Our increasing awareness over the last decade, that reprogramming of cellular energetics is a key hallmark of tumorigenesis, has reinvigorated the search for novel tractable metabolic targets in cancer cells (1). From innovative diagnostics based on the tracer 18F-deoxyglucose in positron emission tomography (FDG-PET) to the development of next-generation antimetabolites built on l-asparaginase, methotrexate, and 5-fluorouracil, there is excitement that a deeper understanding of tumor metabolism will revolutionize patient care (2). Modulating the key substrates glucose and glutamine has great appeal because these provide the essential carbon source for many anabolic cellular processes required during tumor growth (3). However, it has become apparent that there are many other metabolites fueling tumorigenesis. These metabolites range from the rare “oncometabolite” 2-HG in isocitrate dehydrogenase mutant tumors to critical roles for a wider array of amino acids, including serine, glycine, proline, and especially arginine (4–7). Targeting arginine has been the focus of preclinical and promising clinical activity driven by the availability of novel arginine depletors and an appreciation that a range of hematological and solid cancers are defective in urea cycle enzymes, namely argininosuccinate synthetase 1 (ASS1) and argininosuccinate lyase (8, 9). Arginine is a highly versatile amino acid required for the synthesis of proteins, nitric oxide, polyamines, nucleotides, agmatine, creatine, proline, and glutamate. In particular, deficiency of the tumor suppressor ASS1—frequently caused by methylation of the gene promoter—confers a worse prognosis and sensitizes tumors to arginine starvation with pegylated arginine deiminase (ADI-PEG20) and human recombinant arginase (10–13). Arginine withdrawal leads to increased protein turnover—via reduced synthesis and increased breakdown [suppression of mammalian target of rapamycin (mTOR) and proteosomal degradation, respectively]—and triggers caspase-dependent and caspase-independent apoptotic cell death in a cell type-dependent manner (13–15). In PNAS, Changou et al. (16) describe a novel reactive oxygen species (ROS)-mediated mechanism involved in ADI-PEG20–induced cell death in arginine-dependent (auxotrophic) ASS1− prostate cancer cell lines, defined by mitochondrial damage, nuclear DNA leakage, and chromatin autophagy (“self-eating”), termed chromatophagy. This atypical form of autophagic cell death (type 2 cell death) contrasts sharply with their earlier work in prostate cancer cells, showing that early-onset autophagy is a protective mechanism in arginine-auxotrophic tumor cells that otherwise undergo apoptosis via a caspase-independent mechanism (15). The new data by Changou et al. (16) highlight that there is still much to learn about the effects of arginine in cancer cells, and how we should exploit arginine starvation and autophagy in combination with other therapies.
Several groups have studied autophagy induced by arginine depletors and it is clear that many cancer cell types use this homeostatic recycling mechanism to overcome the arginine supply problem. The process begins by enclosing macromolecules and organelles within double membrane-bound vesicles, known as the autophagosomes, which are then fused with lysosomes before disassembly by hydrolases. Work in simple organisms has identified a family of autophagy-related genes (Atg) that are activated following nutrient deprivation stress secondary to loss of the inhibitory effect of mTOR on Atg13, leading to autophagosome formation. Effectively, ADI-induced autophagy buys tumor cells time before additional mechanisms are activated, such as ASS1 reexpression (7, 17). ADI-PEG20–sensitive tumor cell lines exhibit various markers of autophagy, including conversion of cytosolic LC3-1 protein to the lipidated form LC3-II, up-regulation of Beclin 1 (Atg6), Atg genes 5 and 7, appearance of autophagosomes, and degradation of the autophagy substrate protein p62 (18). Moreover, blocking autophagy with the autophagosomal–lysosomal fusion inhibitor, chloroquine, promotes apoptosis and has been suggested as a potential strategy going forward into clinical trial (15, 18, 19). Alternatively, cleavage of Beclin-1 and Atg5 by tumor necrosis factor-related apoptosis-inducing ligand has been shown to enhance apoptosis in ADI-treated melanoma cell lines via caspase-8 (20). Specific pharmacologic inhibitors of the autophagy machinery (e.g., Atg proteins) await further development and may be more effective in combination with arginine depletors. Although autophagy has an undoubted prosurvival function in tumors exposed to various metabolic stresses, studies have revealed paradoxically a role in the regulation of cell death. This finding is best illustrated by genetic studies involving deletion of the autophagic modulators beclin1, atg5, and atg7, which predispose mice to spontaneous tumorigenesis, whereas elimination of p62 suppresses tumorigenesis by preventing mitochondrial and genomic damage that is linked to oxidative stress (17).
