Significance
Proliferating cell nuclear antigen (PCNA) is a homotrimeric DNA sliding clamp that coordinates multiple DNA replication and repair processes by orchestrating the activity of various essential proteins. PCNA can bind up to three partners simultaneously, but despite extensive research, the functional significance of PCNA's trimeric structure remains unclear. We developed a novel approach for the generation of PCNA heterotrimers that contain both wild-type and mutant monomers. Using these heterotrimers, we show that PCNA can efficiently coordinate the activities of the three enzymes involved in Okazaki fragment maturation without binding them simultaneously. In contrast to the previously suggested “toolbelt” model for PCNA function, our results demonstrate sequential binding and release of partners on the PCNA trimer during complex biological processes.
Abstract
The homotrimeric sliding clamp proliferating cell nuclear antigen (PCNA) mediates Okazaki fragment maturation through tight coordination of the activities of DNA polymerase δ (Pol δ), flap endonuclease 1 (FEN1) and DNA ligase I (Lig1). Little is known regarding the mechanism of partner switching on PCNA and the involvement of PCNA's three binding sites in coordinating such processes. To shed new light on PCNA-mediated Okazaki fragment maturation, we developed a novel approach for the generation of PCNA heterotrimers containing one or two mutant monomers that are unable to bind and stimulate partners. These heterotrimers maintain the native oligomeric structure of PCNA and exhibit high stability under various conditions. Unexpectedly, we found that PCNA heterotrimers containing only one functional binding site enable Okazaki fragment maturation by efficiently coordinating the activities of Pol δ, FEN1, and Lig1. The efficiency of switching between partners on PCNA was not significantly impaired by limiting the number of available binding sites on the PCNA ring. Our results provide the first direct evidence, to our knowledge, that simultaneous binding of multiple partners to PCNA is unnecessary, and if it occurs, does not provide significant functional advantages for PCNA-mediated Okazaki fragment maturation in vitro. In contrast to the “toolbelt” model, which was demonstrated for bacterial and archaeal sliding clamps, our results suggest a mechanism of sequential switching of partners on the eukaryotic PCNA trimer during DNA replication and repair.
Proliferating cell nuclear antigen (PCNA) is a central coordinator of genome duplication and maintenance pathways in eukaryotes (1, 2). A member of the conserved sliding clamp family, PCNA is a homotrimeric ring-shaped protein that encircles DNA and serves as a processivity factor for DNA polymerases and a binding platform for many DNA modifying enzymes. PCNA interacts with partners involved in numerous processes, including DNA replication, recombination and repair, chromatin remodeling, and cell-cycle regulation. PCNA recruits these partners to replication forks or other chromosomal locations, enhances their catalytic activities, and orchestrates their cooperation in multistep enzymatic processes. Because most partners interact with the same binding site on PCNA, competition for binding must be tightly regulated during complex PCNA-mediated processes. The switching of partners on the PCNA platform has been shown to be crucial for the proper progression of multiple DNA replication and repair pathways, such as lagging strand replication, translesion synthesis, and mismatch repair (1). In recent years, several regulatory mechanisms, mostly involving posttranslational modifications of PCNA by ubiquitin or small ubiquitin-like modifier, have been shown to affect partner switching on PCNA by favoring the recruitment of specific partners (3–5).
Despite extensive research into the regulation of PCNA-mediated processes, very little is known regarding how PCNA coordinates the activity of several enzymes during sequential processes. Two simple models have been proposed to explain this coordination (1, 2, 6, 7). The first model assumes highly dynamic partner switching on PCNA due to sequential binding and release events on the same or different PCNA monomers (Fig. 1, Upper). This model predicts that a single functional binding site on the PCNA trimer should be sufficient for the coordination of the entire process. In contrast, the second model assumes simultaneous binding of two or three partners to different monomers on the PCNA trimer (Fig. 1, Lower). In this case, the partners are stably associated with PCNA, which acts as a “toolbelt” throughout the process. According to this model, only PCNA trimers with two or three functional binding sites would be able to coordinate the process.
Fig. 1.
Two possible models describing PCNA-mediated Okazaki fragment maturation. (Upper) A dynamic model in which Pol δ, FEN1, and Lig1 are bound and released from PCNA in a sequential manner. (Lower) The toolbelt model in which the three enzymes are simultaneously bound to PCNA using all available PCNA binding sites. The red segments represent the RNA primers; glowing circles represent enzymes currently active on the substrate.
