Significance
The soft shell clam in many areas of the North Atlantic is afflicted with a fatal leukemia-like disease of unknown origin. Leukemic cells from the diseased animals were found to release reverse transcriptase and to express high RNA levels of a previously unknown member of the gypsy family of retroelements, Steamer. The DNA copy number of the element was increased to enormously high levels in diseased cells, mediated by reverse transcription and integration into the host genome. The activation of Steamer expression and transposition may initiate or accelerate the course of leukemia and constitutes a potential diagnostic marker of the disease.
Keywords: retrotransposon, mobile genetic element, integration, disseminated neoplasia, hemic neoplasia
Abstract
Bivalve mollusks of the North Atlantic, most prominently the soft shell clam Mya arenaria, are afflicted with an epidemic transmissible disease of the circulatory system closely resembling leukemia. The disease is characterized by a dramatic expansion of blast-like cells in the hemolymph with high mitotic index. Examination of hemolymph of diseased clams revealed high levels of reverse transcriptase activity, the hallmark of retroviruses and retroelements. By deep sequencing of RNAs from hemolymph, we identified transcripts of a novel retroelement, here named Steamer. The DNA of the element is marked by long terminal repeats and encodes a single large protein with similarity to mammalian retroviral Gag-Pol proteins. Steamer mRNA levels were specifically elevated in diseased hemocytes, and high expression was correlated with disease status. DNA copy number per genome was present at enormously high levels in diseased hemocytes, indicative of extensive reverse transcription and retrotransposition. Steamer activation in M. arenaria is an example of a catastrophic induction of genetic instability that may initiate or advance the course of leukemia.
The soft shell clam Mya arenaria is one of the most primitive species in the animal kingdom to manifest a leukemia-like disease, variously termed hematopoietic, hemic, or disseminated neoplasia (reviewed in ref. 1). The disease is characterized by the presence of abnormal, rounded, rapidly proliferating hemocytes containing large pleiomorphic nuclei and multiple nucleoli. The tumor cells are polyploid or aneuploid (2–4), exhibit abnormal levels and cytoplasmic localization of the p53 tumor suppressor protein (5), and often express a 200-kDa cell surface antigen defined by monoclonal antibody 1e10 (6–9). The increase in DNA content can be detected by flow cytometry, and the severity of the disease can be established using histological methods. As the disease progresses, normal amitotic hemocytes are replaced by proliferating leukemia cells that invade all tissues, with fatal consequences. A similar disease has been described in several species of bivalves, including oysters (Crassostrea virginica, Crassostrea gigas, Ostrea edulis), mussels (Mytilus edulis, Mytilus galloprovincialis, Mytilus trossulus, Mytilus chilensis), cockles (Cerastoderma edule), and clams (Macoma spp., M. arenaria, and Mya trunata) over a wide geographic distribution.
Despite many reports describing some of its characteristics (1, 10), little is known about the onset and etiology of the disease. Stressors such as pollution (1, 11), temperature (12), and overcrowding have been implicated in disease development. There is evidence that the disease can be transmitted from infected to uninfected individuals (13, 14), indicative of an infectious etiology. Unfiltered hemolymph, lysed hemocytes (15), and even filtered hemolymph isolated from BrdU-treated animals (16) were found to induce disease in healthy animals, raising the possibility of a filterable transmissible agent such as a virus. Induction of disease by the retroviral inducer BrdU (17) suggested the possible involvement of an endogenous retrovirus or retrotransposon. Some studies have detected reverse transcriptase (RT) activity in neoplastic clam tissues (14, 18–21), suggesting that a retroelement or retrovirus might be involved in the disease process, but to date searches for viruses and retroviral sequences from leukemic clams have not been successful (22).
Results
Identification of a Novel Retroelement, Steamer.
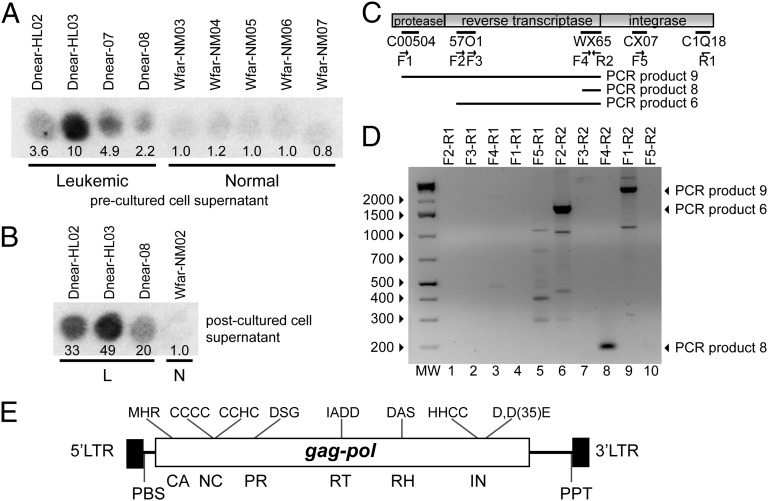
To test for the presence of retroviruses or retroelement virus-like particles, we assayed cell-free hemolymph from diseased and healthy clams for RT activity, using a synthetic homopolymer substrate (23). Hemolymph from diseased clams frequently exhibited high levels of RT activity, 2- to 10-fold above the background activity observed in hemolymph from healthy clams (Fig. 1A). To confirm that the RT activity was released by neoplastic hemocytes rather than other tissues, we cultured the hemocytes and determined the level of RT activity accumulated in the media (postcultured hemolymph). Hemocytes from diseased animals cultured in vitro released high levels of RT into the culture medium, 20- to 50-fold greater than culture medium of hemocytes from healthy animals (Fig. 1B). To identify the potential source of the RT activity, we cultured cells from a diseased clam with high RT activity, isolated total RNA from the culture medium, and used 454 sequencing of cDNAs to generate a database of ∼200,000 sequence reads. Searching these sequences revealed 15 reads with matches to retroviral protease, RT, or integrase sequences. RT-PCR reactions, using different combinations of primers based on these sequences and RNA preparations from cell-free hemolymph of a leukemic clam, yielded three long overlapping DNA fragments (Fig. 1 C and D). The sequence of a complete copy of the retroelement containing these fragments (4,968 bp) was obtained by genome walking using DNA from a healthy animal (Fig. S1). This retroelement was named Steamer for the common name of the host clam and also, by tradition in the transposon field, for a mode of transportation.
