Significance
Quantitative trait loci (QTLs) are major targets for plant breeders. Despite intensive efforts undertaken over the last decades, still little is known how QTLs affect plant resistance to biotic and abiotic stresses and what exact molecular markers (genes) are involved. Here we identified a gene in barley that maps to previously identified QTLs for boron sensitivity, plant height, lodging, stem breaking, days to heading, yield, seed weight, days to maturity, as well as powdery mildew and spot blotch resistance. This gene is identical to a previously described jasmonate-induced protein designated JIP60 that by virtue of its unique structure and processing is capable of reprogramming protein translation for increased stress tolerance and controlled senescence.
Keywords: Hordeum vulgare, jasmonic acid, translational reprogramming
Abstract
Two closely related genes encoding the jasmonate-induced protein 60 (JIP60) were identified in the barley genome. The gene on chromosome arm 4HL encodes the previously identified protein encoded by the cDNA X66376.1. This JIP60 protein is characterized here and shown to consist of two domains: an NH2-terminal domain related to ribosome-inactivating proteins and a COOH-terminal domain, which displays similarity to eukaryotic translation initiation factor 4E (eIF4E). JIP60 undergoes processing in vivo, as a result of which JIP60’s COOH-terminal eIF4E domain is released and functions in recruiting a subset of cellular messengers for translation. This effect was observed for both MeJA-treated and naturally senescing plants. Because the JIP60 gene is in close proximity to several quantitative trait loci for both biotic and abiotic stress resistance, our results identify a unique target for future breeding programs.
Jasmonic acid (JA) and its methyl ester, methyl JA (MeJA), are widespread throughout the plant kingdom. As cyclopentanone compounds, they are involved in a vast variety of developmental processes including flower bud formation, plant fertility, and plant senescence (see refs. 1–3 for reviews). In addition, JA and MeJA accomplish key roles in stress responses and defense. JA causes marked alterations in transcription and translation (1, 2).
In excised leaf tissues of barley, two major jasmonate effects have been observed: (i) the induction of novel abundant proteins designated jasmonate-induced proteins (JIPs), and (ii) the depression of cytoplasmic protein synthesis (summarized in ref. 2). JIP synthesis is controlled transcriptionally. JIP transcripts bind with high efficiency to 80S ribosomes (3). In fact, cytoplasmic polysomes discriminate against photosynthetic transcripts in favor of JIP mRNAs. Previous work identified translational initiation as the point of control (3) and implicated the target of rapamycin pathway as playing a major role (4). Jasmonate-induced proteins comprise JIP5, which is a leaf thionin acting in defense (5); JIP23, which has been implicated in translational control (6, 7); JIP37, a protein of unknown function (8); JIP24 (allene oxide cyclase) (9), JIP54 (allene oxide synthase) (10), and JIP100 (lipoxygenase) (11), all involved in the early steps of jasmonate biosynthesis (summarized in refs. 1 and 2); and JIP60, representing a key player in the translational arrest of cytoplasmic protein synthesis seen for long-term MeJA-treated and stressed plants (12, 13). JIP60 consists of an NH2-terminal domain related to type 1 and type 2 ribosome-inactivating proteins (RIPs) and a COOH-terminal domain, which has similarity to eukaryotic initiation factors of type 4E (eIF4E) (12, 13). Whereas RIPs are extremely potent inhibitors of translation elongation (see below), eIF4Es are part of a multiprotein complex regulating translation initiation. The eIF complex consists of eIF4A, a 46-kDa RNA helicase; eIF4B, a 70-kDa RNA-binding and RNA-annealing protein; eIF4H, a 25-kDa protein that acts with eIF4B to stimulate the helicase activity of eIF4A; eIF4G, a 185-kDa protein that colocalizes with all of the other eIF proteins to the 40S subunit of the ribosome; and eIF4E (14). Joshi et al. (15) identified 411 eIF4E family members from 230 species that display a great variability in their NH2-terminal mRNA-binding domains. For most eukaryotic mRNAs, translation is dependent upon the activity of eIF4E, which binds to the 5′ cap structure of mRNAs and to the initiation factor eIF4G (14). Through the interaction of the eIF4E–eIF4G complex with the ribosome-bound factor eIF3, the 40S ribosomal subunit is positioned at the 5′ end of the mRNA and commences scanning the mRNA for the translational start codon. Then, the binding of the 60S ribosomal subunit leads to the formation of functional initiation complexes (14).
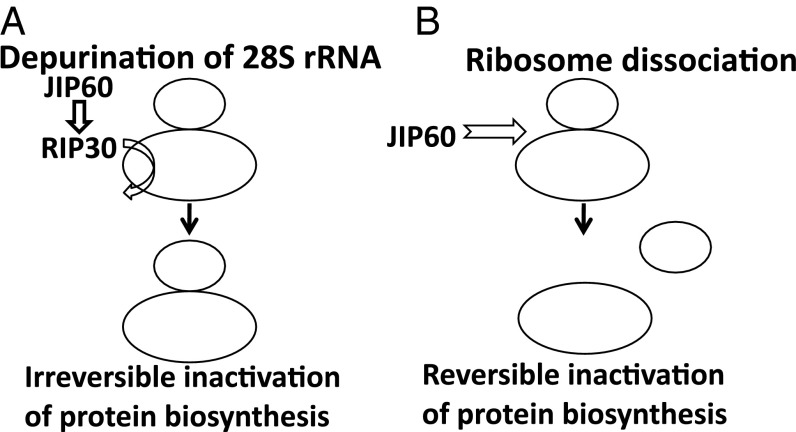
Two models have been proposed to explain JIP60’s role in translation (12, 13) (Fig. 1). The first model suggests JIP60 to be proteolytically processed and its released RIP domain to act as N-glycosidase (12). Classical RIPs catalytically cleave a conserved adenine residue in the α-sarcin/ricin loop of the 28S rRNA of 80S ribosomes and thereby impair binding of elongation factor (EF)2 such that translation is irreversibly inhibited (see refs. 16–18 for reviews). The alternative model assigns JIP60 a role as ribosome dissociation factor (13). Without further processing, JIP60 is supposed to bind to, but not cleave, the highly conserved α-sarcin/ricin loop in the 28S rRNA (13). As a consequence, JIP60 dissociates 80S ribosomes into their large (60S) and small (40S) ribosomal subunits and thereby reversibly inhibits cytoplasmic protein synthesis (13). Both scenarios would have strikingly different effects on cell viability and would lead to either cell death or stress survival (Fig. 1).
Fig. 1.
