SUMMARY
The MYC oncogene regulates gene expression through multiple mechanisms and its overexpression culminates in tumorigenesis. MYC inactivation reverses turmorigenesis through the loss of hallmark features of cancer including autonomous proliferation and survival. Here we report that MYC via miR-17-92 maintains a neoplastic state through the suppression of chromatin regulatory genes Sin3b, Hbp1, Suv420h1, and Btg1, as well as the apoptosis regulator Bim. The enforced expression of miR-17-92 prevents MYC suppression from inducing proliferative arrest, senescence, and apoptosis, and abrogates sustained tumor regression. Knockdown of the five miR-17-92 target genes blocks senescence and apoptosis while it modestly delays proliferative arrest, thus partially recapitulating miR-17-92 function. We conclude that MYC, via miR-17-92, maintains a neoplastic state by suppressing specific target genes.
INTRODUCTIONS
Cancers are often dependent on or addicted to the initiating oncogenes for the maintenance of the malignant phenotype (Chin et al., 1999; Felsher and Bishop, 1999; Huettner et al., 2000; Weinstein, 2002). The inactivation of a single driver oncogene can result in rapid and sustained tumor regression. Oncogene addiction has been exploited clinically in targeted therapies, such as imatinib for BCR-ABL-driven chronic myelogenous leukemia, gefitinib for lung adenocarcinoma with EGFR mutations, and vemurafenib for melanomas with B-RAF mutations (Chapman et al., 2011; Druker et al., 1996; Ladanyi and Pao, 2008). Hence, the targeted inactivation of oncogenes appears to be a generalizable approach for the treatment of many cancers.
The MYC oncogene is overexpressed in over half of human cancers (Dang, 2012). To study the role of MYC in the initiation and maintenance of tumorigenesis, some investigators have used the tetracycline regulatory system (Tet system) to generate reversible models of cancer (Gossen and Bujard, 1992). In these mouse models, the overexpression of a conditional MYC transgene initiates tumorigenesis, and its inactivation results in rapid, complete and sustained tumor regression. MYC inactivation is associated with the loss of many of the hallmark features of tumorigenesis and results in proliferative arrest, apoptosis, differentiation, and senescence, as well as the shutdown of angiogenesis (D'Cruz et al., 2001; Felsher and Bishop, 1999; Hanahan and Weinberg, 2011; Shachaf et al., 2004; Wu et al., 2007).
MYC is a transcriptional regulator of a multitude of genes, but it is unclear if any of these genes are responsible for MYC to maintain a neoplastic state (Dang, 2012). Recently, it has been shown that MYC may regulate gene expression as a general transcriptional amplifier (Lin et al., 2012; Nie et al., 2012). However, it has been pointed out that this would not explain how MYC can suppress gene expression or regulate gene expression in a specific manner (Walz et al., 2013). MYC has also been shown to regulate the expression of several microRNAs, including the polycistronic miR-17-92 cluster (Bui and Mendell, 2010; O'Donnell et al., 2005; Sander et al., 2008). The miR-17-92 cluster is overexpressed in human lymphomas (He et al., 2005). Notably, overexpression of miR-17-92 cooperates with MYC to induce lymphomagenesis, while deletion of miR-17-92 induces the death of lymphoma cells (He et al., 2005; Mu et al., 2009).
We hypothesized that miR-17-92 is causally responsible for at least part of the mechanism by which MYC maintains a neoplastic state (Figure 1A). Here, we found that MYC, through miR-17-92, regulates the expression of specific chromatin regulatory genes, such as Sin3b, Hbp1, Suv420h1, and Btg1, as well as the apoptosis regulator Bim. Upon MYC inactivation, the dowregulation of miR-17-92 and the corresponding induction of these target genes is causally required for the activation of the apoptosis and senescence programs and sustained tumor regression. Hence, MYC suppression of these genes is one of the required mechanisms to maintain a neoplastic state.
