Abstract
Huntington’s disease (HD) is a fatal, progressive neurodegenerative disease with an autosomal dominant inheritance, characterized by chorea, involuntary movements of the limbs and cognitive impairments. Since identification of the HD gene in 1993, tremendous progress has been made in identifying underlying mechanisms involved in HD pathogenesis and progression, and in developing and testing molecular therapeutic targets, using cell and animal models of HD. Recent studies have found that mutant Huntingtin (mHtt) interacts with Dynamin-related protein 1 (Drp1), causing excessive fragmentation of mitochondria, leading to abnormal mitochondrial dynamics and neuronal damage in HD-affected neurons. Some progress has been made in developing molecules that can reduce excessive mitochondrial fission while maintaining both the normal balance between mitochondrial fusion and fission, and normal mitochondrial function in diseases in which excessive mitochondrial fission has been implicated. In this article, we highlight investigations that are determining the involvement of excessive mitochondrial fission in HD pathogenesis, and that are developing inhibitors of excessive mitochondrial fission for potential therapeutic applications.
HD is a fatal, progressive neurodegenerative disease, characterized by involuntary movements, chorea, dystonia, cognitive decline, intellectual impairment and emotional disturbances [1–4]. HD is a midlife disease and mainly found in individuals of Caucasian origin. The prevalence ranges from approximately four to ten individuals in 1000 [5]. A progressive loss of body weight is a major factor in disease progression in patients with HD [6]. Reduced volume of frontal and temporal cortical lobes and an atrophy of striatum were found in HD brains [7,8]. A marked decrease in glucose utilization in the striatum was shown to correlate with several scores in performance-task difficulties in patients with HD, including immediate recall memory, verbal associative learning and executive functions, suggesting that cerebral glucose metabolism is relevant to HD [9,10].
Histopathological examination of brains from patients with HD revealed that several regions of the brain are affected, including caudate and putamen of the striatum, cerebral cortex, hippocampus hypothalamus and subthalamus. The gene for [LM1]causing mutations associated with HD has been identified as an expanded polyglutamine-encoding repeat (or CAG repeat). This mutation is located in exon 1 of the HD gene. In unaffected individuals, polyglutamine repeats are highly polymorphic, whereas in patients with HD, the CAG repeat length ranges from 36 to 120 [5]. The CAG repeat length was found to increase in every generation of male patients with HD who inherited the CAG repeats. This phenomenon, referred to as ‘genetic anticipation’ [5] and CAG repeats, correlates inversely with disease progression in patients with HD.
Htt, a 350-kDa protein, is ubiquitously expressed in the brain and peripheral tissues of patients with HD. Htt has been typically a cytosolic protein. However, a small portion of mHtt as been found in several subcellular organelles, including the nucleus, plasma membrane, mitochondria, lysosomes and endoplasmic reticulum; and the translocated Htt has been found to impair organelle function [11–15]. In addition, mHtt protein aggregates were found in the brains of patients with HD and brain specimens from HD mouse models, mainly in the sites of pathology.
The mechanisms underlying neuronal damage in patients with HD are not well understood. However, the following cellular changes and pathways have been proposed to explain these underlying mechanisms, including: transcriptional dysregulation, expanded polyglutamine repeat protein interactions, calcium dyshomeostasis, defects in axonal trafficking and abnormal mitochondrial dynamics.
Recent studies of HD pathogenesis [16–21] have focused on elucidating impaired mitochondrial dynamics, particularly excessive fragmentation of mitochondria and the subsequent mitochondrial dysfunction, and defective axonal trafficking and synaptic damage in HD-affected neurons. Several groups [17,18] have recently found mHtt interacting with the mitochondrial fission protein Drp1, elevated levels of GTPase Drp1, enzymatic activity, and increased fission and reduced fusion in HD-affected neurons. Furthermore, some progress has been made in identifying molecules that are capable of reducing excessive mitochondrial fission and consequently maintaining healthy mitochondria and neuronal function in HD neurons.
