Significance
Our large-scale survey of genomic nucleotide composition across monocots has enabled the first rigorous testing, to our knowledge, of its biological significance in plants. We show that genomic DNA base composition (GC content) is significantly associated with genome size and holocentric chromosomal structure. GC content may also have deep ecological relevance, because changes in GC content may have played a significant role in the evolution of Earth’s biota, especially the rise of grass-dominated biomes during the mid-Tertiary. The discovery of several groups with very unusual GC contents highlights the need for in-depth analysis to uncover the full extent of genomic diversity. Furthermore, our stratified sampling method of distribution data and quantile regression-like logic of phylogenetic analyses may find wider applications in the analysis of spatially heterogeneous data.
Keywords: plant genome, genome size evolution, Poaceae, phylogenetic regression, geographical stratification
Abstract
Genomic DNA base composition (GC content) is predicted to significantly affect genome functioning and species ecology. Although several hypotheses have been put forward to address the biological impact of GC content variation in microbial and vertebrate organisms, the biological significance of GC content diversity in plants remains unclear because of a lack of sufficiently robust genomic data. Using flow cytometry, we report genomic GC contents for 239 species representing 70 of 78 monocot families and compare them with genomic characters, a suite of life history traits and climatic niche data using phylogeny-based statistics. GC content of monocots varied between 33.6% and 48.9%, with several groups exceeding the GC content known for any other vascular plant group, highlighting their unusual genome architecture and organization. GC content showed a quadratic relationship with genome size, with the decreases in GC content in larger genomes possibly being a consequence of the higher biochemical costs of GC base synthesis. Dramatic decreases in GC content were observed in species with holocentric chromosomes, whereas increased GC content was documented in species able to grow in seasonally cold and/or dry climates, possibly indicating an advantage of GC-rich DNA during cell freezing and desiccation. We also show that genomic adaptations associated with changing GC content might have played a significant role in the evolution of the Earth’s contemporary biota, such as the rise of grass-dominated biomes during the mid-Tertiary. One of the major selective advantages of GC-rich DNA is hypothesized to be facilitating more complex gene regulation.
Deep insights into the genomic architecture of model plants are rapidly accumulating, especially because of advances being made in high-throughput next generation and third generation sequencing techniques (1). However, the genomic constitution of the vast majority of nonmodel plants still remains unknown (2), impeding our understanding of the relationship between particular genomic architectures and evolutionary fitness in various environments. One of the important qualitative aspects of genomic architecture is the genomic nucleotide composition, which is usually expressed as the proportion of guanine and cytosine bases in the DNA molecule (GC content). In prokaryotes, the GC content is a well-studied and widely used character in taxonomy (3), and numerous studies have shown both the impact of GC content on microbial ecology and the influence of the environment in shaping the DNA base composition of microbial communities (4–7). The DNA base composition is also frequently discussed in relation to the evolution of the isochore structure in humans and other homeothermic (warm-blooded) vertebrates (i.e., birds and mammals) (8–10). In contrast, considerably less attention has been paid to the biological relevance of genomic GC content variation in plants (11), with genomic GC contents known only for a limited amount of the total phylogenetic diversity (11–18).
One important feature of the GC base pair is its higher thermal stability compared with the AT base pair, a feature that arises from the stronger stacking interaction between GC bases and the presence of a triple compared with a double hydrogen bond between the paired bases (19). In turn, these interactions seem to be important in conferring stability to higher order structures of DNA and RNA transcripts (11, 20). In bacteria, for example, an increase in GC content correlates with a higher temperature optimum and a broader tolerance range for a species (21, 22). Selection for higher thermal stability has also been suggested to explain the evolution of GC-rich regions in the genomes of homeothermic vertebrates in contrast to their GC-poor homologs found in poikilothermic (i.e., cold-blooded) groups, such as fish and amphibians (9). Nevertheless, other alternative hypotheses have also been proposed to explain GC richness in bacteria and certain regions of vertebrate genomes (7, 8, 11, 23, 24). Two additional important features of the GC base pair are its higher mutability, related to frequent cytosine methylation (25–27), and the higher cost of its synthesis compared with the AT base pair (28). The latter has led to speculation that there will be a tradeoff in the relationship between genomic GC content and genome size (11). Indeed, the higher cost of GC base pairs has been suggested as the reason that explains the lower GC contents observed in giant genomed geophytic plants compared with the species with smaller genomes (16). Nevertheless, it remains unknown whether such observations are limited to species with a geophytic life strategy or a more widespread phenomenon across plants with different life strategies.
To date, the highest GC contents of land plants have been found in grasses (Poaceae) (11, 15, 29–34). Although grasses are reported to have undergone a dramatic spread and evolutionary diversification over the last ∼30 My as the climate has become increasingly arid and cool (35–37), the reasons underpinning their success are controversial given that grasses have extremely desiccation-sensitive (recalcitrant) pollen (38), a feature certainly not well-suited for growth in arid environments (39). The question, therefore, arises as to whether the extremely high GC content might somehow compensate or at least, whether increased GC is also found in other groups with desiccation-sensitive pollen. In contrast to grasses, the lowest GC contents so far reported in plants have been found in several species possessing holocentric chromosomes (i.e., in Cyperaceae and Juncaceae) (15, 17), and this observation raises the question of whether there is an association between genomic GC content and chromosome structure.
The observations that both GC-rich Poaceae and GC-poor Cyperaceae and Juncaceae are closely related (both belong to the monocot order Poales) and that extreme GC contents have also been reported in other monocots (16) make monocots (comprising ∼25% of all angiosperms) an ideal choice to conduct an extensive survey of GC content to provide insights into the extent of its diversity and its possible biological relevance and evolutionary significance. Here, we present the first large-scale analysis, to our knowledge, of GC content variation across 239 monocot species, including representatives of all 11 orders and 70 of 78 families recognized by the Angiosperm Phylogeny Group III (40). By analyzing GC content in relation to several genomic characters, a suite of life history traits, and climatic data within a well-resolved phylogenetic framework, we also explore the possible biological and ecological relevance of GC content variation in monocots and discuss the nature of the driving forces that may have contributed to it.