Fig. 1.
RT activity and isolation of Steamer DNA from hemolymph of leukemic M. arenaria. (A) RT activity in cell-free hemolymph of indicated leukemic or normal M. arenaria animals was determined by assay for incorporation of {32P}dTTP onto a homopolymer substrate (23). Spot intensity reports the yield of labeled DNA synthesized in vitro, quantitated by ImageJ and normalized to the average of normal values. (B) Hemocytes from the indicated animals diagnosed as leukemic or normal (L or N) were cultured in vitro, and RT activity present in the postculture supernatant medium was determined as in A. (C) Alignment of selected sequences obtained by deep sequencing of cDNAs from a leukemic clam with a retroviral pol gene. PCR primers, forward (F) and reverse (R), are indicated. DNAs amplified by various primer pairs are indicated below the element diagram. (D) DNAs amplified in PCR reactions using cDNA obtained from leukemic clams as a template. Major amplified products are indicated by arrows at the right. (E) Schematic of Steamer genome annotated with characteristic retroelement features. The 5′ and 3′ LTR and the locations of the coding sequences for CA (capsid), NC (nucleocapsid), PR (protease), RT, RH (RNaseH), and IN (integrase) domains are indicated. Characteristic sequence features of each domain, and predicted primer binding site (PBS) and polypurine track (PPT) are indicated.
The Steamer element contains a single long ORF with sequence similarity to retroviral Gag and Pol proteins, flanked by 177-bp direct repeats similar to the LTRs of integrated proviral DNAs (Fig. 1E). The region of similarity to Gag includes the major homology region, the most highly conserved motif of retroviral capsid proteins (24), and a nucleocapsid domain with two putative zinc fingers containing CCCC and CCHC motifs. The Pol region includes similarities to the retroviral protease with diagnostic DSG active site motif (25); an RT with a polymerase domain containing an IADD (“YxDD”) box (26) as well as an RNase H domain with a diagnostic DG/AS box (27); and an integrase with a HHCC zinc finger and a characteristic D,D(35)E motif (28). There is no stop codon separating the Gag and Pol ORFs and no ORF similar to an envelope protein. The element contains a primer binding site complementary to the 3′ end of the Leu (CAG codon) tRNA of the purple sea urchin (29) (TGGTGTCAGAAG), suggesting that Leu tRNA likely functions as the primer for minus strand DNA synthesis, and a polypurine tract sequence serving as primer for plus strand DNA synthesis (30). A maximum likelihood phylogenetic tree (31), constructed using representative retrotransposon amino acid sequences (32) and the Gag, protease, RT, and integrase domains of Steamer, indicated that Steamer is a member of the Mag lineage of retrotransposons (33), a subset of the larger family of gypsy/Ty3 elements (32), with closest similarity to the sea urchin retrotransposon SURL (34, 35) (Fig. S2).
Expression of Steamer RNA Is Elevated in Diseased Hemocytes.
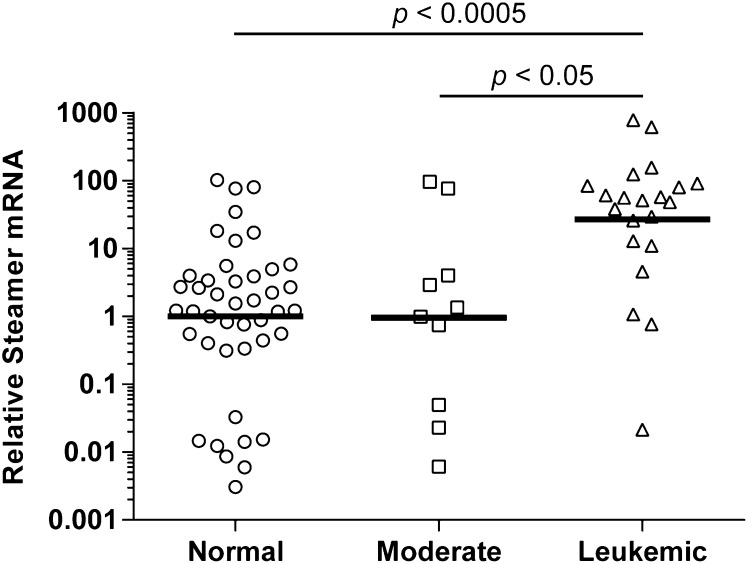
To test for expression of Steamer RNA transcripts, soft shell clams were collected from Prince Edward Island (PEI) in Canada and diagnosed according to hemocyte morphology. Total RNA was isolated from hemocytes of normal (n = 43) and moderately (n = 10) and heavily leukemic (n = 21) individuals, and the levels of Steamer RNA were determined by quantitative RT-PCR (qRT-PCR) and normalized to a housekeeping RNA. Steamer RNA levels were generally low in the normal and moderately leukemic animals, although spanning a large range, and occasional examples were found with high expression (Fig. 2). A large proportion of the highly leukemic samples showed enormously high levels of expression, many fold above the healthy controls. The average level of expression in the diseased animals was ∼27-fold above that in the normal, and the mean levels of Steamer RNA strongly correlated with disease status (P < 0.0005.) The data are consistent with animals showing sporadic induction of RNA at times during the progression of disease, with periods of very high levels of expression occurring with increasing frequency in more advanced disease.
