Significance
Nutritional starvation therapy is under intensive investigation because it provides a potentially lower toxicity with higher specificity than conventional cancer therapy. Autophagy, often triggered by starvation, represents an energy-saving, pro-survival cellular function; however, dysregulated autophagy could also lead to cell death, a process distinct from the classic caspase-dependent apoptosis. This study shows how arginine starvation specifically kills tumor cells by a novel mechanism involving mitochondria dysfunction, reactive oxygen species generation, DNA leakage, and chromatin autophagy, where leaked DNA is captured by giant autophagosomes. These results not only provide insights into the fundamental process of metabolic stress-based cancer therapy but also uncover a new cell-death mechanism.
Keywords: arginine auxotrophy, ADI-PEG20, metabolic stress, cancer therapy, prostate cancer
Abstract
Autophagy is the principal catabolic prosurvival pathway during nutritional starvation. However, excessive autophagy could be cytotoxic, contributing to cell death, but its mechanism remains elusive. Arginine starvation has emerged as a potential therapy for several types of cancers, owing to their tumor-selective deficiency of the arginine metabolism. We demonstrated here that arginine depletion by arginine deiminase induces a cytotoxic autophagy in argininosuccinate synthetase (ASS1)-deficient prostate cancer cells. Advanced microscopic analyses of arginine-deprived dying cells revealed a novel phenotype with giant autophagosome formation, nucleus membrane rupture, and histone-associated DNA leakage encaptured by autophagosomes, which we shall refer to as chromatin autophagy, or chromatophagy. In addition, nuclear inner membrane (lamin A/C) underwent localized rearrangement and outer membrane (NUP98) partially fused with autophagosome membrane. Further analysis showed that prolonged arginine depletion impaired mitochondrial oxidative phosphorylation function and depolarized mitochondrial membrane potential. Thus, reactive oxygen species (ROS) production significantly increased in both cytosolic and mitochondrial fractions, presumably leading to DNA damage accumulation. Addition of ROS scavenger N-acetyl cysteine or knockdown of ATG5 or BECLIN1 attenuated the chromatophagy phenotype. Our data uncover an atypical autophagy-related death pathway and suggest that mitochondrial damage is central to linking arginine starvation and chromatophagy in two distinct cellular compartments.
There is considerable evidence that tumor and normal cells differ in their metabolic requirements. The most prominent examples are the addiction of tumor cells to glucose (i.e., Warburg effect) and to glutamine (1–3). Therapeutics based on selective targeting of these metabolic pathways are under intensive investigation. Starvation therapy generally posts an advantage of having lower toxicity than conventional radiation and chemotherapy. In addition to glutamine, the differential requirement of other amino acids by tumor cells also exists and has been exploited in developing amino acid depletion therapy. The choices, however, are limited, because only 11 amino acids are considered semiessential or nonessential. Nevertheless, recent studies showed that starvation of arginine, asparagine, cysteine, leucine, and glutamine seems to provide preferential killing of tumor cells (4–9). Among them, arginine and asparagine depletion probably are the most advanced in amino acid starvation therapies and have reached clinical trials (10, 11).
Argininosuccinate synthetase (ASS1), a rate-limiting enzyme for intracellular arginine synthesis, was found to have reduced expression in many cancer types including prostate cancer (4, 5, 12–18). As a result, prostate cancer cells become “auxotroph” for and addicted to external arginine. Indeed, in recent publications we showed that depletion of arginine effectively induces cell death of castration-resistant prostate cancer cells but not of normal prostate epithelial cells in vitro and in vivo (18, 19). This was made possible by the availability of pegylated arginine deiminase (ADI-PEG20), a recombinant mycoplasma protein (Polaris), which converts arginine to citrulline and effectively removes extracellular arginine. Previous worldwide clinical trials of ADI-PEG20 showed it is well tolerated, and the FDA has recently approved it for phase III clinical trials in hepatocellular carcinoma (10, 20).
ADI-PEG20 treatment of prostate cancer cells was accompanied by profound autophagy and caspase-independent cell death (18, 19). In this paper, we further demonstrated that prolonged ADI-PEG20 treatment leads to DNA leakage and that this leaked DNA, together with histones and other chromatin-associated proteins, was captured by LC3-containing autophagosomes. We shall refer to it as “chromatophagy.” This nuclear leakage phenomenon was associated with compromised mitochondrial oxidative phosphorylation and increased reactive oxygen species (ROS) and accumulated DNA damage. Our studies revealed a previously unidentified cellular death mechanism associated with therapeutic arginine deprivation.
Results
ADI-PEG20 Targets Cancer Cells via an Atypical Death Pathway with Nuclear DNA Leakage.
