCR. The locations of the primers used are indicated in Figure 2A. The data are shown as means ± standard deviations (SD; n = 3). **P < 0.01 with Student's t-test.
We asked whether this trend would be conserved on a genome-wide scale. We used MACS software to obtain 1775 BmHP1a peaks with a median length of 639 bases. In addition, we estimated the average levels of each histone mark within a 2-kb-long region from the end of each BmHP1a peak (Figure 3A). Enrichments of two euchromatin marks (H3K4me2 and H3K4me3) were clearly detected around the edge of the BmHP1a peak and the H3K4me3 peak was observed immediately external to the BmHP1a-binding region (Figure 3A). In contrast, the heterochromatin marks had not accumulated at all (Figure 3A). These results demonstrate that BmHP1a is closely associated with euchromatic regions throughout the genome. In addition, we counted the number of TSSs that existed within 2 kb of the end of the BmHP1a peak. The histogram representing the frequency clearly showed that a TSS distribution peak was detected at the internal end of the BmHP1a peak (Figure 3B).
Figure 3.
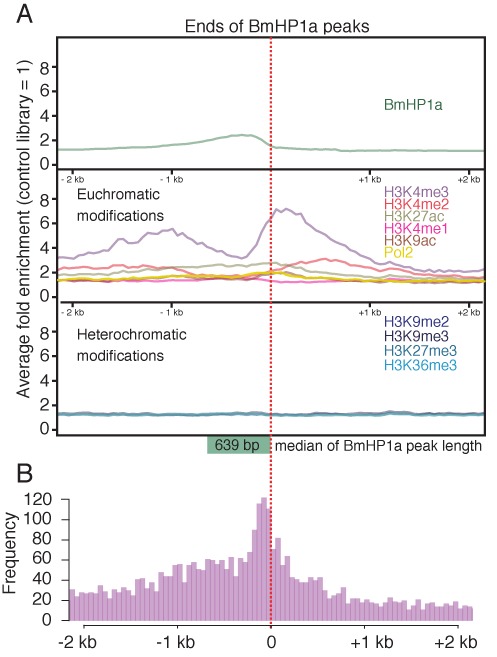
Characterization of the BmHP1a-binding loci. (A) The average levels of each histone mark in a 2-kb-long region from the end of each BmHP1a peak as identified with the MACS program. The red dotted line indicates the ends of the BmHP1a peaks. The green square box indicates the median length of the BmHP1a-binding regions. (B) Distribution of TSSs that existed within 2 kb from the end of the BmHP1a peak. The frequency in each 50-bp window is shown. This figure does not contain directional information for each TSS.
BmHP1a binds the upstream region adjacent to highly euchromatic TSSs
We next investigated the positional relationships between the TSSs and BmHP1 peaks or chromatin modifications. We first calculated the distances from TSSs to each histone modification and found that three euchromatic modifications (H3K4me3, H3K4me2 and H3K27ac) occurred within 1 kb of TSSs (Figure 4, upper panel). Furthermore, we examined the properties of TSSs included within the BmHP1a peaks and observed a marked enhancement of euchromatic modifications among the BmHP1a-associated TSSs (Figure 4, lower panel). In both cases, the heterochromatin marks were not enriched around TSSs (Figure 4). These results indicated that the BmHP1a-binding loci include TSSs that frequently co-occur with the euchromatic histone modifications.
Figure 4.
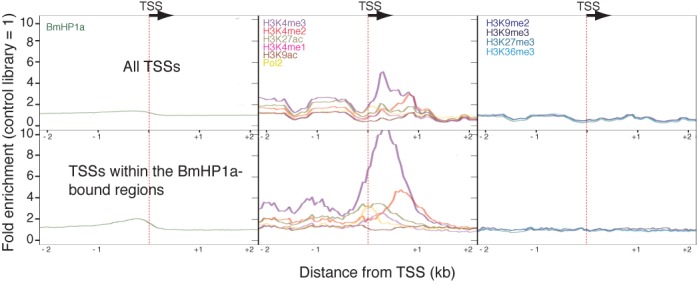
The positional relationships between TSSs and the BmHP1 peaks or chromatin modifications. The average levels of histone modification enrichment in a 2-kb region from the TSSs are shown. The upper panel presents the data for all TSSs; the lower presents the data for TSSs that are included in the BmHP1a-binding regions.