One of the best studied examples of such a multipartner PCNA-mediated process is the synthesis and maturation of Okazaki fragments during lagging strand DNA replication. This process involves the sequential activity of three PCNA binding partners—DNA polymerase δ (Pol δ), flap endonuclease 1 (FEN1), and DNA ligase I (Lig1), which mediate DNA synthesis, flap cleavage, and ligation, respectively (7–9). This is a fast and efficient process that is estimated to take place ∼100,000 times during each yeast cell division with a low tolerance for errors (8). The enzymes involved must cooperate through PCNA in a tightly regulated manner, acting sequentially on the same substrate while repeatedly exchanging access to it (Fig. 1). In particular, removal of the initiator RNA requires several rapid iterative switches between Pol δ and FEN1 (7). This PCNA-dependent cooperation is particularly important to ensure that flaps will not become too long for processing by this short-flap pathway (7, 10, 11).
To directly examine the mechanism of partner switching on PCNA and the functional significance of its homotrimeric structure, we developed a novel approach for the generation of PCNA heterotrimers that contain both wild-type (WT) and mutant monomers that are unable to bind different partners. We used these heterotrimers to determine whether simultaneous binding of more than one partner to a PCNA trimer is necessary to coordinate PCNA-mediated nick translation and Okazaki fragment maturation. Contrary to the toolbelt model, our findings indicate that simultaneous binding is not required, and sequential switching of partners on a single monomer of PCNA is sufficient to coordinate Okazaki fragment maturation. Our findings suggest that PCNA can efficiently orchestrate complex processes by regulating sequential binding and release events of several partners without binding them simultaneously.
Results
Generation of PCNA Heterotrimers.
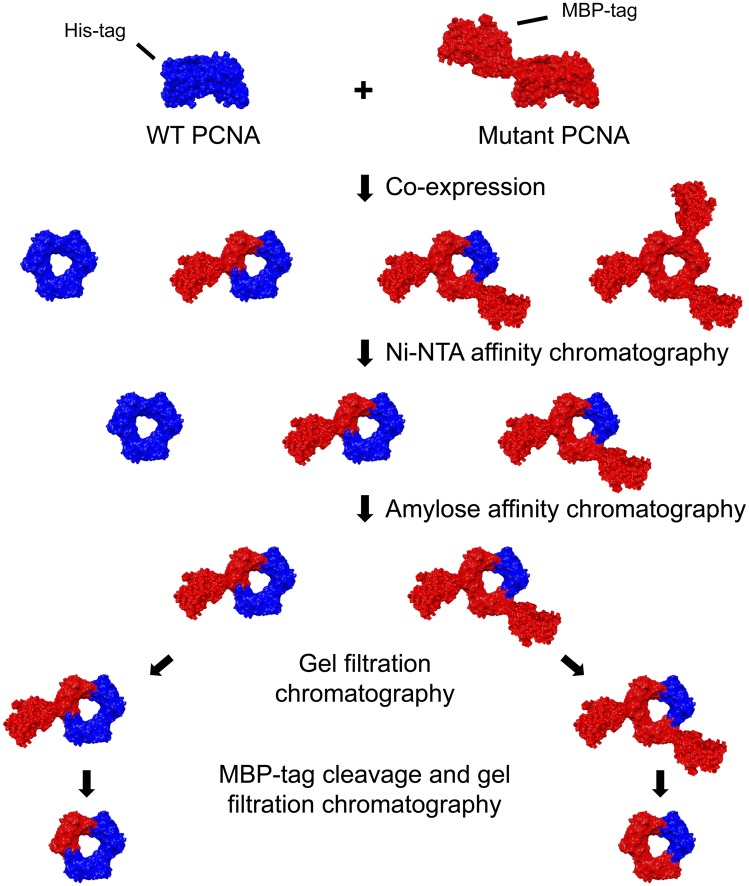
To generate and purify PCNA heterotrimers with native tertiary structure containing both WT and mutant monomers, we coexpressed both monomers in Escherichia coli fused to two affinity tags of considerably different sizes (Fig. 2). We used N-terminal His tag (<1 kDa) and maltose binding protein (MBP) tag (∼40 kDa) for WT and mutant PCNA, respectively. Due to random trimerization of PCNA during coexpression, four trimer species spontaneously form in vivo (Fig. 2). Tandem affinity purification steps using nickel-nitrilotriacetic acid (Ni-NTA) followed by amylose chromatography enabled the isolation of the two heterotrimeric species, containing at least one His tag and one MBP tag. These species were then separated by size exclusion chromatography, due to a difference of ∼40 kDa in their molecular weight (Fig. S1A). Finally, the large MBP tag was removed using site-specific tobacco etch virus (TEV) protease cleavage and the resulting MBP-free heterotrimers were isolated using a second gel filtration step. The contents of these highly purified heterotrimers were verified by SDS/PAGE (Fig. S1B) and MALDI-TOF mass spectrometry (Fig. S1C). The presence of both WT and mutant monomers in the same trimers was further validated by covalent cross-linking of neighboring monomers (Fig. S1D).
Fig. 2.