Fig. 2.
Elevated expression of Steamer RNA correlates with disease status. RNA was extracted from hemocytes obtained from normal (n = 43), moderate (n = 10), and heavily leukemic (n = 21) individuals collected from different sites in the North Atlantic. Steamer RNA levels were measured using qRT-PCR and the relative standard curve method. The results are expressed as relative levels compared with EF1 mRNA and are shown on y axis log scale. Each circle, square, and triangle represents RNA from a single individual animal. The geometric mean values, indicated by the horizontal line, were compared by two-tailed t test.
Steamer DNA Copy Number Is Massively Elevated in Diseased Hemocytes.
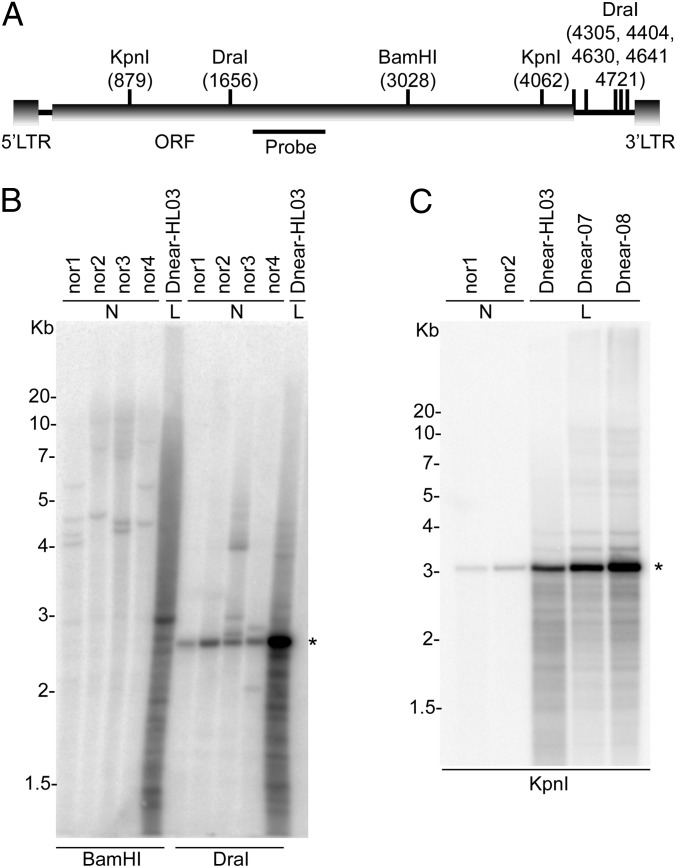
The high levels of Steamer RNAs in leukemic hemocytes raised the possibility that retroelement-encoded gene products with RT and integrase functions might be available to mediate active reverse transcription and transposition of Steamer DNAs. To test for the presence of reverse-transcribed DNAs, we examined total DNA from normal and leukemic clams for Steamer sequences by Southern blotting. Restriction digests of DNA from hemocytes of several healthy clams with BamHI to produce 5′ junction fragments of Steamer (Fig. 3A) revealed a small number of bands (2–4) of uniform intensity and varying sizes, suggestive of a low copy number of elements per genome present at highly polymorphic sites (Fig. 3B). DNA from hemocytes of a leukemic animal revealed an intense smear of heterogeneous fragments, indicative of many new, randomly integrated copies. Digests of normal DNA with DraI predicted to release an internal Steamer fragment yielded a single major product of the expected size with only a few other fragments, indicating that most of the copies were intact and homogeneous. Digestion of leukemic DNA yielded an intense band at the expected size, as well as a number of other fainter fragments, suggesting that most of the newly acquired copies were also intact. Additional digests of DNAs from two normal and three diseased animals with KpnI, again predicted to release an internal fragment, were examined with similar results (Fig. 3C). The patterns are consistent with the presence of a low copy number of elements endogenous to the genome of healthy animals, and the appearance of a large number of newly integrated Steamer DNAs in diseased cells. Digests performed with additional enzymes confirmed these conclusions (Fig. S3). DNA fragments expected for unintegrated or episomal DNAs were not detected, although low levels of such DNAs cannot be ruled out.
Fig. 3.
