Significance
Understanding the mechanisms of the cell cycle remains challenging. The cell cycle is regulated by the underlying gene regulatory networks. We uncovered the underlying Mexican hat landscape of a mammalian cell cycle network. Three local basins of attraction along the cell cycle loop emerge, corresponding to three distinct cell cycle states: the G1, S/G2, and M phases. Two barriers along the loop characterize G1 and S/G2 checkpoints, respectively, of the cell cycle, which provide a physical explanation for cell cycle checkpoint mechanisms. The cell cycle is determined by two driving forces: curl flux and potential barriers. We uncovered the key gene regulations determining the progression of cell cycle, which can be used to guide the design of new anticancer tactics.
Keywords: cell cycle phases, cell cycle checkpoints, landscape, flux
Abstract
Cell cycles, essential for biological function, have been investigated extensively. However, enabling a global understanding and defining a physical quantification of the stability and function of the cell cycle remains challenging. Based upon a mammalian cell cycle gene network, we uncovered the underlying Mexican hat landscape of the cell cycle. We found the emergence of three local basins of attraction and two major potential barriers along the cell cycle trajectory. The three local basins of attraction characterize the G1, S/G2, and M phases. The barriers characterize the G1 and S/G2 checkpoints, respectively, of the cell cycle, thus providing an explanation of the checkpoint mechanism for the cell cycle from the physical perspective. We found that the progression of a cell cycle is determined by two driving forces: curl flux for acceleration and potential barriers for deceleration along the cycle path. Therefore, the cell cycle can be promoted (suppressed), either by enhancing (suppressing) the flux (representing the energy input) or by lowering (increasing) the barrier along the cell cycle path. We found that both the entropy production rate and energy per cell cycle increase as the growth factor increases. This reflects that cell growth and division are driven by energy or nutrition supply. More energy input increases flux and decreases barrier along the cell cycle path, leading to faster oscillations. We also identified certain key genes and regulations for stability and progression of the cell cycle. Some of these findings were evidenced from experiments whereas others lead to predictions and potential anticancer strategies.
The cell cycle is a series of events that take place in a cell leading to its replication and division. Studying the cell cycle process is essential for understanding cell growth, proliferation, development, and death (1–4). Cell cycle comprises several distinct phases: G1 phase (resting), S phase (synthesis), G2 phase (interphase), and M phase (mitosis). Activation of each phase is dependent on the proper progression and completion of the previous one, which can be monitored by cell cycle checkpoints. It is now believed that all proliferation, differentiation, and cell death processes are controlled by the underlying gene regulatory networks (5), which often involve many complex feedback loops (6). The complexity of the large regulatory networks for the cell cycle makes it difficult to understand the global natures and connections between the underlying network and the cell cycle process. Furthermore, in the cell, the intrinsic fluctuations from the finite number of molecules and extrinsic fluctuations from dynamical and inhomogeneous environments (7, 8) coexist. Therefore, stochastic approaches are often required to explore the nature of the underlying network of chemical reaction soups (9–14). The challenge is how to understand the global picture and physical principles for the stability and function of the cell cycle from the underlying gene regulatory network. For example, how do the seemingly many possible gene expression patterns lead to limited specific functions?
Here, we suggest a new global view of the mammalian cell cycle in terms of probability landscape and flux to address the challenge. The probabilistic landscape approach may provide a possible answer, as the significance of each state can thereby be distinguished according to its associated weight. Functional states may correspond to higher probability of occurrence and occupy lower potential valleys (15–20). The state space of gene regulatory networks includes states with different gene expression patterns in the cell, which further determine different cellular functions. Using the landscape concept (15–20), cell types can be represented by the basins of attraction on the landscape reflecting the probability of appearance. By quantifying the topography of the potential landscape in terms of barrier heights between basins, we can explore the global stability and function of the cell cycle. Furthermore, the dynamics of the nonequilibrium system is determined by both landscape gradient and curl flux (16).
We found that the landscape of the cell cycle network depicts a Mexican hat shape, which guarantees the stability of the cell cycle process. Both the landscape and the curl flux determine the dynamics of oscillation (16). Landscape attracts the cell to the oscillation ring, and the curl probability flux drives the coherent oscillations along the ring path. We found the emergence of several local basins of attraction on the potential landscape and saddle points along the limit cycle trajectory. Here the three local basins of attraction on the potential landscape characterize the different phases (G1, S/G2, M) of cell cycle progression. The saddle points (or barriers) between different local basins of attraction on the landscape characterize the checkpoints (G1 checkpoint and S/G2 DNA replication checkpoint) in different stages of the cell cycle, thus providing an explanation for the checkpoint mechanism of cell cycle from the physical perspective. Landscape topography based on the barrier heights separating different basins of attraction provides a quantitative measure of the global stability of the complex network system. We quantify three barriers in the landscape picture. One of them characterizes the global stability of the whole system. The other two reflect the major barriers that the cell needs to overcome along the cycle path to complete the cell cycle. They also characterize the different cell cycle checkpoints. Therefore, the cell cycle can be facilitated in two ways. The first is by enhancing the curl flux as the driving force, and the other is by lowering the potential barriers along the cell cycle path. We found that both the entropy production rate and energy per cell cycle increase as the growth factor increases. This reflects that cell growth and division are driven by energy or nutrition supply. More (less) energy input increases (decreases) flux and decreases (increases) barriers along the cell cycle path, leading to faster (slower) oscillations. Using global sensitivity analysis for the key genes and wirings on landscape topography in terms of barrier heights, we identified certain key elements or wirings that control the stability and the progression of cell cycle. Some of our results were evidenced through experiments, and in some cases lead to predictions and potential anticancer strategies.