Scheme describing the PCNA heterotrimer purification strategy. WT and mutant PCNA are coexpressed in E. coli fused to 6× His- and MBP-fusion tags, respectively, leading to the spontaneous formation of four trimer species. Tandem Ni-NTA and amylose affinity chromatography steps isolate the two heterotrimeric species. These two species are then separated by gel filtration chromatography, owing to the size difference between the two fusion tags (Fig. S1A). Following site-specific cleavage of the MBP tag by TEV protease, a second gel filtration step is used to obtain pure PCNA heterotrimers. Images are schematic models for illustration purposes, created using University of California San Francisco chimera, based on Protein Data Bank entries 1plq and 1anf for PCNA and MBP, respectively.
We specifically constructed heterotrimers of Saccharomyces cerevisiae PCNA, including mutant monomers that are deficient for partner binding while retaining the ability to assemble into stable trimers. Three different mutants were used: the interdomain connector loop (IDCL) mutant pcna-79 (I126A,L128A), which is particularly deficient in stimulation of Pol δ activity (12), the C-terminal mutant pcna-90 (P252A,K253A), which is particularly deficient in stimulation of FEN1 activity (12, 13), and the double mutant, which we designate pcna-7990 (I126A,L128A,P252A,K253A), which we expect to be deficient in stimulation of both Pol δ and FEN1 activities. We denote, for example, a PCNA trimer containing two WT monomers and one monomer of pcna-7990 as WT2:79901. Whereas pcna-79 and pcna-90 are known to be efficiently loaded onto DNA by the clamp loader replication factor C (RFC) (12), we validated that this is also the case for the combined mutant pcna-7990 and heterotrimers containing this mutant, by measuring the PCNA-dependent ATPase activity of RFC in the presence of a suitable DNA effector (Fig. S2).
Heterotrimer Stability.
The PCNA heterotrimers generated using our approach are self-assembled and maintain a native monomer–monomer interface. Consequently, these heterotrimers may dissociate into monomers and randomly reassemble into different trimer species after purification. To assess the kinetics of trimer reassembly, we purified WT1:79902 heterotrimers without removing the MBP tag, incubated them at different temperatures, and examined them by gel filtration chromatography. We found that significant reassembly of the trimers is only observed following prolonged incubations at elevated temperatures (Fig. S3 A and B). Because the in vitro experiments detailed in this report are performed at a maximal temperature of 30 °C for a maximal period of ∼15 min, the extent of reassembly is expected to be negligible under the assay conditions. We also verified that the loading of PCNA onto DNA by RFC does not promote reassembly of the heterotrimers (Fig. S3C).
Activities of Pol δ and FEN1 Separately.
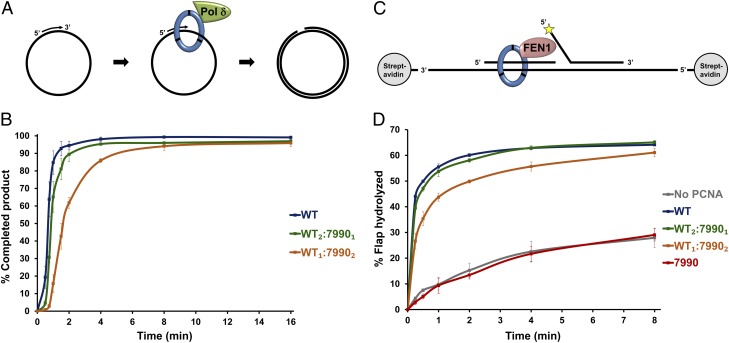
We first examined how Pol δ activity is stimulated by PCNA heterotrimers. This was measured using an in vitro replication assay, in which PCNA is loaded onto primed single-stranded DNA by RFC and ATP, and the kinetics of processive DNA synthesis by Pol δ are analyzed by resolving replication products using gel electrophoresis (Fig. 3A). We found that heterotrimers of WT2:79901 and WT1:79902 display decreased rates of Pol δ activity relative to the WT homotrimer (Fig. 3B, example gel in Fig. S4A), suggesting that the ability of PCNA to stimulate Pol δ is partially dependent on the number of functional binding sites on the PCNA trimer. Nevertheless, heterotrimers containing even one WT monomer significantly stimulate Pol δ activity compared with the pcna-7990 homotrimer, which displays no activity. This indicates that a single functional binding site on the PCNA trimer is sufficient to support processive DNA synthesis by Pol δ.
Fig. 3.