Results
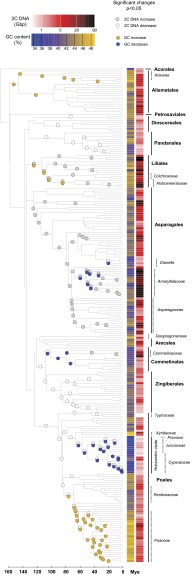
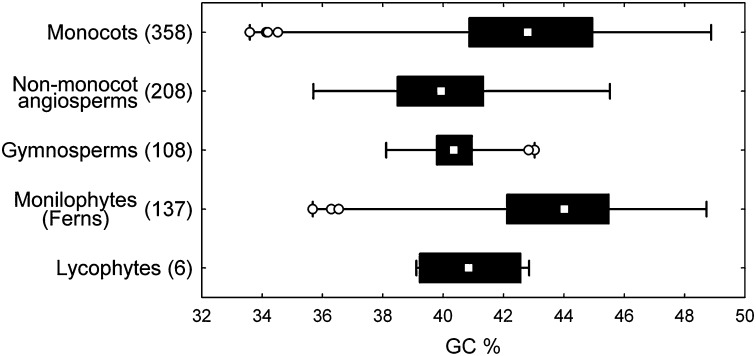
GC content varied from 33.6% in Juncus inflexus to 48.9% in Triticum monococcum (Figs. 1 and 2, Figs. S1 and S2, and Dataset S1, Tables S1 and S2) and showed a strong phylogenetic signal (Pagel λ = 0.919, P < 0.001). Several orders of monocots (i.e., Poales, Liliales, and Alismatales) contained species with GC contents that exceeded those reported for any other group of vascular plants (Fig. 2). Indeed, overall, the range of GC contents in monocots is greater than that encountered in nonmonocot angiosperms, gymnosperms, or lycophytes and broadly similar to the values reported in monilophytes (ferns) (Fig. 2).
Fig. 1.
Reconstructing the evolution of GC content and 2C genome size across the phylogenetic tree of monocots. Significant increases and decreases (P ≤ 0.01) in each character are marked with circles inserted at the appropriate branches of the phylogenetic tree. The names of all monocot orders are shown together with the names of important families and genera with significant shifts in either GC content or genome size. Greater detail of particular branches and species is in Figs. S1 and S3; greater detail for Poaceae is shown in Fig. 3.
Fig. 2.
Variation in GC content measured with flow cytometry across vascular plants. Box plots show the minimum-to-maximum range (whiskers), interquartile range (black boxes), median (white squares),and outliers (empty circles). Numbers in parentheses after the group names indicate the numbers of species with known GC content. The figure is based on our own data (11, 16) and the work by Barow and Meister (12). All of the data were recalculated based on the standards used in this paper, and where multiple values were available for a species, those values estimated for this work were selected.
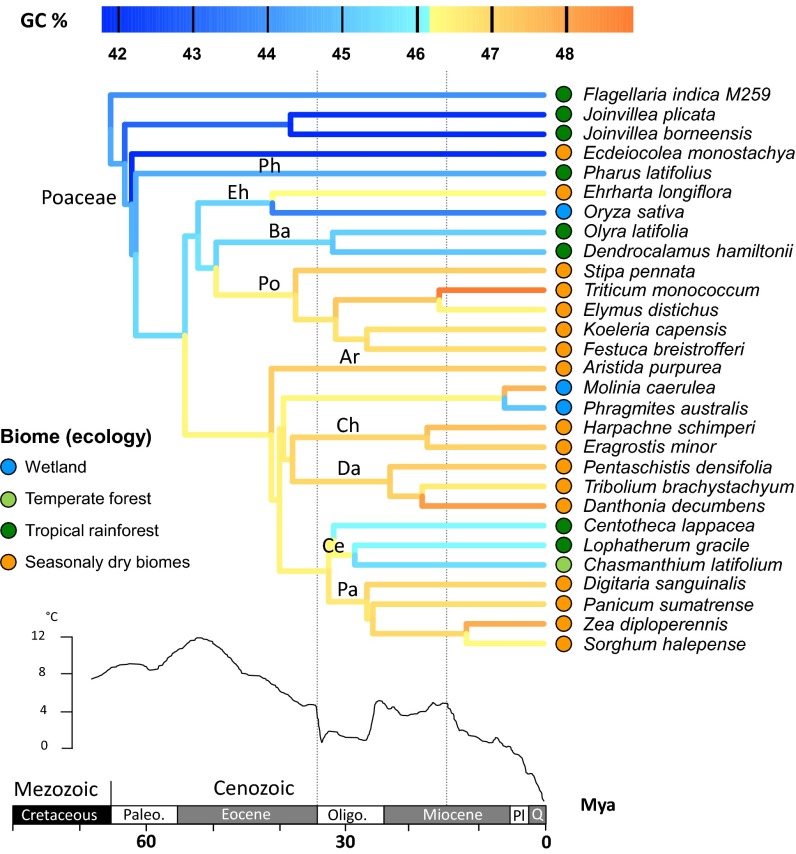
The highest GC contents were found within Poales, especially in the grasses (Poaceae) and Xyris (Xyridaceae) (Fig. 1 and Figs. S1 and S2). In grasses, the increase in GC content was reconstructed to have occurred at the Mezozoic/Cenozoic boundary (68 Mya) (Figs. 1 and 3), when grasses and related families (i.e., Flagellariaceae, Joinvilleaceae and Ecdeiocoleaceae) diverged from Restionaceae. Additional significant increases were reconstructed on many internal branches of Poaceae throughout the Tertiary, mostly in association with the ability to grow in open and seasonally dry habitats (Figs. 1 and 3).
Fig. 3.