Leukemic hemocytes have high copy numbers of Steamer DNA. The presence of Steamer in genomic DNA of four normal (N) and a leukemic (L) soft shell clam was analyzed by Southern blotting. (A) Schematic representation of the Steamer retrotransposon. LTRs at the 5′ and 3′ ends, Gag-Pol ORF, sites for digestion by the indicated restriction enzymes, and location of the 32P-labeled probe are indicated. Nucleotide positions are relative to the first nucleotide of the U3 portion of the 5′ LTR. (B) Genomic DNA of four normal (Nor1-4) and one heavily leukemic animal (Dnear-HL03) were digested with restriction enzymes BamHI, releasing left-junction fragments, or with DraI, releasing an internal fragment, and analyzed by Southern blot. DNA loadings were equal as judged by ethidium stain (not shown). (C) Genomic DNA from two normal individuals (Nor1-2) and three leukemic individuals (Dnear-HL03, Dnear-07, and Dnear-08) were digested with KpnI, releasing an internal fragment, and analyzed by Southern Blot. The migration of the DNA molecular markers is indicated at the left of the panels, and major fragment recognized by the probe is indicated by *.
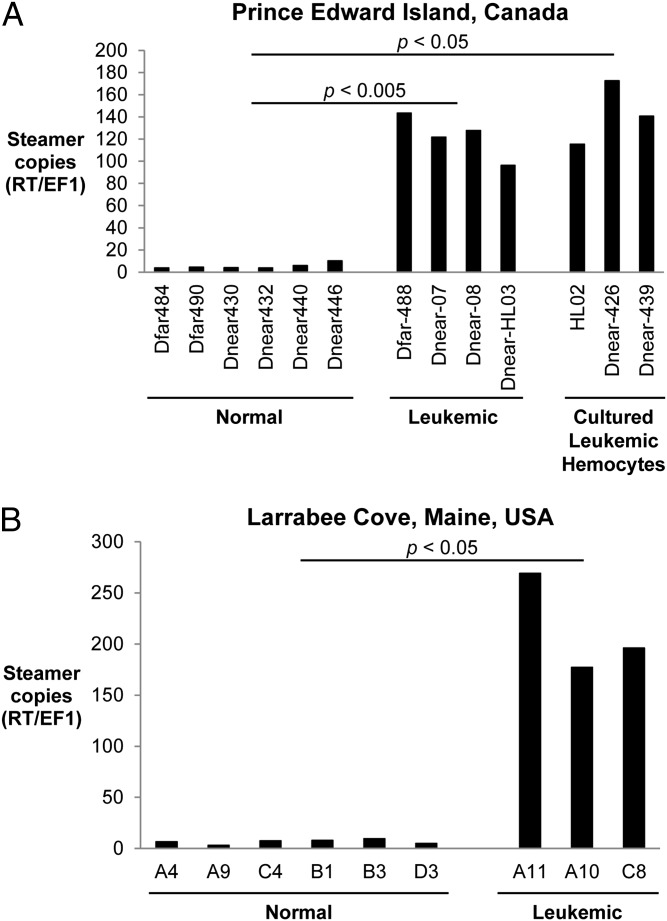
To quantify the Steamer DNA copy number, we carried out qPCR reactions with genomic DNA, normalizing to a single-copy gene, elongation factor 1 (EF1). DNA from hemocytes of six healthy clams from PEI gave a signal of ∼3–10 copies per haploid genome, consistent with the findings from the Southern blots. DNAs from hemocytes of diseased animals, assayed either as primary cells (n = 4) or after culturing (n = 3), yielded copy numbers ranging from 100 to 200 (Fig. 4A). Additionally, clams from an independent population of M. arenaria from Larrabee Cove, ME were assayed for diseased status, and hemocyte DNA was analyzed for Steamer copy number. This isolated population confirmed the strong association between Steamer DNA copy number in hemocytes and disease, with 3–10 copies in normal animals and 150–300 copies in diseased hemocytes (Fig. 4B). The combined Southern and qPCR data suggest that Steamer is an extraordinarily active retrotransposon in diseased animals and undergoes massive expansion and integration into the soft shell clam genome in tumor cells.
Fig. 4.
High copy number of Steamer in leukemic clams from two different populations. Copy number of the Steamer element in genomic DNA was determined by qPCR comparing the ratio of Steamer RT and the single-copy gene EF1. (A) Hemocyte genomic DNA was analyzed from normal and leukemic clams from PEI, Canada and from cultured hemocytes from several leukemic animals. (B) Hemocyte genomic DNA was also analyzed from normal and leukemic clams from Larrabee Cove, ME. Values were compared using a two-tailed t test.