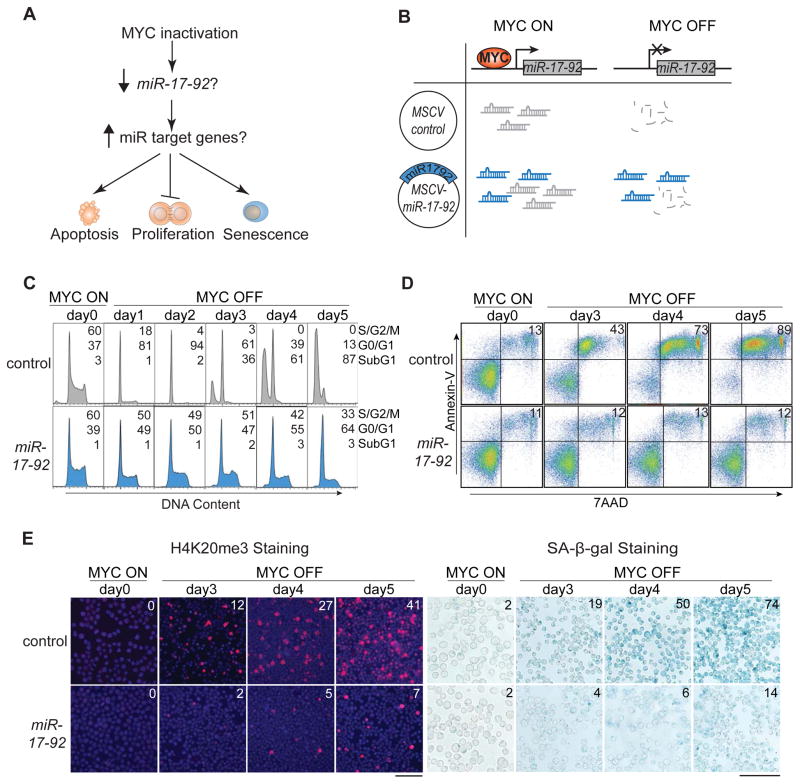
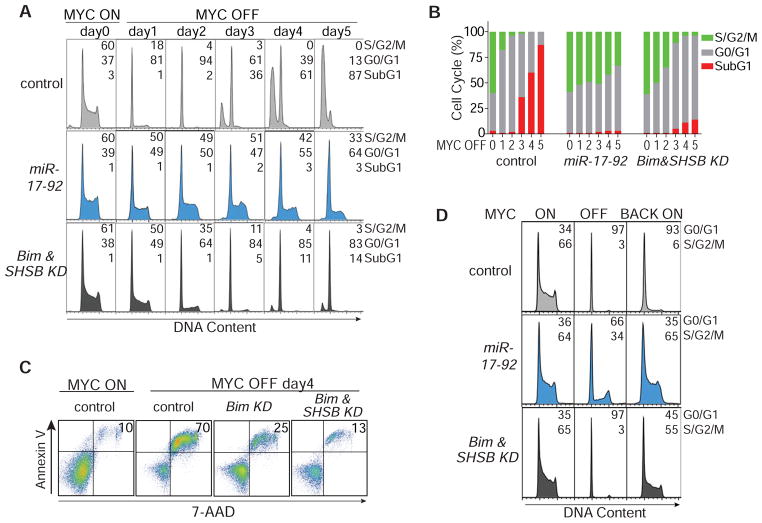
Figure 1. miR-17-92 Expression In Vitro Rescues MYC Oncogene Addiction by Sustaining Proliferation and Blocking Apoptosis and Senescence.
(A) Diagram of cellular changes upon MYC inactivation in MYC-driven tumors. (B) Experimental strategy to sustain miR-17-92 with retroviral expression. The endogenous miR-17-92 is colored grey while the exogenous MSCV-miR-17-92 is colored blue. (C) Cell cycle analysis of tumor cells over a 5-day time course with flow cytometry after propidium iodide staining. Numbers indicate percentage of cells in different phases of cell cycle. The experiments were repeated three times with similar results. (D) Annexin V/7-AAD staining showing apoptotic cells over a 5-day time course of MYC inactivation. Numbers in upper right quadrant indicate percentage of apoptotic cells. The experiments were repeated three times with similar results. (E) H4K20me3 and SA-β-g al staining of tumor cells after MYC inactivation for five days. The control cells are kept alive with Bim shRNA knockdown and Bcl-xL overexpression. Numbers in upper right quadrant indicate percentage of cells stained positive. Cell nuclei in the H4K20me3 panel were stained with DAPI. The experiments were repeated twice with similar results. Scale bar = 50μm. See also Figure S1.
RESULTS
Sustained miR-17-92 Expression Rescues MYC Addiction In Vitro and In Vivo
We examined the level of several microRNAs known to be regulated by MYC using real-time quantitative PCR in three lymphoma cell lines from Eμ-tTA/tet-O-MYC mice (O'Donnell et al., 2005; Sander et al., 2008). Upon MYC inactivation with doxycycline treatment, all members of the miR-17-92 cluster were downregulated, while miR-15/16 and miR-26 were upregulated in a time-dependent manner (Figure S1A). Similarly, in MYC-driven hepatocellular carcinoma derived from LAP-tTA/tet-O-MYC transgenic mice, miR-17-92, but not miR-15/16 and miR-26, was downregulated upon MYC inactivation (Kota et al., 2009; Shachaf et al., 2004) (Figure S1B). Thus, MYC generally regulates the expression of miR-17-92 in tumor cells.