Reconstruction of the evolution of genomic GC content in Poaceae and their closest evolutionary relatives. Within the deep sea paleotemperature curve (58) in Lower, the vertical dotted lines indicate the onset of the two major Cenozoic aridification events (i.e., the beginning of the Oligocene and the Monterey Transition in the mid-Miocene). The highest GC contents are found in modern grassland-forming tribes dominating various seasonally dry ecosystems (savannah, temperate grassland, and Mediterranean-type vegetation). Correspondingly, lower GC content is found in forest-dwelling and wetland grasses experiencing all-year humid conditions. Ar, Aristidoideae; Ba, Bambusoideae; Ce, Centothecoideae; Ch, Chloridoideae; Da, Danthonioideae; Eh, Ehrhartoideae; Oligo, Oligocene; Pa, Panicoideae; Paleo, Paleocene; Ph, Pharoideae; Pl, Pliocene; Po, Pooideae; Q, Quaternary.
Beyond Poales, phylogenetic analyses also identified a significant increase in GC content at the base of Alismatales and within Araceae as well as at the base of Liliales (namely Colchicaceae and Alstroemeriaceae).
At the other end of the scale, the lowest GC contents were found in the holocentric clade [i.e., Prionium with Cyperaceae and Juncaceae; mean GC = 36.9%], sharply contrasting with the high GC contents found in Xyris (mean GC = 48.5%), which is in the sister clade (both within Poales). It is notable that the repeated decreases in GC content within the holocentric clade coincided with significant decreases in genome size (Fig. 1 and Figs. S1 and S3). In addition to the holocentric clade, significant decreases in GC content were also identified at the base of the Commelinales, the large genome-sized family Amaryllidaceae (Asparagales), and the Dianella clade in Xanthorrhoeaceae (Asparagales) (Fig. 1 and Figs. S1 and S3).
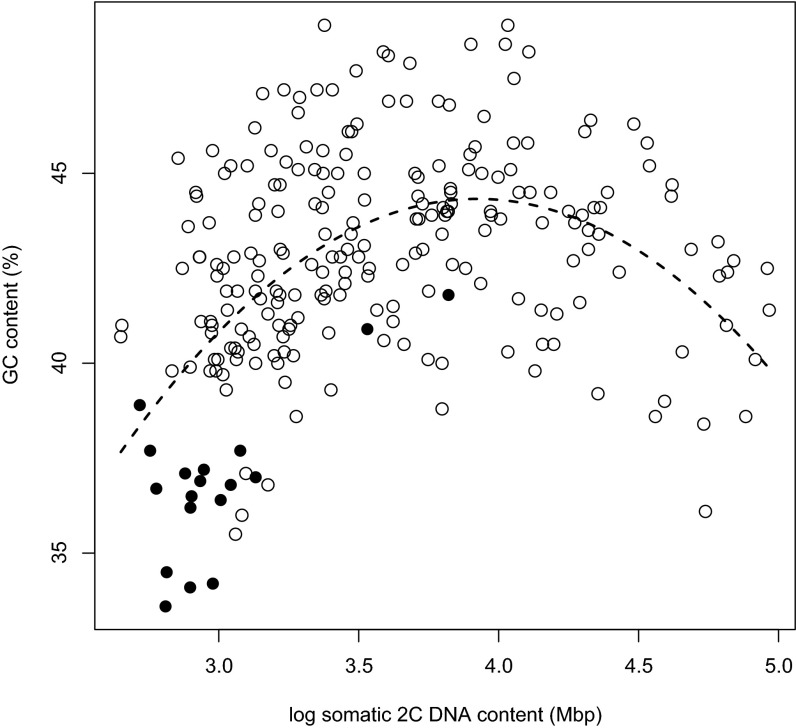
Among the several traits and climate data shown to be significantly associated with changes in GC content in the phylogenetic analyses (Table 1 and Dataset S1, Tables S3 and S4), the strongest relationship was with genome size (with both absolute 2C genome size and 1Cx monoploid genome size, which remove the impact of polyploidy on genome size). In general, GC content increased with increasing genome size, although at both lower and higher genome sizes, there was a tendency for GC content to decrease, making the relationship between GC content and genome size quadratically curved (phylogenetic generalized least squares procedure; P < 0.001) (Fig. 4 and Table 1).
Table 1.
ANOVA showing the final phylogenetic generalized least squares model that explains the observed variation in GC content in monocots
| Character of relationship | F value | P value | Model AIC | Explained residual variance (%) | |
| 2C data* | |||||
| log 2C | Positive | 33.95 | <0.0001 | 1,014.80 | 9.36 |
| (log 2C)2 | Negative | 38.96 | <0.0001 | 986.65 | 10.74 |
| Holocentrics | Negative | 22.56 | <0.0001 | 969.29 | 6.22 |
| BioClim 6† | Negative | 16.89 | <0.0001 | 955.70 | 4.65 |
| Bulb geophyte | Negative | 7.54 | 0.0065 | 950.39 | 2.08 |
| Mediterranean | Positive | 7.13 | 0.0081 | 945.27 | 1.97 |
| Recalcitrant pollen | Positive | 4.77 | 0.0299 | 942.38 | 1.32 |
| 1Cx data‡ | |||||
| log 1Cx | Positive | 13.56 | 0.0003 | 776.57 | 5.44 |
| (log 1Cx)2 | Negative | 19.84 | <0.0001 | 762.16 | 7.98 |
| BioClim 6§ | Negative | 22.36 | <0.0001 | 743.75 | 9.00 |
| Bulb geophyte | Negative | 8.27 | 0.0045 | 737.59 | 3.33 |
| Recalcitrant pollen | Positive | 4.51 | 0.0351 | 734.99 | 1.82 |
Dataset S1, Tables S3 and S4 shows the results of variables not incorporated into the final model. Degrees of freedom equal one in all variables. AIC, Akaike information criterion; BioClim, bioclimatic.
Model with 2C absolute genome size data (n = 239).