In this article, we highlight recent developments in HD research, with a particular focus on mitochondria and mHtt. We also discuss recent advances in developing therapeutic molecules that inhibit excessive mitochondrial fission.
Mitochondrial abnormalities
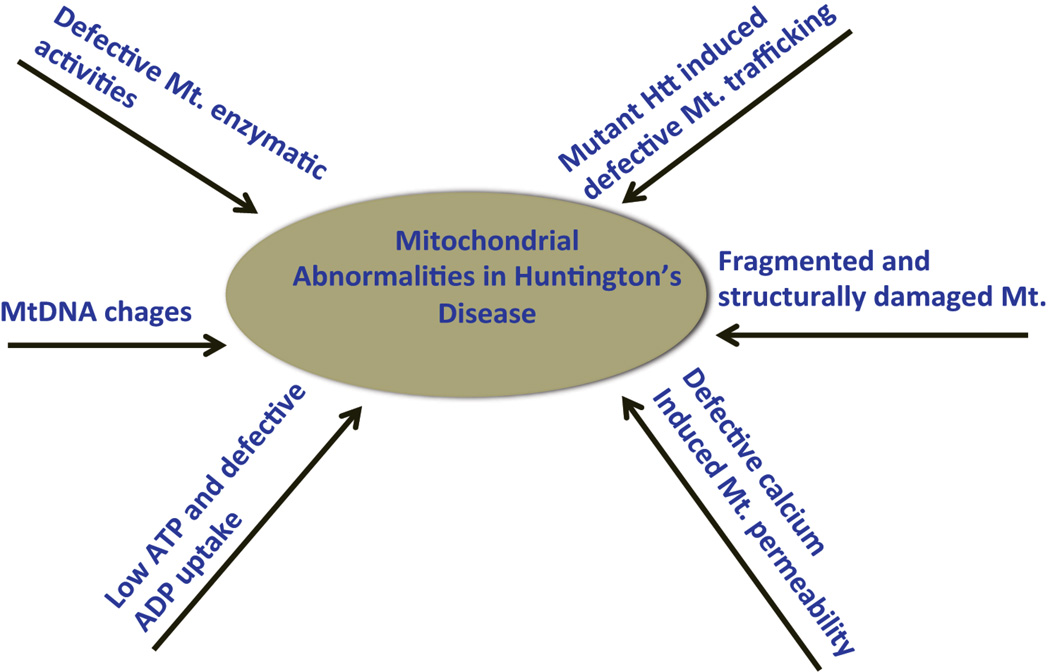
Recent research has revealed multiple alterations in mitochondria, in HD progression and pathogenesis, including: (i) reduced enzymatic activity in several components of oxidative phosphorylation, including complexes II, III and IV of the electron transport chain, in HD postmortem brains and HD mouse models [22–24], suggesting that mitochondria are involved in HD pathogenesis; (ii) low mitochondrial ATP and decreased mitochondrial ADP uptake in HD knock-in striatal cells and lymphoblasts from patients with HD, revealing expanded polyglutamine repeats [25]; (iii) defective calcium-induced mitochondrial permeability in HD cell lines and HD mice (reviewed in [26]); (iv) mHtt-induced defective mitochondrial trafficking in HD primary neurons [15,17,18,27]; (v) age-dependent mitochondrial (mt)DNA damage and mtDNA deletions in HD-affected neurons [28,29]; and (vi) biochemical, confocal and electron microscopy studies revealed structurally damaged mitochondria with broken cristae, and small and round mitochondria in HD-affected neurons [18–21] (Figure 1).
Figure 1.
[LM4]Illustration showing mitochondrial abnormalities involved in the disease process of Huntington’s disease. Abbreviations: Htt, Huntingtin; Mt., mitochondrial; mtDNA, mitochondrial DNA.