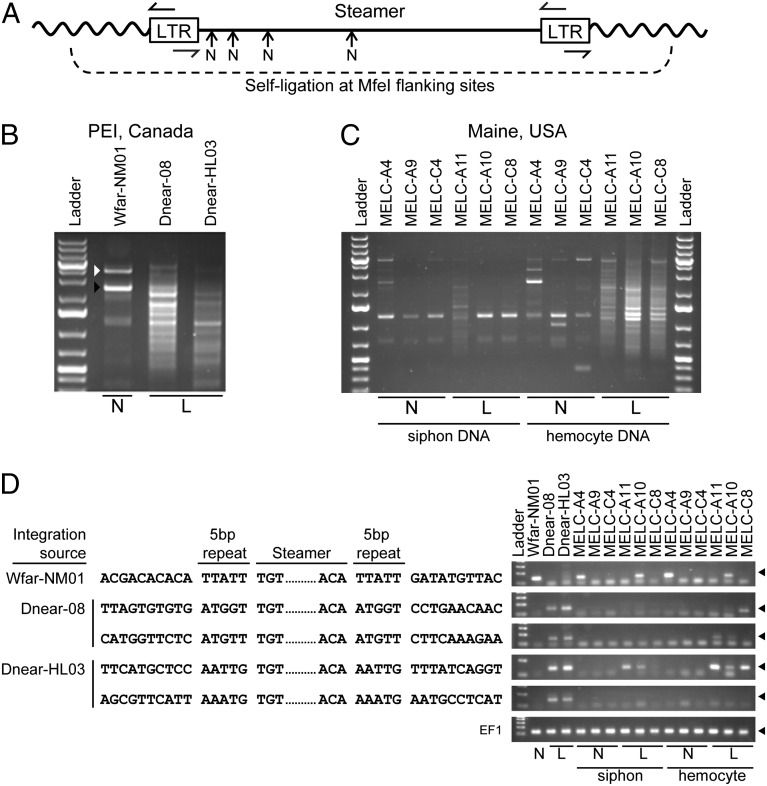
To determine the structure of the Steamer DNAs in more detail, we used inverse PCR to amplify the Steamer integration sites (Fig. 5A). DNA of a healthy clam yielded a single major and some minor PCR products (Fig. 5B). The DNA sequence of the major product revealed integration site junctions corresponding to the predicted LTR 5′ and 3′ ends, and a 5-bp direct repeat flanking the integration site (Fig. 5D). This specific element was cloned using flanking primers, yielding a full-length Steamer retrotransposon (4,968 bp) with an intact Gag-Pol reading frame. The original cDNA products obtained by RT-PCR were nearly identical to the corresponding regions of the genomic sequence (2,453 identical out of 2,457 bp). We subsequently searched the original 454 sequence reads for matches to the genomic sequence and found 63 fragments spanning 3,409 bp of the genome with 99% identity. This full-length sequence was thus selected as defining the prototypical Steamer endogenous retrotransposon (GenBank accession no. KF319019; Fig. S1).
Fig. 5.
Identification of Steamer integration sites in normal and diseased clams. Inverse PCR was used to clone and sequence integration sites of the Steamer retroelement. (A) Schematic of inverse PCR methodology: genomic DNA was digested with MfeI (cleaving only in the flanking DNA), circularized by ligation, and redigested with NsiI at internal sites (N), and finally PCR was performed with outward-directed LTR primers. (B) Inverse PCR was performed using mantle tissue DNA of one normal animal (N), and hemocyte DNA of two heavily leukemic animals (L) from PEI, Canada, and the PCR products were analyzed by agarose gel electrophoresis. For WfarNM01, the white arrowhead marks amplification of the internal Steamer sequence (due to incomplete NsiI cleavage) and the black arrowhead marks the junction product of a single Steamer copy. The leukemic samples yielded a large number of heterogeneous junction products. (C) Inverse PCR was performed using DNA from both siphon tissue and hemocytes for normal and leukemic animals from Maine, USA. (D) Representative DNA sequences of individual cloned integration sites from normal (one site) and leukemic DNAs (two sites from each leukemic PEI animal) are shown. The genomic DNA flanking sequences, the 5-bp duplicated repeats, and the Steamer termini are shown. The presence of the integration sites in the source DNAs and in DNA from other animals was determined for each of the sequences shown by a diagnostic PCR using a forward primer in the Steamer LTR and a reverse primer in the flanking genomic DNA (Right; products are ∼150 bp and marked with a filled triangle). Amplification of EF1 is included as a control.
Inverse PCR of two diseased animals from PEI amplified a large number of integration sites (Fig. 5B), and several were cloned and sequenced from each animal (examples shown in Fig. 5D). Inverse PCR was also carried on DNA from both hemocytes and siphon tissue, of both normal and diseased clams from Maine. Normal animals showed a small number of integration sites in both tissues, and diseased animals showed small numbers in siphon, but a large increase in integration sites can be seen specifically in the leukemic hemocyte DNA (Fig. 5C). Some siphon samples of leukemic animals with higher copies are probably due to contamination with hemocytes. These finding show that the amplification is specific to the diseased hemocytes and not due to diseased individuals inheriting a high number of endogenous copies. To characterize specific integration sites in multiple animals from both PEI and Maine, PCR was performed using primers in the Steamer LTR and in the flanking genomic sequence of each of five selected integration sites (Fig. 5D). The prototypical Steamer integration site identified from the normal PEI animal was present in not all animals but was identified in one normal and one diseased animal from Maine, indicating that it is polymorphic and not associated with disease. In contrast, four integration sites identified in the leukemic PEI animals were not found in any normal animal but were found in both of the two leukemic PEI animals, and several of these same integration sites were also found in hemocyte DNA from diseased animals from Maine (Fig. 5D, rightmost lanes). Thus, Steamer has inserted at multiple new sites in genomic DNA of leukemic clams, and independent tumors often carry Steamer elements at common integration sites. These findings suggest that Steamer may exhibit a preference for selected sites, the transformed phenotype may select for particular insertion sites, or the leukemias may be the result of transmission of a tumor cell line with common Steamer integration sites initiating disease in independent animals at many locations.