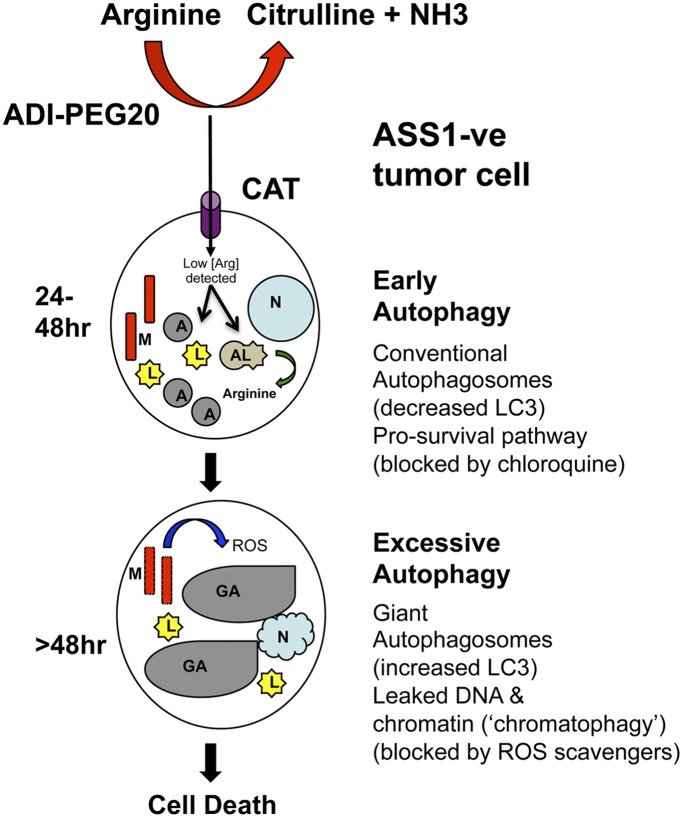
It is has been argued that many of the techniques and tool compounds lack specificity in the analysis of autophagic cell death, and that progress will depend on using novel methodologies and modulating multiple Atg members throughout the process of cell death (21). The biphasic autophagic response reported by Changou et al. (16) in arginine-depleted tumor cells, namely an early prosurvival stage (within 24–48 h) followed by delayed chromatophagy (beyond 48 h) linked to cell death opens up new areas of study for the field of amino acid deprivation (Fig. 1). The formation of giant autophagosomes sequestering leaked nuclear DNA in association with chromatin proteins following arginine withdrawal is striking—20-fold the size of standard vacuoles—and suggestive of a regulated process. Changou et al. (16) supplemented 4D fluorescence microscopic work with transmission electron microscopic studies, revealing giant autophagosomes fused to the nuclear membrane. Moreover, there was a specific lack of lamin A/C proteins, which maintain nuclear membrane integrity, thus pointing to chromatophagy as a dynamic process involved in capturing the leaked DNA. Notably, Atg5 and Beclin-1 short-hairpin RNAs suppressed the giant autophagosomes, arguing for a major role for these proteins in modulating sensitivity to ADI-PEG20. Similarly, the DNA leakage and chromatophagy was inhibited using the autophagy suppressors 3-methyladenine (via blockade of the phosphatylinositol 3-kinase, Vps34) and bafilomycin A1 (a vacuolar-type H+-ATPase antagonist that prevents autophagasomal-lysosomal fusion). Interestingly, in contrast to the prostate cancer cells that exhibit caspase-independent cell death and chromatophagy, the DNA leakage was also observed in ASS1− tumor cells that undergo caspase-dependent cell death when exposed to ADI-PEG20. Further efforts with more specific inhibitors and genetic deletions of the autophagy machinery will be needed to dissect out these differences and assess the significance of the DNA leakage on the respective cell-death pathways.
Fig. 1.
Biphasic autophagic response following arginine deprivation in ASS1-ve tumor cells: prosurvival phase followed by excessive autophagy leading to cell death. A, autophagosome; AL, autolysosome; CAT, cationic amino acid transporter; GA, giant autophagosome; L, lysosome; M, mitochondrion; N, nucleus. ASS1-ve tumor cells are sensitive to the arginine-depletor ADI-PEG20 and exhibit caspase-dependent or -independent apoptosis associated with DNA leakage.
This brings us to the fundamental mechanism initiating the atypical autophagic cell death described by Changou et al. (16). Although the arginine-sensing mechanism per se is obscure, Changou et al. showed that mitochondrial ROS regulated chromatophagy, which was partly reversible with N-acetyl cysteine, a known ROS scavenger. Arginine deprivation via ADI-PEG20 damaged mitochondria, reducing their oxygen consumption rate and the mitochondrial membrane potential, consistent with prior results in malignant mesothelioma cell lines (10).
New data by Changou et al. highlight that there is still much to learn about the effects of arginine in cancer cells.