BmHP1a binds the upstream regions of highly expressed genes
We next examined the relationship between BmHP1a binding and the transcriptional levels of BmHP1a-associated genes. We estimated the distances from the first nucleotide of each gene to the end of each neighboring BmHP1a peak. As shown in Figure 5A, a distinct distribution peak was located within 1 kb of the adjacent BmHP1a-binding loci. We mapped the RNA-seq data from BmN4 cells (17) to the silkworm genes and represented the expression level of each gene as a histogram; thus, showing that the expression levels were distributed across a wide range (Figure 5B). We classified all genes into eight groups according to their expression levels (Figure 5B) and examined the distance between each gene and its adjacent BmHP1a peak. The results clearly indicated that most of the BmHP1a-binding loci were associated with highly expressed genes (RPKM >10, 91%), and were rarely observed in the regions that contained genes expressed at low levels (Figure 5C). We further investigated the expression levels of the gene groups that had been classified according to the distance between each gene and the nearest BmHP1a peak. We noted a clear correlation between the expression level and proximity of the adjacent BmHP1a locus to TSS when BmHP1a bound the upstream region of the gene (Supplementary Figure S5). These results demonstrated that BmHP1a strongly associates with highly expressed genes.
Figure 5.
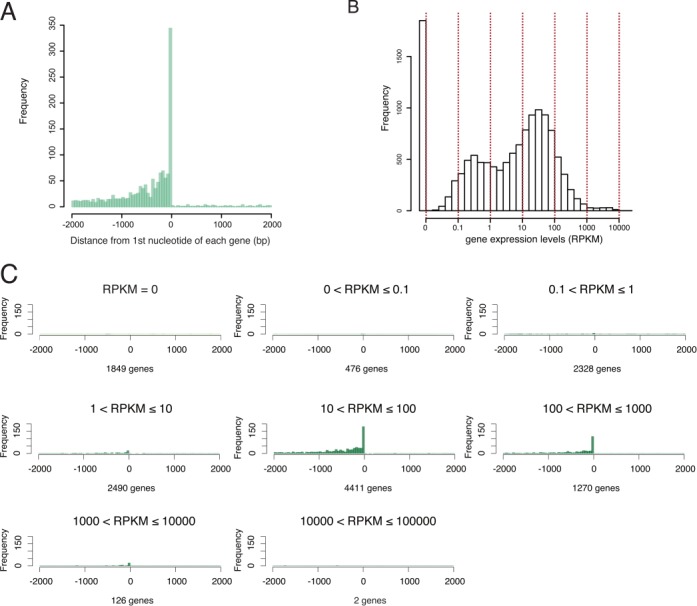
The relationship between BmHP1a-binding and the transcriptional levels of BmHP1a-associated genes. (A) The distances from the first nucleotide of each gene to the end of neighboring BmHP1a peak. (B) The distribution of the transcribed genes in BmN4 cells. The genes were divided into eight groups according to the expression level. (C) The data from Figure 5A were divided into eight groups according to the expression level. The numbers of genes per group are indicated.