Models of the action of JIP60 and its RIP30 domain. (A) After processing of JIP60, the released RIP30 is considered to act as N-glycosidase and to catalytically remove a conserved adenine residue in the α-sarcin/ricin loop of 28S rRNA, leading to an irreversible arrest of translational elongation in the cytosol. (B) JIP60 itself and without further processing is thought to bind to 80S ribosomes and cleave them into their 60S and 40S ribosomal subunits. This effect is dose-dependent and would lead to a reversible block in protein synthesis.
Functional studies on the role of JIP60 and its eIF4E and RIP domains in planta have thus far only been reported by Görschen and coworkers, who expressed the cDNA for barley JIP60 in transgenic tobacco and barley plants (19–21). Western blot analyses suggested processing of JIP60 into an ∼30-kDa fragment in the generated transgenic tobacco plants (19). However, no results were presented whether the ∼30-kDa fragment represents the NH2-terminal RIP or COOH-terminal eIF4E domains of JIP60, thus not permitting drawing definite conclusions on the role of JIP60 and its two domains in planta. On the other hand, JIP60- and RIP-related genes are not ubiquitously distributed in the plant kingdom, and seem to occur only in monocots (Fig. S1). In the model plant Arabidopsis thaliana, for example, no JIP60- and RIP-related genes could be found in the genome (Fig. S1).
In the present work, we mapped JIP60-related genes in the barley genome and discovered two closely related forms. The gene on chromosome 4H most likely encodes the JIP60 mRNA identified previously (clone X66376) (6). For the latter, a functional analysis was carried out using in vivo and in vitro approaches to pinpoint the function of JIP60 in barley. Evidence is provided for a unique mechanism of resistance activation to biotic and abiotic stresses in which JIP60 and its eIF4E domain, but not RIP domain, are involved. Because JIP60 and JIP60-like genes underlie QTLs for biotic and abiotic stresses, detecting and validating associations between novel JIP60 alleles and the traits of interest will find implications in future marker-assisted breeding efforts.
Results
Chromosomal Assignment and Comparative Mapping of JIP60 Genes.
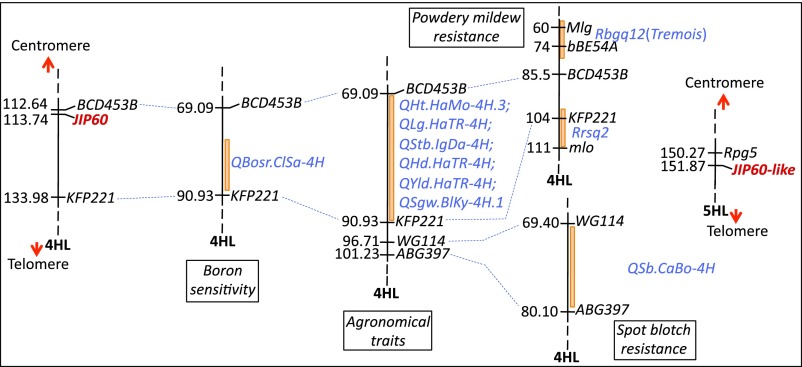
No efforts have been made to map the JIP60 gene since its first description (6). Availability of genetically anchored barley genomic DNA sequences provided the unique opportunity to perform a whole-genome survey for similar sequences and to assign virtually any gene to its respective location in the genome (22). The similarity search of barley genomic DNA sequences with the JIP60 cDNA sequence (X66376.1) identified two copies of the JIP60 gene in the barley genome, one copy on the long arm of chromosome 4H at 113.74 cM and the other copy on the long arm of chromosome 5H at 151.87 cM (Fig. 2). The former gene exhibited 97% similarity to the cDNA sequence, and hence has been designated JIP60. The latter gene, which showed 86% similarity, was called JIP60-like. The observed sequence differences between both JIP60 genes and the cDNA may be due to varietal differences (single-nucleotide polymorphisms or insertions/deletions) and sequencing errors.
Fig. 2.
Chromosome assignment of JIP60 and JIP60-like genes on the long arms of barley chromosome 4H and chromosome 5H, respectively. Genes and QTLs were placed onto the barley consensus map developed by the International Barley Genome Sequencing Consortium as indicated. The maps are not drawn to scale (for details, see SI Materials and Methods).
For comparative mapping, CMap, a web-based application for map alignment and display, was used to order different barley maps (http://wheat.pw.usda.gov/cmap). These maps were developed using various mapping populations, and earlier were used to assign genes and quantitative trait loci (QTLs) for a number of agronomical and stress-related traits (see SI Materials and Methods for details). Common DNA markers identified as flanking the JIP60 gene on the genetically anchored physical map of barley were used to align different maps. As a result, several QTLs mapping close to the JIP60 region on chromosome arm 4HL were identified (Fig. 2). Among them, the QTLs for boron sensitivity, resistance to powdery mildew (presence of resistance genes Mlg and mlo), and resistance to spot blotch caused by Bipolaris sorokiniana are noteworthy (Fig. 2).
Virtual Transcript Profiling of JIP60 Genes.
Both JIP60 genes showed close similarities to expressed sequence tags (ESTs), and thus it could be inferred that these genes are transcriptionally active. Furthermore, the virtual expression profiling of the JIP60 gene based on the abundance of homologous ESTs in tissue-specific cDNA libraries suggested highest JIP60 expression in the culm, followed by root, pericarp, leaf, and seed (Fig. S2A). A more detailed expression analysis was performed using the Genevestigator tool (23) on the available microarray (22K Barley1 GeneChip; Affymetrix; Array content was derived from 350,000 high-quality ESTs, and 1,145 barley gene sequences from the National Center for Biotechnology Information nonredundant database) data obtained under defined experimental conditions from various tissue samples. The analysis suggested highest expression levels for the JIP60 gene in radicle and mesocotyl, followed by leaf blade and pericarp (Fig. S2B). In terms of the developmental stage, JIP60 expression peaked at the time of germination (Fig. S2C). Previous studies also showed abundance of this and other JIPs in germinating barley embryos (24).
Processing of JIP60 in Vivo and in Vitro.