We speculated that miR-17-92 was responsible for maintaining at least some of the hallmark features of cancer in MYC-induced tumors (Figure 1A). To determine whether constitutive expression of miR-17-92 could rescue any of the effects of MYC inactivation, we retrovirally infected MYC-induced lymphomas with Murine Stem Cell Virus containing miR-17-92 (MSCV-miR-17-92), then confirmed that expression of miR-17-92 was maintained even after MYC inactivation (Figure 1B and S1C). Retroviral miR-17-92 expression abrogated the induction of proliferative arrest, apoptosis, and senescence, which we previously described as consequences of MYC suppression (Felsher and Bishop, 1999; Wu et al., 2007). The proliferation was sustained over a 5-day time course in miR-17-92-expressing cells after MYC inactivation as shown by the S/G2/M population in the flow cytometric analysis of cell cycle distributions (Figure 1C, S1D). The induction of apoptosis by MYC inactivation was blocked by miR-17-92 as shown by the subG1 population and the 7-AAD/Annexin V double positive population (Figure 1C–D, S1D). Furthermore, retroviral miR-17-92 expression blocked the induction of cellular senescence in a sustained manner as measured by senescence-associated β-galactosidase (SA-β-g al) staining (Figure 1E, S1E), histone H4 lysine 20 trimethylation (H4K20me3) staining (Figure 1E, S1F), and quantification of trimethylated histone H3 lysine 9 (H3K9me3) (Figure S1G). Similarly, in MYC-induced hepatocellular carcinoma and osteosarcoma (Jain et al., 2002; Shachaf et al., 2004), retroviral expression of miR-17-92 abrogated the induction of cellular senescence upon MYC suppression as shown by SA-β-g al staining (control versus miR-17-92: 19-fold versus 3-fold induction in hepatocellular carcinoma, 10-fold versus 3-fold induction in osteosarcoma; Figure S1H-I). In contrast, miR-17-92 expression in three BCR-ABL-driven B-cell leukemia cell lines failed to rescue proliferative arrest or apoptosis upon BCR-ABL inactivation (Figure S1J). Hence, miR-17-92 specifically rescues the proliferative arrest, apoptosis, and senescence upon MYC inactivation.
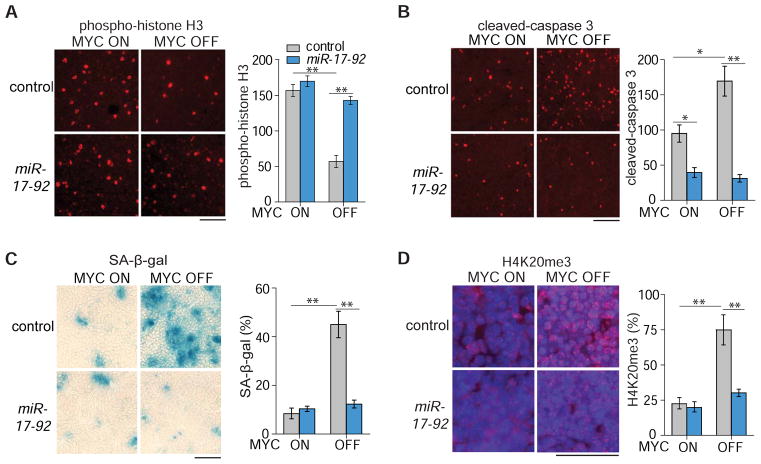
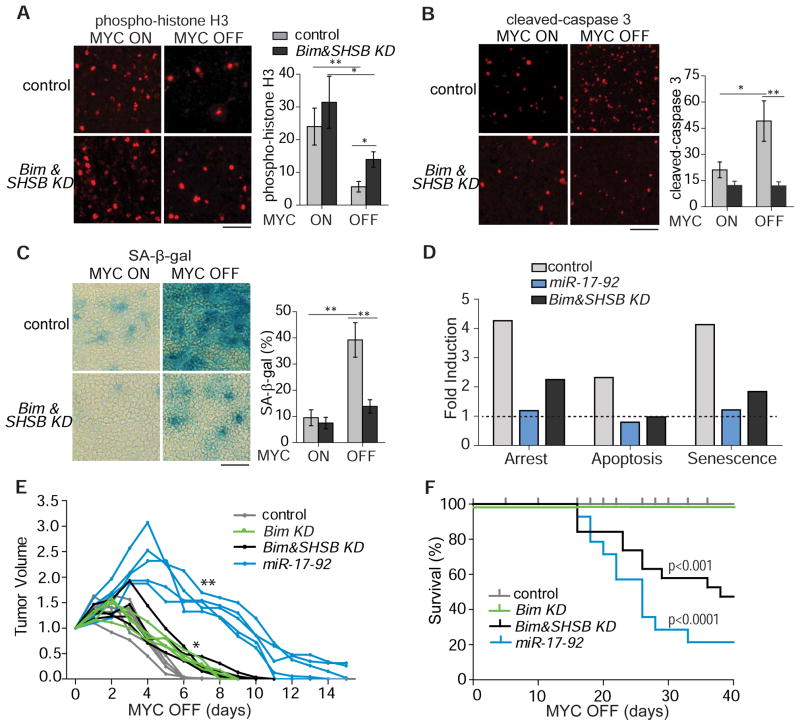
Next, we examined in vivo whether miR-17-92 expression rescues the phenotypes of MYC inactivation. MYC-induced lymphoma cells expressing either empty control vector or MSCV-miR-17-92 were subcutaneously transplanted into syngeneic FVB/N hosts. Tumor cells were allowed to grow in vivo for about 2 weeks before MYC inactivation by doxycycline administration in the drinking water. Tumors were collected before and after MYC inactivation for examination of apoptosis, proliferation, and senescence (Fig. 2A–D). Phospho-histone H3 and Ki67 staining was used to measure mitotic and proliferative cells, respectively. Cleaved-caspase 3 staining was used for apoptotic cells and SA-β-g al and H4K20me3 staining was used for senescent cells. Upon MYC inactivation, in control versus lymphomas with miR-17-92 expression, there was a 67% versus 15% decrease in phospho-histone H3 staining and a 85% versus 25% decrease in Ki67 staining (Figure 2A and Figure S2), a 60% increase versus no change in cleaved-caspase 3 staining (Figure 2B), and a 3-fold increase versus no change in SA-β-g al and H4K20me3 staining (Figure 2C–D). Thus, miR-17-92 expression prevented MYC inactivation from inducing proliferative arrest, apoptosis, and senescence in vivo.