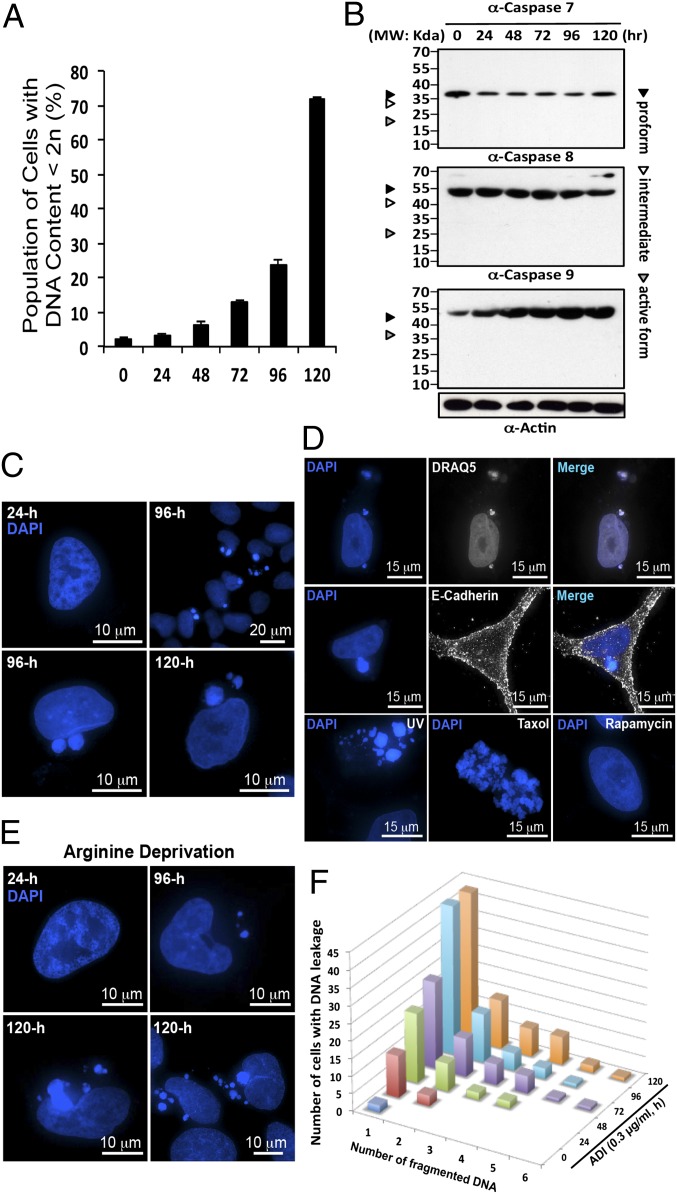
To explore the mechanism underlying the ADI-PEG20- and arginine deprivation-mediated cytotoxic effect, prostate cancer CWR22Rv1 cells were treated with 0.3 μg/mL (IC50) of ADI-PEG20. This treatment resulted in cell death with two unusual features that distinguished it from conventional apoptosis. First, there was a long incubation period (48 h) before the emergence of hypodiploid cells (DNA content <2n) in flow-cytometric analysis (Fig. 1A). Second, the cell death is caspase-independent; caspase 7, 8, and 9 all remained in the respective procaspase form (Fig. 1B). This echoes our earlier finding that treatment of cells with pan-caspase inhibitor (vZAD) did not affect the extent of ADI-PEG20–induced cell death (18). These data prompted us to study whether autophagy is involved in the late phase of cellular killing and to decipher the molecular mechanism underlying ADI-PEG20–induced cell killing.
Fig. 1.
ADI-PEG20 induced both caspase-independent cell death and DNA leakage. (A) CWR22Rv1 cells were treated with ADI-PEG20 and stained with propidium iodide (PI). The percentage of hypodiploid cells began to rise at 48 h and significantly increased after 72 h. (B) Western blotting showing the lack of caspase 7, 8, and 9 activation in cells treated with ADI-PEG20. (C) DAPI staining reveals nuclear DNA before leakage at 24 h (early time points) and after leakage at 96 h and 120 h (late time points) posttreatment. (D) The same samples were stained with another fluorescent DNA dye, DRAQ5. Cells were stained with plasma membrane marker E-cadherin to reveal its boundary. Cells were treated with UV or Taxol (1 nM) to demonstrate the appearance of apoptotic body and mitotic catastrophe, respectively. Rapamycin (2 μM) treatment did not produce the same effect. (E) Prolong incubation with arginine-depleted medium also induced the same phenotype. (F) The bar graph illustrates the positive increment of leaked DNA particles in a population of cells showing the phenotype.
To investigate the mechanism underlying prostate cancer cell death by arginine deprivation, advanced 4D fluorescence microscopy was used to examine the nuclear dynamics. As shown in Fig. 1C, one striking feature was that ADI-PEG20 treatment consistently induced nuclear DNA leakage at the onset of the dying phase (48 h posttreatment) as identified by DAPI-positive particles outside the nucleus. DNA leakage became more apparent after 72 h of treatment, and the size of leaked DNA puncta was notably larger than that of micronuclei and the number was greater (Fig. 1C). To ensure that these particles released from nucleus truly contain DNA (as opposed to RNA), DRAQ5 with higher specificity toward DNA was used. Merged images showed nearly perfect colocalization between DAPI and DRAQ5, confirming the DNA as the major cargo in these particles or puncta (Fig. 1D).
A notable feature of this phenotype, distinguishing it from apoptotic bodies, is that the leaked DNA was not surrounded by plasma membrane and the nucleus was not shattered. In fact, the gross morphology of these cells containing leaked DNA, as indicated by cell membrane marker E-cadherin staining, seemed to be intact (Fig. 1D and Fig. S1A). Moreover, these cells were able to maintain cell adhesion in culture, suggesting that the DNA leakage did not take place after cell death. The leaked DNA structures with largely intact nucleus (Fig. 1C) are distinct from the apoptotic bodies generated by UV or mitotic catastrophe by taxol treatment (Fig. 1D). To ascertain that these observations indeed resulted from arginine depletion, CWR22Rv1 cells were maintained in arginine-free media up to 120 h. DNA leakage was unequivocally observed (Fig. 1E). In addition, a similar phenotype was identified in prostate cancer PC3 cells and pancreatic Mia and L3.3 cells (Fig. S1B); all are lacking ASS1 expression and sensitive to ADI-PEG20–induced cell killing. ADI-PEG20–treated Mia and UMUC3 cells exhibited caspase-dependent cell death (14, 21). Thus, DNA leakage is not limited to cells undergoing caspase-independent death. By contrast, such a phenotype was not found in cells treated with rapamycin alone (Fig. 1D) or subjected to glutamine depletion (20). The number of puncta carrying DNA was at least one, but up to more than six, with an average of 1.92 per cell (n > 300) (Fig. 1F). These data taken together suggest that prolonged arginine deprivation selectively induces DNA leakage.