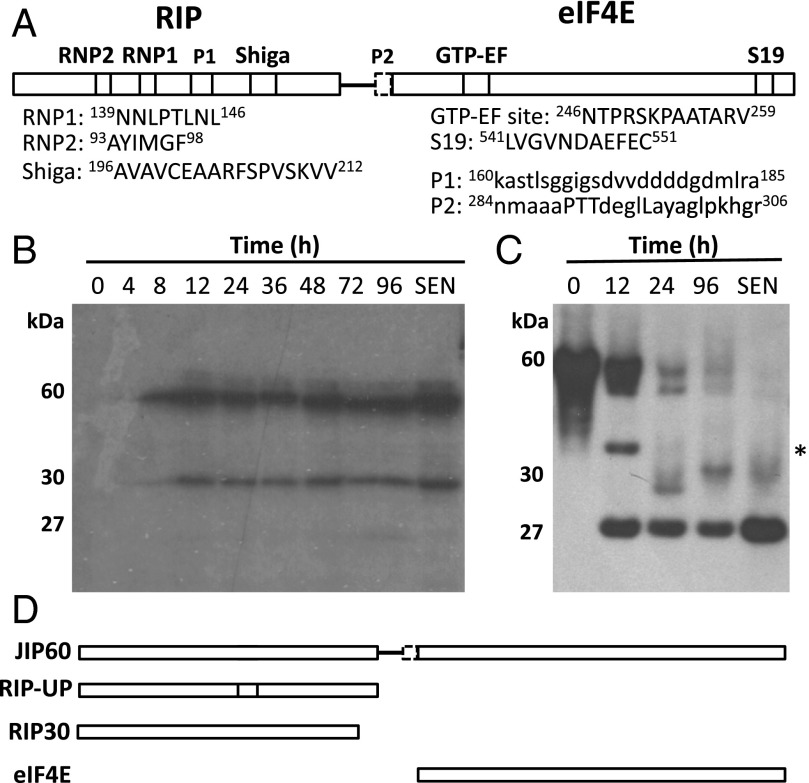
JIP60 shares with other RNA-binding proteins the presence of at least two ribonucleoprotein (RNP) recognition motifs, designated RNP1 and RNP2 (Fig. 3A) (12). In addition, JIP60’s NH2-terminal domain possesses a putative internal processing site, designated P1, as well as the Shiga signature defining the catalytic site of bacterial Shiga toxins and plant RIPs. The COOH-terminal eIF4E domain contains the [(LIVG)GXX(LF)GEXXT] signature of ribosomal protein S19 of the prokaryotic 30S ribosomal subunit (Leu541–Val551) and a [DX4EX3(GC)XT(IV)] site that is similar to the GTP binding site (GTP-EF) of the translation elongation factors (12). The latter motif is flanked by a putative second processing site (P2), linking a hinge region to the NH2-terminal RIP domain (12).
Fig. 3.
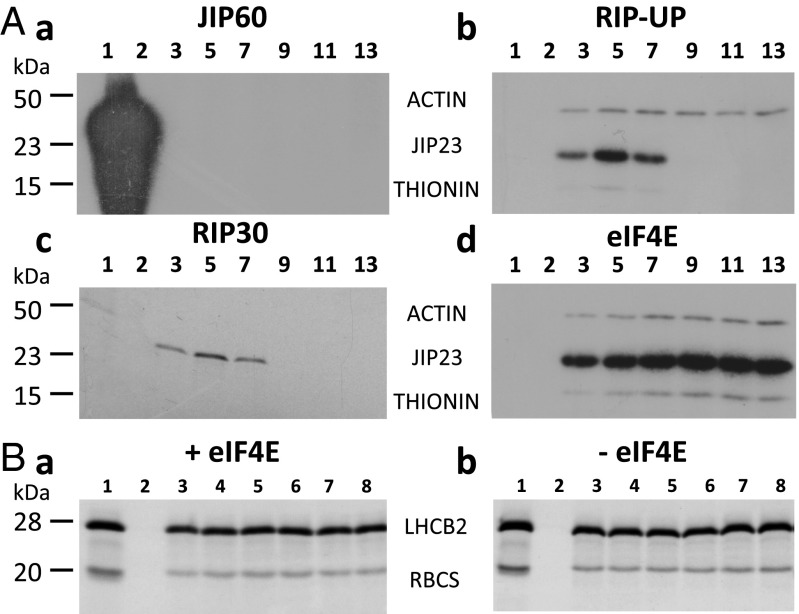
Domain structure and processing of JIP60 in vivo and in vitro. (A) Schematic presentation of JIP60’s overall structure comprising the RIP and eIF4E domains as well as putative processing sites. (B) JIP60 processing in leaf extracts from 7-d-old plants that had been treated with MeJA for the indicated periods and from 56-d-old senescent plants (SEN). Shown are Western blot analyses carried out with antiserum against JIP60. (C) JIP60 processing in vitro using wheat germ-translated, [35S]methionine-labeled JIP60 protein and an aliquot of leaf extract from plants that had been treated with MeJA for 96 h. Processing products were detected by SDS/PAGE and autoradiography. Molecular sizes of JIP60 and its processed domains are indicated. The asterisk marks a presumed processing intermediate of JIP60 that may be identical to the unprocessed RIP domain. (D) Representation of the JIP60 derivatives generated in the present work. eIF4E, COOH-terminal eIF4E domain lacking peptide P2; RIP30, processed NH2-terminal ribosome-inactivating protein domain lacking peptide P1; RIP-UP, unprocessed NH2-terminal ribosome-inactivating protein domain containing peptide P1.
Chaudhry et al. (12) have provided direct evidence for the processing of JIP60 in vivo. To readdress these previous observations, Western blot analyses were carried out with total leaf extracts and the JIP60 antiserum described by Hermann et al. (25). In a time-course experiment, leaf extracts were prepared from 7-d-old plants that had been kept under controlled conditions or treated with MeJA for variable periods. For comparison, 56-d-old senescent plants were used. Results in Fig. 3B underscored the processing of JIP60 in vivo. In addition to JIP60, which remained detectable over the time course of MeJA treatment and also accumulated in 56-d-old senescent plants, a major, ∼30-kDa band and a much weaker, ∼27-kDa band were found. Protein sequencing (26, 27) showed that the ∼30-kDa band is identical to the COOH-terminal eIF4E domain of JIP60, whereas the faint ∼27-kDa band was identical to the NH2-terminal RIP30 domain of JIP60 (Fig. S3). Together, these results suggested processing of JIP60 to occur both in MeJA-treated and senescent plants.
To back up this conclusion, JIP60 was synthesized by coupled in vitro transcription/translation and processing reconstituted with leaf extracts from MeJA-treated and senescent plants. Fig. 3C shows a time course of JIP60 processing with leaf extract from barley plants that had been exposed to MeJA for 96 h. For comparison, the processing of JIP60 was tested with leaf extract from 56-d-old senescent barley plants (Fig. 3C, lane SEN). In both cases, eIF4E and RIP30 accumulated; however, the abundance of RIP30 was much higher than that of eIF4E (Fig. 3C). This finding indicated that RIP30 has a much higher turnover than eIF4E in planta.