Figure 2. miR-17-92 Expression In Vivo Mediates MYC Oncogene Addiction by Sustaining Proliferation and Blocking Apoptosis and Senescence.
(A) Phospho-histone H3 staining showing cells in the metaphase of the cell cycle four days after MYC inactivation. The y-axis denotes number of positive staining cells per 20X magnification field. (B) Cleaved-caspase-3 showing apoptotic cells four days after MYC inactivation. The y-axis denotes number of positive staining cells per 20X magnification field. (C) SA-β-gal staining four days after MYC inactivation. The y-axis denotes percentage of area with positive SA-β-gal staining. (D) H4K20me3 staining four days after MYC inactivation. The y-axis denotes number of positive staining cells per high magnification field. Results are presented as mean +/− SEM. n=4 (A, B, C, D). Student’s t test, *p<0.05. **p<0.01. Scale bar = 50μm. See also Figure S2.
MYC via miR-17-92 Regulates Specific Target Genes
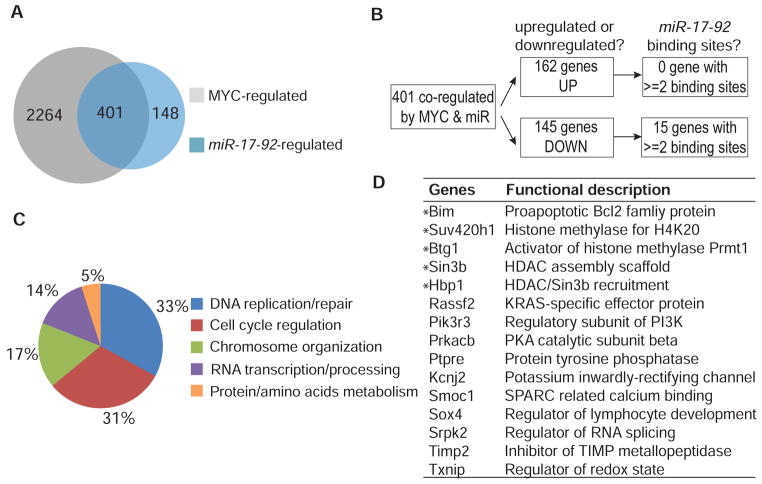
We reasoned that MYC, through miR-17-92, regulates a specific subset of genes responsible for maintaining autonomous proliferation and survival. Since the expression of MYC or miR-17-92 can be turned off independently in our conditional system, this allows for the screening of genes regulated by MYC or miR-17-92 (Figure 1B, S3). The genes that were differentially expressed before and after MYC inactivation in the control lymphoma were categorized as MYC-regulated. The genes that were differentially expressed between control lymphoma and retroviral miR-17-92-expressing lymphoma when MYC was turned off in both populations were defined as miR-17-92-regulated (Figure S3). The specific subset of MYC target genes regulated through miR-17-92 would appear to be co-regulated by both MYC and miR-17-92 (Figure S3 and 3A). By comparing the microarray gene expression profiles of control lymphoma versus miR-17-92-expressing lymphoma upon MYC inactivation, we found that 70% of miR-17-92-regulated genes were also regulated by MYC (Figure 3A). The 401 overlapping genes co-regulated by MYC and miR-17-92 were further separated into the upregulated and downreglated groups (Figure 3B). Among the genes upregulated by both MYC and miR-17-92, there was an enrichment of genes involved in DNA replication, repair, and cell cycle (Figure 3C). Notably, the genes downregulated by both MYC and miR-17-92 had 4.6-fold more miR-17-92 binding sites in their 3’UTR compared with upregulated genes (32% in downregulated versus 7% in upregulated genes, Table S1, S2). We inferred that these downregulated genes are directly regulated by miR-17-92 binding.
