Significance
Small RNAs (sRNAs) are most important regulators in eukaryotes. Although different kinds of sRNAs have been extensively studied in higher eukaryotes, their role remains largely unknown in protozoa. We have systematically investigated in the full genome the sRNAs of Giardia lamblia, the most primitive eukaryote known. Surprisingly, we have found that two major types of sRNAs (i.e., endogenous siRNAs and tRNA-derived sRNAs) are largely encoded in the genome of G. lamblia, whereas canonical microRNAs could not be identified in this parasite. Additional studies showed that both sRNAs might be involved in the differentiation regulation of G. lamblia. This study indicates that these two kinds of eukaryotic sRNAs emerged in the early evolution of eukaryotes.
Keywords: ncRNA sRNA-generating region, development, RNA regulation, single-cell parasite
Abstract
Small RNAs (sRNAs), including microRNAs and endogenous siRNAs (endo-siRNAs), regulate most important biologic processes in eukaryotes, such as cell division and differentiation. Although sRNAs have been extensively studied in various eukaryotes, the role of sRNAs in the early emergence of eukaryotes is unclear. To address these questions, we deep sequenced the sRNA transcriptome of four different stages in the differentiation of Giardia lamblia, one of the most primitive eukaryotes. We identified a large number of endo-siRNAs in this fascinating parasitic protozoan and found that they were produced from live telomeric retrotransposons and three genomic regions (i.e., endo-siRNA generating regions [eSGRs]). eSGR-derived endo-siRNAs were proven to target mRNAs in trans. Gradual up-regulation of endo-siRNAs in the differentiation of Giardia suggested that they might be involved in the regulation of this process. This hypothesis was supported by the impairment of the differentiation ability of Giardia when GLDICER, essential for the biogenesis of endo-siRNAs, was knocked down. Endo-siRNAs are not the only sRNA regulators in Giardia differentiation, because a great number of tRNAs-derived sRNAs showed more dramatic expression changes than endo-siRNAs in this process. We totally identified five novel kinds of tRNAs-derived sRNAs and found that the biogenesis in four of them might be correlated with that of stress-induced tRNA-derived RNA (sitRNA), which was discovered in our previous studies. Our studies reveal an unexpected complex panorama of sRNA in G. lamblia and shed light on the origin and functional evolution of eukaryotic sRNAs.
Small RNAs (sRNAs) with size less than 40 nt are known to be important regulators in all eukaryotes known today (1). They regulate most of the important biologic processes, such as cell apoptosis, division, and differentiation (1, 2). The species, function, and biogenesis mechanism of the sRNAs are diverse. Although related in-depth and fast-paced studies have been carried out for many years, the complexity of sRNAs is largely unknown, and many important questions, such as the origin of various kinds of sRNAs in eukaryotes, are not yet understood.
Argonaut (AGO) family-binding sRNAs, including microRNAs (miRNAs) (3), endogenous siRNAs (endo-siRNAs) (2), and P-element induced wimpy testis (PIWI) protein-interacting RNAs (2, 4), are the most important and well-studied sRNAs in eukaryotes. They can be further divided into DICER-dependent (miRNAs and endo-siRNAs) and -independent, according to whether their biogenesis requires or does not require DICER protein. These AGO-binding sRNAs can regulate gene expression by guiding AGO protein to target mRNAs or DNA regions (2). To date, little is known about non–AGO-binding sRNAs, such as transcription initiation RNAs (5), splice site RNAs (6), and tRNA progenitor-like sRNAs (7), although a large number of them has been identified in various eukaryotes, including humans (1). Preliminary studies have shown that they might also play an important role in the regulation of the normal physiology of eukaryotes (1). It is interesting that many known noncoding RNAs [ncRNAs; for example, tRNAs (8, 9), small nucleolar RNAs (10), and rRNAs (11)] can be further processed into functional sRNAs, and some parts of them are bound to AGO (10). These phenomena add an additional level to the intricacy of sRNA-based gene regulation (12). Although many kinds of sRNAs have been identified, the eukaryotic sRNA repertoire is far from complete, especially in single-cell eukaryotes.
Giardia lamblia is a unicellular parasitic protozoan that causes giardiasis, one of the most common infectious human diseases worldwide (13). The infection causes diarrhea and malnutrition (13). Giardia has a simple two-stage lifecycle consisting of trophozoite and cyst (13), which can be completed in vitro. Giardia cells undergo dramatic biologic changes, including morphological change, DNA replication, and nucleus division, when trophozoites differentiate into cysts. Giardia has a small (∼11.7 M) and compact genome (14). This parasite has been considered the earliest diverging eukaryotic lineage known (14), and it is recognized as an important eukaryotic model, which provides opportunity to gain basic insight into the key pathways that characterize eukaryotic cells (14, 15). However, although several sRNAs have been identified in Giardia (16–18) and the antigenic variation in this parasite has been found to be controlled by RNAi pathway (19), little is known regarding the sRNA regulation system in Giardia. In fact, the sRNA repertoire of Giardia is, at present, very incomplete. A systematic identification of Giardia sRNA is, thus, required to facilitate the functional study of the Giardia sRNA regulation system, which may also provide insight into the origin and evolution of eukaryotic sRNAs.
