Significance
Kinesin-1 is a motor protein central to intracellular transport. Prevailing models of the kinesin mechanochemical cycle—which invoke docking of the neck linker domain upon ATP binding—fail to explain the remarkable processivity of kinesin, which represents a competition between dissociation from the microtubule and continuation of the stepping cycle. We show that kinesin dissociation, which characterizes the end of a processive run, is gated by phosphate release following ATP hydrolysis. The structural change driving kinesin motility, likely neck linker docking, is therefore completed only upon hydrolysis. Our results offer insights into gating mechanisms and necessitate revisions to existing models of the kinesin cycle.
Keywords: single molecule, mechanochemistry, optical tweezers, molecular motors
Abstract
Kinesin-1 is a dimeric motor protein, central to intracellular transport, that steps hand-over-hand toward the microtubule (MT) plus-end, hydrolyzing one ATP molecule per step. Its remarkable processivity is critical for ferrying cargo within the cell: over 100 successive steps are taken, on average, before dissociation from the MT. Despite considerable work, it is not understood which features coordinate, or “gate,” the mechanochemical cycles of the two motor heads. Here, we show that kinesin dissociation occurs subsequent to, or concomitant with, phosphate (Pi) release following ATP hydrolysis. In optical trapping experiments, we found that increasing the steady-state population of the posthydrolysis ADP·Pi state (by adding free Pi) nearly doubled the kinesin run length, whereas reducing either the ATP binding rate or hydrolysis rate had no effect. The data suggest that, during processive movement, tethered-head binding occurs subsequent to hydrolysis, rather than immediately after ATP binding, as commonly suggested. The structural change driving motility, thought to be neck linker docking, is therefore completed only upon hydrolysis, and not ATP binding. Our results offer additional insights into gating mechanisms and suggest revisions to prevailing models of the kinesin reaction cycle.
Since its discovery nearly 30 years ago (1), kinesin-1—the founding member of the kinesin protein superfamily—has emerged as an important model system for studying biological motors (2, 3). During “hand-over-hand” stepping, kinesin dimers alternate between a two–heads-bound (2-HB) state, with both heads attached to the microtubule (MT), and a one–head-bound (1-HB) state, where a single head, termed the tethered head, remains free of the MT (4, 5). The catalytic cycles of the two heads are maintained out of phase by a series of gating mechanisms, thereby enabling the dimer to complete, on average, over 100 steps before dissociating from the MT (6–8). A key structural element for this coordination is the neck linker (NL), a ∼14-aa segment that connects each catalytic head to a common stalk (9). In the 1-HB state, nucleotide binding is thought to induce a structural reconfiguration of the NL, immobilizing it against the MT-bound catalytic domain (2, 3, 10–17). This transition, called “NL docking,” is believed to promote unidirectional motility by biasing the position of the tethered head toward the next MT binding site (2, 3, 10–17). The completion of an 8.2-nm step (18) entails the binding of this tethered head to the MT, ATP hydrolysis, and detachment of the trailing head, thereby returning the motor to the ATP-waiting state (2, 3, 10–17). Prevailing models of the kinesin mechanochemical cycle (2, 3, 10, 14, 15, 17), which invoke NL docking upon ATP binding, explain the highly directional nature of kinesin motility and offer a compelling outline of the sequence of events following ATP binding. Nevertheless, these abstractions do not speak directly to the branching transitions that determine whether kinesin dissociates from the MT (off-pathway) or continues its processive reaction cycle (on-pathway). The distance moved by an individual motor before dissociating—the run length—is limited by unbinding from the MT. The propensity for a dimer to unbind involves a competition among multiple, force-dependent transitions in the two heads, which are not readily characterized by traditional structural or bulk biochemical approaches. Here, we implemented high-resolution single-molecule optical trapping techniques to determine transitions in the kinesin cycle that govern processivity.
Results
Run Lengths Are Asymmetric with Respect to the Direction of External Load.