Figure 3. Identification of miR-17-92 Target Genes by Comparative Analysis of Genes Regulated by MYC and miR-17-92.
(A) Venn diagram of genes regulated by both MYC and miR-17-92. (B) Flowchart showing the analysis of the genes co-regulated by MYC and miR-17-92. Genes were separated into either upregulated or downregulated groups and analyzed for functional annotation and enrichment of miR-17-92 binding sites. (C) Functional categories of genes upregulated by both MYC and miR-17-92 according to DAVID’s Bioinformatic Resource. The percentage refers to the number of genes within a particular category in relation to the total number of genes that have a GO annotation. (D) Candidate target genes with multiple miR-17-92 binding sites. The histone modifiers and Bim were indicated with asterisks. See also Figure S3 and Table S1-3.
Our gene list was further refined by only including genes with at least two miR-17-92 binding sites in their 3’UTR, as predicted by each of three microRNA target scanning programs (miRanda, Targetscan, and miRWalk) (Figure 3B, 3D, and Table S3). Amongst these 15 genes were four chromatin modifiers that have not been previously reported as MYC or miR-17-92 targets (Sin3b, Hbp1, Suv420h1, and Btg1). Also identified was the apoptosis regulator, Bim, that has been reported previously to be a miR-17-92 target (Ventura et al., 2008; Xiao et al., 2008) (Figure 3D). Notably, all of these genes have been associated with proliferative control, senescence, and/or apoptosis (Berthet et al., 2002; David et al., 2008; Roninson, 2003; Swanson et al., 2004; van Oevelen et al., 2010). Thus, we focused our subsequent efforts on assessing whether these MYC/miR-17-92 target genes could contribute to the consequences of MYC suppression in tumors.
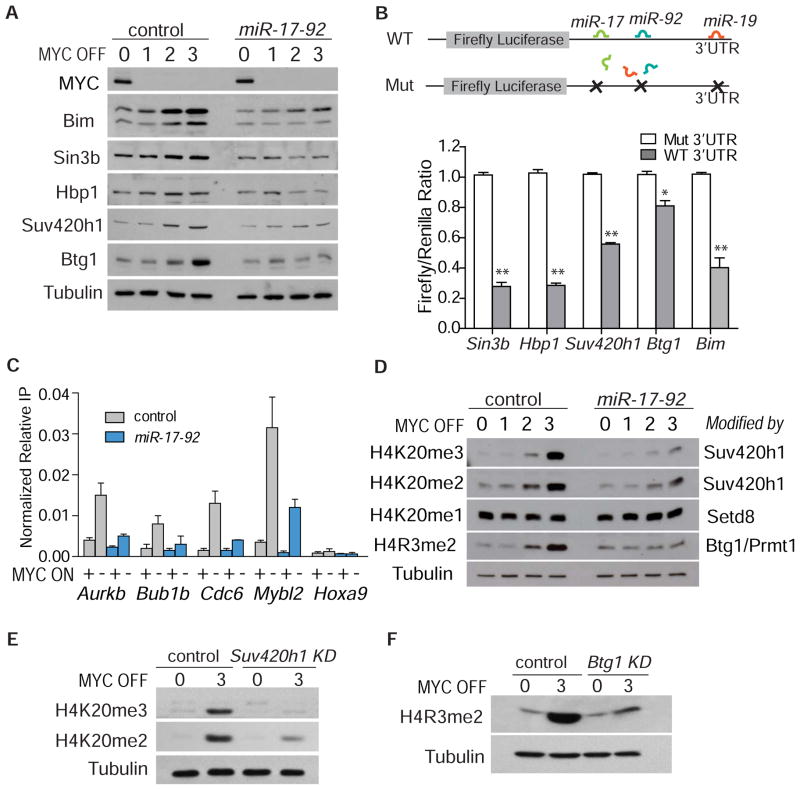
We examined if these miR-17-92 targets were directly regulated by MYC and miR-17-92. First, MYC inactivation induced the protein expression of Sin3b, Hbp1, Suv420h1, Btg1, and all three Bim isoforms in control but not miR-17-92-expressing cells as measured by Western blot analysis (Figure 4A, S4). Second, to validate whether these genes are direct targets of miR-17-92, a dual luciferase assay was performed by cloning 3'UTR fragments of all five genes, with either wild-type or mutant miR-17-92 sites, downstream of the firefly luciferase coding region (Figure 4B, upper panel). Compared with mutant 3'UTRs lacking miR-17-92 binding sites, the wild type 3'UTRs conferred significant repression as shown by the 20–60% lower firefly/renilla ratio (Figure 4B, lower panel). Hence, Sin3b, Hbp1, Suv420h1, Btg1, and Bim are regulated by MYC in a miR-17-92-dependent manner.