In this study, to systematically identify the Giardia sRNA and investigate whether sRNAs are involved in the differentiation of Giardia, we deeply sequenced the sRNA transcriptomes of trophozoites (6- and 24-h encysting trophozoites) as well as cysts of G. lamblia. Through careful analysis of the sequencing data and experimental verification, we have found two kinds of previously unidentified endo-siRNAs, and have identified five novel kinds of tRNA derived sRNAs. All of the sRNAs identified in this work are up-regulated in the differentiation of the parasite, suggesting that they might play important roles in this process. This hypothesis is further supported by the observation that the differentiation ability of G. lamblia was impaired after the knockdown of Giardia DICER (GLDICER), which is essential for the biogenesis of endo-siRNAs.
Results
Deep Sequencing Reveals a Drastic Alteration of the sRNA Transcriptome in the Differentiation of Giardia.
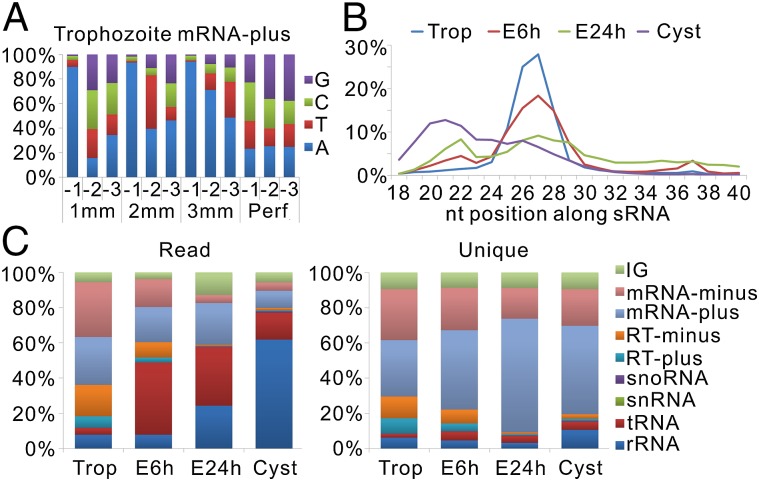
To determine whether the sRNAs are involved in the differentiation of Giardia, we deep sequenced the 18- to 40-nt sRNA libraries constructed from trophozoites and 6- and 24-h encysting trophozoites (encystation 6 and 24 h) as well as cysts, which yielded 13,883,157, 11,843,507, 11,577,787, and 6,350,555 reads, respectively. After mapping these reads to the Giardia genome, we found a strikingly high percentage of mismatches among the last 3 nt at their 3′ ends (Fig. S1) in all four libraries. The annotation results of end-mismatched sRNAs showed that the mismatch occurred at the 3′ ends of all kinds of sRNAs, except for tRNA-associated sRNA (tasRNA; specifically refers to tRNA-derived sRNA with length less than 40 nt) (Figs. S1 and S2 A and B). The 3′-end mismatched nucleotide composition of each kind of sRNA is similar in the four sRNA libraries. An sRNA can have 1, 2, or 3 3′-end mismatched nt, but no matter how many mismatched nucleotides that it has, the −1 mismatched nucleotide (the first mismatched nucleotide from the 3′ end) is preferred to be A, the −2 mismatched nucleotide, if it occurs, is preferred to be A or T, and the −3 mismatched nucleotide, if it exists, has no bias (Fig. 1A and Fig. S2C). These 3′-end mismatched nucleotides might be posttranscriptionally added, which also occurs in many kinds of sRNAs, including miRNAs, and have been found to link to increased or decreased turnover of sRNAs (20).
Fig. 1.
Characteristics of G. lamblia sRNAs. (A) The composition of the last three 3′-end nucleotides of sRNAs with different numbers of untemplated nucleotides; 1, 2, and 3 mm and Perf refer to sRNAs with 1, 2, 3, and 0 untemplated nt. (B) The length distribution of sRNAs. (C) Composition of the sRNA library of four G. lamblia differentiation stages. Trop refers to trophozoite; E6h and E24h refer to encystation 6 and 24 h, respectively.
Because the sRNAs with 3′-end mismatched nucleotides might be functionally relevant, we kept them in the subsequent analysis. All sRNAs, except tasRNAs, with any nucleotide mismatch in the internal sRNA sequence were discarded. We kept the tasRNAs with, at most, 2 mismatched nt, because their mismatch might be caused by the extensive nucleotide modification of mature tRNAs (21). After filtering, the usable reads that remained of four libraries were 9,766,799, 8,040,035, 9,493,416, and 5,290,587 (Dataset S1). Then, we calculated the read percentage (RP) of each sRNA in four libraries based on the total usable read number, which has been widely used to measure the expression level of each sRNA in previous studies (22). The length distributions of sRNAs from the four libraries are substantially different (Fig. 1B and Dataset S2). The 5′ ends of the sRNAs do not have any obvious nucleotide bias (Fig. S2D). The annotation results show that the compositions of the four libraries are remarkably distinct (Fig. 1C and Dataset S1), indicating that the sRNA transcriptome undergoes drastic alteration during Giardia differentiation.