ADI-PEG20 Induces Giant Autophagosome Formation.
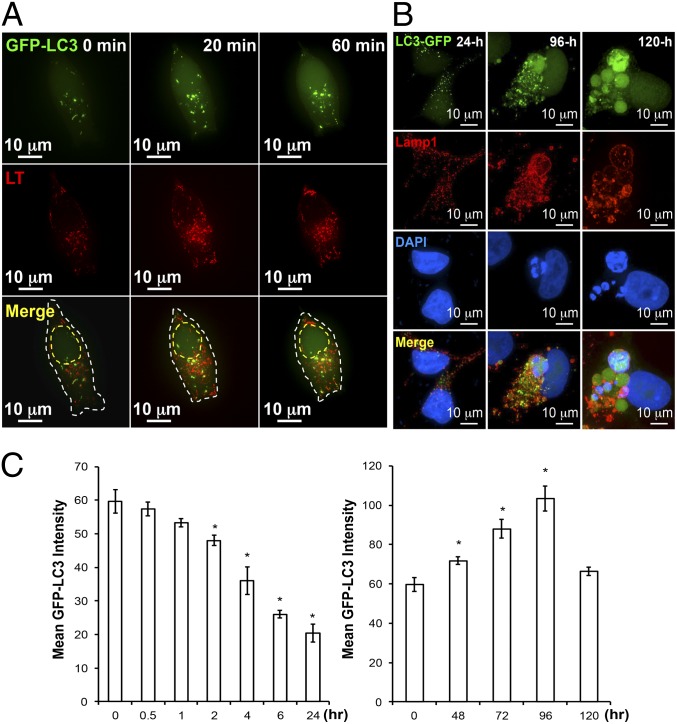
Fig. 2A shows that GFP-LC3, a marker for autophagy, was concentrated at the perinuclear region of the GFP-LC3 stably transfected CWR22Rv1 cells within 20 min of ADI-PEG20 treatment. The number of autophagosome increased as autophagy proceeded. At the same time, lysosomes as marked by LysoTracker Red migrated toward the perinuclear region, where some of them began to fuse with autophagosomes to form autolysosomes (Fig. 2A). This process persisted for at least 24 h in the presence of ADI-PEG20. In contrast to the earlier stage of ADI-PEG20 treatment with conventional-sized autophagosome puncta, a subset of the puncta at the later phase exhibited abnormally large autophagosomes and autolysosomes [as indicated by colocalization of GFP and LysoTracker (Fig. 2B)] or LAMP1 (Fig. S2A). Some of the larger autophagosomes were greater than 20-fold the size of conventional autophagosomes (Fig. S2A). Moreover, Western blot analysis demonstrated that the addition of bafilomycin A1, an inhibitor of vacuolar-type H+-ATPase, caused accumulation of LC3-II and p62 in ADI-PEG20–treated cells (Fig. S2B), but not in cells treated with 3-MA alone, supporting the involvement of lysosomes in this atypical autophagy. Many of these abnormally sized autophagosomes (GFP-C3, green) were colocalized with the LAMP1 (red) and leaked DNA (blue) described above (Fig. 2B and Fig. S2A). Although some of the autophagosomes did not contain leaked DNA particles, we found, with no exception, that all of the leaked DNA particles were contained within autophagosomes by analyzing more than 300 cells with leaked DNA at different time points after ADI-PEG20 treatment. Interestingly, a strong DAPI signal could be found in these particles, suggesting dense DNA material was encapsulated within the autophagosomes.
Fig. 2.
Prolonged ADI-PEG20 treatment can induce giant-autophagosome formation and affect autophagic flux pattern. (A) Representative time-lapse images showing ADI-PEG20 treatment induces autophagy and facilitates the fusion of autophagosome (green) and lysosome (red, LysoTracker) into autophagolysosome (yellow). The outline is shown by a dashed white line. The estimated nucleus location is shown by a dashed yellow line. (B) Prolonged ADI-PEG20 treatment induces abnormally sized autophagosomes (green), which colocalize with lysosome (red) and leaked DNA (blue). (C) ADI-PEG20 treatment induces autophagic flux with distinct kinetics. Mean GFP-LC3 intensity in each group was plotted as a bar graph. Data were collected from three independent experiments and are shown as mean ± SD; *P < 0.05.
Finally, autophagy flux was measured using flow cytometry. As shown in Fig. 2C, Left, during the first 24 h a decrease in GFP-LC3 level was noted, suggesting the progression of autophagic flux to overcome arginine auxotrophy induced by ADI-PEG20, as we previously reported (18). However, the level of GFP-LC3 increased at 48 h after ADI-PEG20 treatment and then tapered off at 120 h (Fig. 2C, Right). The second wave of increased GFP-LC3 coincided with appearance of giant autophagosomes (Fig. 2B). These results suggest that there are two phases of arginine starvation-mediated autophagy induction, with conventional autophagosome morphology at the early phase and giant autophagosomes at the later dying phase.
Nuclear Membrane Remodels During ADI-PEG20–Induced Chromatophagy.