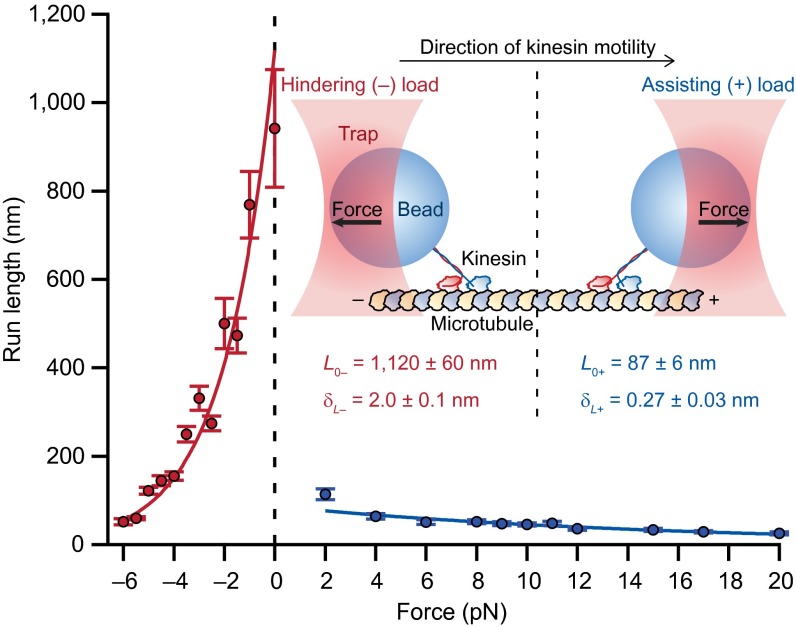
Hindering loads, that is, forces applied against the direction of motion (Fig. 1, Inset, Left), modulate the rates of structural transitions taking place in the MT-bound head (19) and potentially also the binding rates of the tethered (partner) head. Because these separate effects may be confounded during kinesin dissociation from the MT, we instead studied the run length under assisting loads, applied in the direction of kinesin motion using an optical force clamp (Fig. 1, Inset, Right). In this experimental geometry, the docking rate of the NL is not significantly reduced by mechanical force (20). Moreover, the applied load is borne almost exclusively by the bound head via its NL, leaving the tethered head free to undergo diffusive motion in proximity to its next MT binding site. Although kinesin run lengths have been studied extensively under hindering loads, they have not been well characterized in the assisting-load regime. Under assisting loads, single-molecule stepping traces were found to exhibit regular, 8-nm steps (Fig. S1). Measurements of the run length from such stepping traces reveal a dramatic asymmetry with respect to the direction of the applied force (Fig. 1). Under moderate assisting loads, run lengths were an order of magnitude shorter than for corresponding hindering loads, based on the unloaded run length, L0, obtained from exponential fits to the data. Furthermore, the sensitivity of run length to force, as characterized by the distance parameter, δL, was dramatically lower in the assisting-load regime, where run lengths decreased only gradually, out to +20 pN.
Fig. 1.
Kinesin run lengths exhibit an asymmetric dependence on the direction of applied load. Single-molecule kinesin run length measurements (mean ± SE; n = 75‒818) collected under saturating (2 mM) ATP conditions exhibit a clear asymmetry with respect to the direction of the force applied by the optical trap. Characteristic distance (δL) and unloaded run length (L0) parameters (Inset; mean ± SE) were obtained from exponential fits to the hindering load (–, red) and assisting load (+, blue) data. The data point at F = 0 (unloaded run length, obtained from tracking data without the optical force clamp) was included in the fit to hindering loads (0 to −6 pN), but not assisting loads (+2 to +20 pN). (Inset) Illustration of the experimental geometry for the assay under hindering and assisting loads (not to scale).
Run Lengths Are Independent of the Rates of ATP Binding and Hydrolysis.
