Abstract
BACKGROUND
Philadelphia chromosome–like acute lymphoblastic leukemia (Ph-like ALL) is characterized by a gene-expression profile similar to that of BCR–ABL1–positive ALL, alterations of lymphoid transcription factor genes, and a poor outcome. The frequency and spectrum of genetic alterations in Ph-like ALL and its responsiveness to tyrosine kinase inhibition are undefined, especially in adolescents and adults.
METHODS
We performed genomic profiling of 1725 patients with precursor B-cell ALL and detailed genomic analysis of 154 patients with Ph-like ALL. We examined the functional effects of fusion proteins and the efficacy of tyrosine kinase inhibitors in mouse pre-B cells and xenografts of human Ph-like ALL.
RESULTS
Ph-like ALL increased in frequency from 10% among children with standard-risk ALL to 27% among young adults with ALL and was associated with a poor outcome. Kinase-activating alterations were identified in 91% of patients with Ph-like ALL; rearrangements involving ABL1, ABL2, CRLF2, CSF1R, EPOR, JAK2, NTRK3, PDGFRB, PTK2B, TSLP, or TYK2 and sequence mutations involving FLT3, IL7R, or SH2B3 were most common. Expression of ABL1, ABL2, CSF1R, JAK2, and PDGFRB fusions resulted in cytokine-independent proliferation and activation of phosphorylated STAT5. Cell lines and human leukemic cells expressing ABL1, ABL2, CSF1R, and PDGFRB fusions were sensitive in vitro to dasatinib, EPOR and JAK2 rearrangements were sensitive to ruxolitinib, and the ETV6–NTRK3 fusion was sensitive to crizotinib.
CONCLUSIONS
Ph-like ALL was found to be characterized by a range of genomic alterations that activate a limited number of signaling pathways, all of which may be amenable to inhibition with approved tyrosine kinase inhibitors. Trials identifying Ph-like ALL are needed to assess whether adding tyrosine kinase inhibitors to current therapy will improve the survival of patients with this type of leukemia. (Funded by the American Lebanese Syrian Associated Charities and others.)
Acute lymphoblastic leukemia (all) is the most common childhood cancer and a major cause of illness and death in adults.1 ALL encompasses a number of distinct entities characterized by chromosomal rearrangements, structural variations, and sequence mutations that perturb lymphoid maturation, cell proliferation, cell-growth suppression, and epigenetic regulation.2 Our understanding of the genetic basis of ALL has been transformed by genomewide profiling studies that have identified multiple targets of recurring genetic alterations and have defined new subtypes of ALL.
Childhood ALL is more commonly of B-cell than T-cell lineage and includes cases associated with hyperdiploidy, hypodiploidy, and chromosomal rearrangements resulting in chimeric fusion genes, including ETV6–RUNX1, TCF3–PBX1, and BCR–ABL1, as well as rearrangements of MLL and CRLF2. As compared with younger children with ALL, adolescents and adults with ALL have inferior outcomes, partly because of the lower frequency of favorable genetic features such as ETV6–RUNX1 and hyperdiploidy, as well as the higher frequency of BCR–ABL13; however, the genetic basis of ALL in adolescents and adults is poorly defined.
One subtype of precursor B-cell ALL is BCR–ABL1–like, or Philadelphia chromosome–like (Ph-like), ALL. Patients with Ph-like ALL do not have the BCR–ABL1 fusion protein expressed from the t(9;22)(q34;q11.2) Philadelphia chromosome yet have a gene-expression profile similar to that of patients with BCR–ABL1 ALL.4,5 Deletions or mutations of the lymphoid transcription factor gene IKZF1 (encoding Ikaros) are a hallmark of both BCR–ABL1–positive ALL and Ph-like ALL,4,6 and Ph-like ALL in children is associated with poor outcomes.4,5,7–10 Transcriptome sequencing and whole-genome sequencing in 15 children with Ph-like ALL identified chromosomal rearrangements or sequence mutations deregulating cytokine receptor and tyrosine kinase genes in all 15.11 In addition, there have been recent reports of patients with refractory Ph-like ALL and the EBF1–PDGFRB fusion who have a remarkably good response to therapy with tyrosine kinase inhibitors.12,13 Because the full spectrum of kinase-activating genetic alterations in Ph-like ALL, their effect on outcomes in adolescents and young adults, and their potential for therapeutic targeting are unknown, we performed a detailed genomic analysis of 1725 children, adolescents, and young adults with precursor B-cell ALL.
METHODS
STUDY DESIGN
We studied 2013 patients with precursor B-cell ALL, 1725 of whom had material available for microarray gene-expression profiling; 1589 of these 1725 patients had single-nucleotide-polymorphism microarray profiling performed. The cohort included 330 children with National Cancer Institute–classified, standard-risk precursor B-cell ALL (age range, 1 to 9 years; and peripheral-blood leukocyte count at diagnosis, <50,000 per cubic millimeter), 853 children with high-risk precursor B-cell ALL (age range, 10 to 15 years; leukocyte count, ≥50,000 per cubic millimeter; or both), 374 adolescents (age range, 16 to 20 years), and 168 young adults (age range, 21 to 39 years) (Table S1 in Supplementary Appendix 1 and Fig. S1 in Supplementary Appendix 2, available with the full text of this article at NEJM.org). There were few significant differences in the clinical features of patients with gene-expression profiling data available and those without such data available (Table S2 in Supplementary Appendix 2). Samples were obtained from patients enrolled under clinical-trial protocols of St. Jude Children’s Research Hospital, the Children’s Oncology Group, the Eastern Cooperative Oncology Group, the Alliance for Clinical Trials in Oncology (Cancer and Leukemia Group B), and M.D. Anderson Cancer Center. The details of the treatment protocols are provided in Supplementary Appendix 2. Patients, parents, or guardians gave written informed consent for sample collection and research, with assent provided by older children and adolescents. The study was approved by the St. Jude Institutional Review Board. Data from the study have been deposited in the European Genome Phenome archive under accession number EGAS00001000654.