Average minimum temperature of coldest month (90th percentile).
Model with 1Cx monoploid genome size data (n = 186; data for species with holocentric chromosomes are not included in the tested dataset because of their uncertain ploidy-level status).
Average minimum temperature of coldest month (75th percentile).
Fig. 4.
Raw GC contents and 2C genome sizes of the measured species showing a quadratic relationship between the two genomic parameters. The holocentric species are marked with black circles. The quadratic regression fit of the raw data is shown by a dashed line for illustrative purposes. The results of the exact phylogeny-corrected test confirming the existence of the quadratic relationship are shown in Table 1 and Dataset S1, Tables S3 and S4.
After removing the effect of 2C genome size, GC content was shown to be significantly associated with a holocentric chromosomal structure. Species in the holocentric clade had lower GC contents than predicted from their small genomes (Fig. 4) and were generally characterized by possessing the lowest GC contents so far encountered in monocots. After removing the effect of genome size (and holocentrism in the analyses with 2C genome sizes), GC content still remained significantly negatively correlated with the presence of species in Oceania, tropical rainforest biome, mean annual temperature, isothermality (i.e., the proportion of day-to-night to summer-to-winter temperature oscillations), average minimum temperature of coldest month, mean temperature of coldest, warmest, driest, or wettest quarters, annual precipitations, amount of precipitation in wettest month, and wettest, warmest, or coldest quarters and positively correlated with latitude, annual temperature range, and annual temperature seasonality (i.e., coefficient of variation of monthly mean temperatures) (Dataset S1, Tables S3 and S4). These correlations indicate that an increased GC content is associated with the ability of plants to tolerate seasonally dry winter cold regions typical of a continental temperate climate. In the summary explanatory model, these highly intercorrelated variables are best substituted with the 90th percentile of the average minimum temperature of the coldest month (Table 1). Together, in the full 239-species 2C data, genome size, holocentrism, and average minimum temperature of the coldest month were able to explain over 30% of the residual variation in GC content of monocots and caused the most dramatic decrease in the Akaike information criterion of the explanatory model (Table 1). A minor improvement of the model was further achieved by the inclusion of one climatic variable and two life history traits. Specifically, GC content was found to decrease in bulbous geophytes and increase in plants from the global Mediterranean climate biome [only in calculations with the full 2C data] and plants with desiccation-sensitive pollen (Table 1 and Dataset S1, Tables S3 and S4).
Discussion
GC Content and Genome Size.
Our analysis revealed that GC content is closely related with the physical size of the genome. The quadratic nature of the relationship between GC content and genome size (Fig. 4) corroborates previous findings from geophytic (bulbous) plants (16) and suggests that this relationship may hold across the diversity of plants. The positive correlation between GC content and genome size observed for monocot species with small to medium genome sizes reflects a general trend observed in many plant genera (18, 41) as well as bacterial and animal genomes (6, 22, 42). In plants, this correlation might arise simply from the fact that genome growth predominantly arises from increases in the amount of LTR retrotransposons that dominate most plant genomes (43, 44). LTR retrotransposons consist of GC-rich gene regions, making them relatively more GC-rich than other noncoding DNA sequences. Indeed, the expansion of GC-rich retrotransposons may have contributed to the high GC contents observed in some grasses, such as maize (Zea mays; GC = 47.2%) (12), where the extremely GC-rich Huck element (GC ∼ 62%) comprises at least 10% of the genome (45). In general, rapid changes in the abundance of retrotransposons are expected to be the major reason for the differences in GC content observed between closely related taxa that differ sharply in genome sizes (11), like for instance, in the genus Tetraria (Cyperaceae) in our data (Dataset S1, Tables S1 and S2). Here, the three species analyzed range from 36% to 40% GC while possessing genome sizes that vary over fourfold (2C = 793–3,398 Mb). In such cases, alternative mechanisms, such as DNA mutations, are unlikely to operate fast enough to result in the substantial divergence in GC content over such short evolutionary timescales.
The observed quadratic relationship between genome size and GC content (Fig. 4) may point to the presence of a specific mechanism responsible for decreasing the GC percentage when a genome becomes very large. Rocha and Danchin (28) noted that the synthesis of guanine and cytosine (i.e., their deoxyribotriphosphates dGTP and dCTP) is more energetically demanding than dATP and dTTP. It is possible to envisage that a deficiency in dGTP and dCTP during DNA replication (which may be especially pronounced during the replication of large genomes because of the large amounts needed) might result in the misincorporation of less costly dATPs and dTTPs and hence, an overall mutation bias toward AT-rich DNA (11). This hypothesis still remains to be tested experimentally [e.g., by comparing the extent and direction of dNTP misincorporation rate between plants growing under different nutrient regimes and/or between species with weak and strong selection pressures for rapid DNA synthesis (e.g., evergreen perennials and large-genomed annuals, respectively)]. It is also possible that the need for dNTPs economy in large genomes may be coupled with structural constraints, such as the need for compact DNA packing in nuclei, where AT-rich DNA may be favored over GC-rich DNA because of its higher compactness (24).
Decreased GC and Holocentrism.
After genome size, the presence of a holocentric chromosome structure was shown to be the next most significant factor influencing GC content of monocot genomes. Here, the very low GC contents found in species from the holocentric clade (i.e., Prionium, Cyperaceae and Juncaceae) resulted from the combined effects of their small genome size and holocentric chromosome nature (Fig. 1, Table 1, and Fig. S1). In contrast to most plants that have monocentric chromosomes (i.e., the centromere and kinetochore are localized), plants with holocentric chromosomes lack centromeres, and the kinetochore spreads over the whole length of the chromosome (46). One consequence of this type of organization is that holocentric chromosomes are small and rigid, which may in turn, reduce recombination rates (at least during mitosis) (46). If so, this lower recombination might also result in a reduced frequency of repair at heterologous recombination sites. This type of repair preferentially introduces GC bases (47), and it has been suggested to be one of the few mechanisms responsible for maintaining the high GC richness of genes and perhaps, other regions of DNA in the genome (11, 23, 32). However, clearly, more experimental and detailed genomic data are needed from plant and animal species with holocentric chromosomes before attempting any generalization on the relationship between GC content, genome size, and recombination rates.
