Abstract
STUDY QUESTION
Can human spermatogonia be detected in long-term primary testicular cell cultures using validated, germ cell-specific markers of spermatogonia?
SUMMARY ANSWER
Germ cell-specific markers of spermatogonia/spermatogonial stem cells (SSCs) are detected in early (1–2 weeks) but not late (> 6 weeks) primary testicular cell cultures; somatic cell markers are detected in late primary testicular cell cultures.
WHAT IS KNOWN ALREADY
The development of conditions for human SSC culture is critically dependent on the ability to define cell types unequivocally and to quantify spermatogonia/SSCs. Growth by somatic cells presents a major challenge in the establishment of SSC cultures and therefore markers that define spermatogonia/SSCs, but are not also expressed by testicular somatic cells, are essential for accurate characterization of SSC cultures.
STUDY DESIGN, SIZE, DURATION
Testicular tissue from eight organ donors with normal spermatogenesis was used for assay validation and establishing primary testicular cell cultures.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Immunofluorescence analysis of normal human testicular tissue was used to validate antibodies (UTF1, SALL4, DAZL and VIM) and then the antibodies were used to demonstrate that primary testicular cells cultured in vitro for 1–2 weeks were composed of somatic cells and rare germ cells. Primary testicular cell cultures were further characterized by comparing to testicular somatic cell cultures using quantitative reverse transcriptase PCR (UTF1, FGFR3, ZBTB16, GPR125, DAZL, GATA4 and VIM) and flow cytometry (CD9 and SSEA4).
MAIN RESULTS AND THE ROLE OF CHANCE
UTF1, FGFR3, DAZL and ZBTB16 qRT–PCR and SSEA4 flow cytometry were validated for the sensitive, quantitative and specific detection of germ cells. In contrast, GPR125 mRNA and CD9 were found to be not specific to germ cells because they were also expressed in testicular somatic cell cultures. While the germ cell-specific markers were detected in early primary testicular cell cultures (1–2 weeks), their expression steadily declined over time in vitro. After 6 weeks in culture only somatic cells were detected.
LIMITATIONS, REASONS FOR CAUTION
Different groups attempting SSC culture have utilized different sources of human testes and minor differences in the preparation and maintenance of the testicular cell cultures. Differences in outcome may be explained by genetic background of the source tissue or technical differences.
WIDER IMPLICATIONS OF THE FINDINGS
The ability to propagate human SSCs in vitro is a prerequisite for proposed autologous transplantation therapy aimed at restoring fertility to men who have been treated for childhood cancer. By applying the assays validated here it will be possible to quantitatively compare human SSC culture conditions. The eventual development of conditions for long-term propagation of human SSCs in vitro will greatly facilitate learning about the basic biology of these cells and in turn the ability to use human SSCs in therapy.
STUDY FUNDING/COMPETING INTEREST(S)
The experiments presented in this manuscript were funded by a Project Development Team within the ICTSI NIH/NCRR Grant Number TR000006. The authors declare no competing interests.
TRIAL REGISTRATION NUMBER
Not applicable.
Keywords: germ cells, testis, cell culture, fertility, stage-specific embryonic antigens
Introduction
Stem cells are crucial for the maintenance and repair of adult tissues because they serve as a continual source of progenitor cells for specialized cell types that turn over throughout adult life. The stem cells that give rise to spermatozoa are spermatogonial stem cells (SSCs) (Huckins, 1971; Oakberg, 1971). SSCs are a subpopulation of cells known as spermatogonia, which include the true stem cells and progenitor cells that have begun the differentiation process (Bellve et al., 1977; Nakagawa et al., 2007). Like other stem cells, SSCs reside in a niche where they respond to cellular signals arising from neighboring cells, and these cues control SSC fate decisions controlling proliferation and differentiation (Caires et al., 2010). Maintenance of the stem cell pool is necessary for the continual production of spermatozoa throughout the reproductive lifetime of the individual.
Stem cell therapy is an emerging field of medicine with the potential for curing myriad diseases by transplantation of healthy stem cells to restore diseased or damaged tissues. One proposed therapy applies to pre-pubescent boys who risk losing their fertility as a result of chemotherapy or irradiation treatment for cancer; these patients could have their fertility restored by isolating SSCs prior to treatment and transplanting them back into the testes following treatment (Brinster, 2007). Infertility is an important quality of life issue facing an expanding population of childhood cancer survivors as vast improvements in treatment of childhood cancer have occurred in recent decades (Green et al., 2010; Howlader et al., 2010). While the prospect of using SSCs in therapy holds great promise, before cells could be safely reintroduced into a patient one would have to demonstrate that the cells are indeed SSCs and free of cancerous cells (Dovey et al., 2013). Obtaining SSCs in sufficient quantities for molecular characterization and transplantation represents a major hurdle in part because the longevity and quantity of human SSCs maintained in vitro remains limited.
Multiple groups have reported propagating SSCs from human testes in culture for periods ranging from 2 weeks to 6 months (Sadri-Ardekani et al., 2009, 2011; He et al., 2010; Lim et al., 2010; Golestaneh, 2011; Koruji et al., 2012; Piravar et al., 2013). A critical aspect of evaluating the effectiveness of the culture method is the choice of assay(s) for defining which cell types are present. The assays used to identify SSCs in cultures of human testicular cells have varied widely, in part because data are limited on how to define human SSCs properly. Characterization of the cultured cells relies on knowing which mRNAs or proteins/antigens are expressed by human SSCs and also whether those same markers are specific to SSCs or are present in other testicular cell populations (Martin and Seandel, 2013). For instance, ITGA6, ZBTB16 and GPR125 mRNAs have been used to demonstrate that spermatogonia/SSCs are present in cultures of human testicular cells (Golestaneh, 2011; Sadri-Ardekani et al., 2009, 2011). Yet all of these alleged markers of human SSCs have now been found to also be expressed in somatic testicular cells (Dym et al., 2009; Wu et al., 2009; He et al., 2010; Kossack et al., 2013). Hence, it is possible that testicular cell cultures were misinterpreted to contain SSCs when they were composed of somatic cells.
The vast majority of knowledge on the culture and detection of SSCs comes from studies in mice and rats, the model systems in which long-term culturing of SSCs were originally developed (Kanatsu-Shinohara et al., 2003; Kubota et al., 2004a,b; Hamra et al., 2005; Ryu et al., 2005). While the details of culture methods vary, a few key features are shared that lead to successful propagation of rodent SSCs in vitro. First, glial cell line-derived neurotrophic factor (GDNF) is added to the medium; this growth factor is essential for rodent SSC self-renewal in vitro and in vivo (Meng et al., 2000; Kubota et al., 2004a,b; Hofmann et al., 2005; Ryu et al., 2005). Second, while not absolutely required, a feeder layer of mitotically inactivated somatic cells supports optimal SSC propagation (Kanatsu-Shinohara et al., 2003, 2010; Nagano, 2003; Kanatsu-Shinohara et al., 2006; Nagano et al., 2009). Third, during the initiation stage of culturing it is necessary to avoid overgrowth by somatic testicular cells (Oatley and Brinster, 2006; Kubota and Brinster, 2008; Nagano et al., 2009). To overcome this issue SSCs may be enriched either before culturing by immunoselection of spermatogonia/SSCs expressing certain surface proteins (e.g. THY1), or during the first few days of culturing based on differential adherence to certain substrates (e.g. somatic cells bind plastic) (Oatley and Brinster, 2006; Kubota and Brinster, 2008; Nagano et al., 2009). Without an enrichment strategy expansion of testicular somatic cells can quickly overwhelm the primary culture, which contains relatively few SSCs due to their overall scarcity within the testicular cell population. A few weeks after initiating a successful rodent testicular culture, the proliferating germ cells appear as grape-like clusters of spermatogonia, including SSCs (1–10% of cells in rodent cultures) and partly differentiated progenitor cells. Once established such SSC-containing cultures derived from mice can be propagated for years while maintaining stem cell activity (Kanatsu-Shinohara et al., 2003, 2005).
The ability to culture human SSCs similar to the way that one can culture rodent SSCs has been described as a ‘holy grail’ as it will pave the road for future therapies in restoring fertility by autologous cell transplantation (Niederberger, 2012); furthermore, an even wider reaching scope of cell therapeutics is imaginable when one considers the possibility that SSCs could serve as a source of pluripotent cells (de Rooij and Mizrak, 2008). While we share in the enthusiasm for the potential application of SSCs in therapy, we present evidence here indicating that human SSC culture is not as close to translation into therapy as the literature suggests. Our data indicate that some previously described assays for characterizing testicular cultures may not have been reliable. In particular, the ability to distinguish between contaminating somatic cells and spermatogonia/SSCs is of paramount importance. Here we validate multiple assays for characterizing testicular cultures and apply these assays to characterize human testicular cell cultures established using published conditions.