Figure 4. Validation of Target Genes of miR-17-92.
(A) Changes in the expression of Bim and histone modifiers as detected by Western blot three days after MYC inactivation. (B) Top: Wild type (WT) and mutant (Mut) 3’UTR reporter constructs. Bottom: Dual luciferase assay using 3’UTR reporters. The firefly luciferase signals were normalized with internal control renilla luciferase. Results are presented as mean +/− SEM. Student’s t test, *p<0.05. **p<0.01. (C) Chromatin immunoprecipitation with Sin3b antibody showing binding of Sin3b to promoters of four proliferation-related genes upon MYC inactivation. MYC OFF samples were taken at two days after MYC inactivation. PCR with Hoxa9 promoter specific primers was included as a negative control. Data shown are averages of two experiments and are presented as mean +/− SEM. (D) Changes in histone H4 lysine 20 and arginine 3 methylation status upon MYC inactivation. The mono-, di- and trimethylation of H4K20 and dimethylation of H4R3 are shown. MYC off time course includes day 0, 1, 2, and 3. The respective enzymes catalyzing each of the modifications are shown on the left side of the lanes. (E) Western blot analysis of di- and trimethylation of H4K20 in the presence of Suv420h1 knockdown. (F) Western blot analysis of dimethylation of H4R3 in the presence of Btg1 knockdown. See also Figure S4.
Next, we examined if MYC via miR-17-92 was regulating chromatin through each of these gene products. Sin3b and Hbp1 have been shown to be candidate target genes of miR-19 (Mu et al., 2009). Sin3b interacts with Hbp1 and recruits histone deacetylases (HDACs) to repress the transcription of genes related to proliferation, such as Aurkb, Mybl2, Cdc6, and Bub1b (David et al., 2008; Swanson et al., 2004; van Oevelen et al., 2010). Indeed, these genes were upregulated by miR-17-92 and MYC (Figure 3C and Table S1). Upon MYC inactivation, there was a 3 to 8-fold increase versus a 2-fold increase in Sin3b binding to these promoters in control versus miR-17-92-expressing lymphoma according to a chromatin immunoprecipitation assay (Figure 4C). Thus, the induction of Sin3b/Hbp1 upon MYC inactivation may contribute to proliferative arrest and cellular senescence by silencing genes related to proliferation and cell cycle.
Notably, Suv420h1 is a histone methyltransferase that catalyzes dimethylation and trimethylation of histone H4 lysine 20 (H4K20me2 and H4K20me3) (Fraga et al., 2005; Greer and Shi, 2012). H4K20me3 is a marker of heterochromatin formation and senescence, and its loss is a common hallmark of human cancer (Fraga et al., 2005; Greer and Shi, 2012; Nelson, 2012). Upon MYC inactivation, there was an induction of both H4K20me2 and H4K20me3 in control but not miR-17-92-expressing lymphoma cells (Figure 4D). In contrast, the monomethylated H4K20me1, catalyzed by Setd8 (Greer and Shi, 2012), did not increase in either control or miR-17-92-expressing lymphoma cells (Figure 4D), indicating that the induction of Suv420h1 specifically increases the dimethylation and trimethylation of H4K20. Finally, Btg1 is a tumor suppressor that can activate histone methyltransferase Prmt1 to dimethylate histone H4 arginine 3 (H4R3me2) (Berthet et al., 2002; Lin et al., 1996). Upon MYC inactivation, there was an accumulation of H4R3me2 in control but not miR-17-92 expressing cells (Figure 4D). Therefore, MYC inactivation via miR-17-92 regulates the biological function of the chromatin regulatory genes Sin3b, Hbp1, Suv420h1, and Btg1.