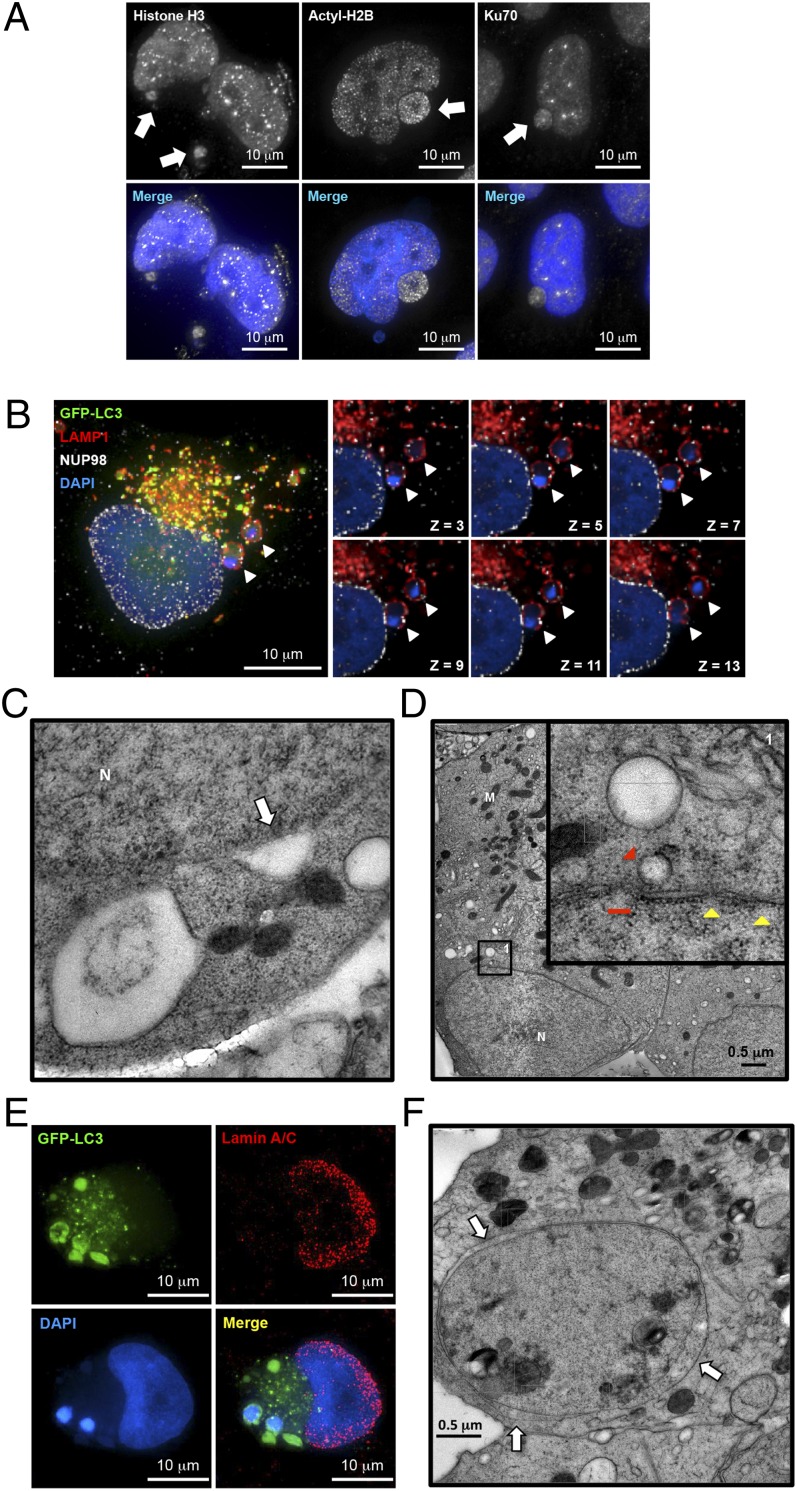
Next we hypothesized that if the leaked DNA was from nucleus it must be sheared from nuclear DNA and therefore could be considered as damaged DNA. To test this hypothesis and to further characterize these DNA-containing autophagosome complexes, we used immunofluorescence to identify proteins known to be associated with chromatin and/or DNA repair. Nuclear DNA/chromatin-bound proteins, both H3 (Fig. 3A and Fig. S3A) and acetyl-H2B (Lys12) (Fig. 3A and Fig. S3B), were found within the DNA-containing autophagosomes. Acetylated histones can be found on the exo-nucleus DNA, indicating destabilized chromatin existed in the autophagosome. Moreover, Ku70, known to be associated with DNA double-strand break (DSB) ends, was identified with leaked DNA (Fig. 3A and Fig. S3C), suggesting that cells might attempt to repair the damaged DNA before it leaked into cytoplasm and was engulfed by autophagosomes.
Fig. 3.
Prolonged arginine deprivation by ADI-PEG20 induces nuclear membrane remodeling. (A) Immunofluorescence analysis showed that histone H3, acetylated H2B, and Ku70 were found associated with the exo-nucleus DNA cluster. (B) Immunofluorescence analysis of NUP98 (white), GFP-LC3 (green), lysosome (red), and DAPI (blue) showed that uniform NUP98 signals were both around and outside of the nucleus and colocalized with lysosome signal (white arrowheads). (C) TEM demonstrated that autophagosomes could potentially fuse with the nuclear membrane (white arrow). (D) Autophagosomes were found in close proximity to the nucleus (red arrowhead, with higher magnification shown in inset). Nucleus membrane also showed partial breakages (red line, ∼150 nm). A typical nuclear pore complex has a thin layer of electron dense disk with an opening about 120 nm; however, it may appear smaller under Cryo-EM (35) (yellow arrowheads, 47 nm ∼ 53 nm). M, mitochondria; N, nucleus. (E) Representative immunofluorescence images showed reduced lamin signals at the site of excessive autophagy. (F) Abnormal nucleus membrane morphology was found in treated cells at later time points. Expansion of intramembrous space was observed between inner membrane and outer membrane (white arrows). One representative cyro-EM image is shown in C, D, and F. n = 3 in A–F.