Endo-siRNAs Are Involved in the Differentiation of Giardia.
mRNA-derived sRNAs and retrotransposon-derived sRNAs are the two most abundant sRNAs in trophozoites (Fig. 1C). It is interesting that they have similar characteristics. (i) Their length distributions are all unimodal (25–28 nt) in the trophozoite library and gradually change to bimodal (20–22 and 25–28 nt) when trophozoites differentiate into cysts (Fig. 2B and Fig. S2E). (ii) A large portion of these two kinds of sRNAs contains posttranscriptionally added 3′-end untemplated nucleotides (Fig. 2B, Fig. S3B, and Dataset S2), and they have length distributions concentrated around 25–28 nt in all of four libraries (Figs. S2E and S3F). Therefore, we first analyzed these two kinds of sRNAs.
Fig. 2.
Characteristics of SGR- and retrotransposon-derived sRNAs. The length and 3′-end nucleotide distribution of (A) SGR-derived sRNAs and (B) retrotransposon-derived sRNAs. (C) Northern blot analysis of SGR- and retrotransposon-derived sRNAs. (D) The composition of retrotransposon-derived sRNAs in four sRNA libraries. (E) The sRNA distribution pattern of GilT, GilM, and GilD. Trop refers to trophozoite. E3h, E6h, E12h, E24h, and E48h refer to encystation 3, 6, 12, 24, and 48 h, respectively. SGR-sRNAs and RT-sRNAs refer to SGR-derived sRNAs and retrotransposon-derived sRNAs, respectively. RT, retrotransposon.
Identification of Three sRNA-Generating Regions in Giardia Genome.
We aligned all mRNA-derived sRNAs to each mRNA and analyzed the characteristics of sRNAs derived from each mRNA (Fig. S3C). We found two types of mRNAx that produced sRNAs with totally different characteristics. One type of mRNAs (type I mRNAs) produced most of the mRNA-derived sRNAs (95.4%) in the trophozoite sRNA library, and both of their strands can produce sRNAs. These sRNAs have two characteristics listed above. In contrast, sRNAs derived from the other mRNA type (type II mRNA) do not have all of these features; they are mainly produced from the plus strand of mRNAs (Fig. S3D and Dataset S2) and have a broad length distribution in the four libraries (Fig. S2F). Also, they do not have posttranscriptionally added 3′ untemplated nucleotides (Fig. S3D and Dataset S2). All of these characteristics indicate that they are not the degradation products of mRNAs. Intriguingly, we found that all type I mRNAs were clustered in the genome and form three clusters (Fig. S3A). Most of them (34 of 39; 87.1%) were in one cluster (Dataset S2), which spans a broad region of 40.55 kb. Notably, the entire region, including the intergenic region of these three mRNA clusters, produced sRNAs with the same characteristics as type I mRNA-derived sRNAs. We named these three regions sRNA generating region I (SGRI), SGRII, and SGRIII. We got the SGR-derived sRNA according to the workflow shown in Fig. S3C. The overall characteristics of SGR-derived sRNA are the same as type I mRNA-derived sRNA (Fig. 2A and Fig. S3B). SGRI and SGRII produced high abundances of sRNAs (Fig. 2A and Fig. S3 C and E), whereas SGRIII only produced a few sRNA. It is interesting that almost no mRNA signatures [referred to as reads sequenced by high throughput mRNA sequencing technology (mRNA-Seq)] could be detected in SGRI and SGRII by a deep coverage (about 1,000×) sequencing of mRNA, which was performed by Franzén et al. (23), whereas high levels of mRNA signatures could be shown in SGRIII.
We tried to validate the expression of SGR-derived sRNAs using Northern blot. However, we failed to detect the SGR-derived sRNA with the highest RP (0.04%), indicating a relatively low abundance of single SGR-derived sRNA. Thus, we tried a mixed probe, which contained 15 probes designed to target the top 15 (Dataset S3) most abundant SGR-derived sRNAs, and succeeded in detecting the expression of SGR-derived sRNAs using Northern blot. We found that, although the RP of SGR-derived sRNAs was gradually dropped in the differentiation process of Giardia (Fig. 1C), their expression levels measured by Northern blot were gradually up-regulated in this process (Fig. 2C). The elevated expression level but dropped RP of SGR-derived sRNAs in encysting and cyst libraries might be caused by the substantial up-regulation of other sRNAs, such as tasRNAs (Figs. 1C and 4D and Fig. S4D), during the differentiation of Giardia.
Fig. 4.
Characteristics of tRNA-derived sRNAs. (A) tasRNA undergoes a larger expression change than endo-siRNAs in the Giardia differentiation process. (B) The length distribution of tasRNAs in four sRNA libraries. (C) The composition of tasRNAs in four sRNA libraries. (D) Northern blot analysis of nine tasRNAs derived from three tRNAs. (E, Upper) Northern blot analysis of 5EsRNA. E, Lower indicates the distribution of sRNA across pseudotRNA-Gln (TTG). (F) Two cleavages in mature tRNAs. (G) Five kinds of sRNAs derived from mature tRNAs. Trop refers to trophozoite. E3h, E6h, E12h, E24h, and E48h refer to encystation 3, 6, 12, 24, and 48, respectively. 3tasRNA, tRNA 3′ end-associated sRNA.