NEXT-GENERATION SEQUENCING
A total of 154 patients with Ph-like ALL underwent detailed genomic analysis, 147 of whom underwent one or more of the following types of next-generation sequencing: transcriptome sequencing (136 patients), whole-genome sequencing (42), and whole-exome sequencing (12) of tumor and matched remission DNA (Table S1 in Supplementary Appendix 1).14 Next-generation sequencing was not performed for 7 patients, who instead underwent reverse-transcriptase polymerase-chain-reaction analysis. Transcriptome sequencing was also performed for 160 patients with non–Ph-like ALL (Table S3 in Supplementary Appendix 2). Details of Ph-like ALL classification, sequencing, and analysis are provided in Supplementary Appendix 2.
MICROARRAY PROFILING AND FUNCTIONAL AND CYTOGENETIC ASSAYS
The details of gene expression and single-nucleotide-polymorphism microarray profiling, fluorescence in situ hybridization, cell-line proliferation and tyrosine kinase inhibitor assays, protein expression, and xenograft experiments are provided in Supplementary Appendix 2.
STATISTICAL ANALYSIS
Associations between categorical variables were examined with the use of Fisher’s exact test. Associations between Ph-like ALL status and treatment outcome (event-free survival and overall survival) were examined with the use of the Kaplan–Meier estimator, with Peto’s estimator of standard deviation and the log-rank test, in each patient cohort (children, adolescents, and young adults).4,15,16 An event was defined as a failure to achieve remission, a relapse after remission, or the development of a second malignant neoplasm. A multivariable analysis of event-free and overall survival was performed with the Cox proportional-hazards regression model.17 Analyses were performed with the use of Prism software, version 6.0 (GraphPad Software), R software (www.r-project.org),18 and SAS software, version 9.1.2 (SAS Institute).
RESULTS
CLINICAL CHARACTERISTICS AND OUTCOMES
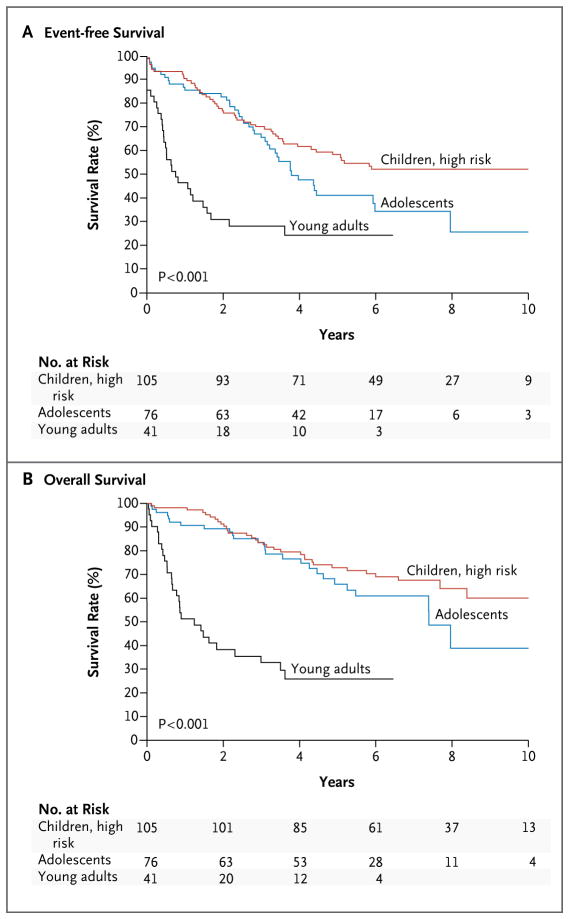
Overall, 264 of 1725 precursor B-cell ALL cases (15.3%) were identified as Ph-like ALL (Table S4 in Supplementary Appendix 2). The prevalence of Ph-like ALL significantly increased with age, from 10% among children with standard-risk ALL and 13% among those with high-risk ALL to 21% among adolescents with ALL and 27% among young adults with ALL (P<0.001 for the comparisons of children with adolescents and children with young adults). Furthermore, patients with Ph-like ALL had higher leukocyte counts at presentation than did patients with non–Ph-like ALL, both overall (106,000 vs. 59,000 per cubic millimeter, P<0.001) and in the different age cohorts (Table S5 in Supplementary Appendix 2). Ph-like ALL was more common among males than among females (Table S5 in Supplementary Appendix 2) and was associated with elevated levels of minimal residual disease at the end of induction therapy in the Children’s Oncology Group trials (Table S6 in Supplementary Appendix 2). Among patients with Ph-like ALL, the median (±SD) 5-year event-free survival rates for children with high-risk ALL, adolescents, and young adults was 58.2±5.3%, 41.0±7.4%, and 24.1±10.5%, respectively, and the 5-year overall survival rates were 72.8±4.8%, 65.8±7.1%, and 25.8±9.9% (Fig. 1). Across all age groups, these survival rates were inferior to those among patients with non–Ph-like ALL (P<0.001 for both comparisons) (Fig. S2 in Supplementary Appendix 2). The presence of Ph-like ALL was an independent prognostic factor in all age groups (Table S7 in Supplementary Appendix 2).
Figure 1. Kaplan–Meier Estimates of Event-free and Overall Survival among Patients with Philadelphia Chromosome–like Acute Lymphoblastic Leukemia (Ph-like ALL).
Panel A shows the median (±SD) rates of event-free survival for young adults, adolescents, and high-risk children with Ph-like ALL (24.1±10.5%, 41.0±7.4%, and 58.2±5.3%, respectively). Panel B shows rates of overall survival in the three age groups (25.8±9.9%, 65.8±7.1%, and 72.8±4.8%, respectively).