The nuclear membrane is composed of lipid bilayer and its integrity is essential for cell survival. To examine whether the nuclear membrane integrity might have been compromised, resulting in DNA leakage, arginine-starved cells were stained with nuclear outer membrane marker nucleolin (NUP98). A series of Z-section fluorescence images revealed that portions of the outer membrane might have fused with autophagosome (Fig. 3B, white arrowheads), suggesting that (i) the generation of these complexes involves nuclear–autophagosome membrane fusion immediately adjacent to the nucleus and (ii) nuclear outer membrane protein nucleolin could provide a docking site for autophagosomes. Moreover, to achieve higher spatial resolution of nucleus and autophagosome junctures, ADI-PEG20–treated cells were subjected to transmission electron microscopy (TEM) analyses. Indeed, TEM image showed autophagosome seemingly fused with nuclear membrane (Fig. 3C, white arrow) and also showed partial nuclear membrane breakage (Fig. 3D, Inset, red line), which is different from a typical nuclear pore complex (Fig. 3D, Inset, yellow arrowheads). Some autophagosomes were also found in close proximity to nucleus, and interestingly enough, the size of the vesicle (Fig. 3D, Inset, red arrowhead) is similar to the opening next to it (Fig. 3D, Inset, red line), suggesting there might be an increased activity of vesicle trafficking between the two. Together with results shown in Fig. 4, autophagosome formation might serve a previously overlooked function related to DSBs in the nucleus and was capable of fusing with nuclear membranes, both of which were critical for DNA leakage.
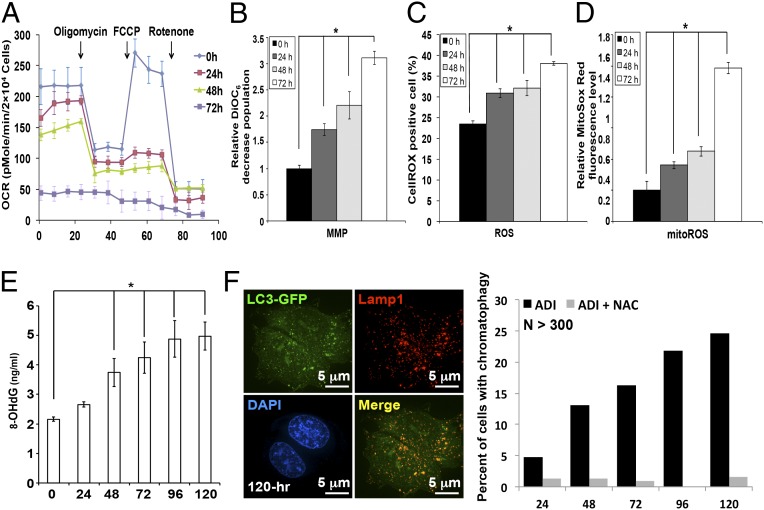
Fig. 4.
ADI-PEG20 treatment impairs mitochondrial function and induces ROS production. (A) ADI-PEG20 treatment suppresses basal OCR and reserve capacity. The tracing revealed the basal OCR decreased in a time-dependent manner with the incubation of ADI-PEG20. (B–D) ADI-PEG20 impairs mitochondrial membrane potential (MMP) and elevates ROS. Respective MMP (B) CellROX oxidation (C), and MitoSox Red oxidation (D) were determined by flow cytometry after staining with DiOC6, DCFDA, and MitoSOX Red. Data are collected from three independent experiments and shown as mean ± SD; *P < 0.05. (E) The increase of 8-OHdG after ADI-PEG20 treatment. Data were collected from three independent experiments with mean ± SD; *P < 0.05. (F) ROS scavenger NAC attenuated DNA leakage phenotype (Right; n > 300) and formation of giant autophagosomes (Left).
Next, we examined the inner membrane structure of nucleus using lamin A/C as marker. The detailed 3D information shows (i) a discontinuity of lamin A/C signals peripherally surrounding the nucleus and (ii) lamin A/C signals displaying a caved-in shape at the site of autophagy (Fig. 3E). Interestingly, stacked sections revealed that lamin A/C signals were markedly reduced at the interface between autophagosomes and nucleus showed discontinuity of lamins (Fig. 3E). Clearly, the lamin A/C signal was also severely reduced in many cells exhibiting the chromatophagy phenotype, suggesting that compromised (or rearranged/remodeling of) nuclear envelope integrity might partake in DNA leakage. In addition, parts of the nuclear membrane also showed some defects and/or breakage (Fig. 3D, Inset, red line). Additional images also showed a separation between the inner and outer membranes (Fig. 3F, white arrows), indicating nuclear membrane integrity might have been compromised. The lack of lamin A/C structure in the area facing autophagosomes (Fig. 3E) implies that autophagy likely engaged in capturing the damaged DNA at the interface.
ADI-PEG20 Impairs Mitochondrial Function and Induces ROS Generation.