All Alive but Not Dead Telomeric Retrotransposons Produce Large Numbers of sRNAs.
There are three retrotransposons in the Giardia genome: two retrotransposons (GilT and GilM) are located in the telomeric region, whereas the third retrotransposon (GilD) is dead (24). Ullu et al. (16) claimed that GilT but not GilM could produce 26-nt sRNAs with unclear function. Through analysis of our sRNA libraries, we found that both GilT and GilM could generate a large number of sRNAs, which occupied 24.2% of reads in the trophozoite library. This kind of sRNA was the second highest abundance sRNA in that library (Fig. 1C and Fig. S3E). The abundance of GilT-derived sRNAs is much higher than that of GilM-derived sRNAs (Fig. 2D). The dead retrotransposon GilD does not generate any sRNA (Fig. 2E). The sRNAs were uniformly distributed throughout the whole regions of GilT and GilM, and their minus strands generated slightly more sRNAs than the plus strands (Fig. 2E). Because the RP of single retrotransposon-derived sRNA is similar to that of single SGR-derived sRNA, we also used a mixed probe (Dataset S3) to verify the expression of retrotransposon-derived sRNA using Northern blot. The Northern blot result showed that retrotransposon-derived sRNAs were also up-regulated in the differentiation of Giardia, which is similar to that of SGR-derived sRNAs (Fig. 2C).
SGR- and Retrotransposon-Derived sRNAs Are both Endo-siRNAs and Might Be Involved in the Differentiation Process of Giardia.
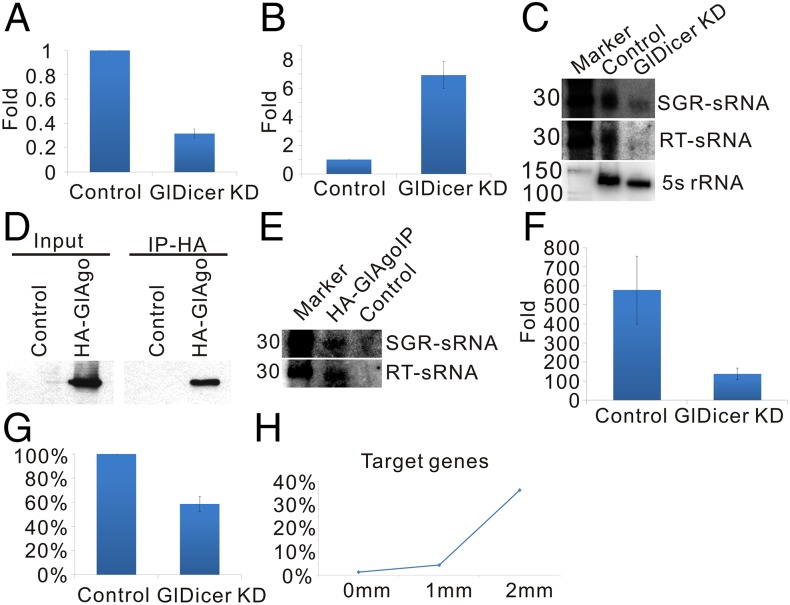
The length distribution of SGR- and retrotransposon-derived sRNAs is similar to that of the in vitro processing products of GLDICER (Fig. 2 A and B) (25), indicating that they might be endo-siRNAs. To verify this hypothesis, we knock downed the expression of GLDICER by constitutive expression of part of its antisense transcripts in trophozoites (19). The substantial down-regulation (68.4% decrease) of GLDICER (Fig. 3A) and up-regulation of variant-specific surface proteins (VSPs) (Fig. 3B), which were previously reported targets of the RNAi pathway, confirmed the successful construction of the GLDICER knockdown strain. The expression levels of both retrotransposon- and SGR-derived sRNAs were dramatically decreased when GLDICER was knocked down (Fig. 3C). We further constructed a 3xHA-GLAGO (Giardia AGO) overexpressing strain and investigated whether these sRNAs were bound to 3xHA-GLAGO using Northern blot. The Northern blot results showed that both SGR- and retrotransposon-derived sRNAs were bound to 3xHA-GLAGO (Fig. 3 D and E). The GLDICER-dependent biogenesis and GLAGO binding of SGR- and retrotransposon-derived sRNAs proved that both of them are endo-siRNAs. Therefore, SGR should be renamed endo-SGR (eSGR).
Fig. 3.
