T cell clonality is a common finding in patients with myelodysplastic syndrome (MDS), but is currently thought to be a reactive phenomenon (van Lom et al, 1995; Epling-Burnette et al, 2007). Recent evidence points to a stem or multipotent progenitor cell as the MDS-initiating cell in some patients, suggesting that the lymphoid lineage may also be involved in the disease. Clonal circulating myeloid and lymphoid precursor dendritic cells have been detected in patients with MDS (Ma et al, 2004) and a high percentage of monosomy 7 in pluripotent stem cells, B cell progenitors and T/Natural Killer (NK) progenitor cells was reported in three of four MDS patients analysed (Miura et al, 2000). In a series of MDS cases that progressed to T cell acute lymphoid leukaemia (T-ALL), the MDS karyotypic aberration was also detected in the T-ALL cells (Disperati et al, 2006).
Many genome-wide studies in MDS have used CD3+ cells from the same patient to represent a patient normal control in order to distinguish between constitutional and acquired variants (Starczynowski et al, 2008). The present study investigated whether T cells are derived from the malignant MDS clone using DNA microarrays in 40 MDS patients.
CD34+ and CD3+ cells were selected from marrow or peripheral blood by immunomagnetic separation (Stem Cell Technologies, Vancouver, BC, Canada). Genomic DNA was extracted with the AllPrep DNA/RNA Mini Kit (QIAGEN, Valencia, CA, USA). Normal reference DNA was purchased as a pool of either male or female genomic DNA (Novagen, Madison, WI, USA).
Details of whole genome array comparative genomic hybridization (aCGH) including DNA extraction, labelling and hybridization as well as image analysis have been described previously (Shah et al, 2006; Coe et al, 2007; Starczynowski et al, 2008). A region was considered altered when a minimum of two overlapping consecutive clones showed the change. The multiplex polymerase chain reaction protocol and primers used for T cell receptor gamma (TRG@) gene analysis followed the standardized BIOMED-2 protocols, followed by analysis on an ABI3730 capillary electrophoresis instrument (van Dongen et al, 2003). Intracellular immunohistochemical staining of marrow/peripheral blood cells with anti-CD3cytoplasmic antibody (Dako North America Inc., Carpenteria, CA, USA) followed by AlexaFluor 594 (Molecular probes Inc., Eugene, OR, USA) preceded the fluorescence in situ hybridization (FISH) procedure with FISH probes (20q or 8 or 11q)(Abbott/Vysis, Downers Grove, IL, USA).
aCGH was performed on 40 DNA samples of matched CD34+ stem/progenitor cells and CD3+ T cells from patients with MDS or MDS/myeloproliferative neoplasm (MPN). Fourteen patients had known cytogenetic abnormalities as identified by conventional karyotyping (Table I). Of these 14 patients, two male patients had a deletion of the Y chromosome. In 11 of the remaining 12 MDS patients the cytogenetic abnormalities could be detected in the CD34+ cells using aCGH. In one patient (Patient 33) with trisomy 8 (4/20 metaphases by conventional karyotyping) and deletion 5q23.1-q31.2 (11/20 metaphases by conventional karyotyping), only the deletion 5q was detectable by aCGH, consistent with a detection threshold of 25–30% aberrant cells (Coe et al, 2007). Additionally, aCGH detected deletion of chromosome 20q11.21-13.33 in this patient. Patient 32 showed deletion of chromosome 11q14.1-q23.1 as well as amplification of 11q12.3-13.4. In five patients (Patients 29, 30, 34, 39 and 40) conventional karyotyping failed or was not performed. One of these patients (Patient 39) revealed partial trisomy 9 from q33.3 to q34.3 as well as trisomies 19 and 22 by aCGH in the CD34+ cells.
Table I.