Increased GC Content and Response to Cold and Dry Climates and Desiccation Stress.
Our study confirmed a significant relationship between GC content and the ecology and distribution of monocot species, particularly their tolerance to temperature extremes. However, in contrast to bacteria (6), where higher GC content correlates with increased thermotolerance (likely under selection because of the higher thermal stability of the DNA molecule) (21, 22), in monocots, higher GC content was associated with increased tolerance and ability to grow in regions of extremely cold winters or experiencing at least some seasonal water deficiency (i.e., biomes characterized by seasonal drought). Such observations suggest that the reasons underlying higher GC contents in plants are different from those in bacteria. These contrasting observations may result from fundamental differences in the structural and regulation complexity of plants compared with prokaryotic (bacterial) genomes as well as the generally lower temperature and environmental extremes that plants experience compared with extreemophilous bacteria.
An inability to cope with low (extreme) temperatures and frequent freezing is likely to restrict the distribution of many vascular plant lineages, especially those that evolved in the humid warm (tropical) climates of the Mesozoic and Early Cenozoic (48). Indeed, plant lineages that are able to establish in regions of seasonally cold climates must have developed a series of physiological adaptations to improve their ecological response to cold and limit the risk of incidental frost damage (49). Adaptations to cold hardiness are principally similar to those for drought, because the major danger of cold temperatures—the freezing of water in living plant tissues—may result in damaging cell dehydration (50). The role of these physiological adaptations is to substitute intracellular water that freezes easily with sugars and substitute water molecules used to stabilize the structure of biomolecules with protective structure-stabilizing proteins (50–52).
Many plants also prevent incidental frost or desiccation damage by the controlled senescing of aboveground tissues (49), with perennials surviving unfavorable climatic periods in the form of renewal organs (buds, rhizomes, and bulbs) protected from the extremely low temperatures or droughts by hiding underground or under a buffering cover of snow. Typically, this type of adaptation is found in true bulbous geophytes, where a need to develop intrinsic cold tolerance adaptations might be of lesser evolutionary advantage than in other life forms. Indeed, it is perhaps not so surprising that, although cold tolerance is generally associated with higher GC contents, this relationship is not so significant in bulbous geophytes, because they had lower GC contents compared with other plants in the explanatory model (Table 1).
Given that freezing and drought stress can be matched by similar physiological and ecomorphological adaptations, their importance might seem particularly pronounced in the Mediterranean climate regions experiencing incidental frosts together with long periods of summer drought. The increased GC content found in plants typical of the global Mediterranean biome supports the interpretation of the above view of a putative function of the increased GC content as a genomic adaptation to increased levels of desiccation stress. The increased GC content found in plants with desiccation-sensitive (recalcitrant) pollen (Table 1) also lends support to this idea. Desiccation-sensitive pollen typically lacks specific pollen wall structures and apertures (38, 39) that prevent uncontrolled water loss and desiccation (53). As a consequence, the viability of desiccation-sensitive pollen is highly dependent on the air humidity when the pollen is shed (39, 54). In grasses, for example, the pollen remains viable for only a very short time (a few minutes to a few hours), even under the most favorable environmental conditions (54, 55). This short period of viability forces plants with desiccation-sensitive pollen to carefully restrict pollen release to humid periods of the day (39, 56). Such plants would certainly benefit from any intrinsic adaptation (possibly associated with increased GC content) that would assure pollen viability at lower water potential. It is, thus, notable that the extremely high GC contents in grasses correspond well with the observation that grasses are the only large monocot group with desiccation-sensitive pollen that dominate cold and drought-stressed environments. We suggest that these hypotheses can effectively be tested, for instance, by measuring the response to incidental frost in tropical plants with different GC contents that have never experienced freezing temperatures or comparing the decrease in pollen viability in plants with the same pollen type but different GC contents.
Adaptations to growth in seasonally cold and/or dry environments (e.g., autumnal cold-hardening, development of dormant organs, or programmed tissue loss) pose a significant physiological and regulatory challenge for plants, requiring complex genome regulation. These challenges are considerably greater than those faced by tropical floras, which experience year-round favorable climate conditions, supporting continuous growth. Current studies of the effect of GC richness on gene function indicate that these complex physiological responses may, indeed, be facilitated by the presence of GC-rich genes and genomes (24, 57). Because this evidence comes from studies of grass genes, the possible mechanisms are discussed below in the context of the evolutionary success of GC-rich grasses.
Tertiary Climate Cooling and the Rise to Dominance of GC-Rich Grasses.
Grasses are among the most spectacular group of monocots showing consistently high genomic GC contents. The timing of the major GC increases (Fig. 3) coincides with the origin and diversification of the modern grassland-forming tribes that then underwent additional diversification, possibly in response to the global cooling and aridification events in the Oligocene (34–23 Mya) and more recently, the mid-Miocene (∼15 Mya) (35, 36, 58, 59). Today, the grasses that can grow in seasonally stressed (dry or cold) climates and especially, those dominating the grassland biomes (i.e., in the Aristidoideae, Danthonioideae, Chlorideoideae, Panicoideae, and Pooideae tribes) have the highest GC contents (mean GC percentage = 47.2) of all monocots and are clearly GC-richer than their forest dwelling relatives in the tribes Pharoideae, Bambusoideae, and Centothecoideae (mean GC percentage = 45.4) or the wetland grass lineages experiencing all-year humid conditions (Fig. 3). For example, the GC content is higher in Ehrharta longiflora (GC percentage = 46.2), which is typical of the Mediterranean-type ecosystems of the Southern Hemisphere, compared with Oryza sativa (GC percentage = 43.6) growing in tropical wetlands (Fig. 3).