Endo-siRNAs in Giardia differentiation process. The expression of (A) GLDICER and (B) VSP evaluated by quantitative RT-PCR in control and GLDICER knockdown strain. Note that VSP quantitative RT-PCR primers can amplify multiple VSPs, because their sequences are highly homologous. (C) Northern blot analysis of SGR- and retrotransposon-derived sRNAs in control and GLDICER knockdown strain. (D) Western blot analysis of immunoprecipitation product of 3xHA-GLAGO. (E) Northern blot analysis of SGR- and retrotransposon-derived sRNAs in sRNAs binding to 3xHA-GLAGO. (F) The fold change of cyst wall protein 1 in control and the GLDICER knockdown strain when they were induced to differentiation for 24 h in vitro. Cyst wall protein 1 expression was evaluated by quantitative RT-PCR. (G) The percentage of the cyst number of the GLDICER knockdown strain relative to that of the control strain when they were induced in vitro for 24 h. (H) Potential trans target gene number of eSGR-derived endo-siRNAs. Error bars indicate SD (n = 3). SGR-sRNAs and RT-sRNAs refer to SGR-derived sRNAs and retrotransposon-derived sRNAs, respectively. IP, immunoprecipitation; KD, knockdown; RT, retrotransposon.
The gradual up-regulation of endo-siRNAs in the differentiation of Giardia suggested that they might have roles in regulation of this process. To verify this hypothesis, we induced the differentiation of trophozoites with GLDICER knocked down to cysts in vitro. We found that the transcription activity of cyst wall protein 1 gene, which is usually used as a marker of Giardia differentiation (13), is much lower in the GLDICER knockdown strain than the control one (Fig. 3F), and the cyst formation ability was obviously impaired in the GLDICER knockdown (Fig. 3G). These evidences supported our hypothesis that endo-siRNAs are involved in the differentiation of Giardia. To investigate how endo-siRNAs regulate Giardia differentiation, we tried to predict the potential targets of endo-siRNAs. The prediction mainly focused on the eSGR-derived endo-siRNAs, because the retrotransposon-derived endo-siRNAs in other eukaryotes always target the transcripts of retrotransposon or retrotransposon-encoding DNA regions. We found that the expression of mRNAs located in the eSGRIII region was not affected by the GLDICER knockdown (Fig. S4B), indicating that eSGR-derived endo-siRNAs might regulate gene expression predominantly in trans. We then predicted the targets of endo-siRNAs through directly aligning them to Giardia mRNAs. It is interesting that, when allowing two mismatches, eSGRs-derived endo-siRNAs could target ∼36.2% of the Giardia genes (Fig. 3H). Gene ontology analysis on potential target genes with single mismatch revealed that eSGR derived endo-siRNAs may regulate the expression of genes with various functions (Fig. S4C).
Six Kinds of tRNA-Derived sRNAs Are Specifically Induced in the Differentiation of Giardia.
The deep sequencing data showed that tasRNAs undergo a far more dramatic expression change than endo-siRNAs (Figs. 1C and 4 A and D and Fig. S4D) when Giardia differentiated from trophozoites to cysts, indicating that they might also play an important role in the differentiation of Giardia.
Identification of Four Kinds of Giardia tasRNA.
Interestingly, the length distributions of tasRNAs have four peaks (Fig. 4B) around 20–22, 24–26, 28–30, and 36–38 nt, indicating that there might be four distinct categories of tasRNAs. We mapped the tasRNA sequences to the mature tRNA sequences and found that 20–22-, 24–26-, and 28–30-nt peaks corresponded to tasRNAs derived from the middle region (most of the anticodon stem loop), 3′ end, and 5′ end of mature tRNAs, respectively (Fig. 4B), which were named tRNA anticodon stem loop-associated sRNAs (actasRNAs), tRNA 3′ end-associated sRNAs, and tRNA 5′ end-associated sRNAs (5tasRNAs), respectively. The 36–38-nt peak corresponded to one sRNA that covers the whole 5′-end exon of pseudotRNA-Gln (TTG) (Fig. 4E). This sRNA was named as 5′-end exon of pseudotRNA-Gln (TTG) derived sRNA (5EsRNA). Other than these four kinds of sRNAs, there are also other kinds of tRNA-derived sRNAs in all of four libraries, which might be the degradation products of tRNAs, because their abundances are low and do not have obvious length distribution pattern. The RPs of these tRNA degradation products gradually drop during the differentiation of Giardia (Fig. 4C).
We then used Northern blot to verify the expressions of 15 tasRNAs with various abundances from 5 tRNAs (Fig. 4D and Fig. S4D). We found that the expression patterns of tasRNAs determined by Northern blot were not quite consistent with those determined by RP value, suggesting that RPs do not faithfully reflect the expression levels of tasRNAs (Fig. 4D and Fig. S4 D and E). To obtain the overall expression patterns of tasRNAs, we used the Northern blot data of eSGR-derived endo-siRNAs, in which the expressions were up-regulated at a relatively lower rate throughout the differentiation of Giardia than that of tasRNAs (Fig. 2C) as an inner control to roughly normalized abundance of tasRNAs (details are in Materials and Methods). After normalization, the deep sequencing data were shown to be consistent with Northern blot results (Fig. 4D and Fig. S4 D and E), and we found that all tasRNAs were gradually up-regulated in the differentiation of Giardia (Fig. 4D and Fig. S4 D–G). It is worthy to note that, although all three kinds of tasRNAs of most tRNAs could be detected by deep sequencing (Fig. S4E), the RP of part of them was low and could not be detected by Northern blot (Fig. 4D and Fig. S4D). The variation of expression levels of different tasRNAs (Fig. S4E) implies that their expressions might be under stringent regulation. The expression pattern of 5EsRNA was also verified by Northern blot (Fig. 4E). It is interesting that we could not detect the mature tRNA of pseudotRNA-Gln (TTG) and could only detect its precursor. Unlike tasRNAs, 5EsRNA are highly expressed in trophozoite and rise substantially in cysts.