Stimulation of Pol δ and FEN1 by heterotrimeric PCNA. (A) Schematic illustration of Pol δ assay. PCNA was loaded on a primed single-stranded plasmid by RFC and Pol δ was added to initiate replication. (B) Pol δ activity assays in the presence of three different trimer species, analyzed by agarose gel electrophoresis and autoradiography. The completed 2.9-kb product was quantified as a percentage of the maximum product observed in the assay. Results with pcna-7990 homotrimers are not shown, because no activity was observed. Results shown are averages of four independent assays (see example gel in Fig. S4A); error bars represent SEM. (C) Schematic illustration of FEN1 assay. PCNA was loaded on a radioactively labeled oligonucleotide substrate, FEN1 was added and the reaction was allowed to proceed for the indicated times. FEN1 activity results in cleavage of the 5′ flap portion of the oligonucleotide. Yellow star denotes radioactive label at 5′ end of the flap. (D) FEN1 activity assays in the absence of PCNA or in the presence of four different PCNA trimer species. Reactions were analyzed by urea-PAGE and autoradiography, and the percentage of substrate cleaved by FEN1 was quantified. Results shown are averages of three independent assays; error bars represent SEM.
To further validate that Pol δ stimulation originates from the presence of heterotrimeric PCNA rather than minute quantities of WT homotrimer due to reassembly, we examined Pol δ activity with mixtures of WT and pcna-7990 homotrimers at different ratios (Fig. S3D). Even at a ratio that resembles full reassembly of the WT1:79902 trimer, Pol δ activity was lower than observed with the actual heterotrimer, indicating that a small degree of reassembly into WT homotrimers cannot explain our results.
FEN1 activity is also stimulated by PCNA (13, 14). We measured the kinetics of 5′-flap cleavage by FEN1 on an oligonucleotide-based model substrate (Fig. 3C). Pcna-7990 homotrimers did not stimulate FEN1 activity over the background levels, whereas PCNA heterotrimers containing one or two pcna-7990 monomers significantly stimulated FEN1 but with slightly lower efficiency relative to the WT homotrimer (Fig. 3D). These results indicate that FEN1 stimulation by a single functional PCNA binding site occurs, with additional binding sites improving kinetics, as observed with Pol δ. Essentially similar results were obtained in a K+-containing as in a Na+-containing buffer system (Fig. S5C). We also examined a double-flap rather than a 5′-flap substrate, but in contrast to previous studies performed using archaeal proteins (15, 16), we could not detect any PCNA-dependent stimulation of FEN1 on this substrate (Fig. S5D).
Nick Translation by Pol δ and FEN1.
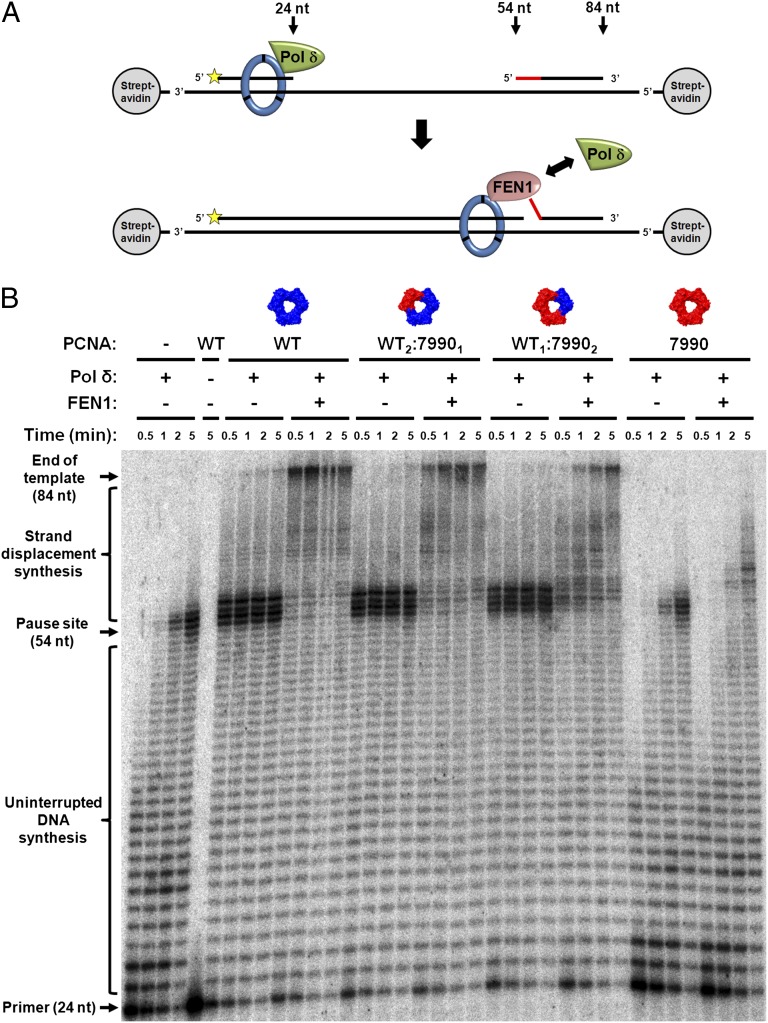
Having observed the activities of Pol δ and FEN1 separately, we next examined whether the cooperation between these two enzymes during Okazaki fragment maturation requires multiple binding sites on the PCNA trimer, as posited by the toolbelt model. Nick translation, the result of iterative, sequential strand displacement synthesis by Pol δ and flap cleavage by FEN1, requires rapid and efficient cooperation between the enzymes in the presence of PCNA (7, 10, 11). We examined the kinetics of nick translation on an oligonucleotide-based model substrate in the presence of different PCNA trimers. In this assay, PCNA-dependent cooperation of Pol δ and FEN1 will result in rapid progression of replication through a downstream RNA–DNA blocking oligo (9, 10). In the absence of FEN1, Pol δ will stall for a relatively long period and only slowly succeed in completely displacing the blocking oligo (Fig. 4A). Hence, a comparison between Pol δ strand displacement activity in the presence and absence of FEN1 provides a measure of the degree of PCNA-dependent cooperation between the two enzymes.