Edwards et al. (36) postulated that the advantageous traits that enabled the rapid expansion of grassland biomes during the mid-Tertiary evolved early (during the shady history of grasses) and before the demise of Tertiary forests and the advent of the C4 photosynthetic pathway in numerous modern grass clades. However, the nature of such traits has remained elusive. Given the timing and the trend in GC content evolution within grasses that we have reconstructed here (i.e., initial increase in GC content in the early diverging and forest dwelling tribes with additional significant increases in the clades, which subsequently gave rise to the modern grassland-forming tribes) (Fig. 3), we propose that such advantageous traits include adaptations at the genome level associated with shifts to higher GC contents.
Advantages of GC-Rich Grass Genomes Under Seasonally Cold and Dry Climate Regimes.
The most notable feature of the GC-rich grass genomes is the presence of extremely GC-rich genes, which mostly represent paralogs of GC standard genes (57, 60). A similar bimodality in the GC composition of genes has also been observed in other plant groups with GC-rich genomes, such as some green algae and ferns (11). It seems, therefore, that understanding the origin and function of GC-rich genes may play a key role in understanding the forces driving the evolution of high genomic GC contents in plants.
Compared with standard genes, GC-rich genes in grasses are characterized by fewer or no introns, a much higher GC content in the 5′ region of the gene, more methylatable CpG dinucleotides in the leading strand, and a higher frequency of regulatory TATA boxes in their promoter regions (57, 60). These findings, together with overrepresentation of the GC-rich paralogs in certain functional groups of genes, have led to the suggestion that GC-rich genes facilitate a plant’s response to environmental stress (57). Hypothetically, an improved response to cold and drought typical of biomes characterized by thermal (warm/cold) and precipitation (summer dry or winter dry) seasonal climates might also be facilitated by GC-rich genes.
Another advantage of GC-rich DNA may arise from the different conformation changes in DNA that are possible in GC-rich compared with GC-poor DNA, because these conformations may also contribute to enabling more complex genome regulation (24). DNA can adopt various conformational states, known as A-DNA, B-DNA, and Z-DNA. A-DNA is considered to be an inactive conformation state, whereas B-DNA is associated with metabolically active DNA, and Z-DNA has been linked to regulation of DNA transcription and gene expression, perhaps affecting the binding of transcription factors (61). When a cell desiccates, the removal of DNA-stabilizing water molecules forces the native B-DNA to adopt different conformations (62), with GC-poor B-DNA forming metabolically inactive A-DNA and GC-rich B-DNA sequences tending to form Z-DNA (63, 64). Furthermore, a positive correlation exists between GC content and the ability of DNA to undergo B→Z conformational transitions in genes of humans and other model vertebrates and plants (24). Given these observations, it is perhaps easy to envisage how GC-rich DNA could be advantageous for cell regulation and survival in plants during cold hardening or as a consequence of tissue freezing or desiccation (50–52). Hypothetically, formation of Z-DNA instead of A-DNA might allow DNA to retain some minimum metabolic activity, even at decreased intracellular water contents, which could be important for the regulation and/or resurrection of frozen or drought-dehydrated tissues. In this way, enabling the formation of a partly functional DNA conformation (i.e., Z-DNA) caused by high GC content might be seen as an additional genomic adaptation along with other physiological cold or drought stress responses to minimize the effect of water loss on the structure and functionality of biomolecules.
Such a hypothesis could be tested, for example, by comparing the GC content of key genes responsible for retaining the functioning of frozen and dehydrated cells or those expressed during cell rehydration. Still, understanding the link between nucleotide composition, DNA conformation, and regulation of gene expression in determining how a plant responds to cold (freezing) or increased drought still poses a significant challenge to cell biologists. Clearly, additional research in this field is essential if we are to improve our understanding of how long-term changes in the environment may have influenced the evolution and composition of plant genomes and the genomic determinants, which shape a plant’s response to climate change.
Methods
GC contents and 2C DNA contents were measured using flow cytometry in 239 species covering all 11 orders and 70 of 78 currently recognized monocot families (40) (Fig. S2 and Dataset S1, Table S1). The measurements of GC content were based on comparison of nuclei fluorescence stained with two different fluorochromes [the DNA intercalating propidium iodide (measuring the absolute 2C genome size) and AT-selective DAPI (measuring the AT fraction of the genome)] using the protocols by Šmarda et al. (14, 15). The chromosome numbers for measured species were taken from the literature or estimated by us in 16 species (Dataset S1, Table S1) to enable monoploid genome size (1Cx) to be calculated (1Cx = 2C genome size divided by the ploidal level) (65). Data on selected biologically important life history traits (life form, pollination strategy, and pollen desiccation sensitivity) as well as information on species distribution and their habitat preferences (including geographic distribution on continents, extent of distribution area, presence in biomes, moisture requirements, or ability to grow in open, sun-exposed habitats) were collected from available floras and taxonomic literature (Dataset S1, Table S2). The geographical distribution data were extracted from the Global Biodiversity Information Facility portal (www.gbif.org) and the South African National Floristic Database (http://bgis.sanbi.org). The geographical data were resampled using a novel spatial data stratification algorithm based on heterogeneity-constrained random resampling (66), which was devised to remove the effect of uneven data sampling (SI Methods, Dataset S2, and Fig. S5). Nineteen bioclimatic variables and altitude were extracted for each selected location from the WorldClim database (67) (Dataset S1, Table S2).