Identification of a Novel Kind of Stress-Induced tRNA-Derived RNA.
Intriguingly, four probes targeting to the 5′ end of tRNAs could also detect a novel kind of sRNAs with a slightly longer length (∼50 nt) than sitRNAs (stress-induced tRNA-derived RNAs) (∼46 nt), which were identified by our previous work and could not be detected by probe target to the tRNA 5′ end (17). Notably, the length of this novel sRNA (∼50 nt) is approximately equal to the sum of the lengths of 5tasRNAs (∼29 nt) and actasRNAs (∼21 nt), suggesting that this novel sRNA shares the same 3′ end with actasRNAs. The simultaneous detection of this sRNA and sitRNA by the probe used to detect actasRNAs supports this assumption (Fig. 4D and Fig. S4D). We named this kind of sRNA RNA derived from the 5′ end of mature tRNAs (5sitRNA). It is the opposite of the previously reported tRNA 3′ end-derived sitRNAs (3sitRNAs). In contrast to 3sitRNA, 5sitRNA is preferentially expressed in the late stage of Giardia differentiation.
tasRNAs and sitRNAs Might Be Produced from Two Endonuclease Cleavage Sites in Mature tRNA.
The tasRNAs might be produced by the cleavage of the mature tRNAs by one or more unknown endonucleases. We found that the cleavage mainly occurred at two sites in mature tRNA: one site is at the 5′ end of the anticodon stem [cleavage site I (CSI)], and one site is at the 5′ end of the TΨC stem (CSII). The cleavage site and frequency did not change during the differentiation of Giardia. Interestingly, the CSI is the cleavage site used to generate the 3sitRNAs by Giardia, which we reported years ago (17), and the 3′ end of 5sitRNA is located at CSII, implying a connection between the biogenesis of tasRNAs and sitRNAs. It is possible that the endonuclease cleavage of CSI and CSII in the mature tRNAs was activated in the differentiation of Giardia and produced five sRNA products simultaneously (Fig. 4 F and G). It is worth noting that, although the expression levels of five tRNA cleavage products were up-regulated in the differentiation, the expressions of the various tRNAs were stable in this process (Fig. 4D and Fig. S4D), indicating that the production of various tasRNAs and sitRNAs were the result of a purposively controlled process but not the degradation of tRNAs.
In summary, our data revealed that extensive tRNA cleavages were involved in the differentiation process of G. lamblia and that cleavages produce six kinds of tRNA-derived sRNAs, with expression patterns indicating that they might be involved in the differentiation of Giardia.
Discussion
sRNAs are among the most important regulators in eukaryotic species. They provide sequence specificity for regulating gene expression (2) or act as ligands of protein (26) in higher eukaryotic individuals. However, their characteristics, biogenesis, and functions in lower eukaryotes are far from known. In this study, we have systematically and deeply studied the sRNAs of G. lamblia through deep sequencing of sRNAs in pure trophozoites, trophozoites induced to cyst formation during 6 and 24 h (these two stages might contain encysting trophozoites, proliferating trophozoites, and even cysts), and pure cysts and identified endo-siRNAs and various kinds of tRNA-derived sRNAs, which are both widely distributed in eukaryotes, including humans. The conservation of these two kinds of sRNAs suggests that they may play critical roles in eukaryotic cells. It is well-known that the Giardia genome encodes three key proteins of the RNAi pathway [i.e., GLAGO, GLDICER, and Giardia RNA-dependent RNA polymerase (GLRDRP)] (19). Because GLAGO and GLDICER are also the two key proteins of the miRNA pathway, almost all previous studies on sRNA of Giardia were focused on the identification of Giardia miRNA in trophozoite (18, 27–33). As far as we know, 166 miRNAs were reported from trophozoite of G. lamblia by different laboratories through analysis of homology searching, in silico prediction, or deep sequencing (Table S1) (18, 27–33). However, by very careful analysis and experimental verification (Figs. 2 and 3), we surprisingly found that most sRNAs (90.8%) in trophozoite are endo-siRNAs (Fig. 1C and Fig. S3E); only 24% (40) of these reported miRNAs could be detected by our high deep sequencing. Among 40 Giardia miRNAs found in our trophozoite sRNA library, 12.5% (5) of them were shown from known ncRNAs, whereas 50% (20) of them were derived from the eSGR region (see below). We consider, therefore, that these eSGR-derived miRNAs might be endo-siRNAs but not miRNAs. Both plus and minus strands of the whole precursor region of these miRNAs were covered by a large number of eSGR-derived sRNAs (Fig. S4A). As a matter of fact, the sRNA distribution pattern is significantly different from the canonical miRNA precursor, which only has its plus strand of the mature miRNA region covered by sRNAs (34). Unfortunately, Friedländer et al. (34), who reported the miRNAs from G. lamblia, did not provide convincing evidence to prove whether these inferred miRNA precursors had a real hairpin structure. The total reads number of 15 expressed canonical miRNAs only accounts for 0.0048% of all reads in the trophozoite sRNA library (Table S1), which is much less than the abundance of eSGR-derived endo-siRNA (60.9%) (Fig. S3E). Therefore, in our opinion, although the Giardia genome can really encode canonical miRNAs, they may play a less important function given their extremely low abundance in this parasite.