Results and Discussion
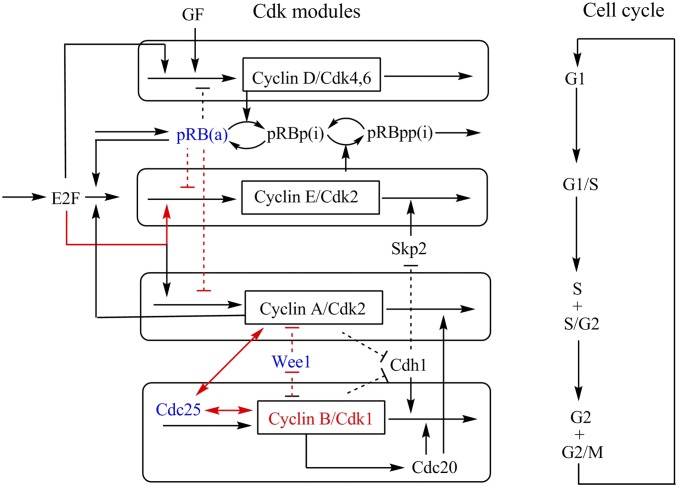
Great efforts have been devoted to investigating cell cycle regulatory wirings (1–3, 17, 21–23). A mammalian cell cycle network wiring has been constructed (Fig. 1) (21–23). It includes the main regulations in the mammalian cell cycle. The network involves four major complexes formed by cyclins and cyclin-dependent kinases (cyclin/CDks), centered on cyclin D/Cdk4-6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1, which together determine the cell cycle dynamics. The mutual repression regulations between the tumor suppressor retinoblastoma protein (pRB) and the trascription factor E2F control the cell cycle progression.
Fig. 1.
The diagram for the mammalian cell cycle model (see SI Appendix, Fig. S11, for a more detailed diagram). Arrows represent activation, and dotted lines with short bars represent repression. The model includes four major cyclin/Cdk complexes centered on cyclin D/Cdk4-6, cyclin E/Cdk2, cyclin A/Cdk2, and cyclin B/Cdk1. The opposite effects of pRB and E2F control the cell cycle progression. The combined effects of four modules determine the cell cycle dynamics of oscillation. Red colors represent the key genes and regulations found by global sensitivity analysis. Blue colors represent the key genes found by global sensitivity analysis, which are consistent with experiments.
At the beginning of the cell cycle, the growth factor (GF) promotes the synthesis of cyclin D and further cyclin D/Cdk4-6 (Module 1). The active forms of cyclin D/Cdk4-6 and cyclin E/Cdk2 (Module 2) ensure the progression in G1 and elicit the G1/S transition by inhibiting pRB. The deactivation of pRB ensures the activation of E2F, which leads to cell cycle progression by promoting the synthesis of G1 cyclins (Module 2). During S and G2 (Module 3), cyclin A/Cdk2 inhibits the Cdh1 that promotes the degradation of cyclin B. The negative feedback loops exerted, via Cdc20 activation, by cyclin B/Cdk1 on itself and cyclin A/Cdk2 (Module 4), and the negative feedback loop exerted by cyclin A/Cdk2 on E2F (Module 3), allow the reset of the cell cycle and the start of a new round of oscillations. Inhibitory phosphorylation by Wee1 and activating dephosphorylation by the Cdc25 regulate the activity of Cdk1 and Cdk2.
Based on mass action or Michaelian kinetics, the dynamics of the model is described by a set of nonlinear ordinary differential equations (44 ODE; see SI Appendix, ODE for 44 Variable Mammalian Cell Cycle Model for detailed equations and SI Appendix, Table S2, for parameter values).
Landscape and Flux of the Mammalian Cell Cycle System.
For nonequilibrium dynamical systems such as cell cycle networks, the main driving force for the dynamics is determined by both the landscape and the flux. The landscape reflects directly the steady state probability distribution, giving the weight of each state, and therefore can be used to quantify the global stability and behavior. On the other hand, the flux has the curl nature, which breaks the detailed balance and time reversal symmetry (see details in SI Appendix, Landscape and Flux Decomposition).