To address the biochemical states contributing to processivity, we performed experiments under moderate assisting loads (+4 pN). This load regime minimizes the effects of forces on known mechanical transitions between nucleotide binding and hydrolysis (19), thereby providing ideal conditions for elucidating force-insensitive steps in the mechanochemical cycle. We observed no difference in the run length scored under low-ATP conditions relative to that in the presence of saturating ATP levels (Fig. 2). This result confirms that the nucleotide-free head remains strongly bound in the ATP-waiting state, and that the run length is not set by transitions in this point of the cycle. To a first-order approximation, the run length is governed by a competition between the rate of tethered-head binding to the next site, which results in a step, and the rate of premature release of the bound head, which results in dissociation. An increase in run length could therefore arise by increasing the rate of tethered-head binding, decreasing the rate of bound-head release, or both. Most conventional models of kinesin mechanochemistry propose that ATP binding induces NL docking (2, 3, 10–17), which repositions the tethered head in the forward direction and enables binding to the MT at the next site. This framework has been taken to imply that the tethered head is generally able to bind to the MT immediately following ATP binding by its partner. One prediction from this model is that reducing the rate of ATP hydrolysis should increase the run length, because it would give the tethered head additional time to bind before the bound head completes its hydrolysis and detaches. To test this hypothesis, we examined the kinesin run length under assisting forces in the presence of saturating levels of ATPγS, a slowly hydrolyzable ATP analog. Contrary to expectation, the run length turned out to be nearly identical to that measured under saturating ATP conditions (Fig. 2), implying that the branch point for MT release occurs subsequent to hydrolysis. A corollary of this finding is that the tethered head remains largely unbound, and that the transition that allows for productive head binding (stepping) is only completed upon hydrolysis.
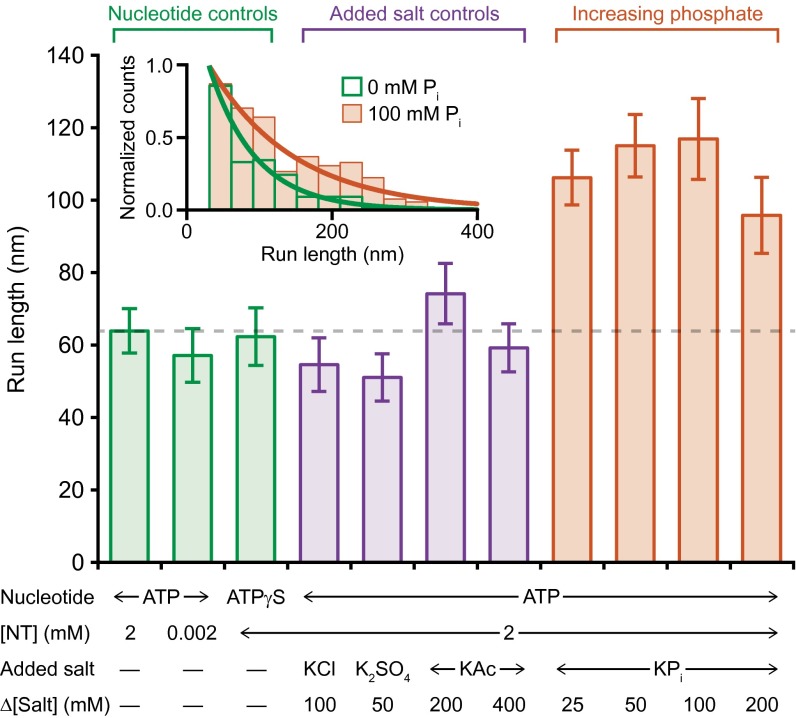
Fig. 2.
Kinesin processivity is enhanced by added phosphate. Run lengths (mean ± SE; n = 93‒345) measured under a +4-pN assisting load in the presence of nucleotide (NT) analogs (green), added salt (purple), or potassium phosphate (orange), at the concentrations indicated. Run lengths scored under various nucleotide conditions (green) or in the presence of nonphosphate salts (purple) showed no statistically significant differences. However, the addition of phosphate (orange) increased the run length by as much as a factor of 2. The increase in mean run length with added phosphate (in 2 mM ATP) was significant (P ≤ 0.01; t test) compared with the baseline run length (dashed line). (Inset) Histograms of normalized run lengths without added salt (green; n = 170) and with 100 mM added phosphate (orange; n = 187), together with exponential fits.
Addition of Phosphate Enhances Kinesin Processivity.
The discovery that decreasing the rate of ATP hydrolysis fails to enhance processivity suggests that tethered-head binding may instead compete with transitions in the mechanochemical cycle that take place subsequent to hydrolysis. One such candidate transition is phosphate (Pi) release. To test the hypothesis that Pi release is critical to the head unbinding transition, we measured run lengths at increasing concentrations of potassium phosphate. We found that run lengths increased by nearly a factor of 2 in the presence of added Pi (Fig. 2). These results were further validated by a series of salt controls (Fig. 2), which confirmed that the observed increase was specific to Pi and not simply due to the increase in ionic strength. Pi release therefore constitutes a key gating mechanism for processivity.