Fig. 4.
Nick translation by Pol δ and FEN1 in the presence of heterotrimeric PCNA. (A) Schematic illustration of the assay. PCNA was loaded on a radioactively labeled oligonucleotide substrate by RFC; Pol δ and FEN1 were added and the reaction was allowed to proceed for the indicated times. Pol δ elongates the primer until reaching the blocking oligo, generating a 54-nucleotide product. Then, strand displacement synthesis through the blocking oligo proceeds with or without FEN1, generating a final product of 84 nucleotides. Yellow star denotes radioactive label at 5′ end of the primer. The red segment represents the RNA portion of the blocking oligo. (B) Results of nick translation assay in the absence of PCNA or in the presence of four different PCNA trimer species. Pol δ and FEN1 were added where indicated. Reactions were analyzed by urea-PAGE and autoradiography.
We found that both heterotrimers allowed FEN1 to significantly stimulate the progression of Pol δ through the blocking oligo (Fig. 4B). These results indicate that Pol δ and FEN1 efficiently cooperate during nick translation even if only one functional binding site is present on the PCNA trimer, suggesting that simultaneous binding of both enzymes to PCNA is not required. To further examine whether simultaneous binding may pose some advantage to this cooperation, we examined the kinetics of nick translation in the presence of heterotrimers containing the pcna-79 and pcna-90 mutants (Fig. S6). As controls, we measured Pol δ and FEN1 activities separately, in the presence of heterotrimers containing pcna-79 and pcna-90 mutants, respectively (Fig. S5 A and B). Because pcna-79 is only partially deficient in stimulation of FEN1 (Fig. S5B) (13), we expect that defects in nick translation that may occur in the WT1:79902 trimer would be alleviated in the WT1:792 trimer, as FEN1 should be able to bind the pcna-79 monomers of this trimer. We found that both pcna-79 and pcna-7990 heterotrimers exhibit the same level of cooperation between Pol δ and FEN1 (Fig. S6, compare samples 5 and 11), indicating that in case simultaneous binding of these enzymes to PCNA does take place, it does not provide a significant functional advantage under these conditions.
Okazaki Fragment Maturation.
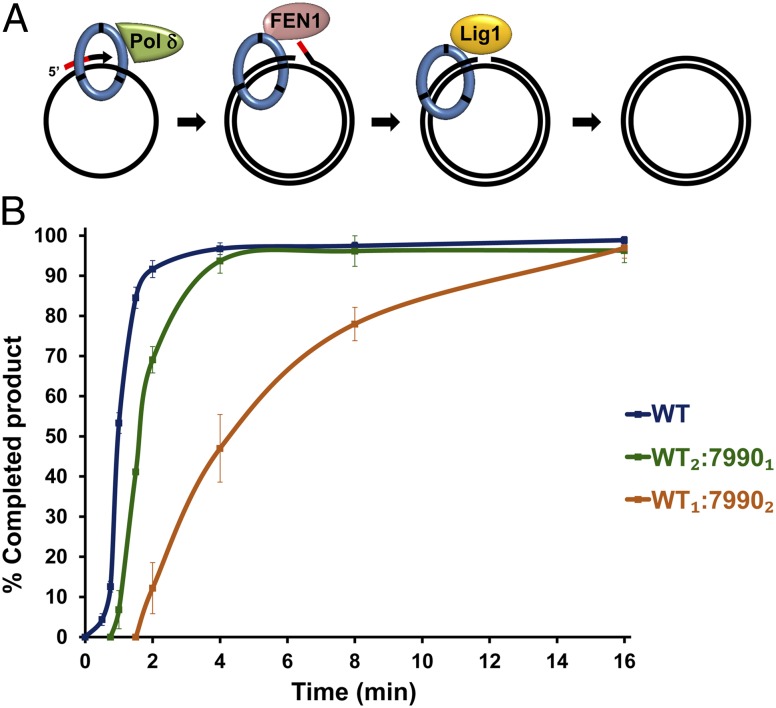
Finally, we examined the ability of the heterotrimers to coordinate the entire Okazaki fragment maturation process in the presence of Pol δ, FEN1, and Lig1. An in vitro assay, which quantifies the product of the concerted activity of all three enzymes was used (Fig. 5A) (9). In this assay, an RNA–DNA primer is annealed to circular single-stranded DNA. Following loading of PCNA by RFC and ATP, Pol δ replicates the plasmid. When reaching the 5′ end of the primer, Pol δ performs nick translation synthesis together with FEN1, removing the RNA portion of the primer. Finally, ligation of the nicked double-stranded plasmid by Lig1 yields a covalently closed plasmid only following RNA removal (17).