Production of JIP60 Derivatives and Tests of Their Biological Activities.
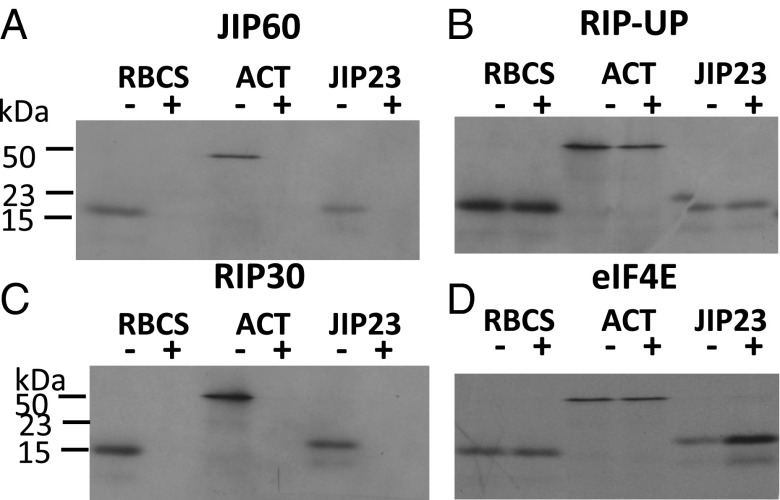
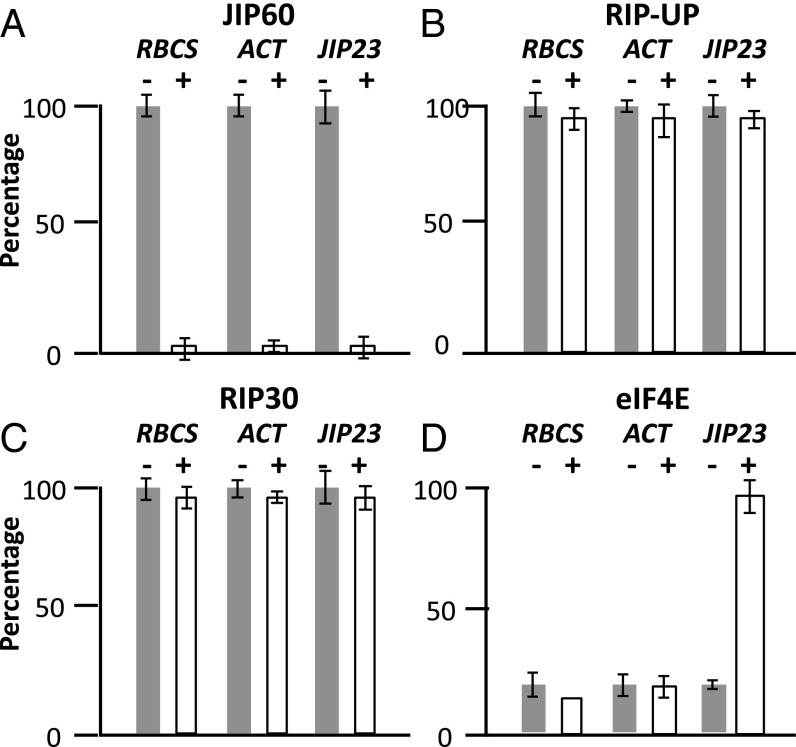
A polymerase chain reaction (PCR)-based approach (28) was used to generate cDNAs encoding the eIF4E and RIP30 domains containing amino-terminal or carboxyl-terminal hexahistidine [(His)6] tags (Fig. 3D). For comparison, cDNAs were constructed for the predicted unprocessed RIP30 (RIP-UP) and the full-length JIP60 (Fig. 3D). The final cDNAs were subcloned into the pQE30 vector and expressed in Escherichia coli. Protein was purified to apparent homogeneity by affinity chromatography on Ni-NTA agarose. Then, equimolar amounts were added to rabbit reticulocyte lysate that had been programmed with model transcripts such as RBCS (ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit) (3) and ACTIN (29), encoding photosynthetic and housekeeping proteins, respectively. In a parallel assay, the mRNA for JIP23 (6) was used. In vitro protein synthesis was conducted in the presence of [35S]methionine. Equimolar amounts of the produced JIP60, RIP-UP, RIP30, and eIF4E proteins were added to the translation mixture, and the levels of translated RBCS, ACTIN, and JIP23 proteins were determined by SDS polyacrylamide gel electrophoresis (SDS/PAGE) and autoradiography. Whereas the full-length JIP60 and RIP30 proteins completely abolished translation of all three model proteins, RIP-UP was largely inactive (Fig. 4). The eIF4E protein did not visibly affect RBCS and ACTIN translation but promoted JIP23 synthesis (Fig. 4).
Fig. 4.
Effect of JIP60 (A) and its RIP-UP (B), RIP30 (C), and eIF4E (D) domains on translation of RBCS, ACTIN, and JIP23 transcripts. A rabbit reticulocyte lysate translation system was programmed with RBCS, ACTIN (ACT), and JIP23 model transcripts and supplemented with (+) or without (−) JIP60 and its derivatives as indicated. After translation, the amount of [35S]methionine-labeled proteins was determined by SDS/PAGE and autoradiography. Molecular sizes of RBCS, ACTIN, and JIP23 are indicated.
In principle, differences in the rate of translation can be explained by alterations in the rates of initiation, elongation, and termination (30). To illuminate the role of JIP60 and its eIF4E as well as its RIP-UP and RIP30 domains, polysomes were recovered by Mg precipitation from rabbit reticulocyte lysates translating the RBCS, ACTIN, and JIP23 model transcripts and subjected to centrifugation on discontinuous step gradients of sucrose. Then, the amounts of polysome-bound versus unbound/free transcripts were determined by Northern hybridization using gene-specific probes or Western blotting with antiserum against (i) ACTIN from soybean (29) (Fig. S4) or (ii) RBCS and JIP23 from barley (3, 8).
Fig. 5 summarizes the results of the Northern blot hybridizations. They revealed a drastic depression of ribosome binding of the RBCS, ACTIN, and JIP23 transcripts in the case of the JIP60-incubated assays. All three transcripts were largely depleted from the polysomal fraction (Fig. 5). For RIP30 and its unprocessed form, no drastic reductions in the binding of RBCS, ACTIN, and JIP23 transcripts to polysomes were observed. For the eIF4E-supplemented assays, four- to fivefold stimulations of JIP23 transcript binding to polysomes were found in three independent experiments (Fig. 5), consistent with the detected increase in the incorporation of [35S]methionine into JIP23 protein (Fig. 4).