IDENTIFICATION OF KINASE ALTERATIONS IN PH-LIKE ALL
A total of 123 of 264 patients with Ph-like ALL had high CRLF2 expression, with the frequency ranging from 24% among children with standard-risk ALL to 60% among adolescents with ALL. Among patients with high CRLF2 expression, we identified P2RY8–CRLF2 in 45 patients and IGH–CRLF2 in 61 patients; for 17 patients, there was insufficient material for analysis. Sixty-eight patients (55%) with CRLF2 rearrangement had concomitant Janus kinase mutations, most commonly in JAK2 (Fig. S3 in Supplementary Appendix 2).
To identify the spectrum of kinase-activating alterations in the remaining patients with Ph-like ALL, we performed next-generation sequencing in 30 patients with CRFL2 rearrangement and 124 patients without CRLF2 rearrangement (Tables S8 and S9 in Supplementary Appendix 1). The analyses of gene-expression levels from transcriptome sequencing recapitulated those from microarray expression data, with clustering of Ph-like and BCR–ABL1–positive cases (Table S10 in Supplementary Appendix 1 and Fig. S4 and Fig. S5 in Supplementary Appendix 2).
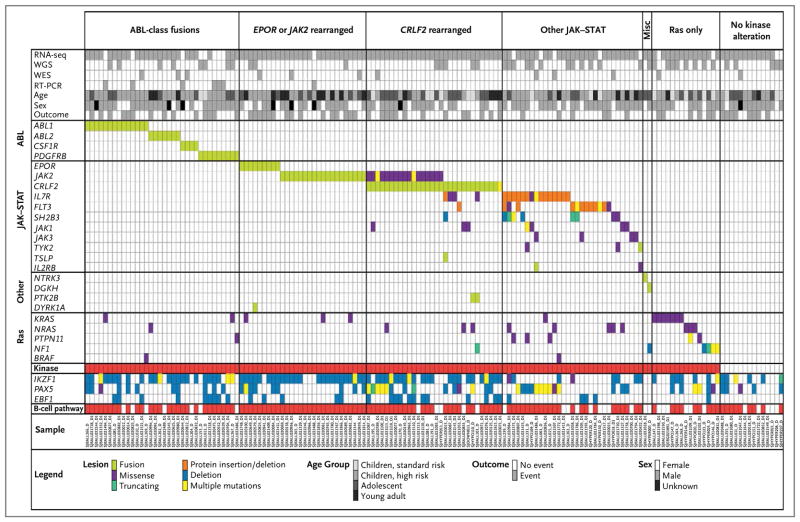
Genomic alterations activating kinase signaling were identified in 91% of patients with Ph-like ALL and were divided into distinct subgroups of kinase and cytokine receptor genes (Fig. 2, and Fig. S6 in Supplementary Appendix 2). These included fusions predicted to respond to ABL1 inhibitors (involving ABL1, ABL2, CSF1R, or PDGFRB) (12.6% of cases); rearrangements of EPOR (3.9%) or JAK2 (7.4%); rearrangements of CRLF2 (49.7%); genetic alterations of IL7R, FLT3, SH2B3, JAK1, JAK3, TYK2, and IL2RB (shown under “Other JAK–STAT” in Fig. 2; 12.6%); Ras pathway mutations, several of which were associated with hypodiploidy (4.3%); and uncommon fusions (e.g., involving NTRK3 or DGKH; 0.9%). A minority of patients (4.8%) were not found to have a kinase-activating alteration on transcriptome sequencing analysis, and suitable material for analysis was not available for 3.9% of patients. The frequencies of these subgroups varied with age. Notably, ABL-class rearrangements were more common among children, and JAK2 rearrangements were more frequent among young adults (Fig. S7 in Supplementary Appendix 2).
Figure 2. Recurring Kinase Alterations in Ph-like ALL.
Data are shown for 154 patients with Ph-like ALL who underwent detailed genomic analysis, including transcriptome sequencing (RNA-seq), whole-genome sequencing (WGS), whole-exome sequencing (WES), and reverse-transcriptase polymerase chain reaction (RT-PCR). The cohort is divided into patients with ABL-class fusions (ABL1, ABL2, CSF1R, PDGFRB) responsive to dasatinib, EPOR or JAK2 rearrangements, CRLF2 rearrangements, other JAK–STAT–activating mutations (IL7R, FLT3, SH2B3, JAK1, JAK3, TYK2, IL2RB, and TSLP), other kinase fusions (miscellaneous group, including NTRK3 and DGKH), alterations in the Ras pathway (KRAS, NRAS, PTPN11, NF1, and BRAF), and no kinase alteration. For details of specific alterations, see Tables S9 and S12 in Supplementary Appendix 1 and Table S20 in Supplementary Appendix 3.
In the transcriptome sequencing analysis, we identified 223 gene rearrangements in 116 of 136 patients (mean, 1.6 per patient; range, 0 to 12) (Table S11 in Supplementary Appendix 1); 115 of these were either chimeric in-frame fusions or rearrangements resulting in deregulated gene expression. Across the entire cohort, rearrangements activating kinase signaling were identified in 96 of 154 patients (62%), including 35 different rearrangements (16 of which were recurrent) in 13 kinase, cytokine, or cytokine-receptor genes: JAK2 (10 fusion partners), ABL1 (6), PDGFRB (4), ABL2 (3), CRLF2 (2), EPOR (2), PTK2B (2), CSF1R (1), DGKH (1), IL2RB (1), NTRK3 (1), TSLP (1), and TYK2 (1) (Table 1 and Fig. 2, and Table S12 in Supplementary Appendix 1 and Table S13 in Supplementary Appendix 2). All kinase fusions retained an intact tyrosine kinase domain (Fig. S8 in Supplementary Appendix 2) and were found by means of fluorescence in situ hybridization to be present in the predominant clone at diagnosis (Fig. S9 in Supplementary Appendix 2).