Abnormal mitochondrial dynamics
Mounting evidence suggests that structural and functional abnormalities in mitochondria are involved in HD pathogenesis [16,18–21]. In neurons that express mHtt, an imbalance between fission and fusion was found to lead to abnormalities in mitochondrial structure and function, and to damaged neurons. Several studies have reported such abnormal mitochondrial dynamics in patients with HD [16–20], HD mouse models [17,18], damaged HD lymphoblasts, HD cell lines and primary neurons that express mHtt [17–19,21].
Recently, we studied abnormal mitochondrial dynamics in tissues from postmortem brains of patients with HD3 (symptomatic with 80% neuronal loss [1]) or HD4 (advanced stage with over 90% neuronal loss [1]) [16] and primary neurons from BACHD transgenic mice [18]. In the 2011 study of postmortem HD brains, we found increased levels of Drp1 and Mitochondrial fission 1 (Fis1) in HD4 rather than in HD3, in HD-affected brain regions but not in HD-unaffected brain regions [16]. We also found reduced expressions of Mitofusin-1 and 2 (Mfn1 and Mfn2) and optic atrophy 4 (Opa1) in HD4 rather than in HD3, in HD-affected brain regions but not in HD-unaffected brain regions. Taken together, these findings suggest that abnormal mitochondrial dynamics are related to HD [16].
Using BACHD mice that express the full-length human Htt gene with 97 CAA and CAG mixed repeats, we studied mHtt, and mitochondrial and synaptic genes [17]. We found significantly increased mRNA levels of Drp1, Fis1 and Cyclophilin D (CypD), and decreased levels of Mfn1 and Mfn2 in 2-month-old BACHD mice relative to age-matched wild type mice, suggesting that abnormal mitochondrial dynamics is an early event in HD progression [17].
Furthermore, to determine whether Drp1 interacts with mutant Htt, we performed co-immunoprecipitation (IP) of Drp1 and of mHtt from HD postmortem brains [17]. We found an 82-kDa and a 40-kDa mHtt protein in IP elutes from patients with HD3 or HD4. We also found Drp1 interacting with wild type Htt in brain specimens from the patients with HD3 or HD4 patients, but to lesser extent than in the control subjects, suggesting that mHtt, interacting with Drp1, is specific and is related to HD progression. Using cortical protein lysates from BACHD mice and wild type mice, we also conducted co-IP analysis. Our results were similar to results from our IP analysis of Drp1 from HD brains. We found Drp1 interacting with mHtt in HD neurons, suggesting that Drp1 participates in mitochondrial fission and impaired mitochondrial biogenesis. We also measured GTPase Drp1 enzymatic activity in brain specimens from patients with HD and from BACHD mice to determine whether increased Drp1, which interacts with mHtt, enhances GTPase activity and results in excessive mitochondrial fission. We found increased levels of Dp1 enzymatic activity in the cortex but not in the cerebellum of patients with HD3 or HD4 relative to Drp1 enzymatic activity in control subjects. We also found elevated levels of Drp1 enzymatic activity in the cerebral cortex and striatum from the BACHD mice relative to the levels of Drp1 enzymatic activity in the cerebral cortex and striatum specimens from wild type mice. Furthermore, using primary neurons from BACHD mice and wild type mice, live-cell imaging techniques and DsRed-mito transfections, we studied mitochondrial transport along the axonal projections of primary neurons from BACHD and wild type mice. We found significantly decreased anterograde movement of the mitochondria in the primary neurons from the BACHD mice relative to the wild type neurons [17].
Song and colleagues [18] studied the effects of mHtt on the fission–fusion balance in mitochondria in HD pathogenesis. They found that mHtt triggers mitochondrial fission in vitro, in rat neurons and in fibroblasts from patients with HD; and in vivo, in a mouse model of HD. These events occurred before the development of neurological deficits and mHtt aggregates. mHtt interacted abnormally with Drp1 in mice and in humans with HD, and this interaction, in turn, stimulated the enzymatic activity of GTPase Drp1. Furthermore, a reduction of Drp1 GTPase activity, in association with the dominant-negative Drp1 K38A mutant, appeared to rescue mHtt-mediated mitochondrial fission, defects in anterograde and retrograde mitochondrial transport, and neuronal cell death [18].