Fig. 5.
Polysome binding of RBCS, ACTIN, and JIP23 transcripts in rabbit reticulocyte lysates translating the RBCS, ACTIN, or JIP23 model transcripts. The translation mixtures were supplemented with either JIP60 (A) or its RIP-UP (B), RIP30 (C), and eIF4E (D) domains (+) or buffer (−). Then, polysomes were Mg2+-precipitated, and RNA present in the pellet and supernatant fractions was extracted with phenol and precipitated with ethanol. Quantification of mRNAs was made by Northern hybridization with gene-specific probes. Percentages refer to total transcript levels found in incubation mixtures lacking JIP60, RIP-UP, RIP30, and eIF4E, respectively, set as 100. Error bars represent the mean ± SD of three independent experiments.
We next simultaneously programmed the reticulocyte lysate with the RBCS, ACTIN, and JIP23 messengers and also included transcripts for THIONIN and LHCB2 (light-harvesting chlorophyll a/b-binding protein 2) as examples of JIPs and photosynthetic proteins, respectively. Messenger recruitment by ribosomes was assessed by sucrose density gradient centrifugation. In each case, the amount of polysome-bound transcript was deduced from the amount of protein synthesized in the different polysomal fractions obtained after centrifugation. This approach allowed scoring both the amount and functionality of each polysome-bound messenger. Whereas accumulation of ACTIN, JIP23, and THIONIN was simultaneously followed on the same Western blots, that of the similarly sized JIP23 (23 kDa), RBCS (21 kDa), and LHCB2 (28 kDa) was followed on replicate Western blots.
Incubation of rabbit reticulocyte lysates translating the RBCS, ACTIN, JIP23, THIONIN, and LHCB2 transcripts with JIP60 caused ribosome dissociation (Fig. 6A, a). This is evident from the colocalization of the ACTIN, JIP23, and THIONIN transcripts in 40S ribosomal subunits. By contrast, no comparable effect was detectable for the RIP-UP–incubated sample (Fig. 6A, b). For the RIP30-treated sample, a drastic reduction in synthesis of JIP23 and the other model proteins was observed. Because the distribution of JIP23 transcript across the sucrose gradient separating the RNP material did not change significantly in the presence or absence of RIP30 (Fig. 6A, c and Fig. S5), we concluded that RIP30 inhibited translation elongation (30). This result was in agreement with the previously documented inhibitory effect of RIPs on translation elongation (summarized in refs. 16–18). For the eIF4E-supplemented assays, an increase of JIP23 and THIONIN transcript binding to polysomes was observed (Fig. 6A, d). By contrast, no comparable effect was found for ACTIN (Fig. 6A, d) as well as the RBCS and LHCB2 model transcripts in the eIF4E-incubated assays (Fig. 6B, a and b). Furthermore, calculation of the ribosome:polysome ratios (P/T values) and rates of protein synthesis (Table S1) confirmed that the full-length JIP60 protein lowered both parameters, and thus exerted its effect by dissociating 80S polysomes into their ribosomal subunits. By contrast, the RIP30 domain operated as a classical RIP and inhibited translation at the elongation step, as evidenced by the only moderate decrease in the P/T value but tremendous reduction in protein synthesis (Table S1). Last but not least, the eIF4E domain stimulated translation initiation at JIP transcripts and thereby caused higher rates of JIP23 protein synthesis (Table S1).
Fig. 6.
Ribosome binding of ACTIN, JIP23, THIONIN, RBCS, and LHCB2 transcripts in rabbit reticulocyte lysates supplemented with JIP60, RIP-UP, RIP30, and eIF4E. (A) Synthesis of ACTIN, JIP23, and THIONIN proteins in JIP60- (a), RIP-UP– (b), RIP30- (c), and eIF4E-incubated (d) reticulocyte lysate. Fraction 1 obtained after sucrose gradient centrifugation contains the 40S ribosomal subunit, fraction 2 contains the 60S ribosomal subunit, and fractions 3–8 contain cytoplasmic polysomes of increasing size. Protein was detected by Western blotting using respective monospecific antisera. Note that, to detect minor traces of polysome-bound messengers, 50-fold–higher levels of RNP material were loaded onto the sucrose gradient in a compared with b–d. (B) Synthesis of RBCS and LHCB2 in eIF4-incubated (a) and mock-incubated (b) reticulocyte lysates, as assessed by Western blotting.
Identification of eIF4E-Bound Transcripts in Vivo.
The processing pattern of JIP60 shown in Fig. 3 suggested a dynamic equilibrium of ribosome dissociation and reengagement in which JIP60 and its eIF4E domain are involved. To identify mRNAs that are bound to either protein, monospecific antisera were raised against JIP60 and its eIF4E domain (Fig. S6) and used for carrying out coimmunoprecipitations. For comparison, an antiserum against human eIF4E was used. mRNAs bound to JIP60 or eIF4E were extracted with phenol/chloroform, purified by oligo-dT cellulose chromatography, and used for Northern hybridization with gene-specific probes. The latter included probes for (i) JIPs, such as JIP60 (12, 13), allene oxide synthase 1 (AOS1) and AOS2 (collectively referred to as AOS in Fig. 7) (10), allene oxide cyclase (AOC) (9), and JIP23 (6); (ii) photosynthetic proteins, such as RBCS and LHCB2 (3); and (iii) housekeeping proteins, such as ACTIN and TUBULIN (3, 29). Moreover, an aliquot of the purified mRNA was translated into [35S]methionine-labeled protein using a cell-free wheat germ system. In addition to the antiserum against the JIP60-derived, plant-type eIF4E, a human-type eIF4E antibody was used.