Fig. 5.
Stimulation of Okazaki fragment synthesis and maturation by heterotrimeric PCNA. (A) Schematic illustration of the assay. PCNA was loaded on a primed single-stranded plasmid; Pol δ, FEN1, and Lig1 were added and the reaction was allowed to proceed for the indicated times. Following replication of the plasmid by Pol δ, flap processing is performed by the coordinated activity of Pol δ and FEN1, thereby removing the RNA portion of the primer (represented by red segment). Ligation of the nicked plasmid is performed by Lig1, resulting in a covalently closed double-stranded plasmid that migrates faster on an agarose gel in the presence of ethidium bromide. (B) Okazaki fragment maturation assays in the presence of three different PCNA trimer species, analyzed by agarose gel electrophoresis and autoradiography. The fully replicated and ligated product was quantified as a percentage of the maximum product observed in the assay. Results with pcna-7990 homotrimers are not shown, because no activity was observed. Results shown are averages of three independent assays (see example gel in Fig. S4B), error bars represent SEM.
In agreement with the nick translation experiments, we found that heterotrimers containing only one WT monomer still promoted rapid Okazaki fragment maturation by coordinating the activity of all three enzymes (Fig. 5B, example gel in Fig. S4B). As in the previous assays, differences in kinetics can be observed between PCNA trimers with one, two, or three wild-type binding sites. However, these differences are likely due to the differences in the kinetics of the individual enzymes rather than defective cooperation between them. Such defective cooperation between maturation enzymes would be expected to cause an accumulation of fully replicated nicked plasmids, but no significant accumulation is visible (Fig. S4B). To examine this process under conditions that more closely mimic those found in vivo, where the concentration of PCNA is significantly higher than the concentrations of Pol δ, FEN1, and Lig1 (18), we repeated the assay with a 5- or 10-fold excess of PCNA over the enzymes and observed no difference in the extent of cooperation between the enzymes (Fig. S7).
Discussion
To directly examine whether binding of partners to PCNA during complex processes is sequential or simultaneous (Fig. 1), we generated novel PCNA heterotrimers, which combine WT monomers with monomers that are structurally similar but deficient in partner binding. These heterotrimers allowed us to determine the minimal number of functional binding sites on the PCNA trimer required for the proper progression of PCNA-mediated Okazaki fragment maturation. Our results showing that heterotrimers containing a single functional binding site can coordinate nick translation and complete Okazaki fragment maturation provide to our knowledge the first direct evidence that partner recruitment to PCNA is sequential. Although we cannot rule out the possibility that simultaneous binding to the PCNA trimer does exist, we show that it is not strictly necessary for Okazaki fragment maturation. Moreover, we demonstrate efficient cooperation between enzymes in both nick translation and Okazaki fragment maturation assays in the presence of PCNA heterotrimers. Whereas the overall kinetics of these processes are slower in the presence of PCNA with only one functional binding site, the results indicate that these differences can mostly be attributed to lower stimulation of each individual enzyme because of the reduction in active binding sites on PCNA, rather than defective cooperation. Therefore, we conclude that simultaneous binding to PCNA, if possible, is not functionally advantageous in vitro.
Our results point toward a model in which Pol δ and FEN1 can repeatedly dissociate and reassociate with PCNA, rapidly replacing each other on a single PCNA monomer (Fig. 1, Upper). We speculate that during processive replication, Pol δ is tightly bound to PCNA, perhaps through several contact points with different PCNA monomers. During strand displacement synthesis, which is considerably slower, the binding of Pol δ to PCNA may be reduced. Indeed, reduced polymerase-clamp interactions upon encountering structural blocks have been documented in the analogous T4 replication system (19). This partial dissociation of Pol δ would allow FEN1 to replace Pol δ by affinity competition. Such a sequential model can be facilitated by changes in the structure of the DNA substrates—i.e., the creation of a flap by Pol δ may increase FEN1's affinity to the PCNA–DNA complex. It may also be facilitated by a rotation of PCNA around a kink in the DNA induced by the partner enzymes, as previously suggested (15). Examination of the in vivo activity of PCNA heterotrimers is difficult due to the random assembly of WT and mutant PCNA monomers into mixtures of homotrimeric and heterotrimic forms. However, our examination of Okazaki fragment maturation at protein concentrations, which more closely mimic the cellular concentration ratio (18), highlights that PCNA may coordinate partners through a sequential mechanism in the cell (Fig. S7). This may allow for higher flexibility in partner switching, considering PCNA’s numerous cellular partners (1).