Lipton’s group [30,31] reported that S-nitrosylation of Drp1 caused mitochondrial fission in brain neurons from patients with AD and HD. Cho et al. studied S-nitrosylation of Drp1 in AD neurons. They found increased S-nitrosylation of Drp1 and GTPase Drp1 activity in AD neurons. They also prevented the nitrosylation of Drp1 by introducing a cysteine mutation in the cDNA of Drp1, which reduced S-nitrosylation of Drp1 and abrogated neurotoxic events in the AD neurons [30].
Huan et al. [31] sought to determine whether S-nitrosylation of Drp1 contributes to HD pathogenesis. They found that, in primary neurons, the mHtt protein triggered a significant increase in nitric oxide (NO). Consistent with this result, increased levels of S-nitrosylation in Drp1 were found in the striatum of an HD transgenic mouse as well as in the neurons from patients with HD. Using specific fluorescence markers, they demonstrated that S-nitrosylation of Drp1 induced excessive mitochondrial fission, which was followed by a loss of dendritic spines, indicating synaptic damage. They then either transfected cDNA with nonnitrosylatable mutant Drp1 (C644A) or blocked the production of NO using a NO synthase inhibitor. They found that both of these therapies significantly reduced neurotoxic events. These findings suggest that S-nitrosylation of Drp1 is a key mediator of mHtt toxicity. Based on findings from their HD studies, Haun et al. [31] proposed that NO, produced in response to an increase in mHtt, might be a key mediator of disease progression, mitochondrial fission, synaptic loss and neuronal damage, in part via the S-nitrosylation of Drp1.
Findings from these studies indicate that mHtt selectively interacts with Drp1, fragmented mitochondria, defective axonal transport and mitochondrial dysfunction in HD-affected neurons. These studies suggest that inhibitors of mitochondrial fission are targets for reducing Drp1 and GTPase activity in patients with HD.
Mechanisms underlying mitochondrial fragmentation
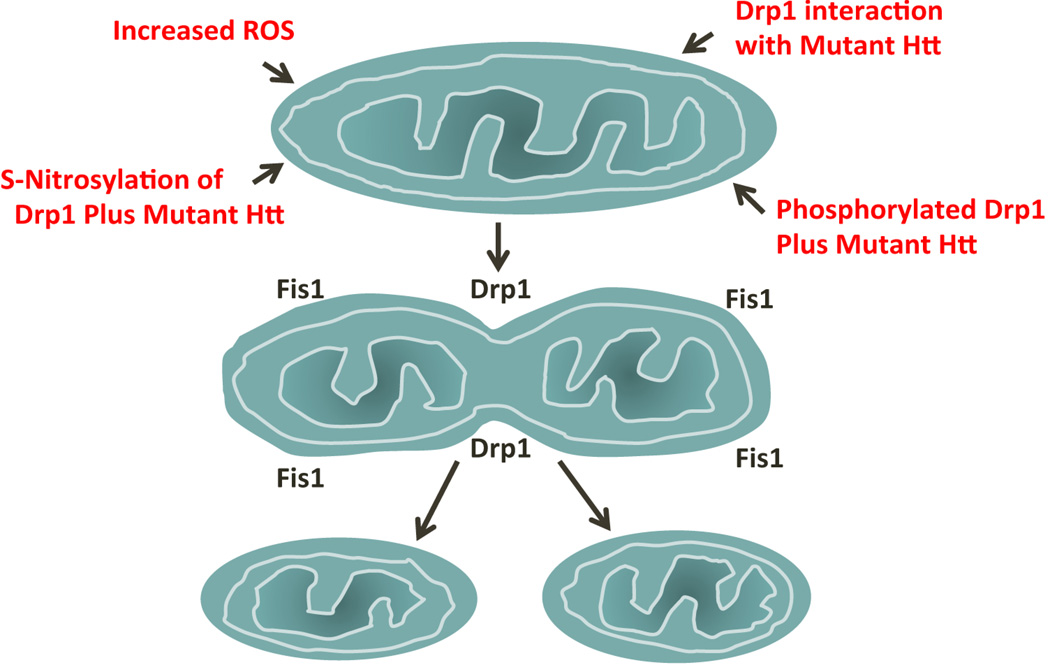
Recent evidence is revealing that several mechanisms are likely to be involved in excessive mitochondrial fission in HD-affected neurons, including the evidence that: (i) excessive production of ROS activates fission proteins and increases GTPase Drp1 enzymatic activity; (ii) mHtt interacts with Drp1 and increases GTPase Drp1 enzymatic activity, which in turn increases mitochondrial fission and creates an imbalance in mitochondrial dynamics [17,18]; (iii) S-nitrosylation of Drp1 in relation to mHtt enhances GTPase Drp1 activity, causing excessive mitochondrial fission [31]; and (iv) phosphorylated Drp1 at Ser 616, Ser 585 and Ser 637 sites elevates GTPase activity, causing an increase in mitochondrial fission [32–36] (Figure 2).