Table 1.
Kinase Fusions Identified in Ph-like Acute Lymphoblastic Leukemia.
| Kinase Gene | Tyrosine Kinase Inhibitor | Fusion Partners | Patients | 5′ Genes |
|---|---|---|---|---|
| number | ||||
| ABL1 | Dasatinib | 6 | 14 | ETV6,11 NUP214,11 RCSD1,11 RANBP2,11 SNX2,19 ZMIZ120 |
| ABL2 | Dasatinib | 3 | 7 | PAG1,* RCSD1,* ZC3HAV1* |
| CSF1R | Dasatinib | 1 | 4 | SSBP2* |
| PDGFRB | Dasatinib | 4 | 11 | EBF1,11–13 SSBP2,* TNIP1,* ZEB2* |
| CRLF2 | JAK2 inhibitor | 2 | 30 | IGH,21 P2RY822 |
| JAK2 | JAK2 inhibitor | 10 | 19 | ATF7IP,* BCR,11 EBF1,* ETV6,23 PAX5,11 PPFIBP1,* SSBP2,24 STRN3,11 TERF2,* TPR* |
| EPOR | JAK2 inhibitor | 2 | 9 | IGH,11 IGK* |
| DGKH | Unknown | 1 | 1 | ZFAND3* |
| IL2RB | JAK1 inhibitor, JAK3 inhibitor, or both | 1 | 1 | MYH9* |
| NTRK3 | Crizotinib | 1 | 1 | ETV625–27† |
| PTK2B | FAK inhibitor | 2 | 1 | KDM6A,* STAG2* |
| TSLP | JAK2 inhibitor | 1 | 1 | IQGAP2* |
| TYK2 | TYK2 inhibitor | 1 | 1 | MYB* |
Thirty patients with CRLF2 rearrangement were studied with the use of transcriptome sequencing and whole-genome sequencing (Fig. 2, and Fig. S10 in Supplementary Appendix 2). We found additional alterations activating JAK–STAT in 5 of 11 patients who had CRLF2 rearrangement but no JAK mutations; these alterations included IL7R mutations (4 patients), an FLT3 mutation (1 patient), and a deletion of SH2B3, which encodes the JAK2 negative regulator LNK (1 patient). One patient had an IQGAP2–TSLP fusion; this resulted in overexpression of TSLP, which encodes thymic stromal lymphopoietin, the ligand for CRLF2. Two patients had fusions that deregulated protein tyrosine kinase 2 β (PTK2B, or FAK2). Thus, multiple kinase-activating lesions are likely to cooperate with CRLF2 rearrangement in leukemogenesis.
Sequence mutations and focal deletions activating JAK–STAT signaling, including in IL7R, FLT3, SH2B3, JAK1, and JAK3, were identified in 31 patients without CRLF2 rearrangement or other kinase fusions. Fifteen patients had alterations in the Ras pathway only, including NRAS, KRAS, PTPN11, NF1, and BRAF (Fig. 2, and Fig. S11 in Supplementary Appendix 2). Eleven patients had mutations in multiple genes, with evidence of subclonality in 7 of the 8 patients who underwent whole-genome or whole-exome sequencing (Table S14 and Fig. S12 in Supplementary Appendix 2). Details of non-kinase fusions and sequence mutations are provided in the Results section of Supplementary Appendix 2.
On analysis of gene-expression data from transcriptome sequencing, patients with ABL-class, EPOR, or JAK2 rearrangements clustered separately from those with other JAK–STAT or Ras pathway alterations (Fig. S13 in Supplementary Appendix 2). We also found differences in outcome between the Ph-like ALL subgroups, with patients who had rearrangements of JAK2 or EPOR having the worst outcome (Table S15 and Fig. S14 in Supplementary Appendix 2).
As previously described,4,5,11 we found a higher frequency of IKZF1 alterations (deletion or point mutation) among patients with Ph-like ALL than among patients with BCR–ABL1–negative non–Ph-like ALL (166 of 244 [68%] vs. 204 of 1241 [16%], P<0.001) (Table S16 in Supplementary Appendix 2). IKZF1 alterations were more common in patients with Ph-like ALL who had kinase fusions (140 of 180 [78%]) than in those with a sequence mutation (14 of 43 [33%], P<0.001) (Fig. S6 in Supplementary Appendix 2). Furthermore, patients with Ph-like ALL who had an IKZF1 alteration had inferior median (±SD) 5-year event-free survival, as compared with patients who had Ph-like ALL without an IKZF1 alteration; these survival differences were seen in children with high-risk ALL (48.6±7.0 vs. 71.7±8.0 years, P<0.001) and in young adults (18.5±11.8 vs. 42.9±18.7 years, P<0.001) (Fig. S15 in Supplementary Appendix 2). When IKZF1 alterations were considered in combination with mutations in other lymphoid transcription factors (ETV6, EBF1, ERG, PAX5, and TCF3), 86% of patients with Ph-like ALL (209 of 244) were found to have alterations affecting lymphoid development, as compared with 61% of patients with non–Ph-like ALL (762 of 1241, P<0.001).