The phylogenetic tree for all measured taxa, except grasses, was obtained by pruning the recent large-scale dated angiosperm phylogeny by Zanne et al. (49) (Fig. 1, SI Methods, and Figs. S1 and S3). This phylogeny contains directly ∼70% of studied species, whereas many of the remaining species studied by us were sufficiently closely related to species studied by Zanne et al. (49) that the latter could be used as surrogates for our species to provide insights into their phylogenetic relationships. For grasses, we adopted the phylogenetic tree of the Grass Phylogeny Working Group II (37) and used maximum likelihood dating with two fossil calibration points (Dataset S3). Significant episodes in the evolution of GC content and genome size were detected on the tree using generalized least squares and tip values reshuffling randomization calculated using the ape package (68) in R (69) (Fig. 1 and Figs. S1, S3, and S4, and Dataset S4). We compared GC contents with genome size, life history traits, and climatic niche data by applying multiple regressions using phylogenetic generalized least squares calculated in the caper package of R (70) and built an explanatory model for GC content variation, including six nonredundant variables (Table 1). For the calculation, we used different (10th, 25th, 50th, 75th, and 90th) percentiles of climatic variables to account for multifactor control of species occurrences using a similar testing logic as in quantile regression. Full methods and associated references are included in SI Methods.
Supplementary Material
Acknowledgments
The authors thank numerous colleagues and botanical gardens, namely M. Dančák (Palacký University), V. Rybka (Prague Botanical Garden), M. Tupá and M. Chytrá (Botanical Garden of the Masaryk University), and C. Berg (Botanical Garden of Graz, Karl Franzens University) for providing fresh plant material (Dataset S1, Table S1) and O. Hájek, P. Veselý, I. Lipnerová, A. Veleba, and J. Šmerda for technical assistance. CapeNature and the Department of Environment and Conservation are acknowledged for the permits for plant samplings in the area of Western Cape and in Western Australia, respectively. The Commonwealth Department of Sustainability, Environment, Water, Population, and Communities provided the relevant export permits. We thank the South African National Biodiversity Institute for providing access to floristic distribution data. L.M. thanks the University of Western Australia, Iluka Chair for logistic support. Czech Science Foundation Grants GACR 206/08/P222, GACR506/11/0890, and GACR13-29362S provided financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.R.G. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321152111/-/DCSupplemental.
References
- 1.Flagel LE, Blackman BK. The first ten years of plant genome sequencing and prospects for the next decade. In: Wendel JF, Greilhuber J, Doležel J, Leitch IJ, editors. Plant Genome Diversity. Vol 1. Vienna: Springer; 2012. pp. 1–15. [Google Scholar]
- 2.Galbraith DW, Bennetzen JF, Kellogg EA, Pires JC, Soltis PS. The genomes of all angiosperms: A call for a coordinated global census. J Bot. 2011 doi: 10.1155/2011/646198. [DOI] [Google Scholar]
- 3.Stackebrandt E, Liesack W. Nucleic acids and classification. In: Goodfellow M, O’Donnell AG, editors. Handbook of New Bacterial Systematics. London: Academic; 1993. pp. 151–194. [Google Scholar]
- 4.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 5.Foerstner KU, von Mering C, Hooper SD, Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;6(12):1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann S, Chen YP. Bacterial genomic G+C composition-eliciting environmental adaptation. Genomics. 2010;95(1):7–15. doi: 10.1016/j.ygeno.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, Zhang Z, Hu S, Yu J. On the molecular mechanism of GC content variation among eubacterial genomes. Biol Direct. 2012;7:2. doi: 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2(7):549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- 9.Bernardi G. The neoselectionist theory of genome evolution. Proc Natl Acad Sci USA. 2007;104(20):8385–8390. doi: 10.1073/pnas.0701652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costantini M, Cammarano R, Bernardi G. The evolution of isochore patterns in vertebrate genomes. BMC Genomics. 2009;10:146. doi: 10.1186/1471-2164-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Šmarda P, Bureš P. The variation of base composition in plant genomes. In: Wendel F, Greilhuber J, Doležel J, Leitch IJ, editors. Plant Genome Diversity. Vol 1. Vienna: Springer; 2012. pp. 209–235. [Google Scholar]
- 12.Barow M, Meister A. Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry. 2002;47(1):1–7. doi: 10.1002/cyto.10030. [DOI] [PubMed] [Google Scholar]
- 13.Meister A, Barow M. In: Flow Cytometry with Plant Cells. Analysis of Genes, Chromosomes, and Genomes. Dolezel J, Greilhuber J, Suda J, editors. Weinheim, Germany: Wiley-VCH; 2007. pp. 177–215. [Google Scholar]
- 14.Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. Genome size and GC content evolution of Festuca: Ancestral expansion and subsequent reduction. Ann Bot (Lond) 2008;101(3):421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Šmarda P, Bureš P, Šmerda J, Horová L. Measurements of genomic GC content in plant genomes with flow cytometry: A test for reliability. New Phytol. 2012;193(2):513–521. doi: 10.1111/j.1469-8137.2011.03942.x. [DOI] [PubMed] [Google Scholar]
- 16.Veselý P, Bureš P, Šmarda P, Pavlícek T. Genome size and DNA base composition of geophytes: The mirror of phenology and ecology? Ann Bot (Lond) 2012;109(1):65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipnerová I, Bureš P, Horová L, Šmarda P. Evolution of genome size in Carex (Cyperaceae) in relation to chromosome number and genomic base composition. Ann Bot (Lond) 2013;111(1):79–94. doi: 10.1093/aob/mcs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veleba A, et al. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol. 2014;203(1):22–28. doi: 10.1111/nph.12790. [DOI] [PubMed] [Google Scholar]
- 19.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34(2):564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biro JC. Correlation between nucleotide composition and folding energy of coding sequences with special attention to wobble bases. Theor Biol Med Model. 2008;5:14. doi: 10.1186/1742-4682-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishio Y, et al. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 2003;13(7):1572–1579. doi: 10.1101/gr.1285603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musto H, et al. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 2006;347(1):1–3. doi: 10.1016/j.bbrc.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 23.Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics. 2001;159(2):907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinogradov AE. DNA helix: The importance of being GC-rich. Nucleic Acids Res. 2003;31(7):1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer GP. Mutagenesis at methylated CpG sequences. In: Doerfler W, Böhm P, editors. DNA Methylation: Basic Mechanisms. Berlin: Springer; 2006. pp. 259–281. [Google Scholar]