Materials and Methods
Ethical approval and human testes procurement and processing
Testes were obtained from brain dead organ donors through the Indiana Organ Procurement Organization (IOPO). IOPO obtained written approval from the next of kin and confirmed negative serology for HepB, HepC and HIV in donors prior to testes procurement. Next of kin were asked if the donor sired children and if the donor was known to have any medical history involving abnormalities in the testes such as undescended testes or testicular cancer. Upon cross clamping, donors were perfused with HTK (histidine-tryptophan-ketoglutarate solution). Testes were procured after other organs were removed for transplant recipients. Testes were packed in sterile, chilled saline and transported in a cooler with ice by courier from Indianapolis to Bloomington. The time from cross clamp until receipt in Bloomington ranged from ∼2 to 4.5 h (Table I). Upon receipt in Bloomington the testes were dissected out of the tunica albuginea, revealing the seminiferous tubules, and most of each testis was cut into portions of ∼0.5–0.7 g for cryopreservation in 10% dimethyl sulphoxide (MP Biomedicals, USA), 10% Dulbecco's Modified Eagle Medium with 4500 mg/l glucose (DMEM; Hyclone, USA) and 80% fetal bovine serum (FBS; Hyclone or Life Technologies, USA) (He et al., 2010). Additionally, two portions from each testes were fixed in formalin and two portions in 4% (v/v) paraformaldehyde (Electron Microscopy Sciences, USA) in phosphate-buffered saline (1× PBS) by rocking at 4°C overnight. Formalin fixed tissue was embedded in paraffin, cut into 5 μm sections and stained with hematoxylin/eosin using standard procedures.
Table I.
Donor data.
| Designation | Age (years) | Biological children | Cause of death | Time in transit |
|---|---|---|---|---|
| Hu2 | 21 | Yes | Drug overdose | 2 h 50 min |
| Hu3 | 30 | No | MVAa/head trauma | 1 h 50 min |
| Hu4 | 30 | Yes | SI-GSWb to head | 2 h 40 min |
| Hu5 | 28 | Yes | MVA/head trauma | 4 h 30 min |
| Hu6 | 13 | No | Diabetes/CVAc | 4 h |
| Hu7 | 40 | No | CVA | 4 h 20 min |
| Hu8 | 39 | Yes | Drug overdose | 3 h 55 min |
| Hu9 | 41 | No | MVA | 3 h 10 min |
aMotor vehicle accident.
bSelf-inflicted gunshot wound.
cCerebrovascular accident.
Primary testicular cell culture
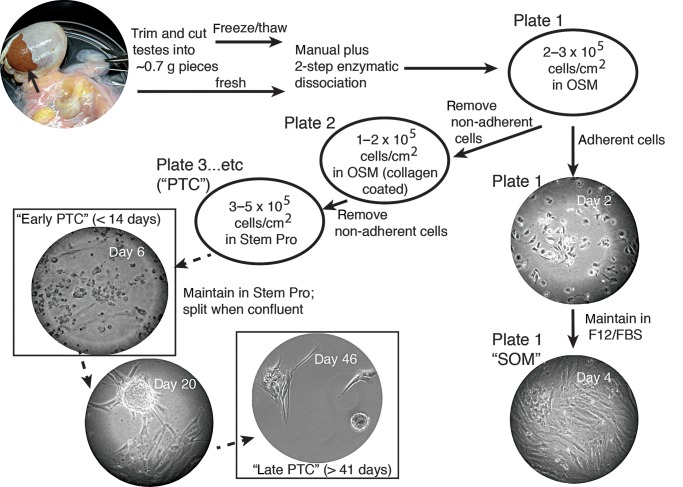
We generated primary testicular cultures using procedures very similar to Sadri-Ardekani et al. (2009); see Fig. 1 for a summary. A weight of fresh or frozen/thawed tissue of 0.5–2 g was used in each experiment and volumes of dissociation enzymes were scaled according to the wet weight of tissue used. Tissue was mechanically disrupted by pulling apart tubules in chilled Hanks Balanced Salt Solution without calcium or magnesium (HBSS; Hyclone, USA). Sequential enzymatic digestion was performed according to Ogawa et al. (1997): we used 1 mg/ml Collagenase Type IV (Sigma, USA)/0.7 mg/ml DNase (Sigma, USA) in HBSS and then 0.25% (w/v) trypsin/1 mM EDTA/0.7 mg/ml DNAse in HBSS in a 37°C water bath with periodic rocking to obtain single cells (Ogawa et al., 1997). Tubules were washed between and after the two digestion steps. Undigested material that remained was removed with a 40 μm mesh strainer.
Figure 1.
Summary of methods used to establish primary testicular cell cultures. See Materials and Methods for full description.
Cells were suspended in overnight selection medium (OSM) consisting of DMEM with 20% (v/v) FBS, 1% (v/v) non-essential amino acids (Hyclone, USA), 1% (v/v) penicillin/streptomycin (Hyclone, USA), 10 μM 2-mercaptoethanol (Sigma, USA) and 10 ng/ml GDNF (Peprotech, USA) and incubated overnight on standard (uncoated) tissue culture plate(s) at a concentration of 2–3 × 105 cells/cm2 (Lim et al., 2010). The next day floating cells were harvested by gentle washing and in most cases a collagen binding step was performed by plating cells in OSM in a 6-well plate (BD Biocoat Collagen I, Rat Tail; BD Biosciences, USA) at 1–2 × 105 cells/cm2 cells for 4 h (Lim et al., 2010). Floating cells were harvested and plated at 3–5 × 105 cells/cm2 in germ cell maintenance medium formulated as described (Sadri-Ardekani et al., 2009). Specifically, the medium was a Stem Pro-34 serum free medium base (Life Technologies, USA) containing all the additives originally described by Kanatsu-Shinohara et al. (2003) except with 1% (v/v) antibiotic/antimycotic (Life Technologies, USA) and knockout serum replacement (Life Technologies, USA) replacing FBS; it contained four recombinant human growth factors: 10 ng/ml GDNF, 10 ng/ml LIF (Peprotech, USA), 20 ng/ml EGF (Peprotech, USA) and 10 ng/ml FGF2 (Life Technologies or Peprotech, USA). Cells cultured in germ cell maintenance medium were termed ‘PTC’ (primary testicular cells). When PTC were confluent, the floating and bound cells were harvested by trypsinization and replated at a ratio to achieve half the original cells:surface area.
Cells that remained bound to the initial plate(s) after the first overnight binding step were subsequently maintained in F12/FBS (Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham (Sigma, USA) with 1.2 g/l sodium bicarbonate (Sigma, USA), 10% (v/v) antibiotic/antimycotic and 10% (v/v) FBS); this fraction of cells was termed ‘SOM’ (somatic).
Immunofluorescence analysis of cultured cells
Cells were washed two times with phosphate buffered saline (1× PBS), fixed for 7.5 min on ice in 4% (v/v) paraformaldehyde, washed with 1× PBS, permeabilized for 15 min with 0.1% (v/v) Triton X-100 in 1× PBS (PBT) and blocked in 1× Blocking Reagent (Roche) in 1× PBS for 1 h. Antibodies were diluted in PBT and 1 μg/ml 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) was added with the secondary antibodies for visualization of DNA. Cells were washed in PBT after each antibody incubation. Primary antibodies included: rabbit anti-deleted in azoospermia-like (DAZL; 1:1000 Ab34139, Abcam, USA), rabbit anti-spalt-like transcription factor 4 (SALL4; 1:500 Ab29112, Abcam, USA), goat anti-GATA binding protein 4 (GATA4; 1:250 SC1237, Santa Cruz, USA), mouse anti-VIMENTIN (VIM; 1:500 VI-10, Pierce, USA), mouse anti-undifferentiated embryonic cell transcription factor 1 (UTF; 1:1000 5G10.2, Millipore, USA), mouse anti-actin, alpha 2, smooth muscle, aorta (ACTA2; 1:2000 1A4; Millipore, USA) and biotin conjugated mouse anti-stage-specific embryonic antigen 4 (SSEA4; 1:400 MC-813–70, Biolegend, USA). Secondary antibodies were diluted 1:500 and included: Cy3 conjugated donkey anti-goat and Alexa Fluor 488 or 594 conjugated goat anti-rabbit or goat anti-mouse (Life Technologies, USA) and Streptavidin conjugated Alexa Fluor 488 (Jackson Immuno, USA). The immunofluorescence data shown are representative of at least two experiments performed using the following donors: Hu2, Hu4 and Hu5.
Immunofluorescence analysis of paraffin sections
Five micrometer sections of formalin-fixed and paraffin-embedded tissue were dewaxed in xylene and rehydrated through a graded series of decreasing concentrations of ethanol. Microwave-mediated antigen retrieval was performed by maintaining slides in a solution of 10 mM sodium citrate with 0.05% (v/v) tween-20 (pH 6.0) at just below boiling temperature for 10 min. Tissue was permeabilized for 15 min with 0.2% (v/v) triton X100 in 1× PBS and blocked in 3% (w/v) BSA (Jackson Immuno, USA) in 1× PBS for 1 h. The same antibodies were used as described for cell staining; however, antibodies were diluted in 1× PBS and washes were in 1× PBS. The immunofluorescence data shown are representative of at least two experiments performed using the following donors: Hu2, Hu5, Hu6 and Hu7.