ACTIVITY OF TYROSINE KINASE INHIBITORS IN PH-LIKE ALL
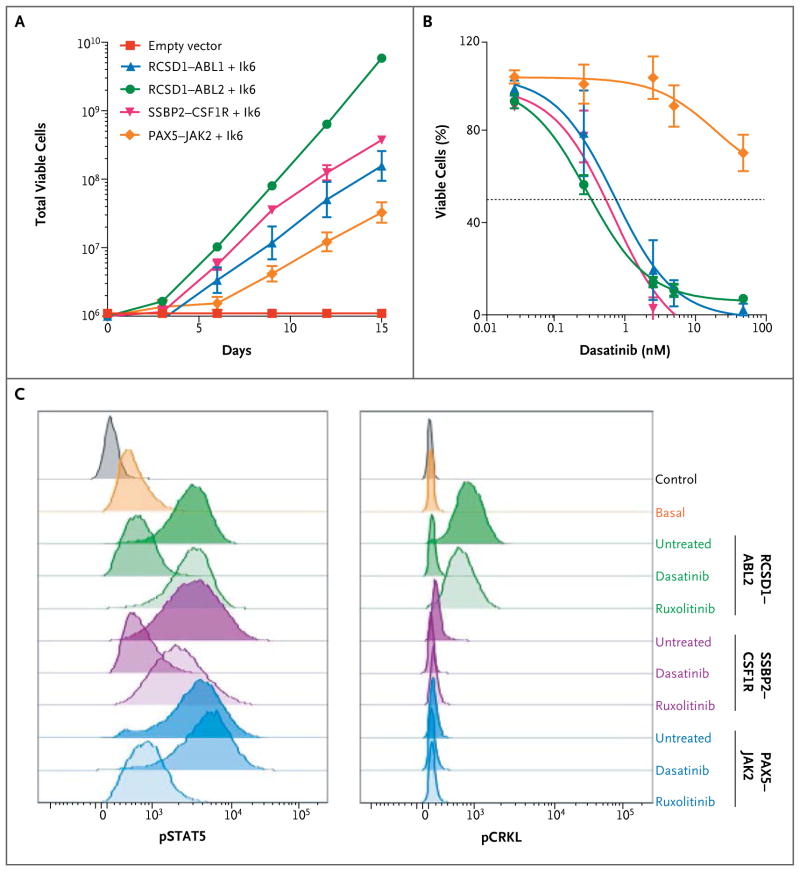
To determine the transforming properties of the kinase fusions, we assessed their ability to induce cytokine-independent proliferation in mouse interleukin-3–dependent Ba/F3 cells and interleukin-7–dependent Arf−/− pre-B cells30,31 expressing the dominant negative isoform of Ikaros, Ik6.6 Expression of all fusions tested (RCSD1–ABL1, RANBP2–ABL1, ZMIZ1–ABL1, RCSD1–ABL2, SSBP2–CSF1R, and PAX5–JAK2) conferred cytokine-independent proliferation (Fig. 3A, and Fig. S16 in Supplementary Appendix 2). The ABL1, ABL2, and CSF1R fusions were sensitive to the ABL-class inhibitors imatinib (50% inhibitory concentration [IC50], 135 to 900 nM) (data not shown) and dasatinib (IC50, 1 to 2 nM), whereas PAX5–JAK2 was not sensitive to these agents (Fig. 3B). Differences in signaling-pathway activation between the fusions were observed. All fusions activated STAT5, which was inhibited by dasatinib in cells expressing ABL1, ABL2, CS-F1R, or PDGFRB fusions,11 and the JAK2 inhibitor ruxolitinib attenuated phosphorylated STAT5 in cells expressing PAX5–JAK2. Phosphorylation of CRKL, a target of ABL1 and ABL2, was seen only in cells expressing these fusions (Fig. 3C). Human leukemic cells harboring ATF7IP–JAK2 or IGH–EPOR were sensitive to ruxolitinib, whereas imatinib had no effect (Fig. S17A in Supplementary Appendix 2). As reported previously in studies of other tumor models,32 cells expressing ETV6–NTRK3 responded to the ALK inhibitor crizotinib (Fig. S17B in Supplementary Appendix 2). The efficacy of dasatinib was also assessed in a xenograft model of ETV6–ABL1 ALL in which human leukemic cells were engrafted into immunodeficient mice. After 4 weeks of treatment, percentages of circulating human CD45+ cells were significantly reduced in the dasatinib-treated mice, as compared with vehicle-treated controls (17% vs. 88%, P<0.001), as was splenic weight (117 vs. 321 mg, P<0.001) (Fig. S17C in Supplementary Appendix 2).
Figure 3. Response to Tyrosine Kinase Inhibitors.
Panel A shows the proliferation of interleukin-7–dependent primary Arf−/− pre-B cells expressing the dominant negative Ikaros isoform (Ik6) with empty vector, RCSD1–ABL1, RCSD1–ABL2, SSBP2–CSF1R, or PAX5–JAK2 in the absence of cytokine. Panel B shows the growth of Arf−/− pre-B cells in increasing concentrations of dasatinib. Panel C shows constitutive phosphorylation of STAT5 and CRKL (pSTAT5 and pCRKL, respectively). Cells expressing ABL2, CSF1R, and JAK2 fusions have enhanced STAT5 activation, which is inhibited by dasatinib in RCSD1–ABL2 and SSBP2–CSF1R and by ruxolitinib in PAX5–JAK2.
The high frequency of kinase-activating lesions in the patients with Ph-like ALL suggests that tyrosine kinase inhibitor therapy is likely to be effective in such patients, as it is in patients with BCR–ABL1–positive ALL.33 To assess this, we studied 34 patients with precursor B-cell ALL who had high-risk clinical features at diagnosis, alterations found on cytogenetic analysis that were suggestive of a tyrosine kinase gene rearrangement, or a poor response to induction chemotherapy (Table S17 in Supplementary Appendix 2). Remarkably, 86% of patients (24 of 28) who underwent testing with a low-density gene-expression array34 had Ph-like ALL, and 22 of these patients had a targetable kinase fusion involving ABL1 (10 patients), ABL2 (3), CRLF2 (3), JAK2 (3), or PDGFRB (3). Notably, we identified 4 patients with induction failure who had EBF1–PDGFRB. Of the 12 patients who began receiving tyrosine kinase inhibitor therapy, 11 with available follow-up data had rapid and sustained responses. Details for each patient are provided in the Results section of Supplementary Appendix 2.