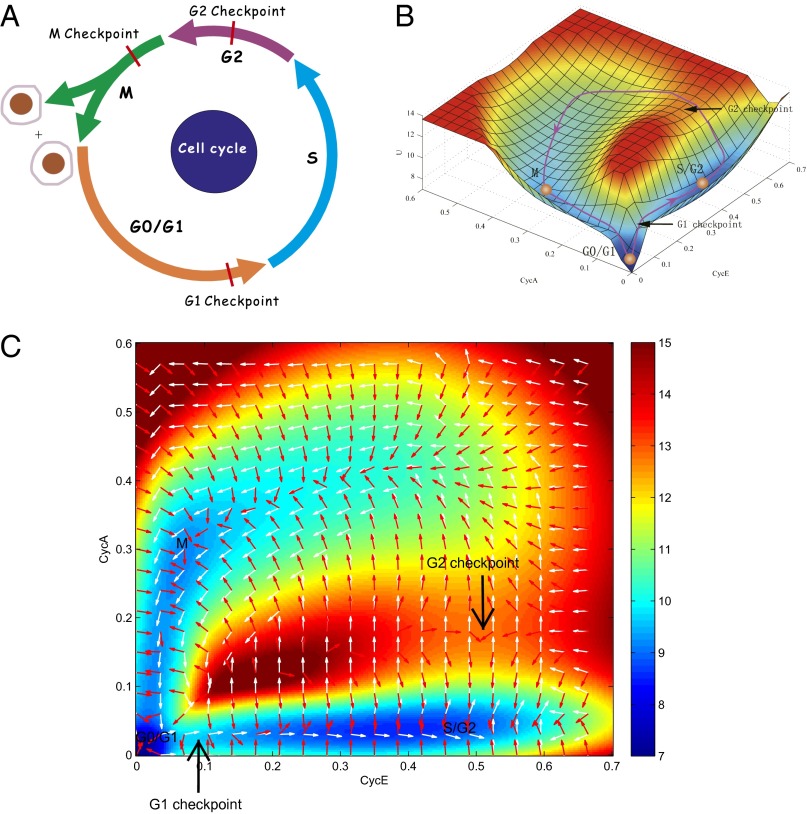
Fig. 2A shows the typical textbook illustration of four phases of cell cycle with three checkpoints: G1, S, G2, and M phase. Based on a mammalian cell cycle network (21–23), we uncovered the underlying landscape and flux. Applying the self-consistent mean field approximation (15, 17, 19), we quantified the steady state probability distribution Pss and the potential landscape U (U = −lnPss) (15–19, 24–26). By projecting a 44-dimensional landscape (in terms of the 44 gene expression variables with each variable representing the expression level of each gene in the gene network) to a two-dimensional state space, we quantified the landscape of the mammalian cell cycle system in terms of two key proteins CycE (cyclin E/Cdk2) and CycA (cyclin A/Cdk2) as shown in Fig. 2 B and C (our new landscape view for cell cycle). The landscape has a Mexican hat shape (SI Appendix, Fig. S1, shows the corresponding landscape using a different coordinate pair CycE and CycB). The blue colored region along the ring represents lower potential or higher probability, which also corresponds to the oscillation trajectory. Inside or outside of the Mexican hat ring (red colored region), the potential is higher and the probability for the system to reach these regions is lower. The Mexican hat shape guarantees the stability and robustness of cell cycle oscillation dynamics. Fig. 2C shows the curl probability flux on the landscape background. White arrows represent the probabilistic flux, and red arrows represent the negative gradient of potential landscape. We found that the landscape and the curl flux are the driving forces of the cell cycle. The force from the negative gradient of potential attracts the cell cycle into the oscillation ring, and the flux drives the cell cycle oscillations along the ring path.
Fig. 2.
(A) The four phases of cell cycle with the three checkpoints: G1, S, G2, and M phase. (B) The three phases (G1, S/G2, and M phase) and the two checkpoints (G1 checkpoint and S/G2 checkpoint) in our landscape view (in terms of gene CycE and CycA). (C) The 2D landscape, in which white arrows represent probabilistic flux, and red arrows represent the negative gradient of potential. The diffusion coefficient D is 0.05.
In Fig. 2B, we labeled the three phases of cell cycle progression along the cell cycle trajectory (magenta trajectory: three basins along the limit cycle trajectory), which separately correspond to biological G1, S/G2, and M phases, and two checkpoints (G1 checkpoint and S/G2 checkpoint). In the cell cycle model we used, the S phase and G2 phase are merged together to become an S/G2 attractor state. Therefore, the four phases become three basins on the landscape and three checkpoints become two (G2 checkpoint and M checkpoint are merged together). We noticed that the cell cycle path is not smooth. Starting the cycle from the G0/G1 basin, the cell needs to overcome two major barriers (two saddle points on landscape) along the cycle path, respectively located between the G1 and S/G2 basins as well as between the S/G2 and M basins, to complete the cell cycle. Here, the landscape from the M basin to the G1 basin is rather flat, i.e., the barrier from M to G1 is not very significant. Therefore, we only consider the two major barriers (the barrier between the G1 and S/G2 basins as well as the barrier between the S/G2 and M basins) along the cell cycle path. This new landscape picture reflects the cell cycle dynamics globally and quantitatively. It provides pictorial yet physical and quantitative explanations for the checkpoint mechanisms of cell cycle. Biologically, cell cycle checkpoints are understood to be the control that ensures the fidelity of cell divisions. It determines whether or not the cell cycle process at its current phase is accurately completed and able to progress into the next phase. In our landscape picture, there are two forces along the cell cycle path determining the progression of the cell cycle. One is the probabilistic flux, which promotes the coherent oscillation (cell cycle forward). The other is the negative potential gradient along the path, which always drives the system to the direction with lower potential.