Figure 2.
Illustration showing the mechanisms of excessive mitochondrial fragmentation in Huntington’s disease. Abbreviations: Drp1, Dynamin-related protein 1; Fis1, Mitochondrial fission 1; Htt, Huntingtin; ROS, reactive oxygen species.
As discussed above, Drp1 has been found to interact with mHtt in cortical tissues from HD brains and brain tissues from BACHD mice [17], and with increased levels of GTPase Drp1 enzymatic activity. Increased Drp1 and Fis1, and reduced Mf1, Mf2 [LM2]and Opa1 in HD brains and BACHD mice. Furthermore, we also found significantly reduced anterograde mitochondrial movements in BACHD primary neurons. These findings suggest that Drp1 interacts with mHtt, elevates GTPase Drp1 activity and causes excessive mitochondrial fragmentation; and that these abnormal interactions disrupt axonal transport of mitochondria in HD neurons. Our findings [17] are supported by Song et al. [18] who also found Drp1 interacting with mHtt in HD mice and in patients with HD, which in turn induced the elevated enzymatic activity of GTPase Drp1, leading to excessive mitochondrial fragmentation in HD-affected neurons.
Recently, Huan et al. [31] reported the involvement of Drp1 nitrosylation in mutant Htt and in increased NO-triggered GTPase Drp1 activity, leading to increased mitochondrial fission in HD brains. It is possible that both events (the interaction of Drp1 with mutant Htt and with nitrosylation) are involved in fragmenting mitochondria in HD neurons.
Inhibitors of excessive mitochondrial fission as therapeutic targets
Progress is being made in developing therapeutic molecules that can reduce excessive mitochondrial fission and still maintain the fission–fusion balance in mitochondria, mitochondrial function and neuronal activity in diseases that are involved with oxidative stress and mitochondrial dysfunction. Several groups have independently screened chemical libraries and have found three different molecules: Mdivi [37], Dynasore [38] and P110 [39]. Among these, the Mdivi molecule has been extensively investigated with experimental rodent models for epilepsy and/or seizures [40,41], ischemia [42,43], oxygen glucose deprivation [44], rhabdomyolysis [45] and conditions such as aggregation of endosomes and vesicle fusion during exocytosis [46]. In all of these diseased states, Mdivi exhibited beneficial effects on affected tissues and cells by reducing excessive mitochondrial fission, and maintaining the fission–fusion balance and the normal functioning of cells.