DISCUSSION
In this study of 1725 children, adolescents, and young adults with precursor B-cell ALL, we defined the frequency and poor outcome of Ph-like ALL, characterized the genetic landscape of alterations activating kinase signaling, and found that the majority of patients with Ph-like ALL have genetic alterations responsive to Food and Drug Administration–approved tyrosine kinase inhibitors. Building on the precedent established for BCR–ABL1–positive ALL,33 these findings provide a strong rationale for testing new therapies to improve the outcome of Ph-like ALL. We studied more than 500 adolescents and young adults with ALL, for whom treatment outcomes are substantially inferior to those of children with ALL. The frequency of Ph-like ALL is higher than 25% among young adults with ALL. Because BCR–ABL1–positive ALL represents more than 20% of precursor B-cell ALL cases in this age group, about half of young adults with precursor B-cell ALL are candidates for tyrosine kinase inhibitor therapy. The frequency of Ph-like ALL among older adults with ALL is unknown but merits investigation in view of the poor treatment outcome in this age group.
We identified several subgroups of Ph-like ALL distinguished by the type of cytokine receptor or kinase alteration that was present. The frequencies of CRLF2 rearrangement (47%) and concomitant JAK1 or JAK2 mutation (55% among patients with CRFL2 rearrangement) in the entire cohort are consistent with previous reports,11,35 with the highest frequency observed in adolescent ALL (60%). Previous reports have shown that the JAK2 inhibitor ruxolitinib is active in models of ALL with CRLF2 rearrangement, which suggests that JAK inhibition may be used in the treatment of these high-risk patients.36,37
The second major subgroup of Ph-like ALL is characterized by fusions involving ABL-class kinase genes. Multiple ABL1 and PDGFRB fusions and new targets of rearrangement were identified, including ABL2 (also known as Abelson-related gene, or ARG) and CSF1R (encoding the macrophage colony-stimulating factor receptor), which are known to respond to the ABL1 inhibitor ima-tinib.38,39 Importantly, we have shown that pre-B cells expressing ABL2 and CSF1R fusions activate oncogenic signaling pathways and cellular proliferation that is potently inhibited by dasatinib at concentrations similar to those for BCR–ABL1, other ABL1 fusions, and EBF1–PDGFRB.11
A striking finding was the high frequency of rearrangements activating JAK2, particularly in young adults. The PAX5–JAK2 fusion activated JAK–STAT signaling and conferred cytokine-independent proliferation that was sensitive to ruxolitinib. We previously identified a single patient with Ph-like ALL who had a cryptic insertion of EPOR, encoding the erythropoietin receptor, into the immunoglobulin heavy-chain locus; the current study shows that the insertion of EPOR into IGH and IGK is a recurring event in Ph-like ALL. Human leukemic cells with EPOR rearrangement exhibit activation of JAK–STAT signaling and are sensitive to JAK inhibitors ex vivo. Thus, JAK inhibition is a widely applicable treatment approach in Ph-like ALL.
We also identified distinct subgroups of patients with Ph-like ALL who had sequence mutations or structural alterations of genes involved in JAK–STAT or MAPK signaling. Alteration of IKZF1 was less common in such patients than in patients with Ph-like ALL who had a kinase rearrangement, and rearrangement of transcriptional regulators and chromatin modifiers was more common (Fig. S20 and Table S21 in Supplementary Appendix 2). Several patients with Ph-like ALL had Ras mutations but no other kinase alterations. Currently, direct therapeutic targeting of oncogenic Ras mutations is challenging, but targeting signaling pathways downstream of Ras may be considered.
The event-free and overall survival rates among patients with Ph-like ALL were markedly inferior to those among patients with non–Ph-like ALL, in all age groups studied. Thus, implementation of therapy directed against the activated kinase is an attractive strategy to improve the outcome for these high-risk patients. Although our data have highlighted the genetic complexity of Ph-like ALL, this entity can be rapidly identified with a low-density gene-expression array,34 and the majority of treatable kinase-activating lesions can be identified with the use of conventional molecular and cytogenetic approaches (Fig. S18 in Supplementary Appendix 2). The importance of identifying Ph-like ALL is highlighted by recent reports of the success of tyrosine kinase inhibitors in the treatment of relapsed or refractory EBF1–PDGFRB Ph-like ALL.12,13 Remarkably, most patients with ALL who had clinical features suggestive of Ph-like ALL and who were tested prospectively in this study were identified as having Ph-like ALL with treatable fusions. Furthermore, all patients treated with the relevant tyrosine kinase inhibitor for whom clinical-response data were available had rapid and durable responses. Together, these findings indicate that Ph-like ALL is common, particularly among adolescents and young adults, and is associated with a high risk of treatment failure. Clinical trials combining kinase inhibitors with chemotherapy in patients with Ph-like ALL, guided by careful use of screening and genomic testing, are warranted.
Supplementary Material
Acknowledgments
Supported in part by funding from the American Lebanese Syrian Associated Charities (to St. Jude Children’s Research Hospital), grants from the National Cancer Institute (CA 21765, to St. Jude Children’s Research Hospital; U01 CA157937-01, to Drs. Willman and Hunger; F32 CA141762, to Dr. Paugh; R37 CA36401, to Dr. Evans; U24-CA114737, U10 CA21115, and CA14958, to Dr. Paietta; U10 CA101140, to Cancer and Leukemia Group B Leukemia Correlative Science; CA145707, to Drs. Will-man and Mullighan; and U10 CA98543, U10 CA98413, and U24 CA114766, to the Children’s Oncology Group), a Stand Up to Cancer Innovative Research Grant and a St. Baldrick’s Foundation Scholar Award (both to Dr. Mullighan), a St. Baldrick’s Consortium Award (to Dr. Hunger), a grant from the Leukemia and Lymphoma Society Specialized Center of Research (to Drs. Carroll, Hunger, and Mullighan), a Leukemia and Lymphoma Society Special Fellow Award (to Dr. Roberts), Alex’s Lemonade Stand Foundation Young Investigator Awards (to Drs. Roberts and Tasian), an Alex’s Lemonade Stand Foundation Scholar in Developmental Therapeutics Award (to Dr. Tasian), and a grant from the National Institute of General Medical Sciences (92666, to Dr. Relling).