Suppression of Sin3b, Hbp1, Suv420h1, Btg1, and Bim Significantly Recapitulates miR-17-92 Function
Our results suggest that suppression of the four epigenetic regulators (Sin3b, Hbp1, Suv420h1, and Btg1, hereafter referred to as SHSB) and the pro-apoptotic protein Bim may contribute to MYC’s ability to maintain tumorigenesis. To examine this, the miR-30-based retroviral short hairpin RNAs (shRNAs) were used to knock down the target genes individually or collectively in MYC-induced lymphoma cells. The target mRNA was knocked down to levels between 6–20% of the scrambled control (Figure S5A). Knocking down the expression of Suv420h1 and Btg1 reduced the levels of H4K20me2/3 and H4R3me2, respectively (Figure 4E–F). After 24 hours of MYC inactivation, lymphoma cells with individual knockdown of the chromatin modifiers exhibited a modest block in proliferative arrest, with 19–30% of cells still remaining in S/G2/M phases compared to only 11% for control cells (Sin3b: 25%; Hbp1: 25%; Suv420h1: 35%; Btg1: 19%; Figure S5B). Concurrently knocking down all five miR-17-92 target genes (Bim and SHSB), modestly delayed the proliferative arrest after MYC inactivation as shown by flow cytometric analysis of the cell cycle distribution (Figure 5A–B). The knockdown of Bim alone reduced the induction of apoptosis following MYC inactivation, whereas the combined knockdown of Bim and SHSB further decreased the rate of apoptosis (Figure 5C, S5C). Hence, these data suggest that miR-17-92 regulation of its target genes Sin3b, Hbp1, Suv420h1, Btg1, and Bim is required for proliferative arrest and apoptosis upon MYC inactivation.
Figure 5. Suppression of Bim and the Chromatin Modifiers Delays Proliferative Arrest and Blocks Apoptosis and Senescence upon MYC Inactivation In Vitro.
(A) Cell cycle distribution of cells with combined knockdowns of SHSB and Bim upon MYC inactivation for 5 days. Numbers indicate percentage of cells in different phases of the cell cycle. (B) Bar graph summarizing data shown in Figure 5A. Different phases of the cell cycle were color-coded. (C) Apoptosis of tumor cells with knockdown of Bim and SHSB upon MYC inactivation for four days. Numbers in upper right quadrant indicate percentage of apoptotic cells. Detailed 5-day time course is shown in Figure S9. (D) Cell cycle distribution of tumor cells upon MYC inactivation and reactivation. MYC OFF samples were taken at four days of MYC inactivation with doxycycline treatment. MYC-Back ON samples were taken at three days after MYC reactivation by removing doxycycline. Events shown are gated on live cells. Numbers indicate percentage of cells in either G1/G0 or S/G2//M phase of the cell cycle. The experiments were repeated three times with similar results. See also Figure S5.
Cellular senescence is characterized by a state of permanent cell cycle arrest (Guney and Sedivy, 2006; Nardella et al., 2011). We have shown previously that even brief suppression of MYC can induce senescence and result in sustained tumor regression (Jain et al., 2002). We tested if cell cycle arrest induced by MYC inactivation is reversible (Figure 5D, ON-OFF-BackON). Upon MYC inactivation, control lymphoma cells progress from cell cycle arrest to high levels of apoptosis, with few viable cells remaining by 4–5 days after oncogene withdrawal (Figure 1C–D). To specifically examine the effect of MYC inactivation on cell cycle arrest, independently from apoptosis, we utilized lymphoma cells with the shRNA-mediated knockdown of Bim. Upon MYC inactivation, lymphoma cells with Bim knockdown persisted and underwent proliferative arrest. Moreover, they remained arrested even after reactivation of MYC (Figure 5D, upper panel). This irreversible cell cycle arrest indicated the induction of senescence by MYC inactivation. In contrast, lymphoma cells expressing miR-17-92 or with the knockdown of Bim and SHSB resumed proliferation upon MYC reactivation (Figure 5D, middle and lower panels). Therefore, the expression of miR-17-92 or suppression of the miR-17-92 target genes Sin3b, Hbp1, Sub420h1, and Btg1 prevents the induction of senescence upon MYC inactivation.
The shRNA-mediated suppression of Bim and SHSB also impeded the in vivo proliferative arrest, apoptosis, and senescence upon MYC inactivation. Tumor cells were transplanted subcutaneously into syngeneic FVB/N hosts and grown for about 2 weeks before MYC inactivation by doxycycline administration in the drinking water. In control lymphoma versus lymphomas with Bim and SHSB knockdown, there was a 4-fold decrease versus one-fold decrease in phospho-histone H3 staining, a 1.5-fold increase versus no change in cleaved-caspase 3 staining, and a 3-fold versus less than one fold increase in SA-β-g al staining (Figure 6A–C), similar to what was observed for miR-17-92 expression (Figure 2 and 6D). Thus, miR-17-92 target genes Sin3b, Hbp1, Suv420h1, Btg1, and Bim are required for the induction of proliferative arrest, apoptosis, and senescence in vivo upon MYC inactivation.