Because it is known that arginine deprivation induces free radical formation and peroxynitrite stress, further studies are warranted in this area, particularly in relation to arginine-dependent cancer cells (22). It is also possible that ammonia, a degradation product of ADI-PEG20 (the other being citrulline, which is readily measured as a pharmacodynamic biomarker), may have an additional modulatory role on the process of autophagy. Recent work has shown that ammonia is released following glutaminolysis in tumor cells and activates autophagy (17). Thus, excess nitrogen-sensing mechanisms involving peroxynitrite or ammonia may play a key role in mediating ADI-induced autophagy.
More broadly, there remain several unanswered questions with regard to ADI-induced chromatophagy that are likely to impact on the success of arginine-depleting agents in the treatment of malignant disease. First, how widespread is the phenotype in arginine-dependent cancers, and is it restricted to arginine alone? Second, how can the process be measured in vivo? The study was restricted to cancer cell lines and thus more definitive conclusions regarding the role of chromatophagy must await studies performed in patients treated with arginine-lowering agents. Third, which agents will be most effective at potentiating the process and how should these be sequenced with conventional as well as newer anticancer agents? Indeed, work performed several decades ago showed that l-asparaginase, the first amino acid depletor, had limited clinical utility as a monotherapy in the treatment of acute lymphoblastic leukemia (2, 9). It was not until it was incorporated into multimodality regimens that the impact of l-asparaginase was observed in the cure of malignant disease. Fourth, will antioxidants suppress chromatophagy and reduce the effectiveness of arginine deprivation therapy? Fifth, how will the chromatophagy play out within the host-tumor arginine metabolome? Understanding the autophagic mechanisms of resistance and cell death of arginine deprivation is gaining momentum. Integrating this data into the hardwiring of the arginine metabolome in cancer cells will be crucial to the success of combination therapies that exploit arginine starvation for a wide spectrum of hard-to-treat malignancies.
Footnotes
The author declares no conflict of interest.
See companion article on page 14147.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG. Targeting cancer metabolism: A therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 3.Lu W, Pelicano H, Huang P. Cancer metabolism: Is glutamine sweeter than glucose? Cancer Cell. 2010;18(3):199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 5.Locasale JW. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109(23):8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delage B, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer. 2010;126(12):2762–2772. doi: 10.1002/ijc.25202. [DOI] [PubMed] [Google Scholar]
- 8.Szlosarek PW, et al. Metabolic response to pegylated arginine deiminase in mesothelioma with promoter methylation of argininosuccinate synthetase. J Clin Oncol. 2013;31(7):e111–e113. doi: 10.1200/JCO.2012.42.1784. [DOI] [PubMed] [Google Scholar]
- 9.Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: Opportunities and challenges. Cancer Res Treat. 2013;45(4):251–262. doi: 10.4143/crt.2013.45.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szlosarek PW, et al. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin Cancer Res. 2006;12(23):7126–7131. doi: 10.1158/1078-0432.CCR-06-1101. [DOI] [PubMed] [Google Scholar]
- 11.Cheng PN, et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67(1):309–317. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 12.Huang HY, et al. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: Aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin Cancer Res. 2013;19(11):2861–2872. doi: 10.1158/1078-0432.CCR-12-2641. [DOI] [PubMed] [Google Scholar]
- 13.Allen MD, et al. Prognostic and therapeutic impact of argininosuccinate synthetase 1 control in bladder cancer as monitored longitudinally by PET imaging. Cancer Res. 2014;74(3):896–907. doi: 10.1158/0008-5472.CAN-13-1702. [DOI] [PubMed] [Google Scholar]
- 14.Morrow K, et al. Anti-leukemic mechanisms of pegylated arginase I in acute lymphoblastic T-cell leukemia. Leukemia. 2013;27(3):569–577. doi: 10.1038/leu.2012.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim RH, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009;69(2):700–708. doi: 10.1158/0008-5472.CAN-08-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Changou CA, et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci USA. 2014;111:14147–14152. doi: 10.1073/pnas.1404171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30(7):671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syed N, et al. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblastoma. Cell Death Dis. 2013;4:e458. doi: 10.1038/cddis.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delage B, et al. Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 2012;3:e342. doi: 10.1038/cddis.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You M, et al. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol Cell Biochem. 2013;374(1-2):181–190. doi: 10.1007/s11010-012-1518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroemer G, Levine B. Autophagic cell death: The story of a misnomer. Nat Rev Mol Cell Biol. 2008;9(12):1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Y, Zweier JL. Superoxide and peroxynitrite generation from inducible nitric oxide synthase in macrophages. Proc Natl Acad Sci USA. 1997;94(13):6954–6958. doi: 10.1073/pnas.94.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]