The molecule Dynasore was identified in 2006 by Macia and colleagues [38] after screening 16 000 small molecules. Dynasore inhibits GTPase activities of Dynamin 1, Dynamin 2 and Drp1, and does not interfere with fusion activity. Gao et al. [47] studied the protective role of Dynasore in ischemia and/or reperfusion injury in mice and observed that mice pretreated with Dynasore exhibited fewer ischemia and/or reperfusion-induced impairments of the heart, suggesting that Dynasore protects heart cells from excessive mitochondrial fission and oxidative stress.
Qi et al. [39] designed a peptide-based Drp1 inhibitor called P110 and, using a cell model of PD[LM3], studied its effect on mitochondrial fission. P110 was found to reduce excessive mitochondrial fission and reactive oxygen species production, and to protect cells from oxidative insults.
Guo and colleagues [48] studied the protective effects of mitochondrial fission inhibitor, P110 using HD fibroblasts, induced pluripotent stem cell-derived HD neurons, HD mouse striatal cells (HdhQ110) and a truncated mouse model (R6/2) of HD. The authors found P110 reduced mHtt-induced excessive mitochondrial fragmentation and corrected mitochondrial function in all cell models and R6/2 mice [48].
Overall, the progress made so far on researching mitochondrial fission inhibitors is encouraging. The Mdivi, P110 and Dynasore molecules need to be tested as a treatment for HD models to protect HD-affected neurons.
Concluding remarks and future directions
Since the discovery of the HD gene in 1993, significant progress has been made in understanding the toxic effects of mHtt and cellular changes that are associated with it. Among multiple cellular mechanisms that might be involved in HD pathogenesis, abnormal mitochondrial dynamics and excessive mitochondrial fission might be crucial conditions and events associated with mitochondrial dysfunction and neuronal damage in HD-affected neurons. Considerable progress has also been made in identifying mechanisms underlying excessive mitochondrial fission in HD, and in identifying molecules, such as Mdivi, Dynasore and P110, that might reduce excessive mitochondrial fission while maintaining normal mitochondrial activity and neuronal function.
Research Highlights.
Mitochondrial dysfunction plays a large role in Huntington’s disease.
Mutant Htt interacts with Drp1 and causes mitochondrial fragmentation.
Drp1-mutant Htt complexes disrupt axonal transport of mitochondria.
Fission inhibitor(s) enhances synaptic activity and neuronal function.
Mdivi, Dynasore and P110 molecules are promising HD therapeutic molecules.
Acknowledgments
This research was supported by National Institutes of Health grants AG028072, AG042178 and RR000163, and a grant from the Medical Research Foundation of Oregon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vonsattel JP, et al. Vulnerability of the basal ganglia. Ann. Neurol. 1985;41:646–653. [Google Scholar]
- 2.Folstein SE. Huntington’s Disease. Johns Hopkins University Press; 1990. [Google Scholar]
- 3.Bates GP. History of genetic disease: the molecular genetics of Huntington disease: a history. Nat. Rev. Genet. 2005;6:766–773. doi: 10.1038/nrg1686. [DOI] [PubMed] [Google Scholar]
- 4.Montoya A, et al. Brain imaging and cognitive dysfunctions in Huntington’s disease. J. Psychiatry Neurosci. 2006;31:21–29. [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy PH, et al. Recent advances in understanding the pathogenesis of Huntington’s disease. Trends Neurosci. 1999;22:248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 6.Aziz NA, et al. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 7.Bäckman L, et al. Cognitive deficits in Huntington’s disease are predicted by dopaminergic PET markers and brain volumes. Brain. 1997;120:2207–2217. doi: 10.1093/brain/120.12.2207. [DOI] [PubMed] [Google Scholar]
- 8.Dierks T, et al. Multimodal imaging of residual function and compensatory resource allocation in cortical atrophy: a case study of parietal lobe function in a patient with Huntington’s disease. Psychiatry Res. 1999;90:67–75. [PubMed] [Google Scholar]
- 9.Berent S, et al. Positron emission tomographic scan investigations of Huntington’s disease: cerebral metabolic correlates of cognitive function. Ann. Neurol. 1998;23:541–546. doi: 10.1002/ana.410230603. [DOI] [PubMed] [Google Scholar]