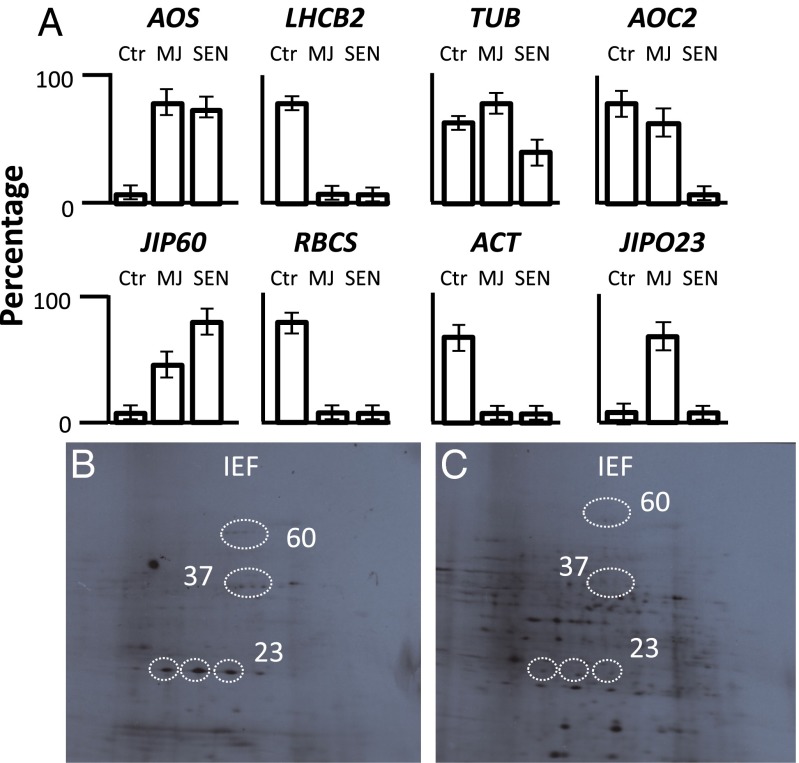
Fig. 7.
Identification of eIF4E-bound RNAs in vivo. Leaf extracts were prepared from 7-d-old freshly harvested barley plants (Ctr; control), 7-d-old barley plants that had been treated with MeJA for 96 h (MJ), and 56-d-old senescent plants (SEN) and subjected to immunoprecipitation using antisera against eIF4E of human (samples from control plants) or the eIF4E domain of JIP60 (samples from MJ and SEN plants). RNA was extracted from the precipitated RNP material with phenol, precipitated with ethanol, and used for either Northern hybridization (A) or in vitro translation (B and C) in a wheat germ extract. (A) Protein synthesis from eIF4E-bound transcripts in control plants versus MeJA-treated and senescent plants. Percentages refer to the ratio of eIF4E-bound messengers to total messengers (eIF4E-bound plus free). Error bars represent the mean ± SD of three independent experiments. (B and C) Two-dimensional patterns including isolelectric focusing (IEF) and SDS-PAGE (from top to bottom) of messenger products translated from the eIF4E-precipitated fractions of MeJA-treated (B) and senescent (C) plants. Circles mark the positions of JIP23, JIP37, and JIP60 messenger products.
Fig. 7A (lane Ctr) indicates that the human eIF4E antiserum coprecipitated the TUBULIN, ACTIN, LHCB2, and RBCS mRNAs from leaf extracts of freshly harvested plants. With the plant-specific eIF4E antiserum, substantial amounts of RNA were recovered from leaf extracts of plants that had been treated with MeJA for 96 h (Fig. 7A, lane MJ). This RNA included mRNAs for JIPs such as AOS, JIP60, AOC, and JIP23 but not mRNAs for photosynthetic proteins such as LHCB2 and RBCS. With leaf extracts from 56-d-old senescent plants, similar results were obtained although only AOS and JIP60 were among the coprecipitated mRNAs (Fig. 7A, lane SEN). Two-dimensional SDS/PAGE of total messenger products translated in wheat germ extracts underscored the presence of only a few common mRNAs in MeJA-treated and senescent plants (Fig. 7 B and C).
Discussion
Despite considerable progress made over the last few years, still not much is known about the nature of QTLs for biotic and abiotic stress resistance and/or other agronomically relevant traits. In the present work, we have identified with JIP60 a putative candidate for such QTLs in barley.
JIP60 is synthesized as an ∼60-kDa protein in planta and is first operative as a ribosome-dissociation factor (Fig. S7). In time-course experiments, this activity was first detectable ∼24 h after the onset of MeJA treatment, where an as-yet unknown mechanism destines 80S ribosomes for disassembly into 40S and 60S ribosomal subunits by the induced JIP60 protein (13). Superimposed on this activity is the processing of JIP60 (ref. 12 and data shown in this work). JIP60’s eIF4E domain then functioned in recruiting stress messengers for translation (Fig. S7). We assume that the released eIF4E of JIP60 bound the 5′ cap structure of capped mRNAs and to the initiation factor eIF4G (14, 15). Together, these effects permitted a rapid reprogramming of translation. Pull-down assays identified JIP60, AOS, AOC, and JIP23 transcripts to be bound to the released eIF4E domain of JIP60 in MeJA-treated plants. By contrast, only a few common messenger products were observed between MeJA-treated and senescent plants, pointing to major distinctions between both senescence types (31, 32).
Alignment of the JIP60 gene on chromosome arm 4HL revealed the presence of several overlapping QTLs, including those for boron sensitivity, plant height, lodging, stem breaking, days to heading, yield, seed weight, days to maturity, and powdery mildew resistance (33–37) (Fig. 2). Two powdery mildew resistance genes, Mlg and mlo, were identified to map proximally and distally to the JIP60-containing region on chromosome 4HL, respectively (38). Moreover, a QTL for spot blotch (B. sorokiniana) resistance was also identified in the region distal to the JIP60 map location (39). Similarly, the JIP60-like gene mapped distally to the Rpg5 locus on chromosome 5HL (40) (Fig. 2).
Interestingly, in an extensive screen of 1,407 spring wheat genotypes, Joshi et al. (41) found an association between the “stay green” trait (delayed senescence) and spot blotch resistance, which indicated that the two traits are coregulated, and selection can be performed simultaneously for resistance to biotic stress and late senescence. This observation also explains colocalization of JIP60 with the QTL for spot blotch resistance.
Biological significance of the colocalization of the JIP60 or JIP60-like gene with the QTLs for a number of agronomically important traits can also be inferred from the results of the earlier microarray/quantitative real-time PCR-based studies, where an increase in transcription levels of JIP23, JIP37, and JIP60 was observed in potassium-starved barley plants (42). Similar induction of JA-related genes was observed as a late response to boron toxicity in barley (43). Dehydration shock and drought stress treatments were also reported to induce expression of JIPs (44–46). Together, these results identify JA as a “master switch” for stress signaling leading to changes in gene expression (Fig. S7) and explain why QTLs for a variety of traits map in the proximity of both JIP60 and the JIP60-like gene.