BmHP1a enhances the expression of BmHP1a-associated, highly expressed genes
We next asked whether BmHP1a would affect the expression levels of genes for which TSSs were closely associated with BmHP1a. We performed RNAi experiments to knock down BmHP1a in BmN4 cells and performed gene expression profiling via RNA-seq. The RNA-seq data plot revealed that BmHP1a depletion did not strikingly affect the global gene expression profile (Figure 6A). Because BmHP1a is closely associated with highly expressed genes, we focused on the expression profiling of genes with RPKMs >100, and selected differentially expressed genes with RPKMs that were 20% higher (red dots in Figure 6A) or lower (blue dots in Figure 6A) in BmN4 cells transfected with BmHP1a siRNA than those in the control cells (EGFP siRNA-transfected cells). Five genes among those that were upregulated (Figure 6B), unchanged (Figure 6C) and downregulated (Figure 6D) in response to treatment with BmHP1a siRNA were validated in RT-qPCR experiments. Similar changes in the expression of these genes were observed in two other silkworm cell lines in which BmHP1a was knocked down (Supplementary Figures S6 and S7), supporting the role of BmHP1a in transcriptional regulation. We also examined the expression of four genes associated with BmHP1a (Figure 2 and Supplementary Figure S4) and verified their downregulation by treatment with BmHP1a siRNA (Supplementary Figure S8). Furthermore, we investigated BmHP1a associations of these differentially expressed genes. Greater BmHP1a binding accumulation was observed on the downregulated genes relative to the upregulated ones (Figure 6E and Supplementary Figure S9). Finally, we examined the tissue distributions of BmHP1a-regulated genes using expressed sequence tags from 15 full-length cDNA libraries (315 854 clones in total; Supplementary Table S2). We found that the upregulated genes were detected in fewer libraries when compared with the downregulated genes (Supplementary Figure S10). Annotation analysis revealed that a number of ribosomal protein and ribosomal RNA-processing protein genes were included in the downregulated group, whereas such a trend was not observed among the upregulated genes (Supplementary Table S3). These results clearly demonstrate that the genes positively regulated by BmHP1a are ubiquitously expressed in silkworm tissues. Taken together with these results, we propose the following mechanism of BmHP1a protein-mediated transcriptional regulation: BmHP1a closely associates with TSSs, and thus positively regulates the transcription of highly and ubiquitously expressed genes (Figure 7).
Figure 6.
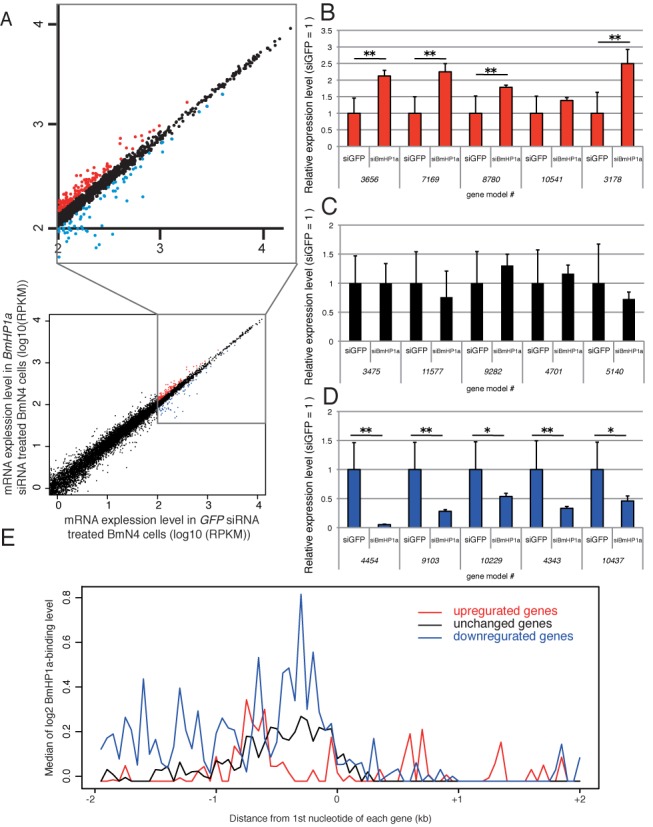
Identification of BmHP1a-regulated genes. (A) Dot plots of the RNA-seq data from GFP siRNA (control) and BmHP1a siRNA-transfected BmN4 cells. x-axis: log10(RPKM in GFP siRNA-transfcetd BmN4 cells); y-axis: log10(RPKM in BmHP1a siRNA-transfected BmN4 cells). Genes for which the RPKMs in control cells were >100 are indicated in the enlarged square. Genes for which the RPKMs were 20% higher or lower in the BmN4 cells transfected with BmHP1a siRNA relative to the control cells are indicated with the red and blue dots, respectively. (B–D) RT-qPCR validation of the RNA-seq experiments. RT-qPCR was performed for five upregulated (B), unchanged (C) and downregulated (D) genes in the BmHP1a-depleted cells. *P < 0.05; **P < 0.01 with Student's t-test. The data are shown as means ± SD (n = 6). (E) The median of BmHP1a-binding levels at 50-base resolution in a 2-kb region from the first nucleotide of the upregulated (red line), unchanged (black line) and downregulated (blue line) genes in the BmHP1a-depleted cells.