Giardia endo-siRNAs consist of two major types: one type is retrotransposon-derived endo-siRNAs, and the other type is eSGR-derived endo-siRNAs. eSGRs are three genomic regions identified in this study where endo-siRNAs are produced. They produced almost all non–retrotransposon-derived endo-siRNAs in trophozoites of G. lamblia. The expression of endo-siRNAs was up-regulated in the differentiation process of Giardia, suggesting that they might be involved in the regulation of differentiation in Giardia. This hypothesis was strongly supported by the observation that the differentiation ability of G. lamblia was impaired after the knockdown of GLDICER, which is essential for the biogenesis of endo-siRNAs. The relatively high expression level of endo-siRNAs in trophozoites indicated that they might also be involved in the regulation of basic biologic processes of Giardia. This assumption was supported by the previous excellent finding that the surface antigenic variation mechanism was disrupted after the knockdown of GLDICER or GLRDRP (19). It is worth noting that, although Prucca et al. (19) speculated that VSP-derived endo-siRNAs were related to the regulation of surface antigenic variation in cis, we only found that low levels of VSP-derived endo-siRNAs express in Giardia trophozoite (Dataset S2). Low expression level of VSP-derived endo-siRNAs might be caused by the strain of Giardia that we used, which expressed several VSPs. It is also possible that low levels of VSP-derived endo-siRNAs can regulate the antigenic variation or that some non–VSP-derived siRNAs may be involved in the regulation of antigenic variation of G. lamblia. Intriguingly, we found that eSGR-derived endo-siRNAs could largely regulate gene expression in trans, because the silencing of GLDICER did not change the expression of mRNA embedded in eSGRIII (Fig. S4B). Therefore, it is possible that VSP mRNAs can be regulated by endo-sRNAs that are not derived from VSP in trans. Recent findings support this hypothesis, because evidence indicated that miRNA-4 is located in eSGRIII (Fig. S4A) (30), and we suggested that it might be an endo-siRNA and able to regulate VSP mRNA in trans.
Endo-siRNAs are not the only sRNA regulators in Giardia differentiation. We found that six different kinds of tRNA-derived sRNAs were specifically expressed during the period of differentiation in this parasite (Figs. 4 and 5), which indicates that tRNA-derived sRNAs may also play a pivotal role in the differentiation of Giardia. Although tRNA-derived sRNAs were proven to be biologically functional in eukaryotes (9, 17), little is known about their biogenesis, characteristics, and classification. Results from this study clearly showed comprehensive tRNA-derived sRNAs in the early evolution of eukaryotes (Fig. 5), and they offer a clue to the biogenesis of various tRNA-derived sRNAs. Among six kinds of tRNA-derived sRNAs, five sRNAs are novel ncRNAs that were identified in this study. Thus, this work has significantly expanded the repertoire of functional sRNAs in G. lamblia. It is hard to figure out how these tRNA-derived sRNAs regulate Giardia differentiation, because each of them has a large number of members, and we currently lack appropriate methods to alter their expressions. The identification of endonucleases responsible for the biogenesis of these sRNAs may overcome this obstacle. In fact, we have identified two potential endonuclease cleavage sites in mature tRNAs of G. lamblia, which may be responsible for the generation of 5tasRNAs, tRNA 3′ end-associated sRNAs, actasRNAs, 5sitRNAs, and 3sitRNAs (Fig. 4F). The features of the sequences and structures of these two cleavage sites may help to find the endonucleases involved in the process of expression of mature tRNAs into various sRNAs.
Fig. 5.
Large numbers of sRNAs are involved in the Giardia differentiation process. RT, retrotransposon.
In somatic cells of higher multicellular eukaryotes, like mammals, sRNAs are also important regulators of many important biologic processes, such as cellular differentiation (2), and the main regulators are miRNAs but not endo-siRNAs or tRNA-derived sRNAs. Our findings suggest that endo-siRNAs and/or tRNA-derived sRNAs might be the main sRNA regulators in early evolution of eukaryotes or if canonical miRNA does not exist. This scenario is reminiscent of tRNA-derived sRNAs and/or endo-siRNAs/Piwi-interacting RNAs, which might be the main regulators in germ cell and very early developmental stages, but miRNAs do not function in these stages (35–37). Therefore, to better understand the functions of siRNAs and miRNAs in the differentiation regulation of protozoan and metazoan, respectively, we will need additional insight into the evolution of these cell differentiation regulators.