Immunofluorescence analysis of whole tubules
Pieces of seminiferous tubules were washed twice in 1× PBS for 15 min. All steps were at 4°C with rocking unless specified otherwise. Permeabilization was in 10% (v/v) methanol + 0.1% (v/v) Triton X-100 in 1× PBS for 1 h. Tubules were blocked for 1.5 h in Blocking Buffer B (1× Blocking Buffer (Roche, USA) + 0.05% (v/v) Triton X-100 in 1× PBS) for 1.5 h. The same antibodies were used as described above for cell staining except anti-SALL4 was used at 1:800 and antibodies were diluted in Blocking Buffer B. Washes were in PBT. The immunofluorescence data shown are representative of at least two experiments performed using the following donors: Hu4 and Hu5.
Oil Red O staining
Cultured cells were fixed in formalin and permeabilized in isopropanol, and 0.18% (w/v) Oil Red O (Santa Cruz Biotechnology, USA) in 60% (v/v) isopropanol was applied to each well for 5 min at room temperature. Type 293 human embryonic kidney (HEK) cells were used as a comparison as an example of a non-lipid rich fibroblast cell type. The Oil Red O staining data shown are representative of at least three experiments performed using the following donors: Hu2, Hu7 and Hu8.
SSEA4 and CD9 analysis and sorting
For analysis of the cell surface markers SSEA4 and CD9 molecule (CD9) by flow cytometry, cultured cells were prepared by trypsinizing both floating and bound cells. Cells were washed twice in PBS/FBS (1× PBS with 2% (v/v) FBS) and incubated with PE conjugated mouse anti-SSEA4 (0.625 μg/ml, Biolegend, USA) or biotin conjugated mouse anti-CD9 (2.5 μg/ml eBioSN4, eBioscience, USA) in 100 μl PBS/FBS on ice for ∼30 min. Cells were washed in PBS/FBS prior to analysis on a FACSCalibur (Becton, Dickinson and Company, USA). In the case of CD9 a secondary antibody was used (1:1000, Streptavidin conjugated Alexa Fluor 488; Jackson Immuno, USA). The SSEA4 and CD9 flow cytometry data shown are representative of at least two experiments performed using the following donors: Hu5 and Hu6.
Quantitative RT–PCR
RNA extraction was performed using RNeasy Mini or RNeasy Micro Plus Kits (Qiagen, USA) and following the manufacturer's instructions. RNAs prepared by RNeasy Mini kit were treated with DNAse (Qiagen, USA). Reverse transcription was performed using qScript reverse transcriptase (Quanta Biosciences, USA). RNA samples were subjected to a negative control test in which reverse transcriptase was omitted. Quantitative PCR was performed using Perfecta SYBR Fast Mix Low Rox (Quanta Biosciences, USA) on a Stratagene MX3000P. PCR primers were either previously published or were designed using NCBI Primer-BLAST tool (Table II). We verified that each primer pair amplified the intended product in multiple ways. First, a dissociation curve was evaluated at the end of each PCR experiment. Second, amplified products were run on an agarose gel and evaluated for purity and size. Finally, for all primer pairs designed in this study and not previously published the sequence of the product was confirmed by Sanger sequencing.
Table II.
Primer design.
| Gene name | Symbol | Sequence | Size | Designb |
|---|---|---|---|---|
| Actin, alpha 2, smooth muscle, aorta | ACTA2 | 5′-ACTGGGACGACATGGAAAAGA-3′ 5′-GGCAACACGAAGCTCATTGTAG-3′ |
59 bp | Kossack et al. (2013) |
| Deleted in azoospermia-like | DAZL | 5′-GCCACGTCCTTTGGTTTTTA-3′ 5′-TGGTTGTGGGCTGCATATAA-3′ |
76 bpa | Wu et al. (2009) |
| Fibroblast growth factor receptor 3 | FGFR3 | 5′-CAAGGTGTACAGTGACGCA-3′ 5′-AAGGAGAGAACCTCTAGCTCC-3′ |
153 bpa | NCBI Primer-BLAST |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 5′-GCTGAGTACGTCGTGGAGTC-3′ 5′-GTGCAGGAGGCATTGCTGA-3′ |
185 bpa | NCBI Primer-BLAST |
| GATA binding protein 4 | GATA4 | 5′-CCTGCGGCCTCTACATGA-3′ 5′-AAGGAGCTGCTGGTGTCTTA-3′ |
123 bpa | NCBI Primer-BLAST |
| G protein-coupled receptor 125 | GPR-125 | 5′-CTTGGCGCAGATGTGATAGA-3′ 5′-TGTCAGAAAAGTTGGCTGCT-3′ |
183 bpa | NCBI Primer-BLAST |
| Undifferentiated embryonic cell transcription factor 1 | UTF1 | 5′-CGACATCGCGAACATCCTG-3′ 5′-CAGGGACACTGTCTGGTCG-3′ |
100 bpa | NCBI Primer-BLAST |
| Vimentin | VIM | 5′-TCTGCCTCTTCCAAACTTTTCC-3′ 5′-CCAGAGGGAGTGAATCCAGATT-3′ |
65 bp | Kossack et al. (2013) |
| Zinc finger and BTB domain containing 16 | ZBTB16 | 5′-GCACATCAAGAGCCACAAA-3′ 5′-CTCAAAGGGCTTCTCACCTG-3′ |
115 bpa | Wu et al. (2009) |
aProduct verified by sequence.
bPrimers were either designed using the NCBI Primer-BLAST tool or were designed by others in previously published work as referenced here.
qRT–PCR reactions were performed in duplicate. Relative quantity was calculated by comparing to a standard curve of input RNA (serial dilutions of human testis RNA). The mean quantity and standard deviation (SD) were calculated for the technical duplicates. For each cDNA reaction a relative quantity of GAPDH was also determined, again by using the standard curve method. The mean GAPDH quantity was used for normalizing the quantity of input RNA for each cDNA reaction. Standard propagation of error calculations were used in determining the error associated with calculating the ratio for each mRNA to GAPDH levels and the error associated with pooling data from biological replicates. For the non-quantitative RT–PCR data shown in Fig. 4 the same RT–PCR protocol was followed and the amplified products were subsequently run on a 1% (w/v) agarose gel.
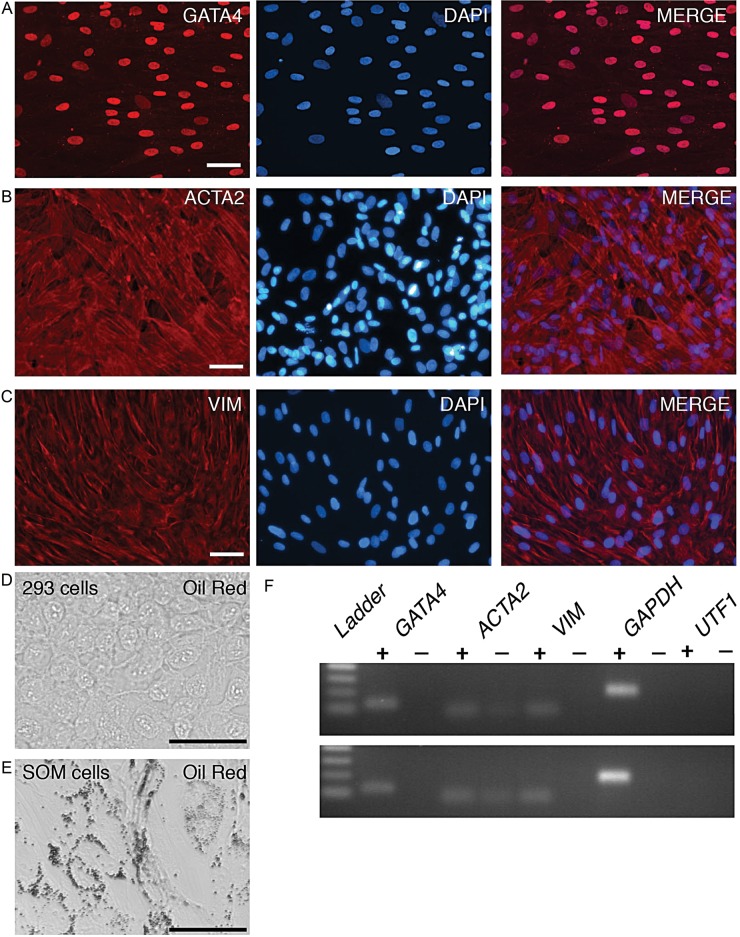
Figure 4.
Immunofluorescence analysis of somatic cell markers in cultured somatic cells. The bound fraction of dissociated testicular cells were cultured in somatic cell maintenance medium (F12/FBS) prior to fixation and immunostaining to detect (A) GATA4, (B) ACTA2 or (C) VIM. DAPI was used to stain the DNA (blue). Bars represent 50 μm. (D and E) Oil Red O staining of 293 human embryonic kidney fibroblasts (D) or SOM cells (E). (F) Agarose gel analysis of the indicated qRT–PCR products using Hu2 SOM RNA (top image) or Hu4 SOM RNA. ‘+’ and ‘−’ refer to the presence or absence of reverse-transcriptase in cDNA reactions. ‘Ladder’ refers to the DNA size markers of 100–400 base pairs.