During the course of analyzing some of the TEM images, abnormal mitochondria were noted (Fig. 3D) in many cells. This led us to speculate that prolonged arginine deprivation has impaired mitochondrial function. To address this possibility, real-time oxygen consumption rate (OCR) was determined in ADI-PEG20–treated CWR22Rv1 cells at indicated time points posttreatment using a Seahorse Extracellular Flux (XF-24) analyzer. Fig. 4A reveals that baseline OCR was suppressed by treatment with ADI-PEG20 in a time-dependent manner starting at 48 h posttreatment. At 72 h following ADI-PEG20 treatment, the basal OCR level showed an ∼80% decrease and failed to respond to the inhibition of oligomycin, an H+-ATP-synthase inhibitor (22). Moreover, ADI-PEG20 impaired the reserve capacity for maximal oxygen consumption, evident by the lack of response to carbonylcyanide p-trifluoromethoxy phenylhydrazone, an uncoupler of the proton gradient across the inner mitochondrial membrane (23). Finally, prolonged ADI treatment showed significant reduction of OCR level before addition of rotenone, a mitochondrial complex I inhibitor. Consistent with the OCR profile (Fig. 4A), the mitochondrial membrane potential level was reduced in a time-dependent manner and the DiOC6-low population was approximately twofold more compared with vehicle-treated control (Fig. 4B). These results suggest that arginine deprivation leads to severe mitochondrial defects. Dysfunctional mitochondria are known to release ROS. As shown in Fig. 4C, the CellROX-positive population is elevated at 24 h after ADI-PEG20 treatment, suggesting an increase of ROS level. MitoSox staining verified an increase of mitochondrial ROS levels to approximate fivefold higher than that in vehicle-treated cells (Fig. 4D). One consequence of elevated ROS is the generation of base damages; 8-hydroxy-2′-deoxyguanosine (8-OHdG) ELISA analyses demonstrated that cellular 8-OHdG levels gradually increased after ADI-PEG20 treatment starting at 48 h posttreatment (Fig. 4E). To determine whether increased ROS contributes to chromatophagy, CWR22Rv1 cells were cotreated with N-acetyl cysteine (NAC, a ROS scavenger, 10 μM) and ADI-PEG20 for 96 h. The addition of NAC essentially abolished the appearance of the DNA autophagosome structure after prolonged ADI-PEG20 treatment (Fig. 4F, Left) and the percentage of cells containing leaked DNA was significantly reduced (Fig. 4F, Right), confirming the involvement of ROS in DNA leakage after prolonged ADI-PEG20 treatment.
Autophagy Is Required for DNA Leakage.
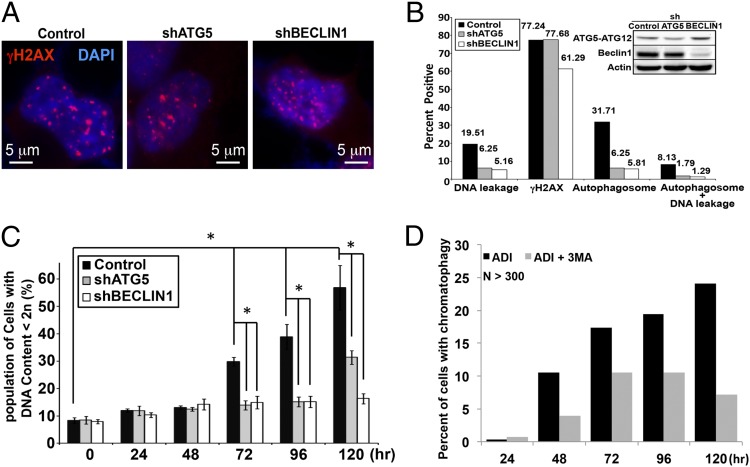
To investigate the potential contribution of autophagy to DNA leakage/DNA damage, we used shRNA to knock down ATG5 or BECLIN1, essential proteins for autophagy initiation and autophagosome formation (24–27) in CWR22Rv1 cells overexpressing GFP-LC3. Western analyses of ATG5-ATG12 and BECLIN1 confirmed that the respective protein expression level was reduced to ∼40 and 10% in the knockdown cells (Fig. 5B, Inset) and that there was reduced autophagy induction in the knockdown cells (Fig. S4A). The reduced autophagy capacity due to ATG5 or BECLIN knockdown did not affect the ADI-PEG20–induced appearance of γH2AX (Fig. 5 A and B), which presumably was a direct result of increased ROS. By contrast, autophagosome formation was extensively inhibited in CWR22Rv1/shATG5 and /shBECLIN1 cells, dropping from 31.7 to 6.3 and 5.8%, respectively (Fig. 5B). The proportion of cells with DNA leakage showed ∼66 and 75% decrease in shATG5 and shBECLIN1 cells (Fig. 5B). Furthermore, the percentage of hypodiploid cells was dramatically reduced in shATG5 and shBECLIN1 cells after 72 h of ADI-PEG20 treatment (Fig. 5C). As a complementary approach, pharmacological inhibitor 3-methyladenine (3-MA) was used to block autophagosome formation (28). As shown in Fig. 5D, such treatment diminished the DNA leakage phenotypes. In addition, bafilomycin A1 was added to prevent maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes. As shown in Fig. S4 B and C, the presence of bafilomycin A1 also reduced the DNA leakage phenotype. Together, the data suggest that autophagosome formation is at least in part involved in the DNA leakage process.
Fig. 5.
Autophagy contributes to ADI-induced DNA leakage. (A) Control, shATG5, and shBECLIN1 stably transfected CWR22Rv1 cells overexpressing GFP-LC3 (green) were treated with ADI-PEG20 for 96 h, followed by staining with anti-γH2AX antibody (red) and DAPI (blue). (B) Approximately 120–150 cells were analyzed for DNA leakage, γH2AX staining, and autophagosome formation. Western blot of ATG5-ATG12 and BECLIN1 demonstrates the expression of these proteins in modified cells. (C) PI staining revealed that knocking down autophagy machinery significantly reduces hypodiploid cell population after 72 h. (D) The bar graph shows the interruption of autophagolysosome formation with 3-MA diminished the DNA-leakage phenotype (n > 250).