Accompanying velocity measurements (Fig. S2) show only a moderate reduction in velocity with salt, with a magnitude dependent on both the identity of the salt and its concentration. The decrease in velocity for increasing Pi concentrations, separate from its effect on processivity, is consistent with a previous report (21).
Discussion
Assisting Loads Facilitate Studies of Kinesin Processivity.
Our results suggest that the presence of additional Pi in solution stabilizes the bound head by increasing the lifetime of the posthydrolysis ADP·Pi state, thereby allowing the tethered head additional time to bind in the forward position before the bound head releases Pi and dissociates from the MT. The fact that the run length remained unchanged under high salt concentrations (Fig. 2) indicates that, under these assisting-load conditions, the rate of tethered-head binding remains largely independent of the ionic strength. Consequently, any changes in the run length arising from additional Pi are attributable to the modulation of biochemical events occurring in the bound head and are not confounded by salt effects. By contrast, run lengths acquired under hindering-load conditions were highly sensitive to ionic strength (Fig. S3). Because assisting loads poise the tethered head in a forward-biased position, this force regime provides a unique window for probing the effect(s) of Pi on processivity while at the same time minimizing any confounding electrostatic effects of added salt on tethered-head binding.
The dramatic asymmetry of run length with respect to direction of load (Fig. 1) has previously been largely unappreciated, but has direct consequences for stochastic models of multimotor transport where individual motors experience either assisting or hindering loads from their interaction with a common intracellular cargo (22). Our data suggest that the unbinding dynamics may vary by an order of magnitude depending on the direction of the effective load on the motor. Such variation in unbinding dynamics would be beneficial for a team of identical motors, where motors lagging behind unbind more readily, and may also influence dynamics in tug-of-war situations involving teams of different motors (23).
ATP Hydrolysis Completes Docking of the Neck Linker.
The finding that processivity is modulated by Pi release indicates that the conformational change responsible for moving the tethered head to a position where it can attach to the next MT binding site must occur subsequent to, or concomitant with, ATP hydrolysis. ATP binding is therefore insufficient to complete the sequence of mechanochemical events necessary for productive tethered-head binding. Although the structural rearrangements associated with ATP binding and hydrolysis may not be restricted to NL docking, extensive previous work (2, 3, 10–17) supports the notion that the NL is central to the conformational changes involved in processive stepping. Because tethered-head binding is gated by ATP hydrolysis, we propose that ATP binding may only incompletely dock the NL. Any such “partial docking” could entail either a shift in a rapid equilibrium between the undocked and fully docked states of the NL, or alternatively, the docking of only a portion of the NL, leaving the remainder unbound. Given the absence of structural evidence for the docking of segments of the NL, we tend to favor the former interpretation. Indirect evidence supporting the notion of partial NL docking comes from previous fluorescence (11, 12) and cryoelectron microscope (16) experiments, which suggest that the NL may be more stably docked in the presence of ADP·AlF4, an analog thought to mimic the posthydrolysis ADP·Pi state (11, 24, 25), than with the nonhydrolyzable analog AMP-PNP, which serves as a proxy for the ATP-bound state (11, 24, 25). Our findings are also consistent with previous work reporting the formation of a cover-neck bundle following NL docking (15). We note that partial NL docking raises the possibility of substeps taking place in the mechanochemical cycle, because separate tethered-head motions are predicted for both ATP-binding and hydrolysis events (with the latter being modulated by [Pi], but not the former). Although there have been previous reports claiming to detect kinesin substeps at different temporal and spatial scales, no such events have been conclusively detected to date, and their existence remains controversial (2). It is possible that any substeps associated with partial NL docking occur on a timescale faster than the experimental bandwidth achieved by most optical trapping work, which is dominated by the viscous damping of trapped beads (2). It is also possible that the displacements arising from kinesin structural rearrangements, or shifts in the equilibrium NL position associated with the substeps, are smaller than the current levels of experimental displacement noise.
A Revised Model of the Kinesin Mechanochemical Cycle.