When the cell begins the cycle from the G0 state, it will encounter the first barrier (BarrierG1/S) characterizing the G1 checkpoint. At this stage, the flux provides the driving force pointing to the forward direction of the cell cycle, whereas the negative gradient of potential points to the opposite direction. At the G1 checkpoint, the combined effects of flux and potential gradient determine the outcome of cell cycle progression. If the driving force from the flux is larger than the backward force from the potential gradient, the cell will go over the G1 checkpoint (BarrierG1/S or saddle point G1/S) and the cell cycle will continue. If the force from the flux is smaller than the backward force from the potential gradient, i.e., if the driving force of the cell cycle is not large enough (biologically, this means that the nutrition supply is not large enough, or the DNA is damaged), the cell will go back to the G0 basin. The cell cycle will then stop at the G0 state until the driving force is large enough (e.g., the GF initiated from the nutrition supply is large enough, or the DNA damage is repaired), which will drive the cell to pass over the G1 checkpoint. In the same way, the second barrier (BarrierG2/M) along the cell cycle path characterizes the S/G2 checkpoint (DNA replication checkpoint), whose function is to make sure that DNA replication is completed before moving to the next phase M. This can be realized with a sufficiently large flux over the landscape gradient at the S/G2 checkpoint.
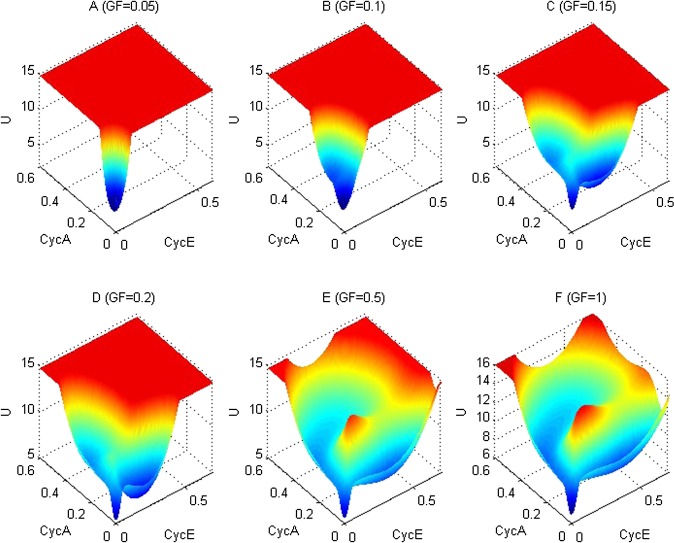
To investigate the influence of GF on the potential landscape, we also show the change of landscape (Fig. 3) when GF is gradually increased (from 0.05 to 1). At small GF, the system exhibits a monostable landscape (a stable steady state corresponding to cell quiescence or G1 state). As the GF grows, the system evolves from a monostable basin at G1 to a stage where local basins of S/G2 and M gradually form (Fig. 3 A−D). As the GF increases further, the central island starts to develop (Fig. 3 E and F) and the system evolves to a robust oscillation stage (corresponding to cell proliferation and division) where the Mexican hat landscape emerges with three local basins.
Fig. 3.
Landscape changes when GF is gradually increased (from 0.05 to 1). At small GF, the system has a stable steady state (monostable landscape), whereas when the GF (representing nutrition supply) is increased, the system evolves to oscillation solutions and the Mexican hat landscape emerges. The diffusion coefficient D is 0.05.
The Effects of the Growth Factor and Other Parameters on Cell Cycle Function.
To quantify the landscape topography, we define three barrier heights for the cell cycle. BarrierG1/S is defined as the barrier from the G0/G1 basin to the saddle point G1/S (G1 checkpoint). BarrierG2/M is defined as the barrier from the S/G2 basin to the saddle point G2/M [the DNA replication checkpoint (27)] along the cell cycle path. BarrierCenter is defined as the potential difference between the global potential minimum and the local maximum inside the cycle ring (the central top of the Mexican hat). BarrierCenter can be used to quantify the global stability of the cell cycle (16). Higher BarrierCenter leads to more robust cell cycle oscillation. Additionally, to quantify the curl probabilistic flux, we define FluxIntLoop as the flux integration along the loop trajectory of the cell cycle oscillation divided by the loop length in gene expression state space (; see SI Appendix for details) (28, 29).
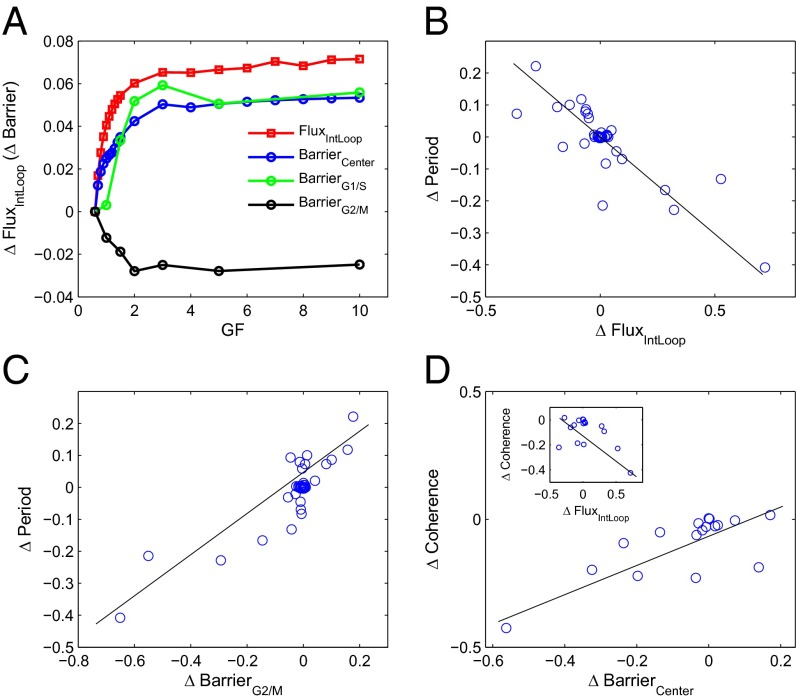
GF, characterizing the initiation signal, induces the expression of cyclins and controls the strength of cell cycle progression. Here, we change the GF to explore the effects of GF on the landscape. Fig. 4A shows the change in the barrier and the flux when GF is changed. ΔFluxIntLoop is defined as the percentage of change for FluxIntLoop relative to the point at GF = 0.6, i.e., ΔFluxIntLoop = (FluxIntLoop − FluxIntLoop(GF=0.6))/FluxIntLoop(GF=0.6). We use this normalization (as well as for the other three curves in Fig. 4A) to display the four curves together and facilitate comparisons. We can see that the BarrierCenter, characterizing the global stability of oscillation system, increases as the GF increases. The GF promotes the cell cycle along the oscillating ring