Discussion
Investigations by our laboratories and others have established that targeting “arginine addiction” could be exploited therapeutically (16, 18, 21, 29, 30), owing to the preferential loss of ASS1 expression in tumor cells. The reduced ASS1 expression renders the cells unable to produce arginine intracellularly and they become addicted to external arginine. In this study, we found that ADI-PEG20 induces autophagy with two distinct phases. Conventional autophagy that protected cells from dying was seen in the first 48 h, and afterward atypical or excessive autophagy and formation of giant autophagosomes, leading to cell death, were detected. This type of cell killing differs from classic type I apoptosis in that it can be caspase-independent, accompanied by DNA leakage from the nucleus, and autophagosomes capture the leaked chromatin, referred to here as chromatophagy. The observation that both Ku70 and histones were also captured suggests that the leaked DNA is of nuclear, but not mitochondrial, origin.
One of the striking findings in this paper is the high level of giant autophagosomes and autolysosomes encapsulating the leaked DNA in the ADI-PEG20–treated dying cells. Nutritional starvation often leads to autophagosome induction, and arginine deprivation is no exception (4, 18, 21, 29, 31 and this paper). However, the average volume of this chromatin-uptaken vesicle, with its diameter ranging from 2 μm to >10 μm, was several-fold greater than the conventional size [700 nm to 1 μm (28)] in the arginine-starved cell (Fig. 2B). These mega-vesicles seemed to be related to the autophagy pathway on the basis of three criteria: (i) As determined by 4D fluorescence microscopy, almost all of them they were GFP-LC3–positive (Fig. 2B); (ii) TEM analyses indicated that many were enclosed by at least two lipid bilayers (Fig. 3 C and D); and (iii) they are ATG5- and BECLIN1-dependent (Fig. 5 B and C). Importantly, 3-MA treatment or ATG5 or BECLIN1 knockdown significantly reduced the number of DNA leakage structures (based on DAPI staining) and accumulation of hypodiploid cell populations (Fig. 5 B–D), suggesting that autophagy contributes to the arginine starvation-induced DNA leakage phenotype and cell death. In principle, the accumulation of giant autophagosomes that we observed in arginine-starved cells could result from an increase in autophagosome formation and/or fusion, or from a decrease in their degradation, or both. These giant autophagosomes could also be found in very close proximity to the nucleus, sometimes seemingly pushing into/budding out of the nucleus (Movie S1).
The correlation of our quantitative data on time-dependent ROS production (Fig. 4 C and D), 8-OHdG accumulation (Fig. 4E), and the ability of NAC to reverse DNA leakage (Fig. 4F) suggest the involvement of dysfunctional mitochondria-released ROS in chronic arginine starvation-induced genome instability. It is tempting to speculate that the intracellular arginine level might affect chromatin structure, thus retarding DSB repair. The leaked DNA could originate from heterochromatin and thus resist repair. However, the presence of acetyl-H2B may suggest otherwise. Given that the accumulation of hypodiploid cells was autophagy-dependent (Fig. 5C), it is likely that these leaked DNAs were irreversibly lost. Although additional studies are warranted to elucidate the mechanisms underlying DNA loss, it is prudent to conclude that chromatinphagy is distinct from previously reported nucleophagy formation (32, 33) and DNautophagy (34) (see SI Discussion for details).
Our observations strengthen and redefine the dynamic role of arginine starvation-induced autophagy in ASS1-lacking cancer cells, which could be the mechanistic foundation of starvation therapy by FDA-approved ADI-PEG20. Application of ADI-PEG20, or arginine starvation, seems to offer a specific therapeutic advantage of starving tumor cells to death without affecting the normal counterpart and using a cell death mechanism distinct from apoptosis-based conventional therapy. To our knowledge, this is the first demonstration of chromatophagy by arginine deprivation and the first example of the concerted damaging of two cellular compartments, mitochondria and nucleus, by metabolic stress.
Materials and Methods
Microscope Image Acquisition and Image Analysis.
For autophagy image acquisition, cells were imaged with wide-field DeltaVision deconvolution microscope (Applied Precision Inc.), equipped with 60×/1.42 N.A. oil-immersion objective lens. Both microscope and camera were controlled by SoftWorX application suite software. Stacks of optical section images, with an image size of 512 × 512 pixels, were collected for all fluorochromes. For live-cell imaging, cells were maintained and visualized by ONIX microfluidic perfusion system (CellASIC) and DeltaVision deconvolution microscope, respectively. For fixed-cell imaging, all target proteins were labeled with appropriate primary antibody (see SI Materials and Methods). Nucleus was visualized with DAPI or DRAQ5 (BioStatus) staining following the manufacturer’s protocol. Lysosomes were labeled with anti-LAMP1 (DSHB) for fixed cells or by LysoTracker Red (Invitrogen) for live cells following the manufacturer’s protocol. Stacks of fluorescence images were deconvolved by using SoftWorX software (Applied Precision Inc.) and later analyzed with VoloCITY software (PerkinElmer). All cell samples were scored for the presence of DNA leakage, and more than one leaked DNA or captured DNA by autophagosomes per cell reflects ADI-induced DNA damage.
TEM.
The ADI-PEG20–treated CWR22Rv1 cells were collected and loaded into flat specimen holders for high-pressure freezing in an electron microscopy PACT HPF station (Leica Microsystems). The samples were freeze-substituted in acetone containing 0.2% glutaraldehyde and 0.1% uranyl acetone at −90 °C for 72 h and then warmed up gradually to −20 °C (AFS; Leica Microsystems) to complete substitution. After dehydration in ethanol of increasing concentration, followed by a series of Lowicryl–ethanol mixtures, samples were infiltrated with pure Lowicryl resin for 16 h. Finally, the resin polymerization was performed at −50 °C under UV light. The sample block was then trimmed and sectioned with a Leica Ultracut microtome. Sections of 100-nm to 120-nm thickness were collected on copper grids for imaging under a JEOL 1230 electron microscope. The electron micrographs were recorded on a F214CCD camera (TVIPS Gauting).