We thank the staff of the Tissue Resources Core Facility, the Hartwell Centre for Bioinformatics and Biotechnology, and the Flow Cytometry and Cell Sorting Core Facility of St. Jude Children’s Research Hospital.
APPENDIX
The authors’ full names and academic degrees are as follows: Kathryn G. Roberts, Ph.D., Yongjin Li, Ph.D., Debbie Payne-Turner, B.S., Richard C. Harvey, Ph.D., Yung-Li Yang, M.D., Deqing Pei, M.S., Kelly McCastlain, B.S., Li Ding, Ph.D., Charles Lu, Ph.D., Guang-chun Song, M.S., Jing Ma, Ph.D., Jared Becksfort, M.S., Michael Rusch, B.A., Shann-Ching Chen, Ph.D., John Easton, Ph.D., Jin-jun Cheng, M.D., Kristy Boggs, Ph.D., Natalia Santiago-Morales, B.S., Ilaria Iacobucci, Ph.D., Robert S. Fulton, Ph.D., Ji Wen, Ph.D., Marcus Valentine, B.A., Cheng Cheng, Ph.D., Steven W. Paugh, Ph.D., Meenakshi Devidas, Ph.D., I-Ming Chen, D.V.M., Shalini Reshmi, Ph.D., Amy Smith, B.S., Erin Hedlund, Ph.D., Pankaj Gupta, M.S., Panduka Nagahawatte, M.S., Gang Wu, Ph.D., Xiang Chen, Ph.D., Donald Yergeau, Ph.D., Bhavin Vadodaria, B.A., Heather Mulder, B.S., Naomi J. Winick, M.D., Eric C. Larsen, M.D., William L. Carroll, M.D., Nyla A. Heerema, Ph.D., Andrew J. Carroll, Ph.D., Guy Grayson, M.D., Sarah K. Tasian, M.D., Andrew S. Moore, M.D., Frank Keller, M.D., Melissa Frei-Jones, M.D., James A. Whitlock, M.D., Elizabeth A. Raetz, M.D., Deborah L. White, Ph.D., Timothy P. Hughes, M.D., Jaime M. Guidry Auvil, Ph.D., Malcolm A. Smith, M.D., Guido Marcucci, M.D., Clara D. Bloomfield, M.D., Krzysztof Mrózek, M.D., Jessica Kohlschmidt, Ph.D., Wendy Stock, M.D., Steven M. Kornblau, M.D., Marina Konopleva, M.D., Elisabeth Paietta, Ph.D., Ching-Hon Pui, M.D., Sima Jeha, M.D., Mary V. Relling, Pharm.D., William E. Evans, Pharm.D., Daniela S. Gerhard, Ph.D., Julie M. Gastier-Foster, Ph.D., Elaine Mardis, Ph.D., Richard K. Wilson, Ph.D., Mignon L. Loh, M.D., James R. Downing, M.D., Stephen P. Hunger, M.D., Cheryl L. Willman, M.D., Jinghui Zhang, Ph.D., and Charles G. Mullighan, M.D.
From the Departments of Pathology (K.G.R., D.P.-T., Y.-L.Y., K. McCastlain, G.S., J.M., S.-C.C., J.C., N.S.-M., I.I., J.W., J.R.D., C.G.M.), Computational Biology and Bioinformatics (Y.L., J.B., M.R., E.H., P.G., P.N., G.W., X.C., J.Z.), Biostatistics (D.P., C.C.), Pharmaceutical Sciences (S.W.P., M.V.R., W.E.E.), and Oncology (C.-H.P., S.J.), the Pediatric Cancer Genome Project (Y.L., L.D., C.L., M.R., J.E., J.C., K.B., R.S.F., E.H., P.G., P.N., G.W., X.C., D.Y., B.V., H.M., M.V.R., W.E.E., E.M., R.K.W., J.R.D., J.Z., C.G.M.), and Cytogenetics Shared Resource (M.V.), St. Jude Children’s Research Hospital, Memphis, TN; the University of New Mexico Cancer Center and School of Medicine, Albuquerque (R.C.H., I-M.C., C.L.W.); the Genome Institute at Washington University (L.D., C.L., R.S.F., E.M., R.K.W.), the Department of Genetics, Washington University School of Medicine (L.D., C.L., R.S.F., E.M., R.K.W.), and Siteman Cancer Center, Washington University (E.M., R.K.W.) — all in St. Louis; Epidemiology and Health Policy Research, College of Medicine, University of Florida, Gainesville (M.D.); the Research Institute at Nationwide Children’s Hospital (S.R., A.S., J.M.G.-F.), the Department of Pathology, College of Medicine, Ohio State University (N.A.H.), and Ohio State University Comprehensive Cancer Center (G.M., C.D.B., K. Mrózek, J.K.) — all in Columbus, OH; the Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas (N.J.W.), Scott and White Hospitals and Clinics and Texas A&M Health Science Center, Temple (G.G.), the University of Texas Health Science Center San Antonio, San Antonio (M.F.-J.), and the Departments of Leukemia and Stem Cell Transplantation, Division of Cancer Medicine, University of Texas M.D. Anderson Cancer Center, Houston (S.M.K., M.K.) — all in Texas; Maine Children’s Cancer Program, Scarborough (E.C.L.); New York University Cancer Institute, New York (W.L.C.), and the Department of Medicine (Oncology), Albert Einstein College of Medicine, Bronx (E.P.) — both in New York; the Department of Genetics, University of Alabama at Birmingham, Birmingham (A.J.C.); the Division of Oncology, Children’s Hospital of Philadelphia, Philadelphia (S.K.T.); Queensland Children’s Medical Research Institute, University of Queensland and Royal Children’s Hospital Brisbane, Herston (A.S.M.), and Cancer Theme, South Australian Health and Medical Research Institute, Adelaide (D.L.W., T.P.H.) — both in Australia; Children’s Healthcare of Atlanta and Emory University School of Medicine — both in Atlanta (F.K.); the Division of Haematology–Oncology, Hospital for Sick Children, and University of Toronto — both in Toronto (J.A.W.); Hunstman Cancer Institute, University of Utah, Salt Lake City (E.A.R.); the Office of Cancer Genomics (J.M.G.A., D.S.G.) and Cancer Therapy Evaluation Program (M.A.S.), National Cancer Institute, and the Children’s Oncology Group TARGET ALL project (M.D., I-M.C., S.R., W.L.C., J.M.G.A., M.A.S., M.V.R., D.S.G., J.M.G.-F., M.L.L., J.R.D., S.P.H., C.L.W., J.Z., C.G.M.), National Institutes of Health, Bethesda, MD; Alliance for Clinical Trials in Oncology Statistics and Data Center, Mayo Clinic, Rochester, MN (J.K.); the University of Chicago Medicine, Chicago (W.S.); the Department of Pediatrics, Benioff Children’s Hospital, and the Helen Diller Family Cancer Center, University of California, San Francisco, San Francisco (M.L.L.); and the University of Colorado School of Medicine and Children’s Hospital Colorado — both in Aurora (S.P.H.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–55. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:314–24. doi: 10.1053/j.seminhematol.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stock W. Adolescents and young adults with acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2010;2010:21–9. doi: 10.1182/asheducation-2010.1.21. [DOI] [PubMed] [Google Scholar]