from two perspectives. First is by increasing the flux, since the FluxIntLoop increases as GF increases (Fig. 4A). Second is by lowering the barrier along the cell cycle oscillation path on the landscape, as we can see that BarrierG2/M decreases as GF increases. We notice that the BarrierG1/S increases as GF increases. However, BarrierG2/M is much larger than BarrierG1/S. BarrierG2/M is, therefore, the most critical barrier (or rate limiting step) for the system to go over along the oscillation trajectory. The increase of the BarrierG2/M thus determines the global cycle behavior, and the cell cycle is promoted. Comprehensively, the landscape picture tells us that the GF controls cell cycle behavior by influencing both flux and BarrierG2/M (landscape topography) along the cycle. This implies that the landscape and flux are both vital to the oscillation dynamics of the cell cycle.
Fig. 4.
(A) The change (percentage) of FluxIntLoop, BarrierCenter, BarrierG1/S, and BarrierG2/M when GF is changed. (B) The correlation between Δflux and Δperiod when parameters (regulation strengths or synthesis rates) are changed (correlation coefficient is −0.839). (C) The correlation between ΔBarrierG2/M and Δperiod when parameters are changed (correlation coefficient is 0.879). (D) The correlation (correlation coefficient is 0.741) between ΔBarrierCenter (Δflux for inner plot; correlation coefficient is −0.553) and Δcoherence when parameters are changed.
The flux results in Fig. 4A are based on the two-dimensional projection of the 44-dimensional system. We also performed the flux integrations along the loop when the entire 44 dimensions are considered (SI Appendix, Fig. S2A). We can see from SI Appendix, Fig. S2A, that for the 44-dimensional case, the trend of the curve for the flux integration along the loop (FluxIntLoop) versus GF is similar to the two-dimensional case (Fig. 4A), being that the flux increases with the increase in GF. This consistency indicates that the flux also works in high-dimensional space and it is reasonable to use the two-dimensional flux projection.
We also investigated the effects of other parameters (regulation strengths or synthesis rates) on the cell cycle. Here, we use the period as a measure for cell cycle progression strength, because the period reflects how fast the cell cycle oscillates and its growth speed. By investigating the correlations among ΔPeriod (period change induced by parameter changes), ΔFluxIntLoop, ΔBarrierCenter, ΔBarrierG1/S, and ΔBarrierG2/M (SI Appendix, Fig. S4), we found that the period has a good anticorrelation with flux (Fig. 4B, correlation coefficient equals −0.839) and a positive correlation with BarrierG2/M (Fig. 4C, correlation coefficient equals 0.879). The flux often takes the form of GF signaling or nutrition supply to the cell, and so characterizes the energy input. Larger flux means more energy input or larger cell cycle driving force and therefore leads to less time to finish cell cycle oscillation (the period is smaller). On the other hand, BarrierG2/M characterizes the major barrier along the oscillation loop that the cell needs to overcome to complete the cell cycle. Therefore, the larger the BarrierG2/M, the more time the cell cycle needs to complete and the larger the period. This also implies that the cell cycle can be promoted in two ways: by enhancing the flux (representing the energy input) or by lowering the barrier (BarrierG2/M as the key barrier) along the cell cycle path.
We also explored coherence as the quality of the cell cycle progression (definition given in SI Appendix, Definition of Phase Coherence). We observed that when the barriers increase (both central barrier and BarrierG2/M), the coherence increases (Fig. 4D and SI Appendix, Fig. S5D). This demonstrates that a stable cell cycle oscillation has better quality of progression. We find that the coherence has a certain anticorrelation with the flux (Inset in Fig. 4D, the correlation coefficient is −0.55). This implies the existence of competition (balance) between stability and speed of a cell cycle (i.e., a faster cycle may disturb the sequential order of the oscillation and lead to lower quality of oscillation, with less coherence or a less stable system). This indicates that a cell needs appropriate cycle speed (to be functional), balanced with appropriate stability (to be stable).