A minimal effective period of photosynthesis is essential for maintaining grain yield in crop plants (47). In fact, under optimal growth conditions, delaying the onset of senescence was shown to be positively correlated with grain yield (48). However, developmentally programmed senescence is essential for remobilization of reserves from source to sink organs. The stay green mutations identified in different crop plants are good examples where yield advantage was reported to be a consequence of delays in senescence (47). However, environmental stresses often trigger early (premature) senescence, which has detrimental effects on grain yield and quality. Thus, it would be beneficial to generate crop plants that are more tolerant to stress and delay the onset of premature senescence. Because JIP60 is involved in both processes and controls translational reprogramming during biotic and abiotic stress as well as leaf senescence (Fig. S7), revealing its exact mechanism of action will have implications for future breeding efforts by engineering or identifying novel JIP60 alleles.
Materials and Methods
Plant Materials.
Seeds of barley (Hordeum vulgare L. cv. Salomé) were germinated on vermiculite at 25 °C under continuous white light illumination provided by fluorescent bulbs (30 W/m2). For treatment with MeJA, 7-d-old primary leaves were cut from the seedlings and placed either onto water or on an aqueous solution of 45 μM MeJA (Firmenich). As a control, leaves from 56-d-old naturally senescing barley plants were used.
Production of JIP60 Derivatives.
JIP60 derivatives were produced by a PCR-based approach (28) essentially as described by Chaudhry et al. (12) using the cDNA described by Becker and Apel (6). Respective clones were subcloned into appropriate expression vectors and sequenced. Protein expression was done in E. coli, giving rise to NH2- or COOH-terminal (His)6-tagged protein variants.
Analysis of Polysomal Messengers and Proteins.
RNA was extracted from the different fractions obtained after sucrose density gradient centrifugation by phenol/chloroform/isoamyl alcohol extraction and analyzed further by in vitro translation and Northern hybridization (3). For further details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank F. Schaller (Ruhr-Universität Bochum) for technical assistance. We are indebted to K. Apel and J. Lehmann for gifts of cDNAs and antisera against THIONIN, LHCB2, JIP23, and JIP60. This work was supported by the Deutsche Forschungsgemeinschaft Bonn (RE 1387/3-1 and 3-2 and FOR222/3-1) and the Life Sciences Discovery Fund (3143956).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415690111/-/DCSupplemental.
References
- 1.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 2007;100(4):681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009;276(17):4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- 3.Reinbothe S, Reinbothe C, Parthier B. Methyl jasmonate-regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv. Salome) J Biol Chem. 1993;268(14):10606–10611. [PubMed] [Google Scholar]
- 4.Reinbothe C, Pollmann S, Reinbothe S. Singlet oxygen signaling links photosynthesis to translation and plant growth. Trends Plant Sci. 2010;15(9):499–506. doi: 10.1016/j.tplants.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Andresen I, et al. The identification of leaf thionin as one of the main jasmonate-induced proteins of barley (Hordeum vulgare) Plant Mol Biol. 1992;19(2):193–204. doi: 10.1007/BF00027341. [DOI] [PubMed] [Google Scholar]
- 6.Becker W, Apel K. Isolation and characterization of a cDNA clone encoding a novel jasmonate-induced protein of barley (Hordeum vulgare L.) Plant Mol Biol. 1992;19(6):1065–1067. doi: 10.1007/BF00040538. [DOI] [PubMed] [Google Scholar]
- 7.Görschen E, Dunaeva M, Reeh I, Wasternack C. Overexpression of the jasmonate-inducible 23 kDa protein (JIP 23) from barley in transgenic tobacco leads to the repression of leaf proteins. FEBS Lett. 1997;419(1):58–62. doi: 10.1016/s0014-5793(97)01433-6. [DOI] [PubMed] [Google Scholar]
- 8.Hause B, Demus U, Teichmann C, Parthier B, Wasternack C. Developmental and tissue-specific expression of JIP-23, a jasmonate-inducible protein of barley. Plant Cell Physiol. 1996;37(5):641–649. doi: 10.1093/oxfordjournals.pcp.a028993. [DOI] [PubMed] [Google Scholar]
- 9.Maucher H, et al. The allene oxide cyclase of barley (Hordeum vulgare L.)—Cloning and organ-specific expression. Phytochemistry. 2004;65(7):801–811. doi: 10.1016/j.phytochem.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome): Tissue specific regulation in seedling development. Plant J. 2000;21(2):199–213. doi: 10.1046/j.1365-313x.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- 11.Vörös K, et al. Characterization of a methyljasmonate-inducible lipoxygenase from barley (Hordeum vulgare cv. Salome) leaves. Eur J Biochem. 1998;251(1-2):36–44. doi: 10.1046/j.1432-1327.1998.2510036.x. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhry B, et al. The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome-inactivating protein. Plant J. 1994;6(6):815–824. doi: 10.1046/j.1365-313x.1994.6060815.x. [DOI] [PubMed] [Google Scholar]
- 13.Reinbothe S, et al. JIP60, a methyl jasmonate-induced ribosome-inactivating protein involved in plant stress reactions. Proc Natl Acad Sci USA. 1994;91(15):7012–7016. doi: 10.1073/pnas.91.15.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhoads RE. eIF4E: New family members, new binding partners, new roles. J Biol Chem. 2009;284(25):16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi B, Lee K, Maeder DL, Jagus R. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol. 2005;5:48. doi: 10.1186/1471-2148-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girbés T, Ferreras JM, Arias F-J, Stirpe F. Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini Rev Med Chem. 2004;4(5):461–476. doi: 10.2174/1389557043403891. [DOI] [PubMed] [Google Scholar]
- 17.Hartley MR, Lord JM. Cytotoxic ribosome-inactivating lectins from plants. Biochim Biophys Acta. 2004;1701(1-2):1–14. doi: 10.1016/j.bbapap.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Stirpe F, Battelli MG. Ribosome-inactivating proteins: Progress and problems. Cell Mol Life Sci. 2006;63(16):1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Görschen E, et al. Expression of the ribosome-inactivating protein JIP60 from barley in transgenic tobacco leads to an abnormal phenotype and alterations on the level of translation. Planta. 1997;202(4):470–478. doi: 10.1007/s004250050151. [DOI] [PubMed] [Google Scholar]