Statistical analysis
Unpaired two-tailed Student's t-test was used to calculate P-values comparing the mean relative expression of each mRNA normalized to mean GAPDH mRNA expression. Means were calculated with the standard curve method as described in the quantitative RT–PCR section of the Materials and Methods.
Results
In this study we obtained cells from testes of organ donors varying in age from 13 to 40 years old (Table I). A portion of each testis was fixed and stained with hematoxylin and eosin to confirm that the donors had grossly normal spermatogenesis based on the presence of expected spermatogenic cells. Additionally, next of kin confirmed that the donors had no known reproductive health issues and several donors were known to have children (Table I). We obtained testicular cells following a standard two-step procedure for tissue digestion of either fresh or frozen/thawed samples. We obtained a mean ± SD of 29 ± 16 million cells per gram of fresh tissue (n = 8). The yield of cells from frozen/thawed samples was significantly lower than that obtained from fresh tissue (Supplementary data, Fig. S1) and varied among donors.
Validation of antibodies by immunofluorescence analysis of testicular sections
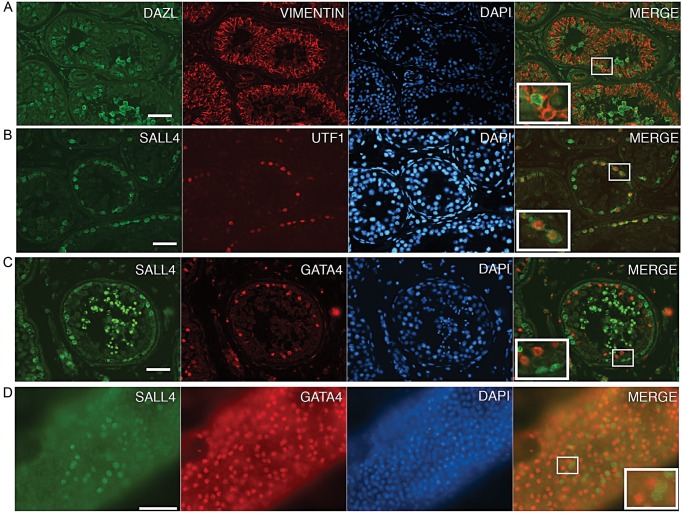
Given that a major challenge while initiating a new culture of SSCs is overgrowth by somatic testicular cells, we sought to establish assays for determining the relative contributions of germ and somatic cell types present. First we validated the use of particular antibodies for the detection of germ and somatic cells by performing co-immunofluorescence experiments on sections of seminiferous tubules from donor tissue. We tested antibodies to detect DAZL, UTF1 or SALL4 in germ cells (Fig. 2A, B and C). DAZL immunostaining was present in spermatogonia and in spermatocytes. UTF1 was present in spermatogonia. SALL4 was also present in spermatogonia, a subset of which were the same population as UTF1-positive spermatogonia; SALL4 immunostaining was also unexpectedly observed in spermatids. We also tested antibodies to detect VIM or GATA4 in somatic cells. As expected, co-immunostaining with DAZL and VIM or GATA4 and SALL4 revealed a non-overlapping staining pattern between the germ cell and somatic cell markers (Fig. 2A and C). Similar results were obtained by co-immunostaining whole tubules to detect GATA4 and SALL4 (Fig. 2D). Our results are consistent with previously described expression patterns for these proteins in testes and validate these particular antibodies for use in detecting somatic and germ cells in the context of germ cell cultures (Miettinen et al., 1985; Reijo et al., 2000; Wu et al., 2009; Eildermann et al., 2012; Valli et al., 2014).
Figure 2.
Immunofluorescence analysis of germ and somatic cell markers in seminiferous tubules. Immunostaining on paraffin sections (A, B and C) or whole tubule pieces (D). Co-immunostaining was performed to detect (A) DAZL (green) and VIM (red), (B) SALL4 (green) and UTF1 (red) or (C) SALL4 (green) and GATA4 (red). DAPI was used to stain the DNA (blue). Merges of green and red show that the germ cell and somatic cell staining were mutually exclusive. SALL4 immunostaining was unexpectedly observed in post meiotic cells, although it may be background staining associated with this lot of polyclonal antibody (Eildermann et al., 2012). Bars represent 50 μm.
Immunofluorescence analysis of primary testicular cell cultures
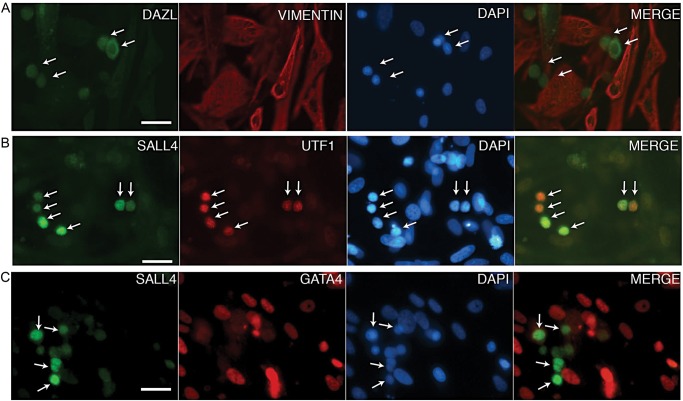
We then used the validated antibodies to detect germ and somatic cells in cultures of primary testicular cells. Specifically, we established a culture using a protocol previously described to support SSC growth in vitro (Sadri-Ardekani et al., 2009). Similar to what has been described by others, we observed clusters of smaller cells, typical of spermatogonia, present on top of a proliferating layer of flat, spread cells, typical of fibroblast-like somatic cells. Co-immunofluorescence analysis of early primary testicular cell cultures (‘PTC’; day 11–13) revealed that the flat monolayer of cells expressed GATA4 and VIM, confirming the prediction that these cells are somatic cells (Fig. 3A and B). We also observed relatively rare groups of DAZL positive and VIM negative or SALL4 positive and GATA4 negative cells (Fig. 3A and B). Also, UTF1-positive cells were detected; some cells expressed both SALL4 and UTF1 while others expressed only SALL4 or UTF1 (Fig. 3C). The DAZL, UTF1 and SALL4 expressing cells had small round nuclei that were distinct from the larger, oblong somatic cell nuclei, consistent with the interpretation that DAZL, UTF1 and SALL4 expression marked the germ cells. These results confirmed the use of DAZL, UTF1 or SALL4 as markers for identifying the germ cell population and VIM or GATA4 for identifying the somatic cell population. These results also established that early primary testicular cell cultures were a heterogeneous mixture of somatic and germ cells.
Figure 3.
Immunofluorescence analysis of germ and somatic cell markers in early primary testicular cell cultures. Primary testicular cells were cultured in germ cell maintenance medium prior to fixation and co-immunostaining to detect (A) DAZL (green) and VIM (red) or (B) SALL4 (green) and UTF1 (red) or (C) SALL4 (green) and GATA4 (red). Merges of green and red are shown. DAPI was used to stain the DNA (blue). Bars represent 50 μm.
qRT–PCR analysis of early primary testicular cell cultures
While the immunofluorescence assay showed that spermatogonia and somatic cells were present in early primary testicular cultures, technical limitations prevented it from being useful for quantitative analyses. As a quantitative assay would be required for optimization of culture conditions we evaluated qRT–PCR as a method for analyzing primary testicular cultures. Given the finding that germ cells were relatively rare cells in the mixed cell population of PTC, qRT–PCR would be expected to provide a sensitive method for detecting rare germ cells. The goal was to identify spermatogonia/SSC associated mRNAs that could be detected in PTC but with minimal or no expression in somatic cells. We compared two types of samples. The first sample type was RNA isolated from early PTC (Day 7) that contained a mixture of somatic cells and clusters of germ cells as described above. For comparison we used RNA isolated from cells that were removed by differential adherence during initiation of the culture and had subsequently been maintained for >11 days in a simple growth medium (F12/FBS, Fig. 1 and Materials and Methods) in which they formed a monolayer of fibroblast-like cells; this cell population appeared relatively homogeneous with cells exhibiting a flat, spread morphology. Many of the cells contained Oil Red staining lipid droplets, and widespread VIM and GATA4 mRNA and protein expression, consistent with the interpretation that the population included Sertoli cells. The population also exhibited widespread expression of ACTA2 (SMA), consistent with the cells having a peritubular myoid origin (Tung and Fritz, 1990). Given that the population had properties of both Sertoli and myoid cells we simply termed the population somatic (‘SOM’) (Fig. 4).