Discussion
We identify a previously uncharacterized retrotransposon, Steamer, and find that its expression and genomic amplification is strongly associated with leukemia in M. arenaria. Transposons, ubiquitous in the genomes of all eukaryotes, are by convention grouped into families according to their sequence similarity. The Steamer element of M. arenaria is a member of the gypsy/Ty3 family of retrotransposons, which are marked by the presence of LTRs and undergo reverse transcription and integration by mechanisms virtually identical to those used by the true retroviruses (36). The single gene product encoded by Steamer contains many of the motifs present on retrovirus Gag and Pol proteins, including those of the capsid, nucleocapsid, protease, RT, RNase H, and integrase. Steamer does not encode an envelope protein. Most gypsy family members do not encode envelope proteins, and most retrotransposition events mediated by these elements are likely to occur intracellularly, by the formation of cytoplasmic virion-like particles that mediate reverse transcription and DNA integration into the genome of the same cell. Those elements that do encode envelope proteins [such as ZAM (37) and gypsy itself (38)] can act as infectious retroviruses and can transmit from cell to cell and from one animal to another, perhaps with the help of cellular vesicle trafficking machinery (37–39). However, such infection events may take place even without the use of the envelope protein encoded by the element (40), and in these cases an envelope-like protein from the cell, or from a complementing retroelement, may provide the functionality in trans. The filter-feeding mollusks are capable of concentrating viruses present at very low concentrations in seawater, and can concentrate even viruses such as human hepatitis A virus that do not replicate in the mollusk, to sufficient levels to allow infection of humans upon ingestion (41). Thus, although Steamer does not contain an envelope gene, it is easily conceivable that virion-like particles could mediate movement of the element horizontally from one animal to another. This process may explain the accounts of transmission of disease by filtered hemolymph or by coculture of healthy animals with leukemic animals (13, 42, 43).
It is possible that the activation of Steamer that we find associated with leukemia may be a consequence rather than a cause of tumor development. A recent study has documented significant changes in the expressed mRNAs of hemocytes from leukemic compared with healthy animals, suggesting alterations in the transcriptional program that could include Steamer activation (44). It is notable that a previous report using subtractive hybridization of cDNA identified a small sequence fragment overexpressed in leukemic hemocytes that matches the Steamer genome, confirming the association between Steamer expression and disease (45). Whether Steamer activation is a passive phenotype of the disease or a causal agent of disease remains to be determined.
Transposons create insertional mutations upon each transposition event and thus can be agents of profound genome instability in cancers (46, 47). The scale of activation of Steamer in leukemic cells seen here is extraordinary, unprecedented in magnitude for an induction of transposition in a natural setting. The introduction of more than 100 new copies of a retroelement per genome is bound to lead to profound genetic changes, and it is very plausible that Steamer activity and amplification is involved as a factor or cofactor in the initial development of the leukemia. There are so many new copies of Steamer DNA per genome in the leukemia cells that it will be hard to determine whether there has been an insertional activation of a critical oncogene, but the leukemias are clearly polyclonal with respect to Steamer insertions and are acquiring new proviruses as the pool of transformed cells expands. One or more of the new insertions could significantly alter the phenotypes of these cells.
The observation that multiple integrations are found at the same sites in independent diseased animals (and not in normal tissue from the same animals) is intriguing. It is possible that Steamer integration events may be occurring in a targeted fashion, or that particular sites are selected as part of the process of oncogenesis. Another possible explanation is that a primary leukemia with a primordial repertoire of integration sites is spread between animals as a transmissible leukemia cell line, similar to a transmissible canine venereal tumor (48), or to a facial tumor disease of the Tasmanian devil (49). Such a mechanism would not explain the transmission of disease by filtered hemolymph or the induction of disease by BrdU.
Endogenous retroviruses and retroelements in mammals are often induced by DNA damaging agents, notably halogenated nucleosides such as BrdU and iododeoxyuridine, and this induction can be enhanced by polycyclic hydrocarbons (50). Thus, exposures to environmental toxins may be triggers for the activation of Steamer and disease. An induction of Steamer either early or late in the course of disease would induce rapid genetic instability and so could accelerate or promote disease progression. This scenario may account for the ability of BrdU to experimentally induce disease in clams (17).
Recent studies have shown that some clam populations are more susceptible than others to induction of disease by DNA damaging agents (16). If Steamer is responsible for the disease, susceptible populations may harbor a higher copy number of Steamer or distinctive copies that are more readily induced for expression. Both inheritance of a high number of endogenous copies of the element and somatic amplification of the element within individuals could contribute to development of disease. The availability of sequence probes for Steamer will allow surveys of the prevalence of the element in various populations, tests of experimental transmission from animal to animal, and further tests for its functional involvement with disease. Because genomes of M. arenaria are highly polymorphic for the Steamer element, the DNA probes should also allow the development of populations of M. arenaria that lack the element entirely through selective breeding, and such element-free populations may be less prone to induction of leukemia by environmental stresses. The identification of Steamer and its dramatic amplification in leukemia provides a potential marker for the disease and a remarkable example of catastrophic genomic instability caused by retroelements.
Materials and Methods
M. arenaria Collection.
M. arenaria were collected from PEI, Canada and Larrabee Cove, ME and diagnosed by microscopic analysis of cell morphology as described in SI Materials and Methods.
Molecular Analyses.