Previous studies have provided inconclusive evidence for a PCNA toolbelt model. In vivo and in vitro data suggest that Pol δ may bind PCNA together with FEN1, whereas Lig1 binding is exclusive (9, 20, 21). In contrast, another in vivo study in mammalian cells has demonstrated that PCNA is stably associated with DNA, whereas its partners are transiently associated (22). In the archaeon Sulfolobus solfataricus, PCNA is a heterotrimer and each monomer specifically binds one of the three Okazaki fragment maturation enzymes (23). It has recently been shown that Okazaki fragment maturation in this archaeon is stimulated by simultaneous binding of all three partners to a single PCNA trimer (24). Simultaneous binding of two different partners has also been demonstrated for the bacterial sliding clamp (25). These analogous systems, however, are thought to possess a considerably lower number of sliding clamp-interacting proteins compared with the eukaryotic system. It has been suggested that the toolbelt model provides a simple solution for the problem of recruiting the correct enzymes to the sliding clamp at the correct time, but only when the number of possible partners is limited (1, 24). Eukaryotes, possessing dozens of PCNA partners, may have evolved more complex regulated mechanisms to drive the sequential recruitment of multiple partners, thus rendering the possibility of simultaneous binding unnecessary.
Interestingly, we found that strand displacement by Pol δ was significantly stimulated by FEN1 even when only one binding site on the PCNA trimer was available (Fig. 4B). Previously, it was shown that the strand displacement activity of the S. solfataricus replicative polymerase is stimulated by the presence of FEN1, as in eukaryotes, but only when the two enzymes bind different PCNA monomers on the same trimer (24). When an alternative polymerase was used, which binds the same archaeal PCNA monomer as FEN1, strand displacement activity was inhibited by the presence of FEN1 due to competition between the two enzymes for PCNA binding. Comparing this study to our findings suggests that eukaryotic PCNA-partner interactions, unlike the archaeal system, are governed by sequential cooperation rather than competition.
It remains unclear why Pol δ and FEN1, when examined separately, exhibit higher stimulation by PCNA trimers with more WT binding sites. There are two main possible explanations for these observations: first, the probability of enzyme recruitment to the DNA substrate may depend on the number of available binding sites on PCNA. Simple mass action collision theory would predict higher activity when tripling the number of functional binding sites on each trimer. A second explanation is that PCNA trimers with more WT binding sites may have an intrinsically superior ability to stimulate partner enzymes. To examine this possibility, we performed true processivity assays (26), which measure the rate of PCNA–Pol δ dissociation after each binding event (Fig. S8). We observed a significant difference in processivity between trimers with one, two, or three WT binding sites, indicating that multiple functional sites on PCNA increase the intrinsic affinity to Pol δ. These results are in good agreement with studies analyzing PCNA–Pol δ interactions. It was recently shown that Pol δ, which is a heterotrimer, possesses multiple PCNA binding motifs that contribute to processive PCNA-dependent DNA replication (27, 28). This suggests that Pol δ may engage different PCNA monomers simultaneously or consecutively during replication. Using our PCNA heterotrimers, we may have limited the number of PCNA–Pol δ contacts leading to a decrease in Pol δ processivity.
We present here a fast, reproducible approach that allows the simple purification of heterotrimers with a native tertiary and quaternary structure. Our method takes place under native conditions and can be extended to incorporate any mutant into heterotrimers, as long as it does not impede natural trimerization. For example, heterotrimers bearing the K164R mutation (3), combined with in vitro ubiquitylation methods (5, 29), may address several fundamental questions regarding the polymerase switching mechanism during translesion DNA synthesis (30). We believe that this can be an effective approach for the mechanistic study of structure–function relationships in PCNA and can be applied for the study of many other homooligomeric proteins that participate in a variety of complex biological processes.
Materials and Methods
Purification of PCNA Heterotrimers.
S. cerevisiae PCNA was cloned with an N-terminal His tag into the first multiple cloning site (MCS) of pETDuet-1 (Novagen). Mutant pcna-79, pcna-90, or pcna-7990 was cloned with an N-terminal MBP tag followed by a TEV protease cleavage site into the second MCS of the same vector. This vector allows the simultaneous overexpression of both proteins at similar levels. Overexpression was performed in E. coli BL21(DE3) cells. Following cell lysis using a French press (Thermo Scientific), the lysate was purified over a Ni-NTA His-bind column (Novagen), and the eluate was pooled and purified over an amylose column (New England Biolabs). The amylose eluate was concentrated and injected into a Superdex 200 16/60 prep grade gel filtration column (GE Healthcare) using the AKTA purifier FPLC system (GE Healthcare). Next, selected 0.5-mL fractions were collected and examined for purity by performing analytical gel filtration on a Superdex 200 10/300 GL column (GE Healthcare). For each heterotrimer species, fractions that contained the desired heterotrimer without significant contamination were selected and pooled. TEV protease was added at 1:100 (enzyme:substrate) molar ratio and incubated overnight at 4 °C. Following cleavage, the samples were again purified by gel filtration on a Superdex 200 10/300 GL column.