Figure 7.
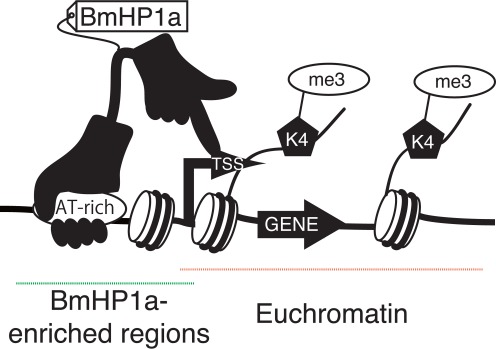
The proposed models of BmHP1a-mediated transcriptional regulation. The model diagram of a novel role for BmHP1a. Here, BmHP1a is closely associated with TSSs via unknown interactions with protein or nucleic acid modifications. Upon association, BmHP1a positively regulates the transcription of highly and ubiquitously expressed genes via an unknown mechanism.
DISCUSSION
In this study, we focused on BmHP1a, one of the two silkworm HP1 proteins, and showed that BmHP1a plays two independent roles in transcriptional regulation. As observed in other organisms, BmHP1a may play a canonical role of the HP1 protein with respect to forming and maintaining telomeric heterochromatin (Figure 1 and Supplementary Figure S2). In addition, we discovered a non-canonical role for BmHP1a as a transcriptional activator (Figure 7). In Drosophila, certain euchromatic genes, such as those that regulate the cell cycle and development, require DmHP1a for stage and/or tissue-specific expression; thus, suggesting a positive role for HP1 proteins in euchromatic gene expression (12,13). Our experimental results clearly indicated that BmHP1a binds near the TSSs of highly expressed euchromatic genes to positively affect their expression (Figure 6). In addition, a bioinformatic analysis of 15 silkworm cDNA libraries revealed that the genes positively regulated by BmHP1a were ubiquitously expressed in silkworm tissues (Supplementary Figure S10 and Table S3). These results suggest that HP1a target genes differ between Drosophila and silkworm and that BmHP1a likely plays a novel role in the transcriptional regulation of euchromatic genes.
Although BmHP1a is recruited to telomeric heterochromatin, presumably by identifying H3K9me2 or H3K9me3 via CD (Figure 1, Supplementary Figure S2 and Figure 7), we did not observe any heterochromatic modifications in euchromatic, transcriptionally active BmHP1a-binding regions (Figure 3). In Drosophila, the HP1a protein can be recruited to promoters and within the bodies of active genes independently of histone H3 Lys 9 methylation (30). The Drosophila promoter sequences in the fourth chromosome and the 2L:31 region, where HP1a is enriched without heterochromatin marks, is known to exhibit a significantly higher A/T content relative to the promoter sequences at other chromosomal locations (30,31). This finding is similar to our observation that A/T contents within BmHP1a-binding regions were relatively high (data not shown), suggesting that these two HP1a proteins bind to the genome via a similar, albeit unknown process in the absence of H3K9 methylation. On the other hand, the chromatin states of the HP1a-enriched loci considerably differ between the two species. Although heterochromatin modifications exist near the HP1a-binding regions in Drosophila, no heterochromatin marks were found to be enriched near the BmHP1a-binding loci (Figure 3). It has been proposed that in Drosophila, the HP1 binding in the absence of H3K9me initiates the spread of heterochromatin modifications near the binding site (30). In the silkworm, this system is likely non-functional because of the lack of heterochromatin modifications near the BmHP1a binding loci. BmHP1a may recruit proteins that activate the transcription of genes with BmHP1a-associated promoters.