- 20.Dunaeva M, Goerschen E. RIP-JIP60 alters conformation of ribosomes in vivo. Biochem Biophys Res Commun. 1999;258(3):572–573. doi: 10.1006/bbrc.1999.0685. [DOI] [PubMed] [Google Scholar]
- 21.Dunaeva M, Goebel C, Wasternack C, Parthier B, Goerschen E. The jasmonate-induced 60 kDa protein of barley exhibits N-glycosidase activity in vivo. FEBS Lett. 1999;452(3):263–266. doi: 10.1016/s0014-5793(99)00645-6. [DOI] [PubMed] [Google Scholar]
- 22.Mayer KF, et al. International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 23.Hruz T, et al. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson L, Henry RJ. Microarray analysis of gene expression in germinating barley embryos (Hordeum vulgare L.) Funct Integr Genomics. 2005;5(3):155–162. doi: 10.1007/s10142-005-0133-6. [DOI] [PubMed] [Google Scholar]
- 25.Hermann G, et al. Species and tissue specificity of jasmonate-induced abundant proteins. J Plant Physiol. 1989;134(6):703–709. [Google Scholar]
- 26.Chang J-Y, Brauer D, Wittmann-Liebold B. Manual micro-sequence analysis of peptides and proteins using 4-N,N-dimethylaminoazobenzene 4′isothio-cyanate/phenylisothiocyanate double coupling method. FEBS Lett. 1978;93:205–214. [Google Scholar]
- 27.Chang J-Y. Manual micro-sequence analysis of polypeptides using dimethylaminoazobenzene isothiocyanate. Methods Enzymol. 1983;91:455–466. doi: 10.1016/s0076-6879(83)91043-1. [DOI] [PubMed] [Google Scholar]
- 28.Innis MA, Gelfand DH, Sninsky JJ, White TJ. PCR Protocols. San Diego: Academic; 1990. [Google Scholar]
- 29.Shah DM, Hightower RC, Meagher RB. Complete nucleotide sequence of a soybean actin gene. Proc Natl Acad Sci USA. 1982;79(4):1022–1026. doi: 10.1073/pnas.79.4.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassart G, Dumont JE, Cantraine FR. Translational control of protein synthesis: A simulation study. Biochim Biophys Acta. 1971;247(3):471–485. doi: 10.1016/0005-2787(71)90034-7. [DOI] [PubMed] [Google Scholar]
- 31.Reinbothe C, Reinbothe S. 2005. Regulation of photosynthetic gene expression by the environment: From seedling de-etiolation to leaf senescence. Photoprotection, Photoinhibition, Gene Regulation, and Environment, eds Demmig-Adams B, Adams WW, Mattoo AK, Advances in Photosynthesis and Respiration, series ed Govidjee B (Springer, Dordrecht, The Netherlands), Vol 21, pp 333–365.
- 32.Jibran R, A Hunter D, P Dijkwel P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol. 2013;82(6):547–561. doi: 10.1007/s11103-013-0043-2. [DOI] [PubMed] [Google Scholar]
- 33.Backes G, et al. Localization of quantitative trait loci (QTL) for agronomic important characters by the use of a RFLP map in barley (Hordeum vulgare L.) Theor Appl Genet. 1995;90(2):294–302. doi: 10.1007/BF00222217. [DOI] [PubMed] [Google Scholar]
- 34.Bezant J, Laurie D, Pratchett N, Chojecki J, Kearsey M. Mapping QTL controlling yield and yield components in a spring barley (Hordeum vulgare L.) cross using marker regression. Mol Breed. 1997;3(1):29–38. [Google Scholar]
- 35.Jefferies SP, et al. Mapping of chromosome regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.) Theor Appl Genet. 1999;98(8):1293–1303. [Google Scholar]
- 36.Tinker NA, et al. Regions of the genome that affect agronomic performance in two-row barley. Crop Sci. 1996;36(4):1053–1062. [Google Scholar]
- 37.Marquez-Cedillo LA, et al. QTL analysis of agronomic traits in barley based on the doubled haploid progeny of two elite North American varieties representing different germplasm groups. Theor Appl Genet. 2001;103(4):625–637. [Google Scholar]
- 38.Aghnoum R, et al. Basal host resistance of barley to powdery mildew: Connecting quantitative trait loci and candidate genes. Mol Plant Microbe Interact. 2010;23(1):91–102. doi: 10.1094/MPMI-23-1-0091. [DOI] [PubMed] [Google Scholar]
- 39.Bilgic H, Steffenson BJ, Hayes PM. Molecular mapping of loci conferring resistance to different pathotypes of the spot blotch pathogen in barley. Phytopathology. 2006;96(7):699–708. doi: 10.1094/PHYTO-96-0699. [DOI] [PubMed] [Google Scholar]
- 40.Brueggeman R, et al. The stem rust resistance gene Rpg5 encodes a protein with nucleotide-binding-site, leucine-rich, and protein kinase domains. Proc Natl Acad Sci USA. 2008;105(39):14970–14975. doi: 10.1073/pnas.0807270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi AK, et al. Stay green trait: Variation, inheritance and its association with spot blotch resistance in spring wheat (Triticum aestivum L.) Euphytica. 2007;153(1-2):59–71. [Google Scholar]
- 42.Davis J, Armengaud P, Newton A, White P, Amtmann A. 2009. Potassium deficiency and JA-dependent responses to biotic stress in barley. Univ of Glasgow Post Graduate Symposium, June 9, 2009, Glasgow, United Kingdom.
- 43.Öz MT, et al. Microarray analysis of late response to boron toxicity in barley (Hordeum vulgare L.) leaves. Turk J Agric For. 2009;33:191–202. [Google Scholar]
- 44.Wierstra I, Kloppstech K. Differential effects of methyl jasmonate on the expression of the early light-inducible proteins and other light-regulated genes in barley. Plant Physiol. 2000;124(2):833–844. doi: 10.1104/pp.124.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreenivasulu N, Varshney RK, Kavi Kishor PB, Weschke W. Functional genomics for tolerance to abiotic stress in cereals. In: Gupta PK, Varshney RK, editors. Cereal Genomics. Dordrecht, The Netherlands: Kluwer Academic; 2004. pp. 483–514. [Google Scholar]
- 46.Walia H, et al. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics. 2006;6(2):143–156. doi: 10.1007/s10142-005-0013-0. [DOI] [PubMed] [Google Scholar]
- 47.Penfold CA, Buchanan-Wollaston V. Modelling transcriptional networks in leaf senescence. J Exp Bot. 2014;65(14):3859–3873. doi: 10.1093/jxb/eru054. [DOI] [PubMed] [Google Scholar]
- 48.Distelfeld A, Avni R, Fischer AM. Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot. 2014;65(14):3783–3798. doi: 10.1093/jxb/ert477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.