RT activity assays, 454 sequencing, genome walking, Southern blot analysis, qRT-PCR, qPCR, and inverse PCR were conducted by standard methods using the specific primers and conditions described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Brian Beal (University of Maine at Machias) for obtaining and providing us with clams from Maine, and Niklas Thalén for assistance in integration site analysis. This work was supported by the Howard Hughes Medical Institute (G.A., M.J.M., and S.P.G.), the Pew Fellows Program (G.A.), Fondo Nacional de Desarrollo Cientifico y Tecnologico Grant 1130852 (to G.A.), National Institutes of Health (NIH) Training Grant T32 CA009503 (to M.J.M.), and awards from the National Science Foundation (to C.R.), Strategic Applications of Genomics in the Environment (to J.S.), the Pesticide Science Fund (to J.S. and C.R.), and NIH (to C.S. and W.I.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KF319019).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409945111/-/DCSupplemental.
References
- 1.Barber B. Neoplastic diseases of commercially important marine bivalves. Aquat Living Resour. 2004;17(4):449–466. [Google Scholar]
- 2.Cooper K, Brown RS, Chang PW. Accuracy of blood cytologial screening techniques for the diagnosis of a possible hematopoetic neoplasm in the bivalve mollusc, Mya arenaria. J Invertebr Pathol. 1982;39(3):281–289. [Google Scholar]
- 3.Lowe DM, Moore MN. Cytology and quantitative cytochemistry of a poliferative atypical hemocytic condition in Mytilus edulis (Bivalvia, mollusca) J Natl Cancer Inst. 1978;60(6):1455–1459. doi: 10.1093/jnci/60.6.1455. [DOI] [PubMed] [Google Scholar]
- 4.Reno PW, House M, Illingworth A. Flow cytometry and chromosome analysis of Softshell clams, Mya arenaria, with disseminated neoplasia. J Invertebr Pathol. 1994;64(3):163–172. [Google Scholar]
- 5.Walker C, et al. p53 superfamily proteins in marine bivalve cancer and stress biology. Adv Marine Biol. 2011;59:1–36. doi: 10.1016/B978-0-12-385536-7.00001-7. [DOI] [PubMed] [Google Scholar]
- 6.Miosky DL, Smolowitz RM, Reinisch CL. Leukemia cell specific protein of the bivalve mollusc Mya arenaria. J Invertebr Pathol. 1989;53(1):32–40. doi: 10.1016/0022-2011(89)90071-2. [DOI] [PubMed] [Google Scholar]
- 7.Reinisch CL, Charles AM, Troutner J. Unique antigens on neoplastic cells of the soft shell clam Mya arenaria. Dev Comp Immunol. 1983;7(1):33–39. doi: 10.1016/0145-305x(83)90052-6. [DOI] [PubMed] [Google Scholar]
- 8.Smolowitz RM, Reinisch CL. A novel adhesion protein expressed by ciliated epithelium, hemocytes, and leukemia cells in soft-shell clams. Dev Comp Immunol. 1993;17(6):475–481. doi: 10.1016/s0145-305x(05)80003-5. [DOI] [PubMed] [Google Scholar]
- 9.White MK, Miosky D, Flessas DA, Reinisch CL. The expression of an adhesion-related protein by clam hemocytes. J Invertebr Pathol. 1993;61(3):253–259. doi: 10.1006/jipa.1993.1049. [DOI] [PubMed] [Google Scholar]
- 10.Muttray A, et al. Haemocytic leukemia in Prince Edward Island (PEI) soft shell clam (Mya arenaria): Spatial distribution in agriculturally impacted estuaries. Sci Total Environ. 2012;424:130–142. doi: 10.1016/j.scitotenv.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Pariseau J, et al. Potential link between exposure to fungicides chlorothalonil and mancozeb and haemic neoplasia development in the soft-shell clam Mya arenaria: A laboratory experiment. Mar Pollut Bull. 2009;58(4):503–514. doi: 10.1016/j.marpolbul.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Schneider KR. Heat stress in the intertidal: Comparing survival and growth of an invasive and native mussel under a variety of thermal conditions. Biol Bull. 2008;215(3):253–264. doi: 10.2307/25470709. [DOI] [PubMed] [Google Scholar]
- 13.Collins CM, Mulcahy MF. Cell-free transmission of a haemic neoplasm in the cockle Cerastoderma edule. Dis Aquat Organ. 2003;54(1):61–67. doi: 10.3354/dao054061. [DOI] [PubMed] [Google Scholar]
- 14.Oprandy JJ, et al. Isolation of a viral agent causing hematopoietic neoplasia in the soft-shell clam Mya arenaria. J Invertebr Pathol. 1981;34(1):45–51. [Google Scholar]
- 15.Walker C, et al. Mass culture and characterization of tumor cells from a naturally occurring invertebrate cancer model: Applications for human and animal disease and environmental health. Biol Bull. 2009;216(1):23–39. doi: 10.1086/BBLv216n1p23. [DOI] [PubMed] [Google Scholar]
- 16.Taraska NG, Anne Böttger S. Selective initiation and transmission of disseminated neoplasia in the soft shell clam Mya arenaria dependent on natural disease prevalence and animal size. J Invertebr Pathol. 2013;112(1):94–101. doi: 10.1016/j.jip.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Oprandy JJ, Chang PW. 5-bromodeoxyuridine induction of hematopoietic neoplasia and retrovirus activation in the soft-shell clam, Mya arenaria. J Invertebr Pathol. 1983;42(2):196–206. doi: 10.1016/0022-2011(83)90062-9. [DOI] [PubMed] [Google Scholar]
- 18.Romalde JL, et al. Evidence of retroviral etiology for disseminated neoplasia in cockles (Cerastoderma edule) J Invertebr Pathol. 2007;94(2):95–101. doi: 10.1016/j.jip.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 19.AboElkhair M, et al. Reverse transcriptase activity associated with haemic neoplasia in the soft-shell clam Mya arenaria. Dis Aquat Organ. 2009;84(1):57–63. doi: 10.3354/dao02038. [DOI] [PubMed] [Google Scholar]