Collectively, our findings support a revised model of the kinesin mechanochemical cycle (Fig. 3A). Starting from a 1-HB ATP-waiting state [1] (26–29), we propose that ATP binding by the bound head triggers partial NL docking, biasing the tethered (ADP-bound) partner head forward [2]. ATP hydrolysis then completes the docking of the NL against the bound head, enabling productive MT binding by the tethered head at the forward binding site [3]. This is the point in the cycle where processivity is gated. If Pi is released prematurely from the bound head before tethered-head binding [3‡], the motor generally dissociates from the MT and the run terminates [4off]. However, if productive MT binding and ADP release by the tethered head can be completed before Pi release by the bound head, the dimer remains attached and is able to proceed through the cycle. Kinesin, now in a strained, 2-HB state [4], releases Pi from its rear head, which unbinds from the MT (30). Once the rear head is released, the motor is once again ready to bind ATP and repeat another stepping cycle [1], having translocated forward by 8.2 nm. The revised model is shown alongside the prevailing model (2, 3, 10, 14, 15, 17) for comparison (Fig. 3 A and B): the key difference is the addition of an intermediate state associated with partial NL docking, before hydrolysis.
Fig. 3.
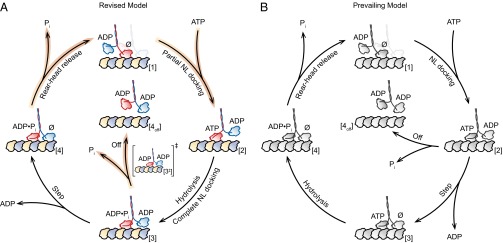
Comparison of the proposed model for the kinesin mechanochemical cycle and the prevailing model. (A) The revised cycle begins in the 1-HB ATP-waiting state [1] with ADP bound to the tethered head and a nucleotide-free, MT-bound head (∅). ATP binding by the bound head triggers partial NL docking [2], and subsequent hydrolysis induces complete NL docking, which induces productive MT binding by the tethered head [3]. At this stage, the kinesin molecule either completes the mechanical step [4], binding to the next MT site and releasing ADP, or prematurely releases Pi [3‡], and dissociates in an off-pathway event [4off]. In the event that a forward step is completed [4], Pi release followed by rear-head unbinding returns the dimer to the ATP-waiting state [1]. The motor is now ready to begin the cycle anew, having translocated one 8.2-nm step toward the MT plus-end. Highlighted arrows (orange) indicate rates decreased by Pi, notably the dimer dissociation rate (Off). (B) The prevailing model of the kinesin mechanochemical cycle (2, 3, 10, 14, 15, 17). The key difference between the revised (A) and prevailing (B) models is whether a step can be completed upon ATP binding.
The Revised Model Is Consistent with Bulk Biochemical Data.
The notion that ATP binding by the MT-bound head induces the attachment of its tethered partner to the MT (10, 31, 32), which is a key feature of the prevailing model (2, 3, 10, 14, 15, 17) (Fig. 3B), is consistent with evidence from biochemical measurements for the release of the fluorescent ADP analog, mantADP, from the tethered head upon nucleotide binding its MT-bound partner (31, 33–35). This release has often been interpreted as reflecting the attachment of the tethered head to the MT in the forward position. However, if mantADP release follows the attachment of the tethered head to the MT, then decreasing the hydrolysis rate—for example, by substituting ATPγS for ATP—would be expected to enhance motor processivity through prolongation of the MT-attached state. However, no such increase in processivity has been found (Fig. 2), suggesting instead that any tethered-head binding generally occurs subsequent to hydrolysis. An alternative interpretation, and one that is consistent with the mantADP release data, is that nucleotide binding to the MT-attached head facilitates ADP release from its tethered partner, but leaves the tethered head free, or weakly associated with the MT, until hydrolysis renders it able to bind the MT. This interpretation, where the tethered head can release ADP while remaining unbound from the MT, reconciles bulk biochemical results (31, 33–35) with single-molecule measurements and is consistent with the model of Fig. 3A.
Phosphate Release Gates Kinesin Processivity.