- 10.Powers WJ, et al. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 12.Choo Y, et al. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 13.Strehlow AN, et al. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum. Mol. Genet. 2007;16:391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- 14.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 15.Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirendeb UP, et al. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum. Mol. Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirendeb UP, et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, et al. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum. Mol. Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa V, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol. Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne SE, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 23.Tabrizin SJ, et al. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann. Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Pandey M, et al. Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3-nitropropionic acid-induced rat model of Huntington’s disease. J. Neurochem. 2008;104:420–434. doi: 10.1111/j.1471-4159.2007.04996.x. [DOI] [PubMed] [Google Scholar]
- 25.Seong IS, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 26.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro . Mol. Cell. Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy PH, et al. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res. Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acevedo-Torres K, et al. Mitochondrial DNA damage is a hallmark of chemically induced and the R6/2 transgenic model of Huntington’s disease. DNA Repair. 2009;8:126–136. doi: 10.1016/j.dnarep.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banoei MM, et al. Huntington’s disease and mitochondrial DNA deletions: event or regular mechanism for mutant huntingtin protein and CAG repeats expansion?! Cell. Mol. Neurobiol. 2007;27:867–875. doi: 10.1007/s10571-007-9206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haun F, et al. S-nitrosylation of Dynamin-Related Protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington’s disease. Antioxid. Redox Signal. 2013;19:1173–1184. doi: 10.1089/ars.2012.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taguchi N, et al. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 33.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 35.Merrill RA, et al. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011;9:e1000612. doi: 10.1371/journal.pbio.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J. Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cassidy-Stone A, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Qi X, et al. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci. 2013;126:789–802. doi: 10.1242/jcs.114439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie N, et al. A selective inhibitor of Drp1, mdivi-1, protects against cell death of hippocampal neurons in pilocarpine-induced seizures in rats. Neurosci. Lett. 2013;545:64–68. doi: 10.1016/j.neulet.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 41.Qiu X, et al. Role of mitochondrial fission in neuronal injury in pilocarpine-induced epileptic rats. Neuroscience. 2013;245:157–165. doi: 10.1016/j.neuroscience.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Zhang N, et al. A selective inhibitor of Drp1, mdivi-1, acts against cerebral ischemia/reperfusion injury via an anti-apoptotic pathway in rats. Neurosci. Lett. 2013;535:104–109. doi: 10.1016/j.neulet.2012.12.049. [DOI] [PubMed] [Google Scholar]
- 43.Park SW, et al. A selective inhibitor of drp1, mdivi-1, increases retinal ganglion cell survival in acute ischemic mouse retina. Invest. Ophthalmol. Vis. Sci. 2011;52:2837–2843. doi: 10.1167/iovs.09-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wappler EA, et al. Mitochondrial dynamics associated with oxygen-glucose deprivation in rat primary neuronal cultures. PLoS ONE. 2013;8:e63206. doi: 10.1371/journal.pone.0063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang WX, et al. Amelioration of rhabdomyolysis-induced renal mitochondrial injury and apoptosis through suppression of Drp-1 translocation. J. Nephrol. 2013 doi: 10.5301/jn.5000268. http://dx.doi.org/10.5301/jn.5000268. [DOI] [PubMed] [Google Scholar]
- 46.Chlystun M, et al. Regulation of mitochondrial morphogenesis by annexin A6. PLoS ONE. 2013;8:e53774. doi: 10.1371/journal.pone.0053774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao D, et al. Dynasore protects mitochondria and improves cardiac lusitropy in Langendorff perfused mouse heart. PLoS ONE. 2013;8:e60967. doi: 10.1371/journal.pone.0060967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X, et al. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease associated neurodegeneration. J. Clin. Invest. 2013 doi: 10.1172/JCI70911. http://dx.doi.org/10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]