A previous study showed that both BmHP1a and BmHP1b function as transcriptional repressors and can form heterodimers (16). In order to determine whether these two HP1 proteins cooperate in transcriptional regulation in BmN4 cells, we examined the expression levels of the genes that were identified in BmHP1a-depleted cells (Figure 6B–D), following treatment with BmHP1b siRNA. Although the upregulated and unchanged groups exhibited very similar patterns in response to either treatment, four of the five downregulated genes observed in the BmHP1a-depleted cells did not exhibit transcriptional changes relative to the control (Supplementary Figure S11). These results clearly demonstrated that the target genes of the two silkworm HP1 proteins differed with respect to transcriptional activation. Taken together with a report stating that BmHP1b possesses a higher level of transcriptional repression activity than BmHP1a in BmN4 cells (16), BmHP1a and BmHP1b presumably cooperate in transcriptional repression through their heterodimerization, whereas BmHP1a can activate the transcription of highly expressed genes irrespective of BmHP1b action. Further experiments are needed to understand the precise role of the BmHP1a-BmHP1b complex in heterochromatin establishment and/or maintenance.
In addition to the five well-known HP1 genes in D. melanogaster (8), five HP1-related genes in D. melanogaster and 21 genes in other Drosophila species were recently identified (33), suggesting that the roles of HP1 proteins have been evolutionarily subdivided or diversified in Drosophila species. In contrast, we along with the others have discovered only three HP1-related genes, BmHP1a, BmHP1b and BmCdp1, in the silkworm genome (16–18). Therefore, silkworm HP1 proteins may play multiple roles during silkworm development. In this study, we demonstrated that BmHP1a plays a canonical role in transcriptional repression and heterochromatin maintenance, thus suggesting that BmHP1a is a functional ortholog of Drosophila HP1a. In addition, expression profiling revealed that all of the silkworm HP1-related genes were expressed at relatively high levels in germline tissues (Supplementary Figure S1). Drosophila HP1d/Rhino and HP1e are predominantly expressed in germline cells (8), and HP1d/Rhino is essential for piRNA production by dual-strand piRNA clusters and piRNA-mediated transposon silencing (32). According to the relatively abundant expression profiles in the gonads, one or more of the three Bombyx HP1-related proteins may possibly contribute to the piRNA pathway in the silkworm germline. Because the BmN4 cell line harbors a complete piRNA pathway (19) and piRNA clusters have been observed in the silkworm genome (17,33), it will be possible to examine the HP1-related silkworm genes that are involved in the piRNA pathway.
In conclusion, we showed that BmHP1a exerts two context-dependent modes of action with respect to transcriptional regulation. It will be of great interest to identify the factors that determine whether BmHP1a acts in canonical or non-canonical pathways and to determine how BmHP1a binds to the euchromatic regions of the silkworm genome to selectively activate highly expressed genes.
ACCESSION NUMBERS
Deep sequencing data obtained in this study are available under the accession number DRA002247 (DDBJ).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Masashi Iwanaga for BmVF cells and National Institute of Agrobiological Sciences (NIAS) for NIAS-Bm-M1 cells, respectively, and the members of our laboratories for their support of this study.