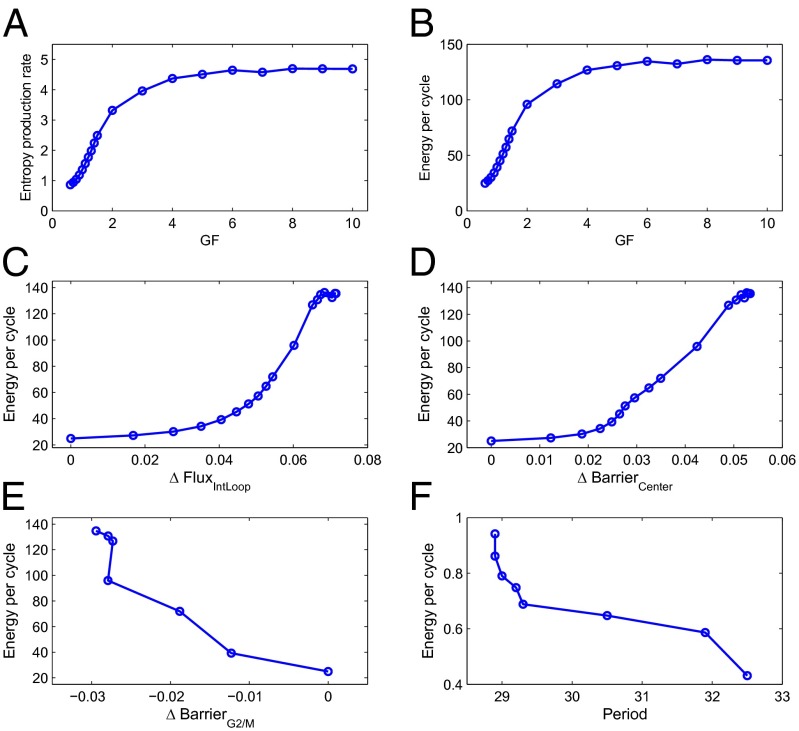
To explore the origin of the flux, we calculated the changes of the entropy production rate (EPR) (Fig. 5A) and the energy per cycle (Fig. 5B) with respect to the GF (see SI Appendix for detailed methods of calculating the EPR). The energy per cycle is quantified by the EPR times the period of oscillation. We see that both the EPR and the energy per cell cycle monotonically increase with the increase of the GF. This reflects that cell growth and division are driven by energy or nutrition supply. From the definition of EPR (see SI Appendix), we can see clearly that the entropy production and energy per cycle are directly related to the flux. Zero flux corresponds to zero entropy production and zero energy per cycle. We can see that the origin of the flux is the chemical energy from the nutrition supply. Fig. 5 C−F individually shows the changes in energy per cell cycle with respect to the Flux, BarrierCenter, BarrierG2/M, and Period. We can see that the energy per cycle increases as the Flux or the BarrierCenter increases, whereas it decreases as the BarrierG2/M or the Period increases. The larger flux costs more energy (see definition of entropy production in SI Appendix). The larger BarrierCenter means more stable system with higher coherence, and it consumes more energy to maintain the stability of the system. BarrierG2/M stands for the major barrier or obstacle along the cell cycle loop. Therefore, larger BarrierG2/M will make the system easier to trap, and cost less energy. Furthermore, a faster oscillation (smaller period) costs more energy, and therefore leads to a larger energy per cycle.
Fig. 5.
(A and B) The change of EPR and energy per cell cycle increase as GF is increased. (C−F) The Flux (C), BarrierCenter (D), BarrierG2/M (E), and Period (F) versus the energy per cell cycle with GF changed. It can be seen that the energy per cycle increases as the Flux and BarrierCenter increase, while it decreases as the BarrierG2/M and Period increase.
Global Sensitivity Analysis of Key Genes and Regulations.
The cell cycle is a process involving many genes and regulations among genes. Therefore, we did a global sensitivity analysis on the function through the oscillation period, and the landscape topography through checkpoints and central island barriers as well as the curl flux (flux integrated over the cell cycle path), to find those key genes or regulatory wirings for the cell cycle (SI Appendix, Fig. S3).
By selecting the top influential parameters (regulation strengths or synthesis rates) in terms of the genes and regulations of the network (those influencing the checkpoints and central island barriers, the flux, and the period most in SI Appendix, Fig. S3), we identified certain key factors or hot spots for the cell cycle progression. SI Appendix, Table S1, provides a summary of major results from our global sensitivity analysis. Some of these have been confirmed by experiments as indicated. For example, pRB serves as a key gene in controlling the G1 checkpoint, as its activation represses the cell cycle and also represses the cancer (30–32). The results from our global sensitivity analysis (SI Appendix, Fig. S3) are consistent with the above findings, where activation of pRB leads to the increase of the period, the decrease of the flux, and the increase of the BarrierG2/M as well as the elongation of the cell cycle (parameter 3 represents the synthesis rate of pRB). We also provide predictions (SI Appendix, Table S1), which can be further tested by experiments. We marked the key sensitivity analysis results (key genes and regulations) in the wiring diagram for the cell cycle network (Fig. 1) in red.
In summary, the driving force for cell cycle dynamics is determined by the landscape topography and the curl flux (from chemical energy pump and, in this case, the nutrition supply). The perturbations in underlying genes (nodes) and regulations (wirings) causing the significant changes in landscape barriers and curl flux (flux integral along the loop) (SI Appendix, Fig. S3) will disturb the function (oscillation period). Therefore, through quantifying the dependence of the two driving forces, we can identify the key genes and wirings for the global stability and function.