- 20.AboElkhair M, et al. Reverse transcriptase activity in tissues of the soft shell clam Mya arenaria affected with haemic neoplasia. J Invertebr Pathol. 2009;102(2):133–140. doi: 10.1016/j.jip.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 21.House ML, Kim CH, Reno PW. Soft shell clams Mya arenaria with disseminated neoplasia demonstrate reverse transcriptase activity. Dis Aquat Organ. 1998;34(3):187–192. doi: 10.3354/dao034187. [DOI] [PubMed] [Google Scholar]
- 22.AboElkhair M, et al. Lack of detection of a putative retrovirus associated with haemic neoplasia in the soft shell clam Mya arenaria. J Invertebr Pathol. 2012;109(1):97–104. doi: 10.1016/j.jip.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: Use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craven RC, Leure-duPree AE, Weldon RA, Jr, Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69(7):4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb DD, Hutchison CA, 3rd, Edgell MH, Farmerie WG, Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J Virol. 1989;63(1):111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuki S, Ishimaru S, Inouye S, Saigo K. Identification of genes for reverse transcriptase-like enzymes in two Drosophila retrotransposons, 412 and gypsy; a rapid detection method of reverse transcriptase genes using YXDD box probes. Nucleic Acids Res. 1986;14(7):3017–3030. doi: 10.1093/nar/14.7.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanaya S, et al. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990;265(8):4615–4621. [PubMed] [Google Scholar]
- 28.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorge J, Hughes SH. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982;43(2):482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 32.Llorens C, et al. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Res. 2011;39(Database issue):D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaille JJ, Mathavan S, Gaillard J, Garel A. The complete sequence of mag, a new retrotransposon in Bombyx mori. Nucleic Acids Res. 1990;18(3):674. doi: 10.1093/nar/18.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Springer MS, Davidson EH, Britten RJ. Retroviral-like element in a marine invertebrate. Proc Natl Acad Sci USA. 1991;88(19):8401–8404. doi: 10.1073/pnas.88.19.8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez P, Lessios HA. Evolution of sea urchin retroviral-like (SURL) elements: Evidence from 40 echinoid species. Mol Biol Evol. 1999;16(7):938–952. doi: 10.1093/oxfordjournals.molbev.a026183. [DOI] [PubMed] [Google Scholar]
- 36.Levin HL. Newly identified retrovtransposons of the Ty3/gypsy class in fungi, plants, and vertebrates. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM; 2002. pp. 684–701. [Google Scholar]
- 37.Brasset E, et al. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8(17):2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 39.Kim A, et al. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91(4):1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalvet F, et al. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18(9):2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruddy SJ, et al. An epidemic of clam-associated hepatitis. JAMA. 1969;208(4):649–655. [PubMed] [Google Scholar]
- 42.Elston RA, Kent ML, Drum AS. Transmission of hemic neoplasia in the bay mussel, Mytilus edulis, using whole cells and cell homogenate. Dev Comp Immunol. 1988;12(4):719–727. doi: 10.1016/0145-305x(88)90047-x. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin SM, Farley CA, Hetrick FM. Transmission studies of sarcoma in the soft-shell clam, Mya arenaria. In Vivo. 1992;6(4):367–370. [PubMed] [Google Scholar]
- 44.Siah A, McKenna P, Berthe FC, Afonso LOB, Danger JM. Transcriptome analysis of neoplastic hemocytes in soft-shell clams Mya arenaria: Focus on cell cycle molecular mechanism. Res Immunol. 2013;3:95–103. doi: 10.1016/j.rinim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siah A, McKenna P, Danger JM, Johnson GR, Berthe FC. Induction of transposase and polyprotein RNA levels in disseminated neoplastic hemocytes of soft-shell clams: Mya arenaria. Dev Comp Immunol. 2011;35(2):151–154. doi: 10.1016/j.dci.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28(11):550–559. doi: 10.1016/j.tig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Solyom S, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22(12):2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen D. The canine transmissible venereal tumor: A unique result of tumor progression. Adv Cancer Res. 1985;43:75–112. doi: 10.1016/s0065-230x(08)60943-4. [DOI] [PubMed] [Google Scholar]
- 49.Pearse AM, Swift K. Allograft theory: Transmission of devil facial-tumour disease. Nature. 2006;439(7076):549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikura H, et al. Enhancement of 5-iododeoxyuridine-induced endogenous C-type virus activation by polycyclic hydrocarbons: Apparent lack of parallelism between enhancement and carcinogenicity. J Natl Cancer Inst. 1977;58(4):1035–1040. doi: 10.1093/jnci/58.4.1035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.