Because kinesin processivity is so tightly controlled by Pi release, we propose that kinesin passes through two states with distinct conformations: a 1-HB state before [2] and after [3] ATP hydrolysis. The conformational changes induced by hydrolysis control the point in the mechanochemical cycle where the tethered head is able to bind the MT efficiently. However, the duration of the posthydrolysis ADP·Pi state [3], which is dictated by the rate of Pi release, determines whether the tethered head binds [4] and completes the stepping cycle, or whether the bound head accesses the weakly bound ADP state [3‡] and dissociates [4off]. We note that this “dissociation branch point” is distinct from both ATP binding and the subsequent, load-dependent mechanical step, thereby ensuring that long runs can be completed even against significant opposing loads, which may be a desirable property for some kinesin motors. The revised kinetic cycle for kinesin-1 suggests new challenges for future work. For example, does the partial NL docking implied by phosphate sensitivity represent a distinct structural state—and therefore a mechanical substep—or a shift in some rapid equilibrium between fully docked and undocked conformations? Cryoelectron microscope structural studies of Eg5, a mitotic kinesin-5, have found hydrolysis-induced shifts in the NL conformation (36, 37), and similar shifts may take place in kinesin-1. Moreover, previous work characterizing the motility of Eg5 has shown that its processivity can be also be enhanced by the addition of Pi (38), suggesting that the Pi release gating mechanism proposed here may not be limited to kinesin-1 motors.
Materials and Methods
Protein Expression and Purification.
A recombinant Drosophila melanogaster kinesin-1 construct was truncated at position 559 and fused to a C-terminal eGFP and 6×His-tag (39). The protein was expressed in Escherichia coli and purified by nickel column chromatography (40), as previously described.
Single-Molecule Optical-Trapping Assay.
Flow cells and incubations with kinesin and 440-nm polystyrene beads (Spherotech) were prepared as previously described (17). Motility buffers consisted of PEM80 [80 mM Pipes (Sigma), 1 mM EGTA (Sigma), 4 mM MgCl2 (Fluka)] at pH 6.9, 2 mM DTT (Sigma), 10 μM Taxol (Sigma), and 2 mg⋅mL−1 BSA (Calbiochem). An oxygen-scavenging system with final concentrations of 50 μg⋅mL−1 glucose oxidase (Calbiochem), 12 μg⋅mL−1 catalase (Sigma), and 1 mg⋅mL−1 glucose (EM Science) was added to motility buffers before introduction into flow cells. Concentrations of ATP (Sigma), ATPγS (Calbiochem), and salt were adjusted for the desired conditions. Potassium phosphate (KPi) (50% monobasic, 50% dibasic; EMD), potassium sulfate (K2SO4) (Sigma-Aldrich), potassium acetate (KAc) (EMD), and potassium chloride (KCl) (EMD) were each dissolved in PEM80 buffer and adjusted to pH 6.9 before addition to the motility buffer.
Optical-trapping experiments were performed under force-clamped conditions, as previously described (17, 41). However, for data collected at +4 pN, the detection scheme (41) was modified to include a second position-sensitive detector for measuring the trapping beam, thereby improving triggering of the force clamp and facilitating increased data throughput. Run length measurements under no load (0 pN) were obtained by video tracking, as before (17).
Data Analysis.
Starting and ending points of kinesin runs were identified by eye in individual records. The average run length, L, for the unloaded (0 pN) data was determined by fitting an exponential to the histogram of run lengths, excluding bins at longer distances containing <6 counts, as well as the initial bin (17). For nonzero assisting loads (other than +4 pN), L was determined using maximum-likelihood estimation, to account for varying starting points for kinesin runs. For data acquired in the hindering-load regime (also for +4-pN assisting loads), L was calculated from the number of runs, N1,2, binned in two different intervals: one inside the detection region, and the other outside. The lower limit of the first bin interval, x1 < x < x2, was set to x1 = 30 nm, to avoid misclassifying extremely short runs. The upper limit of the first bin interval was set to x2 = 150 nm, chosen such that the second bin interval, x > x2, included all runs that reached the boundary of the detection region. This choice minimizes the run length error, σL, propagated from the errors associated with the Poisson-distributed counts, . The expressions used to determine the average run length, , and the estimated run length error, , were derived by assuming an exponential run length distribution. Velocities for individual kinesin runs were obtained from linear fits to the experimental traces; average velocities and the associated SEs were weighted by run length.
The mean run length, L, as a function of hindering (–) or assisting (+) load (Fig. 1) was fit to the exponential form, , where L0 is the unloaded run length, F is the external force applied by the optical trap, δL is a distance parameter that characterizes the load dependence, and kBT is Boltzmann’s constant times the absolute temperature.