FUNDING
Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry; Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry; Grants-in-Aid for Scientific Research on Innovative Areas (‘Functional machinery for non-coding RNAs’) [to S. K.]. Funding for open access charge: Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry [to S. K.].
Conflict of interest statement. None declared.
REFERENCES
- 1.James T.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 2.Aasland R., Stewart A.F. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 4.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 5.Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eissenberg J.C., Elgin S.C. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 8.Vermaak D., Henikoff S., Malik H.S. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smallwood A., Estève P.O., Pradhan S., Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 11.Lu B.Y., Emtage P.C., Duyf B.J., Hilliker A.J., Eissenberg J.C. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cryderman D.E., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 13.De Lucia F., Ni J.Q., Vaillant C., Sun F.L. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005;33:2852–2858. doi: 10.1093/nar/gki584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piacentini L., Fanti L., Negri R., Del Vescovo V., Fatica A., Altieri F., Pimpinelli S. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5:e1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsunobu H., Izumi M., Mon H., Tatsuke T., Lee J.M., Kusakabe T. Molecular characterization of heterochromatin proteins 1a and 1b from the silkworm, Bombyx mori. Insect Mol. Biol. 2012;21:9–20. doi: 10.1111/j.1365-2583.2011.01115.x. [DOI] [PubMed] [Google Scholar]
- 17.Kawaoka S., Hara K., Shoji K., Kobayashi M., Shimada T., Sugano S., Tomari Y., Suzuki Y., Katsuma S. The comprehensive epigenome map of piRNA clusters. Nucleic Acids Res. 2012;41:1581–1590. doi: 10.1093/nar/gks1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoji K., Kiuchi T., Hara K., Kawamoto M., Kawaoka S., Arimura S., Tsutsumi N., Sugano S., Suzuki Y., Shimada T., et al. Characterization of a novel chromodomain-containing gene from the silkworm, Bombyx mori. Gene. 2013;527:649–654. doi: 10.1016/j.gene.2013.06.071. [DOI] [PubMed] [Google Scholar]
- 19.Kawaoka S., Hayashi N., Suzuki Y., Abe H., Sugano S., Tomari Y., Shimada T., Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M.G., Imanishi S., Dohmae N., Nishimura T., Shimada T., Matsumoto S. Establishment of a novel in vivo sex-specific splicing assay system to identify a trans-acting factor that negatively regulates splicing of Bombyx mori dsx female exons. Mol. Cell. Biol. 2008;28:333–343. doi: 10.1128/MCB.01528-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwanaga M., Hitotsuyama T., Katsuma S., Ishihara G., Daimon T., Shimada T., Imanishi S., Kawasaki H. Infection study of Bombyx mori macula-like virus (BmMLV) using a BmMLV-negative cell line and an infectious cDNA clone. J. Virol. Methods. 2012;179:316–324. doi: 10.1016/j.jviromet.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Osanai-Futahashi M., Suetsugu Y., Mita K., Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi H., Okazaki S., Fujiwara H. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 1997;25:1578–1584. doi: 10.1093/nar/25.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanti L., Giovinazzo G., Berloco M., Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 29.Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., Berloco M., Turano C., Ferraro A., Pimpinelli S. HP1 controls telomere capping telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 30.Figueiredo M.L., Philip P., Stenberg P., Larsson J. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. Plos Genet. 2012;8:e1003061. doi: 10.1371/journal.pgen.1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greil F., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B. Distinct HP1 and Su(var)3–9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2925–2938. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klattenhoff C., Xi H., Li C., Lee S., Xu J., Khurana J.S., Zhang F., Schultz N., Koppetsch B.S., Nowosielska A., et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaoka S., Mitsutake H., Kiuchi T., Kobayashi M., Yoshikawa M., Suzuki Y., Sugano S., Shimada T., Kobayashi J., Tomari Y., et al. A role for transcription from a piRNA cluster in de novo piRNA production. RNA. 2012;18:265–273. doi: 10.1261/rna.029777.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