Conclusions
Beyond our previous works on the landscapes for limit cycle systems (16, 17, 24, 28, 33), our current work exhibits the following novel results: (i) We uncovered the underlying Mexican hat landscape of a mammalian cell cycle gene regulatory network. (ii) More importantly, for the first time, we identified and quantified different cell cycle phases as landscape basins on the cell cycle ring; we identified and quantified the checkpoints of the cell cycle as the potential barriers along the cycle ring. This provides an intuitive yet quantitative explanation for the checkpoint mechanism of the cell cycle—a persistent focal problem in cell biology—from the physical perspective. (iii) We identified and quantified the global driving forces for the cell cycle progression—landscape barriers and cycle flux. Especially, we identified and quantified the nutrition supply as the origin of the flux. For the cell cycle progression, the curl flux is responsible for driving the oscillation on the cycle ring, while the potential barriers act to block the oscillation along the cycle path where the different phases and checkpoints of the cell cycle emerge. These illustrate the importance and quantification of barriers and the curl flux along the cell cycle.
In the current work, we focus on the effects of external noises (environment changes) on the system, and the additive noise is applied. We have investigated the landscapes of some biological systems under external noises (16–19, 34) and under intrinsic noises (multiplicative noise is used) (24, 33), previously. In mammalian cells, the number of protein molecules is often abundant; therefore it is expected that the source of fluctuations is mainly from the extrinsic noises, rather than from the intrinsic noises.
Compared with the previous works on cell cycle modeling (1–4, 21–23), which mostly focused on the deterministic dynamics, our results from the landscape view provide new insights and explanations for cell cycle mechanisms. Different from Tyson’s model (1–3), the stochastic model we used does not have periodic time-dependent mass as a driving force variable. For this case, the cell cycle is driven by the nutrition supply (not time dependent) and depends on the threshold of GF. The threshold controls the switching of the behavior from monostable state, to multiple states coexisting, and finally to oscillations on the landscape (Fig. 3). Additionally, Tyson and Novak (4) proposed a “gate control”-like model to explain the cell cycle progression and the cell cycle checkpoints. However, there were no quantitative results for checkpoints, and the physical picture is not completely clear in their work. In this work, we identified local basins as the phases and potential barriers as the checkpoints along the cell cycle ring. These findings provide a physical foundation and quantification of different cell cycle phases and checkpoints. They also give an intuitive yet quantitative explanation for the mechanism of cell cycle checkpoints. We quantified the two cell cycle driving forces: the potential barrier for forming basins/checkpoints of the cell cycle and the flux for driving the cell cycle. Therefore, the cell cycle can be promoted (suppressed), either by enhancing (suppressing) the flux (representing the energy input) or by lowering (increasing) the barrier (BarrierG2/M as the decisive barrier) along the cell cycle path. These can be realized by perturbing the genes or regulations based upon the results of our global sensitivity analysis according to the topography of landscape and flux (SI Appendix, Fig. S3). The key genes and regulations identified from the global sensitivity analysis regarding, especially, the period (fast for cancer and slow for normal cells) of the cell cycle suggest potential anticancer strategies. Our approach is general and can be applied to other gene regulatory networks or protein networks.
Materials and Methods
The statistical nature of the chemical reactions can be captured by corresponding diffusion equations, which describe the evolution of the networks probabilistically. It is hard to solve a diffusion equation due to its inherent huge dimensions. We therefore apply a self-consistent mean field approximation to reduce the dimensionality (15, 17, 19, 35). In this way, we can follow the time evolution and steady state probability of the protein concentrations, and finally map out the potential landscape, which is closely associated with steady state probability distribution.
The state dynamics of the cell cycle can be quantitatively described through a probabilistic evolution dictated by the Fokker−Planck diffusion equation: P(X1, X2, …, Xn, t) where X1, X2,…, Xn are the concentrations of proteins or populations of molecules. We expected to have an N-dimensional partial differential equation, which is not feasible to solve, because if every variable can have M values, then the dimensionality of the system becomes MN. Following a self-consistent mean field approach (15, 17, 19, 36), we split the probability into the products of individual ones: and solve the probability self-consistently. This effectively reduces the dimensionality from MN to M × N and therefore makes the problem computationally tractable (see details in SI Appendix, Self-Consistent Mean Field Approximation).
However, for the multidimensional system, it is still hard to solve the diffusion probability directly. We can start from moment equations and then simply assume specific probability distribution based on physical arguments, i.e., we assume some specific connections between moments (36). In principle, once we know all of the moments, we can construct probability distribution. For example, the Poisson distribution has only one parameter, so we may calculate all other moments from the first moment, i.e., the mean. Here we assume a Gaussian distribution as an approximation (25, 26); then we need two moments, mean and variance.
When the diffusion coefficient D is small, the moment equations could be approximated to (25, 26):
| [1] |
| [2] |
Here, x, σ(t), and A(t) are vectors and tensors, and AT(t) is the transpose of A(t). The matrix elements of A are . According to these equations, we can solve x(t) and σ(t). We consider here only diagonal element of σ(t) from mean field splitting approximation. Therefore, the evolution of distribution for one variable can be obtained using the mean and variance by Gaussian approximation:
| [3] |
We can extend the current results to the multidimensional system by considering the total probability as the product of the individual probability for each variable from the mean field splitting mentioned at the beginning of this section. Finally, once we have the total probability, we can quantify the potential landscape by: U(x) = −lnPss(x).
Supplementary Material
Acknowledgments
C.L. thanks Mr. Dick Goldman and Mr. Peter Soo for help in editing the English text. This work was supported by the National Science Foundation. J.W. acknowledges support from the National Science Foundation of China (Grants 21190040 and 11174105) and the 973 Project of China (2009CB930100 and 2010CB933600).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408628111/-/DCSupplemental.