In summary, we have provided the first comprehensive sRNA transcriptome panorama, to our knowledge, of the primitive eukaryote G. lamblia. The data have revealed an unexpected complex sRNA system used in the regulation of differentiation of this parasite (Fig. 5). Our work provides insight into the function, biogenesis, and evolution of endo-siRNAs and tRNA-derived sRNAs in eukaryotes.
Materials and Methods
G. lamblia isolate WB clone C6 (ATCC 50803) was cultured as described previously (38). Encystation and cysts purification were carried out as reported (38). Total RNAs were isolated using TRIzol. sRNA library preparation and deep sequencing were performed by BGI. Giardia genome assemblage A 2.0 was used throughout this study. Northern blot and quantitative RT-PCR were performed as described (39). All primers used in this study’s analysis are listed in Dataset S3. Trophozoites were transfected using the LONZA electrophoration system. Additional details of materials and methods are supplied in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Barbara J. Davids for providing the protocol for Giardia lamblia culture. We greatly appreciated the critical comments and suggestions of the two reviewers, which were very useful for improving the manuscript. J.-Y.L. thanks the Guangdong Province Key Laboratory of Computational Science and the Guangdong Province Computational Science Innovative Research Team for support. This work was supported by National Natural Science Foundation of China Grants 30870530 (to H.Z.), 31401092 (to J.-Y.L.), 31401975 (to L.-L.Z.) and 31272305 (to Z.-R.L.), Sun Yat-Sen University Grant 12lgjc11 (to Z.-R.L.), and National Basic Research Program (973 Program) Grant 2011CB811300 (to L.-H.Q.).
Footnotes
Data deposition: The sRNA deep sequencing data reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA; accession no. SRP043553).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414394111/-/DCSupplemental.
References
- 1.Jacquier A. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 6.Taft RJ, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010;17(8):1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 7.Yu CH, Liao JY, Zhou H, Qu LH. The rat mitochondrial Ori L encodes a novel small RNA resembling an ancestral tRNA. Biochem Biophys Res Commun. 2008;372(4):634–638. doi: 10.1016/j.bbrc.2008.05.092. [DOI] [PubMed] [Google Scholar]
- 8.Zheng LL, et al. Comparative transcriptome analysis of small noncoding RNAs in different stages of Trypanosoma brucei. RNA. 2013;19(7):863–875. doi: 10.1261/rna.035683.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2(5):E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27(10):422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. Behind the smile: Cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol. 2010;8(6):413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- 14.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317(5846):1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 15.Svärd SG, Hagblom P, Palm JE. Giardia lamblia — a model organism for eukaryotic cell differentiation. FEMS Microbiol Lett. 2003;218(1):3–7. doi: 10.1111/j.1574-6968.2003.tb11490.x. [DOI] [PubMed] [Google Scholar]
- 16.Ullu E, Lujan HD, Tschudi C. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell. 2005;4(6):1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36(19):6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4(11):e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prucca CG, et al. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456(7223):750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- 20.Katoh T, et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23(4):433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24(24):2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin RD, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzén O, et al. Transcriptome profiling of Giardia intestinalis using strand-specific RNA-seq. PLOS Comput Biol. 2013;9(3):e1003000. doi: 10.1371/journal.pcbi.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arkhipova IR, Morrison HG. Three retrotransposon families in the genome of Giardia lamblia: Two telomeric, one dead. Proc Natl Acad Sci USA. 2001;98(25):14497–14502. doi: 10.1073/pnas.231494798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24(1):13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Huang PJ, et al. Identification of putative miRNAs from the deep-branching unicellular flagellates. Genomics. 2012;99(2):101–107. doi: 10.1016/j.ygeno.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YQ, Chen DL, Tian HF, Zhang BH, Wen JF. Genome-wide computational identification of microRNAs and their targets in the deep-branching eukaryote Giardia lamblia. Comput Biol Chem. 2009;33(5):391–396. doi: 10.1016/j.compbiolchem.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Chen XS, Collins LJ, Biggs PJ, Penny D. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol Evol. 2009;1:165–175. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraiya AA, Li W, Wang CC. A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA. 2011;17(12):2152–2164. doi: 10.1261/rna.028118.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Saraiya AA, Wang CC. Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl Trop Dis. 2011;5(10):e1338. doi: 10.1371/journal.pntd.0001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Saraiya AA, Wang CC. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cell Microbiol. 2012;14(9):1455–1473. doi: 10.1111/j.1462-5822.2012.01811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saraiya AA, Li W, Wu J, Chang CH, Wang CC. The microRNAs in an ancient protist repress the variant-specific surface protein expression by targeting the entire coding sequence. PLoS Pathog. 2014;10(2):e1003791. doi: 10.1371/journal.ppat.1003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedländer MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 35.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22(11):1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh N, Blelloch R. Small RNAs in early mammalian development: From gametes to gastrulation. Development. 2011;138(9):1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davids B, Gillin F. Methods for giardia culture, cryopreservation, encystation, and excystation. In: Luján H, Svärd S, editors. Vitro Giardia. Vienna: Springer; 2011. pp. 381–394. [Google Scholar]
- 39.Liao JY, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5(5):e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.