Clinical, chromosomal and TCR rearrangement characteristics of 40 patients with MDS or MDS/MPN analyzed by array CGH for chromosomal changes in T cells..
| Patient number |
Age (years) |
Gender | Diagnosis | Karyotype | Origin of T cells |
Chromosomal abnormality present in T cells |
TRG@ rearrangement |
|---|---|---|---|---|---|---|---|
| 1 | 77 | F | 5q- syndrome | 46,XX, del5q | BM | No | ND |
| 2 | 86 | F | RA | 46,XX | BM | NA | ND |
| 3 | 78 | F | RA | 46,XX | PB | NA | Polyclonal |
| 4 | 78 | M | RA | 46,XY, del20q | BM | Yes | ND |
| 5 | 54 | M | MDS/MPN (RARS-T) | 46,XY, del20q | PB | Yes | Clonal |
| 6 | 78 | M | RA | 46,XY/45,X-Y | BM | NA | ND |
| 7 | 80 | M | RCMD | 46,XY | PB | NA | Clonal |
| 8 | 75 | F | RA | 46,XX | PB | NA | ND |
| 9 | 56 | M | RA | 46,XY | BM | NA | ND |
| 10 | 79 | M | RA | 46,XY | BM | NA | Polyclonal |
| 11 | 69 | F | RA | 46,XX | BM | NA | ND |
| 12 | 63 | M | MDS/MPN | 46,XY | BM | NA | ND |
| 13 | 49 | F | RA | 46,XX | BM | NA | Oligoclonal |
| 14 | 73 | F | RA | 46,X idicXq13 | PB | Yes | Clonal |
| 15 | 84 | M | RA | 45,X-Y | PB | NA | ND |
| 16 | 74 | M | RA | 45,X-Y, del7 | BM | No | ND |
| 17 | 53 | M | RA | 46,XY, del20q | PB | No | Polyclonal |
| 18 | 79 | M | RCMD | 46,XY, del7, +8 | BM | No | Oligoclonal |
| 19 | 83 | M | RARS | 46,XY | BM | NA | ND |
| 20 | 68 | F | RARS | 46,XX | BM | NA | ND |
| 21 | 81 | F | MDS/MPN (RARS-T) | 46,XX | BM | NA | ND |
| 22 | 81 | M | RAEB-1 | 46,XY | BM | NA | Polyclonal |
| 23 | 83 | M | RARS | 46,XY | NA | NA | ND |
| 24 | 60 | M | RARS | 46,XY | BM | NA | Oligoclonal |
| 25 | 81 | M | RARS | 46,XY, +8 | BM | Yes | ND |
| 26 | 33 | F | RARS | 46,XX | BM | NA | ND |
| 27 | 76 | M | RARS | 46,XY | BM | NA | Oligoclonal |
| 28 | 52 | M | RARS | 46,XY | BM | NA | ND |
| 29 | 71 | F | RA | NA | BM | NA | ND |
| 30 | 66 | M | RA | NA | PB | NA | ND |
| 31 | 60 | M | MDS-NOS | 46,XY | BM | NA | ND |
| 32 | 74 | M | RCMD-RS | 46,XY, del11q | BM | Yes | ND |
| 33 | 74 | M | RCMD-RS | 46,XY, del5q, +8 | BM | Yes | Oligoclonal |
| 34 | 61 | M | RAEB-1 | NA | BM | NA | ND |
| 35 | 66 | F | MDS/MPN | 46,XX | BM | NA | Polyclonal |
| 36 | 85 | M | MDS/MPN | 46,XY | BM | NA | ND |
| 37 | 66 | M | RARS | 46,XY, +8 | BM | Yes | Clonal |
| 38 | 68 | F | RARS | 46,X idicXq13 | BM | Yes | ND |
| 39 | 64 | M | MDS/MPN | NA | BM | Yes | ND |
| 40 | 82 | M | MDS-NOS | NA | BM | NA | ND |
F, female; M, male; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; NOS, not otherwise specified; RA, refractory anaemia; RARS(-T), refractory anaemia with ring sideroblasts (in transformation); RCMD(-RS), refractory cytopenia with multilineage dysplasia (with ring sideroblasts); RAEB-1, refractory anaemia with excess blasts, type 1; Del, deletion; Idic, isodicentric; NA, not applicable; PB, peripheral blood; BM, bone marrow; ND, not done.