Supplementary Material
Acknowledgments
We thank A. Chakraborty, C. García-García, D. Hogan, C. Perez, A. Savinov, and other members of the S.M.B. laboratory, as well as S. Rosenfeld (Cleveland Clinic) for helpful comments and discussions. B.M. acknowledges the support of a Stanford Graduate Fellowship. This work was supported by National Institute of General Medical Sciences, National Institutes of Health Grants 5R01GM051453 (to S.M.B.) and 5R01GM076476 (to W.O.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410943111/-/DCSupplemental.
References
- 1.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42(1):39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block SM. Kinesin motor mechanics: Binding, stepping, tracking, gating, and limping. Biophys J. 2007;92(9):2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21(1):59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302(5653):2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 6.Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348(6299):348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 7.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388(6640):386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 8.Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388(6640):390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 9.Kozielski F, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91(7):985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 10.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402(6763):778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld SS, Jefferson GM, King PH. ATP reorients the neck linker of kinesin in two sequential steps. J Biol Chem. 2001;276(43):40167–40174. doi: 10.1074/jbc.M103899200. [DOI] [PubMed] [Google Scholar]
- 12.Asenjo AB, Weinberg Y, Sosa H. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat Struct Mol Biol. 2006;13(7):648–654. doi: 10.1038/nsmb1109. [DOI] [PubMed] [Google Scholar]
- 13.Tomishige M, Stuurman N, Vale RD. Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat Struct Mol Biol. 2006;13(10):887–894. doi: 10.1038/nsmb1151. [DOI] [PubMed] [Google Scholar]
- 14.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134(6):1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil AS, et al. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci USA. 2008;105(49):19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci USA. 2010;107(9):4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18(9):1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365(6448):721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 19.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2(10):718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 20.Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100(5):2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schief WR, Clark RH, Crevenna AH, Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc Natl Acad Sci USA. 2004;101(5):1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klumpp S, Lipowsky R. Cooperative cargo transport by several molecular motors. Proc Natl Acad Sci USA. 2005;102(48):17284–17289. doi: 10.1073/pnas.0507363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller MJ, Klumpp S, Lipowsky R. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc Natl Acad Sci USA. 2008;105(12):4609–4614. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4- Biochemistry. 1995;34(28):8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 25.Phan BC, Cheung P, Stafford WF, Reisler E. Complexes of myosin subfragment-1 with adenosine diphosphate and phosphate analogs: Probes of active site and protein conformation. Biophys Chem. 1996;59(3):341–349. doi: 10.1016/0301-4622(95)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450(7170):750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 27.Asenjo AB, Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc Natl Acad Sci USA. 2009;106(14):5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461(7260):125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proc Natl Acad Sci USA. 2009;106(31):12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klumpp LM, Hoenger A, Gilbert SP. Kinesin’s second step. Proc Natl Acad Sci USA. 2004;101(10):3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37(3):792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- 32.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288(5463):88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 33.Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272(2):724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 34.Farrell CM, Mackey AT, Klumpp LM, Gilbert SP. The role of ATP hydrolysis for kinesin processivity. J Biol Chem. 2002;277(19):17079–17087. doi: 10.1074/jbc.M108793200. [DOI] [PubMed] [Google Scholar]
- 35.Klumpp LM, Mackey AT, Farrell CM, Rosenberg JM, Gilbert SP. A kinesin switch I arginine to lysine mutation rescues microtubule function. J Biol Chem. 2003;278(40):39059–39067. doi: 10.1074/jbc.M304250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goulet A, et al. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem. 2012;287(53):44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goulet A, et al. Comprehensive structural model of the mechanochemical cycle of a mitotic motor highlights molecular adaptations in the kinesin family. Proc Natl Acad Sci USA. 2014;111(5):1837–1842. doi: 10.1073/pnas.1319848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentine MT, Block SM. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys J. 2009;97(6):1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20(10):939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uppalapati M, Huang Y-M, Shastry S, Jackson TN, Hancock WO. Microtubule motors in microfluidics. In: Zahn JD, Lee LP, editors. Methods in Bioengineering: Microfabrication and Microfluidics. Boston: Artech House Publishers; 2009. pp. 311–336. [Google Scholar]
- 41.Valentine MT, et al. Precision steering of an optical trap by electro-optic deflection. Opt Lett. 2008;33(6):599–601. doi: 10.1364/ol.33.000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.