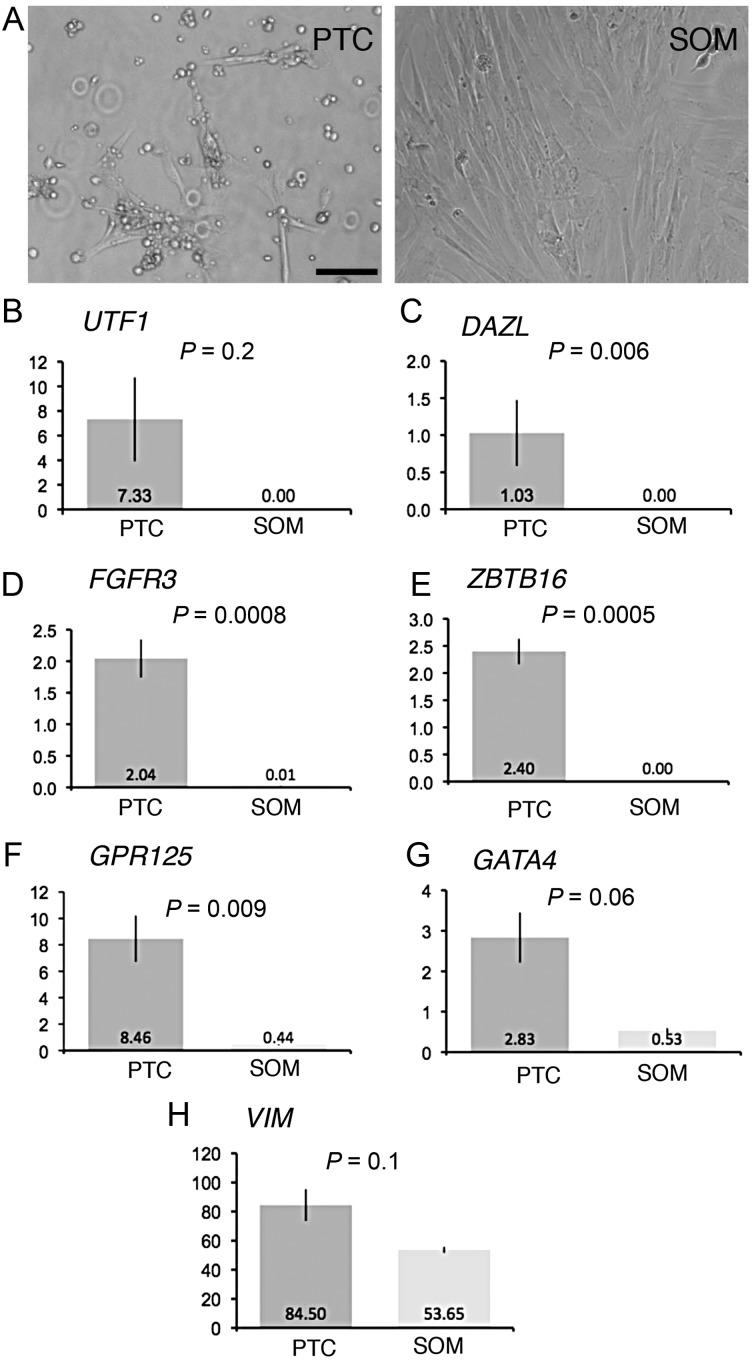
We evaluated the expression of several mRNAs previously described as being expressed in human spermatogonia/SSCs (UTF1, FGFR3, ZBTB16 and GPR125) or more widely expressed in germ cells (DAZL). Our qRT–PCR results showed that UTF1 and DAZL were present in PTC and undetectable in SOM; similarly, FGFR3 and ZBTB16 were highly enriched (>100-fold) in PTC compared with SOM (Fig. 5 and Supplementary data, Fig. S1). In contrast, GATA4 and GPR125 were moderately enriched in PTC versus SOM (5-fold and 19-fold, respectively); VIM was detectable at similar levels in PTC and SOM. These results were consistent with the idea that primary testicular cells comprise a rare but detectable germ cell population and an abundant somatic cell population. Also, the results supported the idea that qRT–PCR analysis of UTF1, FGFR3, ZBTB16 or DAZL mRNAs was an effective tool for detecting rare germ cells in early cultures of primary testicular cells.
Figure 5.
Quantitative RT–PCR analysis of early primary testicular cell cultures. (A) Representative images of PTC on Day 7 and SOM on Day 20 are shown. (B–H) RNA was isolated from cultures of primary testicular cells on Day 7 (n = 2, PTC from Hu2 and Hu4) and somatic cells on Day 20 (n = 3, SOM from Hu2 twice and Hu4 once). Related data from additional culture experiments are presented in Supplementary data, Fig. S2. Graphs depict the mean ± SD of the relative mRNA levels of the indicated mRNAs normalized to GAPDH mRNA levels for each sample. The y-axis represents arbitrary units. The P-values and means are shown. Bar represents 100 μm.
Validation of SSEA4 as a germ cell specific marker in primary testicular cell cultures
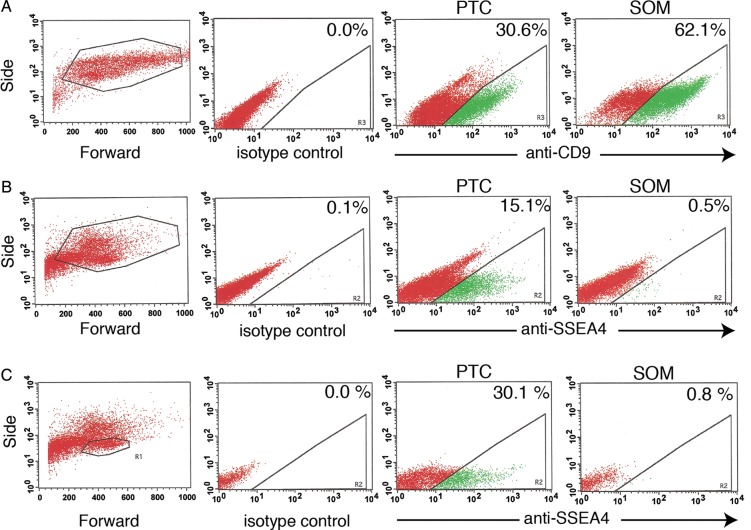
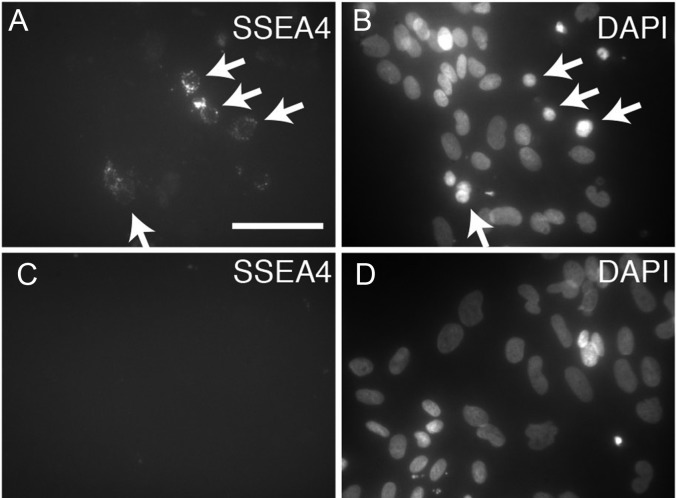
To complement the qRT–PCR assay we sought an assay that could be used to directly count rare spermatogonia from among a small total number of cells. We considered flow cytometry for detection of two cell surface antigens previously described as human spermatogonial markers: CD9 and SSEA4 (Izadyar et al., 2011; Zohni et al., 2012). Again, comparisons were made between PTC and SOM cells. Surprisingly, CD9 expression was abundant in both PTC and SOM cells (Fig. 6). In contrast, a fraction of PTC cells exhibited bright SSEA4 staining whereas bright staining was virtually absent in SOM cells. Dim SSEA4 staining that was observed in a small fraction of SOM cells was defined as background. The SSEA4bright cells were enriched within the smaller, less complex population of the forward/side scatter plot, where germ cells typically appear. Additionally, by using microscopy to observe immunostaining of PTC SSEA4 staining was observed in rare cells with nuclear morphology typical of spermatogonia but not in cultured SOM cells (Fig. 7). Our results suggested that SSEA4bright staining was specific to germ cells.
Figure 6.
Flow cytometry analysis of SSEA4 and CD9 in primary testicular cell cultures. PTC and SOM cells from Day 8 of culture were immunostained with antibodies to CD9 (A) or SSEA4 (B and C) or the corresponding IgG isotype control for each. Representative dot plots depicting the forward/side scatter for SOM (A, left) and PTC (B and C, left) are shown. Dot plots from the same samples are shown in (B) and (C) except a more restrictive forward/side scatter gate is applied in (C). The plots shown in this figure are from an experiment using frozen Hu5 testes. The same results were obtained using cultures derived from frozen Hu6 testes.
Figure 7.
Immunofluorescence analysis of SSEA4 in primary testicular cultures. Primary testicular cells (A and B) or SOM cells (C and D) were cultured for 6 days prior to fixation and immunostaining to detect SSEA4. DAPI was used to stain the DNA (blue). The images shown are from cultures derived from Hu2 and the same results were found with cultures from Hu4 and other donors. Bar represents 50 μm.