References
- 1.Tyson JJ, Novak B. Regulation of the eukaryotic cell cycle: Molecular antagonism, hysteresis, and irreversible transitions. J Theor Biol. 2001;210(2):249–263. doi: 10.1006/jtbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 2.Chen KC, et al. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11(1):369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen KC, et al. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15(8):3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyson JJ, Novak B. Temporal organization of the cell cycle. Curr Biol. 2008;18(17):R759–R768. doi: 10.1016/j.cub.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hejmadi M. Introduction to Cancer Biology. London: Bookboon; 2009. [Google Scholar]
- 6.Qi H, Blanchard A, Lu T. Engineered genetic information processing circuits. WIREs Syst. Biol Med. 2013;5(3):273–287. doi: 10.1002/wsbm.1216. [DOI] [PubMed] [Google Scholar]
- 7.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99(20):12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98(15):8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 10.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295(5560):1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 11.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1996;93(19):10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311(5767):1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 13.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 14.Kar S, Baumann WT, Paul MR, Tyson JJ. Exploring the roles of noise in the eukaryotic cell cycle. Proc Natl Acad Sci USA. 2009;106(16):6471–6476. doi: 10.1073/pnas.0810034106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasai M, Wolynes PG. Stochastic gene expression as a many-body problem. Proc Natl Acad Sci USA. 2003;100(5):2374–2379. doi: 10.1073/pnas.2627987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Xu L, Wang E. Potential landscape and flux framework of nonequilibrium networks: Robustness, dissipation, and coherence of biochemical oscillations. Proc Natl Acad Sci USA. 2008;105(34):12271–12276. doi: 10.1073/pnas.0800579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Li C, Wang E. Potential and flux landscapes quantify the stability and robustness of budding yeast cell cycle network. Proc Natl Acad Sci USA. 2010;107(18):8195–8200. doi: 10.1073/pnas.0910331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Zhang K, Xu L, Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc Natl Acad Sci USA. 2011;108(20):8257–8262. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wang J. Quantifying cell fate decisions for differentiation and reprogramming of a human stem cell network: Landscape and biological paths. PLOS Comput Biol. 2013;9(8):e1003165. doi: 10.1371/journal.pcbi.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ao P. Global view of bionetwork dynamics: Adaptive landscape. J Genet Genomics. 2009;36(2):63–73. doi: 10.1016/S1673-8527(08)60093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gérard C, Goldbeter A. Temporal self-organization of the cyclin/Cdk network driving the mammalian cell cycle. Proc Natl Acad Sci USA. 2009;106(51):21643–21648. doi: 10.1073/pnas.0903827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gérard C, Goldbeter A. Entrainment of the mammalian cell cycle by the circadian clock: Modeling two coupled cellular rhythms. PLOS Comput Biol. 2012;8(5):e1002516. doi: 10.1371/journal.pcbi.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gérard C, Goldbeter A. From simple to complex patterns of oscillatory behavior in a model for the mammalian cell cycle containing multiple oscillatory circuits. Chaos. 2010;20(4):045109. doi: 10.1063/1.3527998. [DOI] [PubMed] [Google Scholar]
- 24.Li CH, Wang J, Wang EK. Landscape and flux decomposition for exploring global natures of non-equilibrium dynamical systems under intrinsic statistical fluctuations. Chem Phys Lett. 2011;505(1-3):75–80. [Google Scholar]
- 25.Van Kampen NG. Stochastic Processes in Chemistry and Physics. 1st Ed. Amsterdam: North-Holland; 1992. pp. 120–127. [Google Scholar]
- 26.Hu G. In: Stochastic Forces and Nonlinear Systems. Hao B, editor. Shanghai: Shanghai Sci Technol Educ Press; 1994. pp. 68–74. [Google Scholar]
- 27.Dart D, Adams K, Akerman I, Lakin N. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J Biol Chem. 2004;279(16):16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Shi H, Feng H, Wang J. The energy pump and the origin of the non-equilibrium flux of the dynamical systems and the networks. J Chem Phys. 2012;136(16):165102. doi: 10.1063/1.3703514. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K, Sasai M, Wang J. Eddy current and coupled landscapes for nonadiabatic and nonequilibrium complex system dynamics. Proc Natl Acad Sci USA. 2013;110(37):14930–14935. doi: 10.1073/pnas.1305604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seville LL, Shah N, Westwell AD, Chan WC. Modulation of pRB/E2F functions in the regulation of cell cycle and in cancer. Curr Cancer Drug Targets. 2005;5(3):159–170. doi: 10.2174/1568009053765816. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Wang E, Wang J. Landscape, flux, correlation, resonance, coherence, stability, and key network wirings of stochastic circadian oscillation. Biophys J. 2011;101(6):1335–1344. doi: 10.1016/j.bpj.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han B, Wang J. Quantifying robustness and dissipation cost of yeast cell cycle network: The funneled energy landscape perspectives. Biophys J. 2007;92(11):3755–3763. doi: 10.1529/biophysj.106.094821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Wolynes P. Stem cell differentiation as a many-body problem. Proc Natl Acad Sci USA. 2014;111(28):10185–10190. doi: 10.1073/pnas.1408561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KY, Wang J. Potential energy landscape and robustness of a gene regulatory network: Toggle switch. PLOS Comput Biol. 2007;3(3):e60. doi: 10.1371/journal.pcbi.0030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.