- 4.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 7.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–9. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh ML, Zhang J, Harvey RC, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood. 2013;121:485–8. doi: 10.1182/blood-2012-04-422691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiyokawa N, Iijima K, Yoshihara H, et al. An analysis of Ph-like ALL in Japanese patients. Presented at the American Society of Hematology Annual Meeting; New Orleans. December 7–10, 2013; p. abstract 352. [Google Scholar]
- 10.Te Kronnie G, Silvestri D, Vendramini E, et al. Philadelphia-like signature in childhood acute lymphoblastic leukemia: the AEIOP experience. Presented at the American Society of Hematology Annual Meeting; New Orleans. December 7–10, 2013; p. abstract 353. [Google Scholar]
- 11.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31(25):e413–e416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- 13.Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98(11):e146–e148. doi: 10.3324/haematol.2013.095372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–31. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–70. [PubMed] [Google Scholar]
- 16.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 17.Cox DR. Regression models and life-tables. J R Stat Soc Series B Stat Methodol. 1972;34:187–220. [Google Scholar]
- 18.R Development Core Team. R: A language and environment for statistical computing. 2009 ( http://www.R-project.org)
- 19.Ernst T, Score J, Deininger M, et al. Identification of FOXP1 and SNX2 as novel ABL1 fusion partners in acute lymphoblastic leukaemia. Br J Haematol. 2011;153:43–6. doi: 10.1111/j.1365-2141.2010.08457.x. [DOI] [PubMed] [Google Scholar]
- 20.Soler G, Radford-Weiss I, Ben-Abdelali R, et al. Fusion of ZMIZ1 to ABL1 in a B-cell acute lymphoblastic leukaemia with a t(9;10)(q34;q22. 3) translocation. Leukemia. 2008;22:1278–80. doi: 10.1038/sj.leu.2405033. [DOI] [PubMed] [Google Scholar]
- 21.Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–98. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 22.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–6. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–12. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 24.Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, Morton CC. Novel SSBP2-JAK2 fusion gene resulting from a t(5;9) (q14.1;p24. 1) in pre-B acute lymphocytic leukemia. Genes Chromosomes Cancer. 2008;47:884–9. doi: 10.1002/gcc.20585. [DOI] [PubMed] [Google Scholar]
- 25.Knezevich SR, Garnett MJ, Pysher TJ, Beckwith JB, Grundy PE, Sorensen PH. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58:5046–8. [PubMed] [Google Scholar]
- 26.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–7. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 27.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 28.Alessandri AJ, Knezevich SR, Mathers JA, Schultz KR, Sorensen PH. Absence of t(12;15) associated ETV6-NTRK3 fusion transcripts in pediatric acute leukemias. Med Pediatr Oncol. 2001;37:415–6. doi: 10.1002/mpo.1222. [DOI] [PubMed] [Google Scholar]
- 29.Eguchi M, Eguchi-Ishimae M. Absence of t(12;15) associated ETV6-NTRK3 fusion transcripts in pediatric acute leukemias. Med Pediatr Oncol. 2001;37:417. doi: 10.1002/mpo.1223. [DOI] [PubMed] [Google Scholar]
- 30.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–93. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–7. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taipale M, Krykbaeva I, Whitesell L, et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol. 2013;31:630–7. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2009;27:5175–81. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey RC, Kang H, Roberts KG, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-Like (“Ph-like” or “BCR-ABL1-Like”) signature for therapeutic targeting and clinical intervention. Blood. 2013;122:826. [Google Scholar]
- 35.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–21. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maude SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–8. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tasian SK, Doral MY, Borowitz MJ, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–42. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer. 2013;13:559–71. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewar AL, Cambareri AC, Zannettino AC, et al. Macrophage colony-stimulating factor receptor c-fms is a novel target of imatinib. Blood. 2005;105:3127–32. doi: 10.1182/blood-2004-10-3967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.