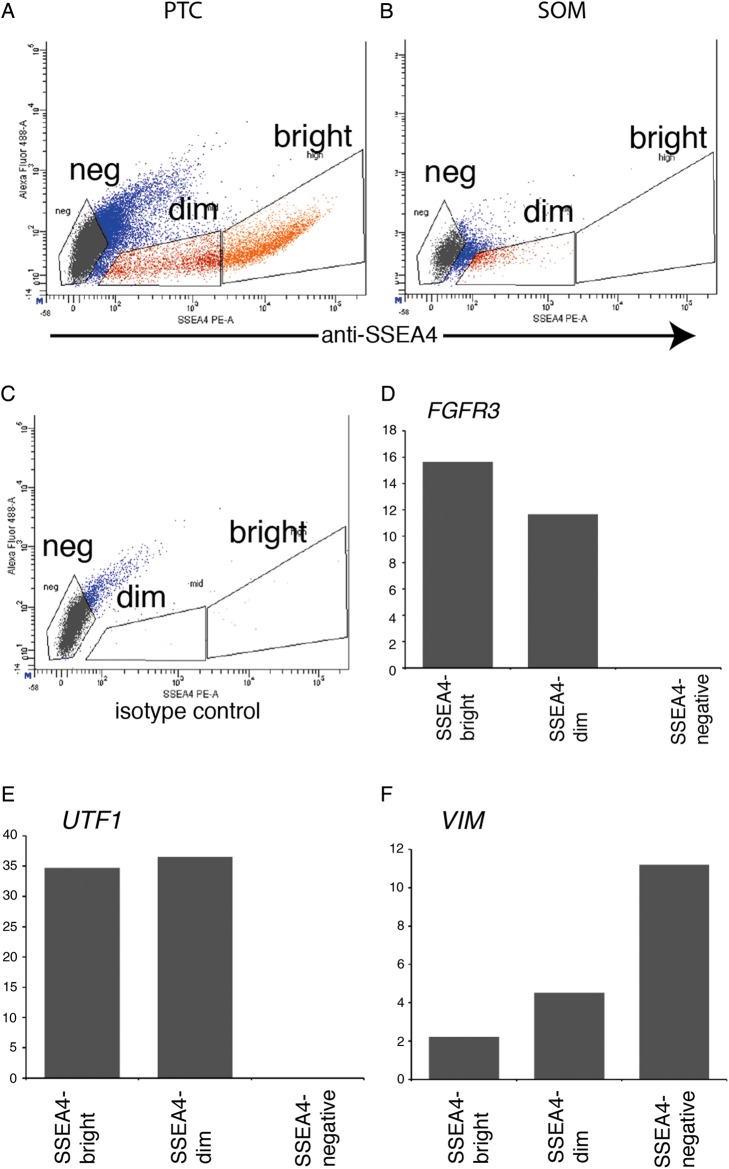
In order to further evaluate the nature of the cells that stained with SSEA4 we performed qRT–PCR analysis on SSEA4bright, SSEA4dim and SSEA4negative cells isolated from primary testicular cell cultures by fluorescence-activated cell sorting (FACS). This experiment used PTC derived from frozen/thawed Hu2 testes. As predicted, SSEA4bright cells were highly enriched in UTF1 and FGFR3 mRNA while SSEA4negative cells were highly enriched in VIM mRNA (Fig. 8). These results indicated that SSEA4 staining could be used to specifically and quantitatively detect and isolate rare spermatogonia/SSCs from the heterogeneous population of primary testicular cultures.
Figure 8.
Quantitative RT–PCR analysis of SSEA4 sorted cells. Dot blots show SSEA4 fluorescence on the x-axis in (A) primary testicular cells or (B) somatic cells on Day 11 of a culture derived from frozen Hu2 testes. (C) Isotype control. RNA was isolated from SSEA4bright, SSEA4dim and SSEA4negative cells. Graphs depict the mean relative mRNA levels of (D) FGFR3, (E) UTF1 or (F) VIM normalized to GAPDH mRNA levels for that sample. The y-axis represents arbitrary units.
Quantitative analysis of primary testicular cell cultures over time in culture
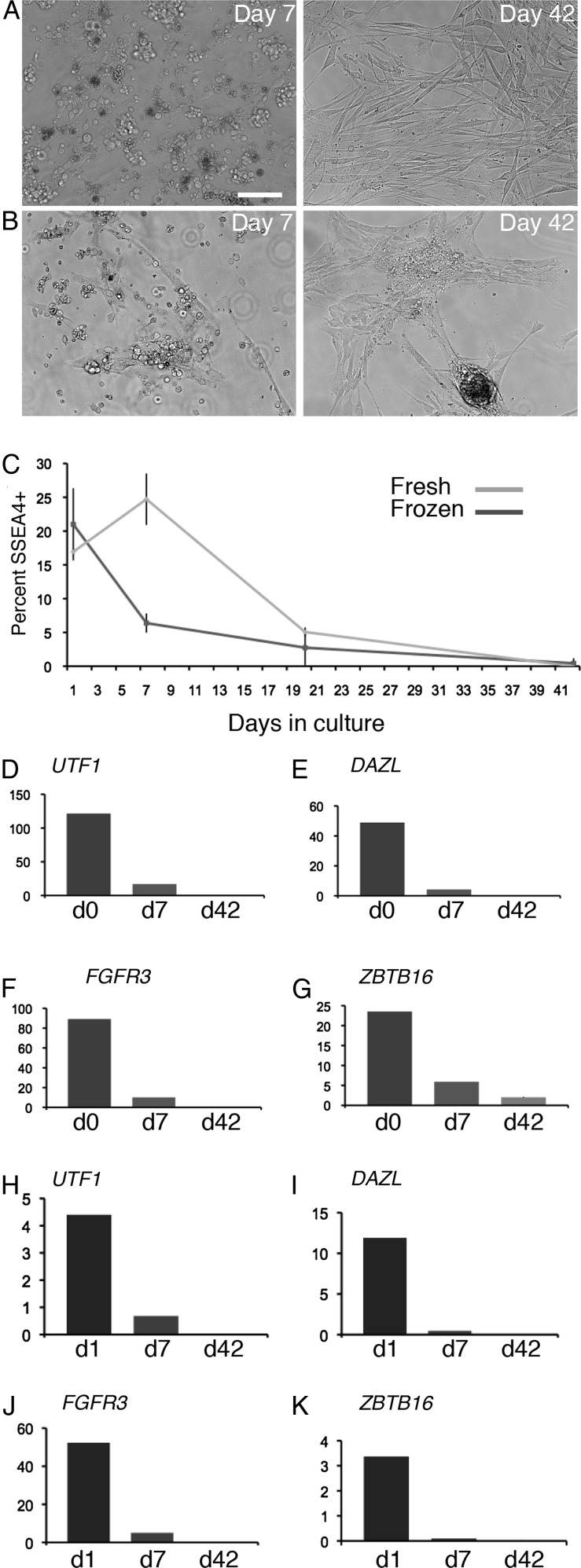
We then used qRT–PCR and SSEA4 assays to evaluate the composition of cells present over an extended time in culture. As in the short-term culturing experiment described above, we used the method of Sadri-Ardekani et al. (2009) wherein somatic cells present in PTC serve as a supporting feeder layer for the germ cells. Primary testicular cell cultures were established from four donors (Hu2 and Hu4 frozen tissue and Hu8 and Hu9 fresh tissue) and analyzed at the initiation of the culture and then at Day 7 and Day 42 (Day 42 analysis was on cultures from Hu2, Hu4 and Hu9 only). In multiple trials at deriving long-term cultures of human SSCs, either from fresh or frozen testes we consistently found a steady decline in the percentage of SSEA4bright cells (Fig. 9). A peak of SSEA4 expression was observed at Day 7 in cultures established from fresh tissue but not frozen tissue; nonetheless, the long-term outcome was the same. The SSEA4 flow cytometry assay was as an efficient method for detecting rare SSCs/spermatogonia (e.g. 0.1%) from among small populations of analyzed cells.
Figure 9.
Quantitative analysis of validated germ cell markers in primary testicular cells over time in culture. (A and B) Representative images of PTC on Day 7 and Day 42 derived from either fresh (A) or frozen/thawed (B) testes. (C) The graph depicts the mean ± SD of the percentage SSEA4bright cells on Day 1 (n = 2: Hu8 and Hu9 fresh; n = 2: Hu2 and Hu4 frozen), Day 7 (n = 2: Hu8 and Hu9 fresh; n = 2: Hu2 and Hu4 frozen) and Day 42 (n = 1: Hu9 fresh; n = 2 Hu2 and Hu4 frozen) of culture. (D–K) Graphs depict the mean of the relative mRNA levels of the indicated mRNAs normalized to GAPDH mRNA levels for each timepoint. Each graph depicts data from a PTC of a single donor with the mean from technical duplicates displayed at each timepoint for either fresh (D–G) or frozen (H–K). The y-axis represents arbitrary units. Similar qRT–PCR results from different donors are shown in Supplementary data, Fig. S3.
