In response to the concerns of Teasdale and Collins (1) regarding of our report (2), our comments are as follows.
First, the fully intact FAM65B (family with sequence similarity 65, member B) spans a polypeptide chain of 1,068 residues. Because the genetic deletion that we identified spans residues 34–86 (Δ34–86), we restricted our structural analysis of FAM65B exclusively to its N-terminal region (residues 1–300) in lieu of the whole protein, as Teasdale and Collins (1) seem to have done. It is unlikely that, using the whole protein as a bait, one would find any sort of hit, no matter what database is searched.
Second, the initial search for proteins that potentially harbored homology to the N-terminal region (residues 1–300) of FAM65B was conducted using the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). Among several proteins identified as potential templates for homology modeling, the phox homology (PX) and bin/amphiphysin/rvs (BAR) module of sorting nexin 33 (SNX33) (PDB ID code 4AKV) ranked the best. Because we demonstrated that FAM65B localizes to cell membranes, we were particularly motivated to use SNX33 as a template to shed structural light into how the Δ34–86 deletion might impair the physiological function of FAM65B.
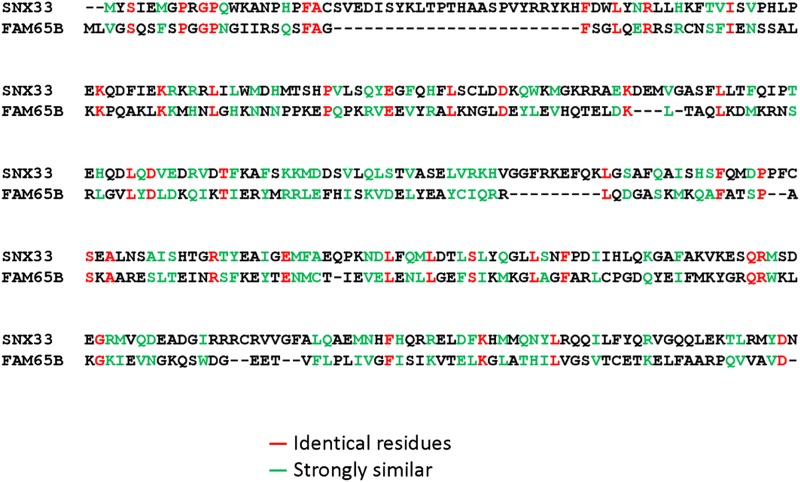
Third, amino acid sequence alignment of the N-terminal region (residues 1–300) of FAM65B with the PX–BAR module of SNX33 used to build our structural model is provided for your reference (Fig. 1). Of these 300 residues, 138 are either identical or strongly similar to the corresponding PX–BAR module of SNX33, in agreement with 40% sequence similarity mentioned in our report (2).
Fig. 1.
Amino acid sequence alignment of PX–BAR module of SNX33 with the N-terminal region (residues 1–300) of FAM65B.
Fourth, although we acknowledge the poor sequence similarity between FAM65B and SNX33, one cannot rule out the presence of the PX–BAR module in FAM65B, particularly in light of its demonstrated potential to localize to plasma membranes. Indeed, many protein domains adopt similar structural folds, despite their low sequence similarities.
Finally, of all known protein structures, our structural analysis merely suggests that the N-terminal region (residues 1–300) of FAM65B bears best similarity to the PX–BAR module of SNX33. Whether it is indeed a bona fide PX–BAR module can only be verified experimentally. We invite Teasdale and Collins to pursue this goal in their future endeavors, as it bears the potential to be a novel PX–BAR module.
Footnotes
The authors declare no conflict of interest.
References
- 1.Teasdale RD, Collins BM. Little evidence that FAM65B belongs to the family of phox homology (PX) and bin/amphiphysin/rvs (BAR) domain-containing proteins. Proc Natl Acad Sci USA. 2014;111:E4064. doi: 10.1073/pnas.1412755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz-Horta O, et al. FAM65B is a membrane-associated protein of hair cell stereocilia required for hearing. Proc Natl Acad Sci USA. 2014;111(27):9864–9868. doi: 10.1073/pnas.1401950111. [DOI] [PMC free article] [PubMed] [Google Scholar]