Additional methods, extended data display items, and discussion are available in Supporting Information.
Supplementary Material
Acknowledgments
We thank Drs. Bor-Wen Wu, John Bomalaski, and Wei-Jen Kung (Polaris, Taiwan) for providing high-quality ADI-PEG20. We also thank GeneTech Biotech (Taiwan) for providing access to the DV microscope. This work was supported in part by National Institutes of Health (NIH) Grants CA165263, CA150197, CA150197S1, NHRI 02D1-MMD0H02 (Taiwan), and DOH102-TD-M-111-102001 (to H.-J.K.), Grant CA150197S1 (to C.A.C.), and Grants DE10742 and DE14183 (to D.K.A.). Y.-R.C. was supported by a City of Hope Women’s Cancers Program Award. This work was also supported by an Isabelle J. McDonald Endowment and Department of Defense Grant W81XWH-08-1-385 (to R.J.B.). The Center for Biophotonics Science and Technology is managed by the University of California, Davis, under Cooperative Agreement PHY 0120999 (to F.Y.S.C.). R.H.C. was supported by NIH Grant AI095382 and Discovery Grant UCDG178969.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 14015.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404171111/-/DCSupplemental.
References
- 1.Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66(18):8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70(3):859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feun L, et al. Arginine deprivation as a targeted therapy for cancer. Curr Pharm Des. 2008;14(11):1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang CC, et al. Arginine deprivation as a new treatment strategy for head and neck cancer. Oral Oncol. 2012;48(12):1227–1235. doi: 10.1016/j.oraloncology.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Müller HJ, Boos J. Use of L-asparaginase in childhood ALL. Crit Rev Oncol Hematol. 1998;28(2):97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 7.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheatley DN, Campbell E. Arginine deprivation, growth inhibition and tumour cell death: 3. Deficient utilisation of citrulline by malignant cells. Br J Cancer. 2003;89(3):573–576. doi: 10.1038/sj.bjc.6601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise DR, Thompson CB. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ascierto PA, et al. Pegylated arginine deiminase treatment of patients with metastatic melanoma: Results from phase I and II studies. J Clin Oncol. 2005;23(30):7660–7668. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- 11.Wetzler M, et al. Effective asparagine depletion with pegylated asparaginase results in improved outcomes in adult acute lymphoblastic leukemia: Cancer and Leukemia Group B Study 9511. Blood. 2007;109(10):4164–4167. doi: 10.1182/blood-2006-09-045351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong H, Zölzer F, von Recklinghausen G, Havers W, Schweigerer L. Arginine deiminase inhibits proliferation of human leukemia cells more potently than asparaginase by inducing cell cycle arrest and apoptosis. Leukemia. 2000;14(5):826–829. doi: 10.1038/sj.leu.2401763. [DOI] [PubMed] [Google Scholar]
- 13.Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res. 2002;62(19):5443–5450. [PubMed] [Google Scholar]
- 14.Bowles TL, et al. Pancreatic cancer cell lines deficient in argininosuccinate synthetase are sensitive to arginine deprivation by arginine deiminase. Int J Cancer. 2008;123(8):1950–1955. doi: 10.1002/ijc.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi E, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9(3):535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 16.Hsueh EC, et al. Deprivation of arginine by recombinant human arginase in prostate cancer cells. J Hematol Oncol. 2012;5:17. doi: 10.1186/1756-8722-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly MP, et al. Arginine deiminase PEG20 inhibits growth of small cell lung cancers lacking expression of argininosuccinate synthetase. Br J Cancer. 2012;106(2):324–332. doi: 10.1038/bjc.2011.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim RH, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009;69(2):700–708. doi: 10.1158/0008-5472.CAN-08-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim RH, Bold RJ, Kung HJ. ADI, autophagy and apoptosis: Metabolic stress as a therapeutic option for prostate cancer. Autophagy. 2009;5(4):567–568. doi: 10.4161/auto.5.4.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin TC, et al. Autophagy: Resetting glutamine-dependent metabolism and oxygen consumption. Autophagy. 2012;8(10):1477–1493. doi: 10.4161/auto.21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syed N, et al. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblastoma. Cell Death Dis. 2013;4:e458. doi: 10.1038/cddis.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shchepina LA, et al. Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21(53):8149–8157. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- 23.Brennan JP, et al. Mitochondrial uncoupling, with low concentration FCCP, induces ROS-dependent cardioprotection independent of KATP channel activation. Cardiovasc Res. 2006;72(2):313–321. doi: 10.1016/j.cardiores.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyo JO, et al. Essential roles of Atg5 and FADD in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita M, et al. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282(9):6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 27.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu F, et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci Signal. 2014;7(319):ra31. doi: 10.1126/scisignal.2004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng PNM, et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67(1):309–317. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 31.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4(5):621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 32.Park YE, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5(6):795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 33.Rello-Varona S, et al. Autophagic removal of micronuclei. Cell Cycle. 2012;11(1):170–176. doi: 10.4161/cc.11.1.18564. [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara Y, et al. Direct uptake and degradation of DNA by lysosomes. Autophagy. 2013;9(8):1167–1171. doi: 10.4161/auto.24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa-Goto K, et al. Microtubule network facilitates targeting of human cytomegalovirus capsid. J Virol. 2003;77(15):8451–8547. doi: 10.1128/JVI.77.15.8541-8547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.