- 27.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18(6):291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 29.Salinas J, Matassi G, Montero LM, Bernardi G. Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res. 1988;16(10):4269–4285. doi: 10.1093/nar/16.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 31.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 32.Serres-Giardi L, Belkhir K, David J, Glémin S. Patterns and evolution of nucleotide landscapes in seed plants. Plant Cell. 2012;24(4):1379–1397. doi: 10.1105/tpc.111.093674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KY. Studies on the base composition of higher plants. 1. Monocotyledons. BMB Rep. 1968;1(2):99–107. [Google Scholar]
- 34.Biswas SB, Sarkar AK. Deoxyribonucleic acid base composition of some angiosperms and its taxonomic significance. Phytochemistry. 1970;9(12):2425–2430. [Google Scholar]
- 35.Stromberg CAE. Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci. 2011;39:517–544. [Google Scholar]
- 36.Edwards EJ, et al. C4 Grasses Consortium The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science. 2010;328(5978):587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 37.Grass Phylogeny Working Group II New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193(2):304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- 38.Franchi GG, Nepi M, Dafni A, Pacini E. Partially hydrated pollen: Taxonomic distribution, ecological and evolutionary significance. Plant Syst Evol. 2002;234(1-4):211–227. [Google Scholar]
- 39.Franchi GG, et al. Pollen and seed desiccation tolerance in relation to degree of developmental arrest, dispersal, and survival. J Exp Bot. 2011;62(15):5267–5281. doi: 10.1093/jxb/err154. [DOI] [PubMed] [Google Scholar]
- 40.Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161(2):105–121. [Google Scholar]
- 41.Bureš P, et al. Correlation between GC content and genome size in plants. Cytometry A. 2007;71A(9):764. [Google Scholar]
- 42.Vinogradov AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: The triangular relationship. Cytometry. 1998;31(2):100–109. doi: 10.1002/(sici)1097-0320(19980201)31:2<100::aid-cyto5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 43.Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot (Lond) 2005;95(1):127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grover CE, Wendel JF. Recent insights into mechanisms of genome size change in plants. J Bot. 2010 doi: 10.1155/2010/382732. [DOI] [Google Scholar]
- 45.SanMiguel P, Vitte C. The LTR-retrotransposons of maize. In: Bennetzen J, Hake S, editors. Handbook of Maize Genetics and Genomics. New York: Springer; 2009. pp. 307–327. [Google Scholar]
- 46.Bureš P, Zedek F, Marková M. Holocentric chromosomes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel J, editors. Plant Genome Diversity. Vol 2. Vienna: Springer; 2013. pp. 187–208. [Google Scholar]
- 47.Brown TC, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- 48.Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends Ecol Evol. 2004;19(12):639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506(7486):89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 50.Pearce RS. Plat freezing and damage. Ann Bot (Lond) 2001;87(4):417–424. [Google Scholar]
- 51.Beck EH, Heim R, Hansen J. Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J Biosci. 2004;29(4):449–459. doi: 10.1007/BF02712118. [DOI] [PubMed] [Google Scholar]
- 52.Beck EH, Fettig S, Knake C, Hartig K, Bhattarai T. Specific and unspecific responses of plants to cold and drought stress. J Biosci. 2007;32(3):501–510. doi: 10.1007/s12038-007-0049-5. [DOI] [PubMed] [Google Scholar]
- 53.Katifori E, Alben S, Cerda E, Nelson DR, Dumais J. Foldable structures and the natural design of pollen grains. Proc Natl Acad Sci USA. 2010;107(17):7635–7639. doi: 10.1073/pnas.0911223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dafni A, Firmage D. Pollen viability and longevity: Practical, ecological and evolutionary implications. Plant Syst Evol. 2000;222(1-4):113–132. [Google Scholar]
- 55.Reddi CS, Raju NSN, Rao MVS. Pollination and seed set in tropical wetland grasses. Nord J Bot. 2010;28(3):354–365. [Google Scholar]
- 56.Franchi GG, Nepi M, Matthews ML, Pacini E. Anther opening, pollen biology and stigma receptivity in the long blooming species, Parietaria judaica L. (Urticaceae) Flora. 2007;202(2):118–127. [Google Scholar]
- 57.Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA. GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics. 2010;11:308. doi: 10.1186/1471-2164-11-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 59.Linder PH, Rudall PJ. Evolutionary history of Poales. Annu Rev Ecol Evol Syst. 2005;36:107–124. [Google Scholar]
- 60.Guo X, Bao J, Fan L. Evidence of selectively driven codon usage in rice: Implications for GC content evolution of Gramineae genes. FEBS Lett. 2007;581(5):1015–1021. doi: 10.1016/j.febslet.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 61.Rich A, Zhang S. Z-DNA: The long road to biological function. Nat Rev Genet. 2003;4(7):566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 62.Saenger W, Hunter WN, Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature. 1986;324(6095):385–388. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- 63.Foloppe N, MacKerell AD., Jr Intrinsic conformational properties of deoxyribonucleosides: Implicated role for cytosine in the equilibrium among the A, B, and Z forms of DNA. Biophys J. 1999;76(6):3206–3218. doi: 10.1016/S0006-3495(99)77472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuller W, Forsyth T, Mahendrasingam A. Water-DNA interactions as studied by X-ray and neutron fibre diffraction. Philos Trans R Soc Lond B Biol Sci. 2004;359(1448):1237–1247. doi: 10.1098/rstb.2004.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann Bot (Lond) 2005;95(1):255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lengyel A, Chytrý M, Tichý L. Heterogeneity-constrained random resampling of phytosociological databases. J Veg Sci. 2011;22(1):175–183. [Google Scholar]
- 67.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25(15):1965–1978. [Google Scholar]
- 68.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 69.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 70.Orme D. 2012. The Caper Package: Comparative Analysis of Phylogenetics and Evolution in R. Available at http://cran.r-project.org/web/packages/caper/vignettes/caper.pdf. Accessed March 23, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.