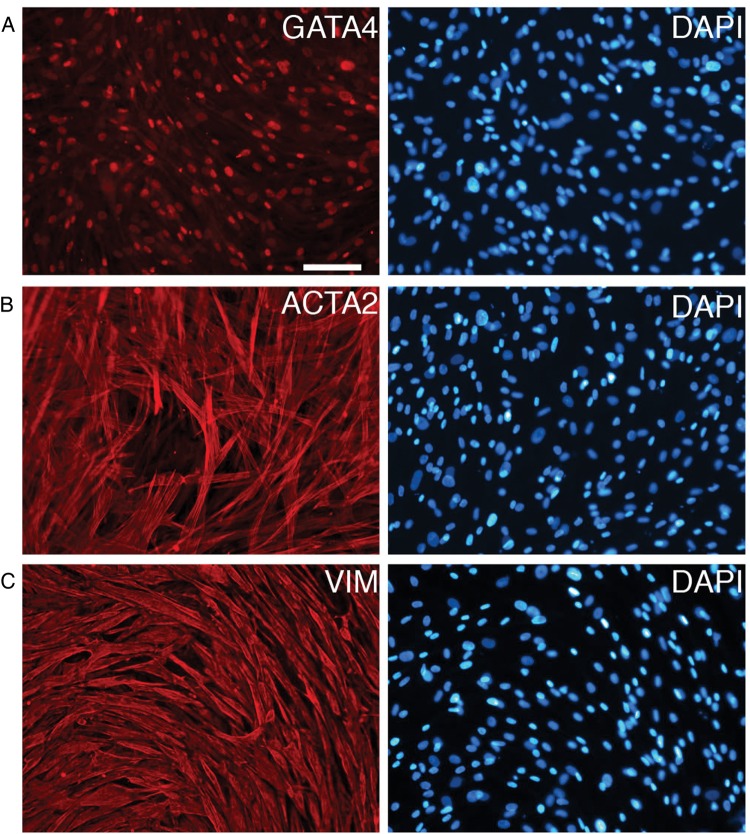
To further characterize the long-term cultures we used qRT–PCR to analyze mRNAs that we validated as germ cell specific. UTF1, FGFR3 and DAZL were detectable during the first week of culture but not after several weeks of culture (3/3 cultures; Fig. 9 and Supplementary data, Fig. S3). ZBTB16 was detected in some long-term cultures (2/3 cultures; Fig. 9 and Supplementary data, Fig. S3). Altogether, our SSEA4 and qRT–PCR analysis indicated that the testicular cells cultured using previously published conditions are likely to contain few, if any, SSCs. Rather, cells present after several weeks of culture had the appearance of somatic cells and expressed the somatic cell markers VIM, ACTA2 and GATA4 (Fig. 10).
Figure 10.
Immunofluorescence analysis of somatic cell markers in Day 48 primary testicular cultures. Primary testicular cells derived from fresh Hu9 testes were cultured for 48 days prior to fixation and immunostaining to detect GATA4 (A), ACTA2 (B) or VIM (C). DAPI was used to stain the DNA (blue). These same results were obtained using cultures from frozen Hu4 testes. Bar represents 100 μm.
Discussion
In this study we validate and apply expression assays that distinguish between human testicular somatic and germ cell populations, allowing the key challenge of somatic versus germ cell growth during culture initiation to be addressed. In particular, we find that FGFR3, UTF1, DAZL and ZBTB16 qRT–PCR reliably and sensitively detect germ cells while VIM can be used to detect somatic cells. Also, we demonstrate that SSEA4 analysis by flow cytometry provides a quantitative and sensitive method for tracking spermatogonia/SSCs over time in culture, thereby facilitating comparison of conditions for human SSC culture. Application of these validated assays to examine primary testicular cells cultured using previously described methods leads to the conclusion that considerable improvement in SSC culture conditions is still needed.
Propagation of human SSCs in vitro is essential for performing autologous transplantation therapies aimed at restoring fertility to survivors of childhood cancer. Availability of a SSC culture system could also provide a useful model to expand our understanding of the molecules regulating the development of human SSCs, which exhibit some similarities but also several differences from their rodent counterpart (Dym et al., 2009; Wu et al., 2009; He et al., 2010; Izadyar et al., 2011). While short-term culture periods of up to a month may be sufficient for some studies, ideally human SSCs could be robustly propagated for several months or even indefinitely, similar to the way that rodent SSCs can be propagated.
To our knowledge, three groups have described long-term (>5 months) culture of human SSCs (Sadri-Ardekani et al., 2009; Lim et al., 2010; Golestaneh, 2011). The culture medium used for maintenance by each group was very similar and consisted of a Stem Pro-34 serum free base medium with numerous supplements plus growth factors that included GDNF, FGF, EGF and LIF. We applied this culture method and characterized cells using similar and additional assays and concluded that following propagation of periods over a month the cells remaining in culture are almost entirely somatic cells. One key difference in our analysis of cultured testicular cells to that performed by other groups was the use of cultured somatic cells as a comparison. Immediately following testicular dissociation we and other groups typically perform a selection step in which cells are plated overnight on standard tissue culture plates to allow some somatic cells to be removed based on differential adherence. Unbound cells (germ and somatic) are washed off and transferred to a new plate containing stem cell medium with GDNF and no serum for establishment of SSC cultures. We also maintain the bound cells (somatic) in a standard fibroblast-type growth medium with 10% FBS so that they can be used as a comparison. The bound cells exhibit several properties of somatic cells including their ability to readily reach confluence without GDNF, growth as a monolayer, presence of lipid droplets characteristic of Sertoli cells and widespread expression of VIM, GATA4, ACTA2 mRNA and protein. Hence, we routinely use these somatic cells as a negative control when characterizing cultures of germ cells. Such analysis led to the realization that several markers that we and others had been using for identifying human spermatogonia/SSCs may not be germ cell specific. Examples include GPR125 mRNA and the cell surface protein CD9.
Consistent with our findings, Kossack et al. (2013) recently published an elegant study comparing gene expression in human testicular samples from normal patients and patients lacking germ cells (e.g. Sertoli Cell Only, SCO). They found that only nine of the eighteen tested mRNAs thought to be markers of germ cells were actually specific to germ cells. The nine non-specific markers included GPR125, ZBTB16, ITGA6 and GFRA1, all of which have been used to test for the presence of SSCs in attempts at developing a method for human SSC culture (Sadri-Ardekani et al., 2009; Lim et al., 2010; Golestaneh, 2011; Koruji et al., 2012); in contrast, FGFR3 and UTF1 mRNAs were among mRNAs that Kossak et al. found to be germ cell specific. Our analysis of FGFR3 and UTF1 mRNA expression in cultured germ cells compared with somatic cells also showed essentially no expression of these two mRNAs in somatic cells; in contrast, GPR125 mRNA could be readily detected in somatic cells in our study. We noticed one interesting difference between their study and our study. ZBTB16 mRNA was not detected in our somatic cell cultures, suggesting that it is a germ cell-specific marker. Yet, in a subset of long-term (Day 42) primary testicular cell cultures we detected low levels of ZBTB16 mRNA even though four other germ cell-specific markers (SSEA4, DAZL mRNA, FGFR3 mRNA and UTF1 mRNA) could not be detected. Based on the currently available data it remains unclear whether the presence of ZBTB16 in some of our long-term cultures reflects the presence of a few germ cells of a specific sub-type, such as partly differentiated spermatogonia, or if the ZBTB16 expression is from a somatic cell type (Valli et al., 2014). Overall, our results support and build upon those of Kossack et al. (2013).
In order to achieve the ultimate goal of long-term human SSC culture it will be necessary to identify conditions that support germ cell proliferation without the concurrent expansion of the somatic testicular population. The issue of somatic cell ‘contamination’ during the establishment of a new SSC culture also presents a challenge for mouse SSC culture, particularly from certain genetic backgrounds and ages of mice. One method for overcoming this hurdle is cell sorting. For example, when establishing a rodent SSC culture from immature mouse testes, where the Sertoli cells are highly proliferative, Thy1pos cells can be isolated using MACS (magnetic associated cell sorting), allowing SSCs to be greatly enriched and somatic cells to be mostly removed (Kubota et al., 2004a,b). Similar attempts at SSC enrichment have been made using human testes. Although THY1 is not ideal for enrichment of human SSCs by MACS, flow cytometry can be used to enrich SSCs by sorting THYdim cells (Valli et al., 2014). Other strategies have recently been used for enriching human SSCs including EPCAMpos cells sorted by FACS, or ITGA6pos cells sorted by FACS or MACS, or SSEA4pos cells sorted by MACS, or the combined phenotype of HLAneg/CD9pos sorted by MACS (Golestaneh, 2011; Izadyar et al., 2011; Zohni et al., 2012; Dovey et al., 2013; Valli et al., 2014). Based on our results, together with other studies assessing the cellular identity of SSEA4pos testicular cells, we propose that SSEA4 could be an excellent marker for enrichment of spermatogonia/SSCs prior to long-term culturing (Izadyar et al., 2011). We note, however, that it may be necessary to use a flow cytometry approach to separate the SSEA4bright and SSEA4dim cells, because our data suggest that even some somatic cells exhibit a SSEA4dim phenotype. The use of MACS may be problematic because it may not distinguish between SSEA4bright and SSEA4dim cells.