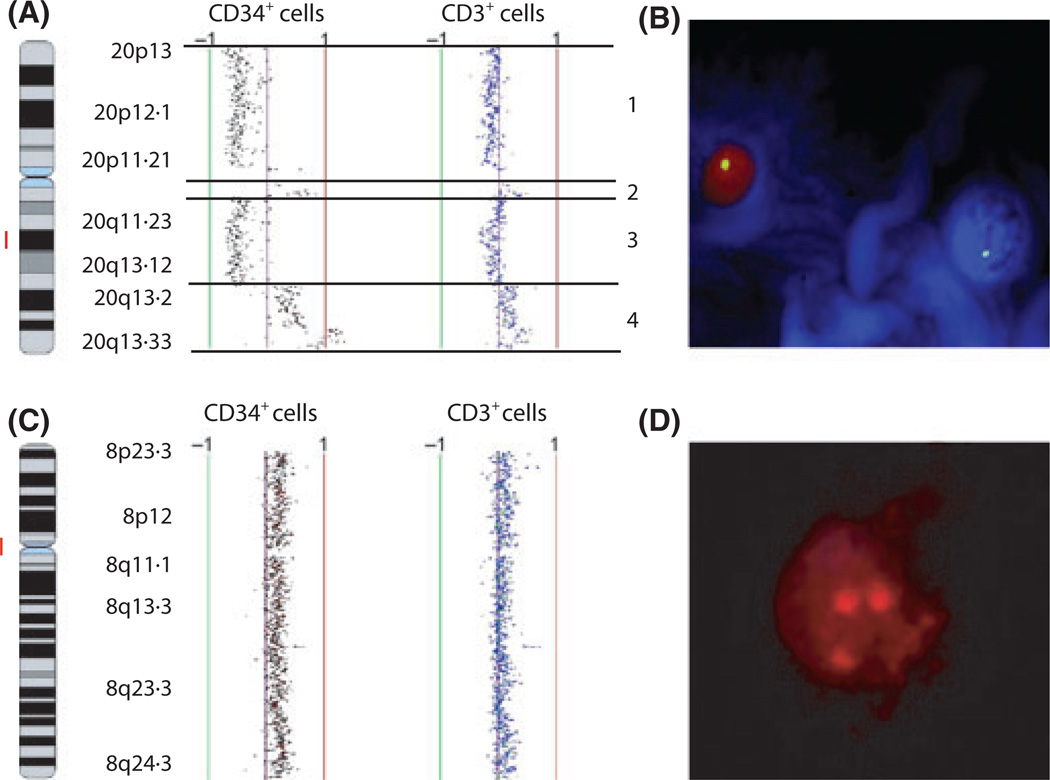
aCGH analysis of CD3+ T cells demonstrated the same cytogenetic abnormalities in nine of 12 MDS patients with karyotypic abnormalities, excluding the two patients with –Y (Table I and Fig 1). Deletion of chromosome 20q (Patients 4 and 5), isodicentric X chromosome (Patients 14 and 38), trisomy 8 (Patients 25 and 37), 11q abnormalities (Patient 32) and deletion 5q (Patient 33), partial trisomy 9 and trisomies 19 and 22 (Patient 39) were detected in the T cells of these patients. Consistent with the findings in the CD34+ cells, deletion of chromosome 5q23.1-q31.2 but not trisomy 8, was identified in the CD3+ cells of Patient 33. The presence of deletion 20q in the T cells from the marrow of Patient 5 (60% of CD3+ cells), trisomy 8 in the marrow T cells of Patient 25 (15% of CD3+ cells) and deletion of chromosome arm 11q in the peripheral blood T cells of Patient 32 (8% of CD3+ cells) was confirmed by FISH (Fig 1, and data not shown).
Fig 1.
Large genomic alterations in marrow CD34+ cells are detected in matched T cells of some patients with MDS. Array comparative genomic hybridization (aCGH) (A) and fluorescence in situ hybridization (FISH) (B) show deletions of chromosome 20 (1 and 3) as well as amplifications of chromosome 20 (2 and 4) in Patient 5, and trisomy 8 by aCGH (C) and FISH (D) in Patient 25. The red bar depicts the locus of the probe used for FISH.
T cell clonality of 14 MDS cases was determined by analysing TRG@ rearrangement (Table I). Six of the 14 patients analysed had karyotypic abnormalities, four of whom had the identical copy number variant identified in both the CD34+ and CD3+ populations by aCGH. These four patients either had clonal (Patients 5, 14 and 37) or oligoclonal (Patient 33) TRG@ rearrangements. In contrast, the two patients without the genetic abnormality in the T cells, showed polyclonal (Patient 17) and oligoclonal (Patient 18) TRG@ rearrangement. Clonal TRG@ rearrangement was detected in only one of the eight patients with a normal karyotype.