Heterotrimer stability assays, RFC ATPase assays, cross-linking assays, and purification of other proteins are described in SI Materials and Methods.
Pol δ Replication Assays and Okazaki Fragment Maturation Assays.
Assays were performed essentially as described previously (9). For consistency, Pol δ assays and Okazaki fragment maturation assays were performed under identical conditions. The template DNA, single-stranded Bluescript SKII(+) plasmid, was obtained as previously described and hybridized with primer SKrc14 (9). Standard 40-µL assays contained 20 mM Tris⋅HCl pH = 7.8, 1 mM DTT, 100 µg/mL BSA, 7.5 mM MgAc2, 0.4 mM ATP, 100 µM each of dCTP, dGTP, and dTTP, 10 µM dATP, 4 nM [α-32P]dATP (3,000 Ci/mmol), 100 mM NaCl, 50 fmol of template plasmid, 10 pmol of replication protein A (RPA), 100 fmol of RFC, 100 fmol of PCNA trimers, and 200 fmol of Pol δ. Okazaki fragment maturation assays also contained 200 fmol each of FEN1 and Lig1. The template plasmid was preincubated with RPA, PCNA, and RFC for 1 min at 30 °C for RPA coating and PCNA loading. The other enzymes were then added in a mix, and the reactions were incubated at 30 °C for the indicated times. Products were analyzed by electrophoresis on a 1% agarose gel in the presence of 0.5 µg/mL ethidium bromide. The gels were dried, exposed to a storage phosphor screen (GE Healthcare), and analyzed on a PhosphorImager (Fuji Film).
Pol δ processivity assays were performed as previously described (26) with slight modifications. Details can be found in SI Materials and Methods.
FEN1 Flap Cleavage Assays.
Oligonucleotide-based FEN1 assays were performed as previously described (13, 14) with slight modifications. Details can be found in SI Materials and Methods.
Nick Translation Assays.
Assays were performed essentially as previously described (7, 9, 10). Details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Carrie Stith and Dan Od-Cohen for protein purification and technical assistance, Enav Drori for assistance with graphics, and Ron Milo for assistance with the calculation of cellular protein concentrations. This work was supported in part by Grant GM032431 from the National Institutes of Health (to P.M.J.B.). A.A. was supported by the European Research Council “Ideas Program” (201177).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.D.S. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321349111/-/DCSupplemental.
References
- 1.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116(Pt 15):3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 3.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 4.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425(6954):188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 5.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102(51):18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann AR. Clubbing together on clamps: The key to translesion synthesis. DNA Repair (Amst) 2006;5(3):404–407. doi: 10.1016/j.dnarep.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J Biol Chem. 2008;283(49):34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgers PMJ. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284(7):4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayyagari R, Gomes XV, Gordenin DA, Burgers PMJ. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278(3):1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 10.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18(22):2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi ML, Bambara RA. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J Biol Chem. 2006;281(36):26051–26061. doi: 10.1074/jbc.M604805200. [DOI] [PubMed] [Google Scholar]
- 12.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol. 1997;17(11):6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19(14):3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270(38):22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 15.Chapados BR, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116(1):39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 16.Craggs TD, Hutton RD, Brenlla A, White MF, Penedo JC. Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates. Nucleic Acids Res. 2014;42(3):1857–1872. doi: 10.1093/nar/gkt1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432(7016):473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 18.de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 19.Hacker KJ, Alberts BM. The rapid dissociation of the T4 DNA polymerase holoenzyme when stopped by a DNA hairpin helix. A model for polymerase release following the termination of each Okazaki fragment. J Biol Chem. 1994;269(39):24221–24228. [PubMed] [Google Scholar]
- 20.Subramanian J, Vijayakumar S, Tomkinson AE, Arnheim N. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics. 2005;171(2):427–441. doi: 10.1534/genetics.105.042861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riva F, et al. Distinct pools of proliferating cell nuclear antigen associated to DNA replication sites interact with the p125 subunit of DNA polymerase δ or DNA ligase I. Exp Cell Res. 2004;293(2):357–367. doi: 10.1016/j.yexcr.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 22.Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33(11):3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell. 2003;11(1):275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 24.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31(6):1556–1567. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19(6):805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Chilkova O, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35(19):6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Netz DJ, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc Natl Acad Sci USA. 2011;108(44):17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103(17):6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedberg EC, Lehmann AR, Fuchs RPP. Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18(5):499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.