Another potential challenge in the pursuit of conditions for human SSC culture may be the availability of normal testes for experimentation. We and others have found that organ donors serve as an excellent source of testes for research (He et al., 2010; Sachs et al., 2014; Valli et al., 2014). Although the time of transfer to the laboratory is difficult to control, and possibly longer than is desirable, a major advantage is that one can obtain a much larger quantity of tissue (∼30 g) from an organ donor than from clinical biopsies (<1 g), another common source of tissue. Since one obtains a substantial quantity of tissue from each donor, we cryopreserved most of the tissue using a cryopreservation medium described by He et al. (2010). The yield of cells was much lower from frozen tissue compared with fresh tissue but we could readily offset the reduced yield by dissociating several pieces of tissue. To maximize cell yield one could alternatively cryopreserve dissociated cells instead of masses of tissue (Valli et al., 2014). It remains to be determined whether human SSCs can be propagated long term using organ donors as a source of testes. We obtained similar overall results whether cultures were established with fresh or frozen tissue. Considering that several markers associated with undifferentiated spermatogonia/stem cells, including SSEA4 and UTF1, were present in early cultures, it is likely that organ donor tissue provides an adequate source of stem cells but that the stem cells were simply lost over time as a result of inadequate culturing conditions.
In future studies we propose that SSEA4bright cells could be isolated, cultured in a variety of conditions, and then SSEA4 analyzed by flow cytometry as an efficient and quantitative means for comparing many conditions for identification of a method for human SSC growth in vitro. Subsequent utilization of a combination of qRT–PCR (UTF1, FGFR3 and VIM) and immunofluorescence (DAZL/VIM and UTF1 or SALL4/GATA4) as described here could provide secondary assays to confirm the identity of cells growing in culture. Application of these assays will allow continued optimization of human SSC culture as we and others continue to seek conditions that permit in vitro expansion of human SSCs with a robustness sufficient for studying the basic biology of human SSCs and eventually cell transplantation therapy.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
Y.Z., C.M.S. and A.T. were involved in acquisition and analysis of data and C.T.D. was involved in conception, design, coordination, acquisition and analysis of data and wrote the manuscript. All authors were involved in critical discussion and approved the final manuscript.
Funding
The experiments presented in this manuscript were funded by a Project Development Team within the ICTSI NIH/NCRR Grant Number TR000006.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We are very grateful to the staff of Indiana Organ Procurement Organization for coordinating the procurement and transfer of donor tissue for research and to Sue Childress and Crystal Heim for technical assistance.
References
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316:404–405. doi: 10.1126/science.1137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caires K, Broady J, McLean D. Maintaining the male germline: regulation of spermatogonial stem cells. J Endocrinol. 2010;205:133–145. doi: 10.1677/JOE-09-0275. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Mizrak SC. Deriving multipotent stem cells from mouse spermatogonial stem cells: a new tool for developmental and clinical research. Development. 2008;135:2207–2213. doi: 10.1242/dev.015453. [DOI] [PubMed] [Google Scholar]
- Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, Chu T, Sanfilippo JS, Orwig KE. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123:1833–1843. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dym M, Kokkinaki M, He Z. Spermatogonial stem cells: mouse and human comparisons. Birth Defects Res C Embryo Today. 2009;87:27–34. doi: 10.1002/bdrc.20141. [DOI] [PubMed] [Google Scholar]
- Eildermann K, Aeckerle N, Debowski K, Godmann M, Christiansen H, Heistermann M, Schweyer S, Bergmann M, Kliesch S, Gromoll J, et al. Developmental expression of the pluripotency factor sal-like protein 4 in the monkey, human and mouse testis: restriction to premeiotic germ cells. Cells Tissues Organs. 2012;196:206–220. doi: 10.1159/000335031. [DOI] [PubMed] [Google Scholar]
- Golestaneh N. Long term culture of human SSEA4 positive spermatogonial stem cells. Stem Cell Res Ther. 2011:1–8. doi: 10.4172/2157-7633.S2-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Kawashima T, Stovall M, Leisenring W, Sklar CA, Mertens AC, Donaldson SS, Byrne J, Robison LL. Fertility of male survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J Clin Oncol. 2010;28:332–339. doi: 10.1200/JCO.2009.24.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL. Self-renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci USA. 2005;102:17430–17435. doi: 10.1073/pnas.0508780102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Kokkinaki M, Jiang J, Dobrinski I, Dym M. Isolation, characterization, and culture of human spermatogonia. Biol Reprod. 2010;82:363–372. doi: 10.1095/biolreprod.109.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol. 2005;279:114–124. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER Cancer Statistics Review 1975–2010. Bethesda, MD: National Cancer Institute; 2010. [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec. 1971;169:533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, Yuen C, Greilach S, Zhao HH, Chow M, et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26:1296–1306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, Shinkai Y, et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Miki H, Ogonuki N, Takehashi M, Morimoto T, Ogura A, Shinohara T. Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol Reprod. 2006;75:68–74. doi: 10.1095/biolreprod.106.051193. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Ogonuki N, Morimoto H, Ogura A, Shinohara T. Serum- and feeder-free culture of mouse germline stem cells. Biol Reprod. 2010;84:97–105. doi: 10.1095/biolreprod.110.086462. [DOI] [PubMed] [Google Scholar]
- Koruji M, Shahverdi A, Janan A, Piryaei A, Lakpour MR, Gilani Sedighi MA. Proliferation of small number of human spermatogonial stem cells obtained from azoospermic patients. J Assist Reprod Genet. 2012;29:957–967. doi: 10.1007/s10815-012-9817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossack N, Terwort N, Wistuba J, Ehmcke J, Schlatt S, Scholer H, Kliesch S, Gromoll J. A combined approach facilitates the reliable detection of human spermatogonia in vitro. Hum Reprod. 2013;28:3012–3025. doi: 10.1093/humrep/det336. [DOI] [PubMed] [Google Scholar]
- Kubota H, Brinster RL. Culture of rodent spermatogonial stem cells, male germline stem cells of the postnatal animal. Methods Cell Biol. 2008;86:59–84. doi: 10.1016/S0091-679X(08)00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod. 2004a;71:722–731. doi: 10.1095/biolreprod.104.029207. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA. 2004b;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JJ, Sung SY, Kim HJ, Song SH, Hong JY, Yoon TK, Kim JK, Kim KS, Lee DR. Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Prolif. 2010;43:405–417. doi: 10.1111/j.1365-2184.2010.00691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LA, Seandel M. Serial enrichment of spermatogonial stem and progenitor cells (SSCs) in culture for derivation of long-term adult mouse SSC lines. J Vis Exp. 2013:e50017. doi: 10.3791/50017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Virtanen I, Talerman A. Intermediate filament proteins in human testis and testicular germ-cell tumors. Am J Pathol. 1985;120:402–410. [PMC free article] [PubMed] [Google Scholar]
- Nagano MC. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol Reprod. 2003;69:701–707. doi: 10.1095/biolreprod.103.016352. [DOI] [PubMed] [Google Scholar]
- Nagano M, Yeh JR, Zohni K. Spermatogonia. In: Masters J, Palsson B, editors. Human Adult Stem Cells (Human Cell Culture) 2009. pp. 157–170. Springer Netherlands. [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Niederberger C. Re: in vitro propagation of human spermatogonial stem cells. J Urol. 2012;187:994. [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol. 2006;419:259–282. doi: 10.1016/S0076-6879(06)19011-4. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Arechaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- Piravar Z, Jeddi-Tehrani M, Sadeghi MR, Mohazzab A, Eidi A, Akhondi MM. In vitro culture of human testicular stem cells on feeder-free condition. J Reprod Infertil. 2013;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, Cooke H, Page DC. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Kubota H, Avarbock MR, Brinster RL. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc Natl Acad Sci USA. 2005;102:14302–14307. doi: 10.1073/pnas.0506970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs C, Robinson BD, Andres Martin L, Webster T, Gilbert M, Lo HY, Rafii S, Ng CK, Seandel M. Evaluation of candidate spermatogonial markers ID4 and GPR125 in testes of adult human cadaveric organ donors. Andrology. 2014;2:607–614. doi: 10.1111/j.2047-2927.2014.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, Hovingh S, de Reijke TM, de la Rosette JJ, van der Veen F, et al. Propagation of human spermatogonial stem cells in vitro. JAMA. 2009;302:2127–2134. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305:2416–2418. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- Tung PS, Fritz IB. Characterization of rat testicular peritubular myoid cells in culture: alpha-smooth muscle isoactin is a specific differentiation marker. Biol Reprod. 1990;42:351–365. doi: 10.1095/biolreprod42.2.351. [DOI] [PubMed] [Google Scholar]
- Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, Chu T, Marshall GR, Orwig KE. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril. 2014;102:566–580. doi: 10.1016/j.fertnstert.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Schmidt JA, Avarbock MR, Tobias JW, Carlson CA, Kolon TF, Ginsberg JP, Brinster RL. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci USA. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohni K, Zhang X, Tan SL, Chan P, Nagano M. CD9 is expressed on human male germ cells that have a long-term repopulation potential after transplantation into mouse testes. Biol Reprod. 2012;87:27. doi: 10.1095/biolreprod.112.098913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.