Here we show that cytogenetic abnormalities, identical to those seen in stem/progenitor cells, are present in the T cells of some MDS patients. We speculate that low numbers of T cells derived from the malignant clone are probably also present in other cases, but that this population may be smaller and thus not detectable by aCGH. Alternatively, the aberrant CD3+ cells may undergo apoptosis in the marrow before entering the circulation, and again may not be detectable in cases in which peripheral blood rather than marrow T cells are examined. This is in agreement with one report, in which a high percentage of monosomy 7 cells was identified in marrow-derived stem cells, B cell progenitors and T/NK progenitor cells but not in peripheral blood B and T cells (Miura et al, 2000). A recent publication has described the presence of TET2 mutations in T cells of a significant number of MDS patients (Smith et al, 2010). We conclude that, in a large proportion of MDS cases, at least a proportion of the T cells are part of the malignant clone, and suggest that CD3+ cells do not represent an appropriate patient normal control for genome-wide studies, but rather a non-haematopoietic cell type should be used.
Acknowledgements
This work was supported by a Canadian Institutes of Health Research (CIHR, MOP 89976) grant to AK, Genome Canada and CIHR grants to WLL. SMV is supported by a Leukemia and Lymphoma Society Clinical Research Fellowship. AK is a Senior Scholar of the Michael Smith Foundation for Health Research.
Footnotes
Author contributions
SMV designed research, analysed data and wrote the manuscript, DTS analysed data, SS, KM, CS and CJ performed experiments and analysed data, HB analysed data, WL provided BAC arrays and expertise in aCGH analysis, AK conceived the idea, designed research, analysed data, and wrote the manuscript.
References
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Disperati P, Ichim CV, Tkachuk D, Chun K, Schuh AC, Wells RA. Progression of myelodysplasia to acute lymphoblastic leukaemia: implications for disease biology. Leukemia Research. 2006;30:233–239. doi: 10.1016/j.leukres.2005.06.011. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- Epling-Burnette PK, Painter JS, Rollison DE, Ku E, Vendron D, Widen R, Boulware D, Zou JX, Bai F, List AF. Prevalence and clinical association of clonal T-cell expansions in Myelodysplastic Syndrome. Leukemia. 2007;21:659–667. doi: 10.1038/sj.leu.2404590. [DOI] [PubMed] [Google Scholar]
- van Lom K, Hagemeijer A, Smit E, Hahlen K, Groeneveld K, Lowenberg B. Cytogenetic clonality analysis in myelodysplastic syndrome: monosomy 7 can be demonstrated in the myeloid and in the lymphoid lineage. Leukemia. 1995;9:1818–1821. [PubMed] [Google Scholar]
- Ma L, Delforge M, van Duppen V, Verhoef G, Emanuel B, Boogaerts M, Hagemeijer A, Vandenberghe P. Circulating myeloid and lymphoid precursor dendritic cells are clonally involved in myelodysplastic syndromes. Leukemia. 2004;18:1451–1456. doi: 10.1038/sj.leu.2403430. [DOI] [PubMed] [Google Scholar]
- Miura I, Kobayashi Y, Takahashi N, Saitoh K, Miura AB. Involvement of natural killer cells in patients with myelodysplastic syndrome carrying monosomy 7 revealed by the application of fluorescence in situ hybridization to cells collected by means of fluorescence-activated cell sorting. British Journal of Haematology. 2000;110:876–879. doi: 10.1046/j.1365-2141.2000.02294.x. [DOI] [PubMed] [Google Scholar]
- Shah SP, Xuan X, DeLeeuw RJ, Khojasteh M, Lam WL, Ng R, Murphy KP. Integrating copy number polymorphisms into array CGH analysis using a robust HMM. Bioinformatics. 2006;22:e431–e439. doi: 10.1093/bioinformatics/btl238. [DOI] [PubMed] [Google Scholar]
- Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, Przychodzen B, Mian SA, Nasser EE, Shooter C, Westwood NB, Strupp C, Gattermann N, Maciejewski JP, Germing U, Mufti GJ. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Vercauteren S, Telenius A, Sung S, Tohyama K, Brooks-Wilson A, Spinelli JJ, Eaves CJ, Eaves AC, Horsman DE, Lam WL, Karsan A. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112:3412–3424. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]