Figure 6. Suppression of Bim and the Chromatin Modifiers Delays Proliferative Arrest, Blocks Apoptosis and Senescence, and Abrogates Sustained Tumor Regression upon MYC Inactivation In Vivo.
(A) Phospho-histone H3 staining showing cells in the metaphase of the cell cycle four days after MYC inactivation. The y-axis denotes number of positive staining cells per 40X magnification field. (B) Cleaved-caspase-3 showing apoptotic cells four days after MYC inactivation. The y-axis denotes number of positive staining cells per 40X magnification field. (C) SA-β-gal staining at four days after MYC inactivation. The y-axis denotes percentage of area with positive SA-β-gal staining. n=3. For panel 6A, 6B, and 6C, the results are presented as mean +/− SEM. Student’s t test, *p<0.05. **p<0.01. Scale bar = 50μm. (D) Comparison of in vivo induction of proliferative arrest, apoptosis, and senescence upon MYC inactivation in control lymphomas, lymphomas with retroviral miR-17-92, and lymphomas with knockdown of Bim and SHSB. To calculate the fold induction of proliferative arrest, we used the MYC ON/MYC OFF ratio for phospho-histone H3 staining. To calculate the fold induction of apoptosis and senescence, we used the MYC OFF/MYC ON ratio of cleaved-caspase 3 and SA-β-g al staining, respectively. The fold induction was computed by combining data presented in Figure 2 and Figure 6A–C. The dashed line indicates no induction. (E) In vivo regression of transplanted tumors in SCID mouse host. n=4-6. For miR-17-92 versus control, the double asterisk (**) indicates p<0.05 for day 3-14. For Bim KD or Bim&SHSB KD versus control, the single asterisk (*) indicates p<0.05 for day 5-8. The comparisons were made with the two-tailed Student’s t test. (F) In vivo tumor reoccurrence upon MYC inactivation in syngeneic wild type FVB/N mice. Tumor bearing mice were monitored for six weeks of doxycycline administration in drinking water. Mice were euthanized when the diameters of the relapsed tumors reach 1 cm. n= 10-16. Logrank test. p<0.001 for tumors with Bim&SHSB KD. p<0.0001 for tumors with miR-17-92 expression. See also Figure S6.
Suppression of Sin3b, Hbp1, Suv420h1, Btg1, and Bim Abrogates Sustained Tumor Regression upon MYC Inactivation
Since miR-17-92 expression and knockdown of miR-17-92 target genes significantly block the induction of proliferative arrest, apoptosis, and senescence both in vitro and in vivo, we examined their impact on sustained tumor regression upon MYC inactivation. MYC inactivation in lymphoma induced rapid tumor regression within 6 days (Figure 6E), without evidence of lymphoma recurrence even after six months of continuous observation (Figure 6F). In contrast, the lymphomas with enforced retroviral miR-17-92 expression regressed only after 14 days and 80% of tumors reoccurred within 6 weeks (Figure 6E–F). Compared with the dramatic delay in tumor regression with miR–17-92 expression, the knockdown of either Bim or Bim combined with SHSB modestly delayed the kinetics of tumor regression (Figure 6E). Interestingly, although Bim knockdown was not associated with any tumor recurrence, the combined knockdown of Bim and SHSB was associated with the recurrence of 50% of tumors (Figure 6F). After prolonged MYC inactivation, the recurrent tumors eventually regained high levels of MYC expression, similar to what we have described previously (Figure S6) (Choi et al., 2011). Hence, the expression of miR-17-92 or the suppression of miR-17-92 target genes, Sin3b, Hbp1, Suv420h1, Btg1, and Bim prevented sustained tumor regression upon MYC inactivation.
DISCUSSION
We have found that MYC through miR17-92 directly suppresses the expression of chromatin regulatory genes Sin3b, Hbp1, Suv420h1, Btg1 and pro-apoptotic gene Bim. The suppression of these defined factors is causally required to maintain survival, autonomous proliferation, and self-renewal. Our results have general implications for how MYC maintains a neoplastic state.
MYC is known to globally regulate gene and protein expression (Dang, 2012). Many studies have identified hundreds of genes associated with MYC overexpression and tumorigenesis (Kim et al., 2008; Schlosser et al., 2005; Zeller et al., 2006). The expectation is that a similar multitude of genes would be required by MYC to initiate and maintain a neoplastic state. Surprisingly, we found that a single microRNA cluster, miR-17-92, amongst thousands of genes controlled by MYC, can maintain a neoplastic state in MYC-induced tumors by sustaining autonomous proliferation and survival. We found that this mechanism was specific to MYC and unique to miR-17-92. Furthermore, the function of miR-17-92 can be partially attributed to the suppression of a small number of target genes, such as the chromatin regulatory genes Sin3b, Hbp1, Suv420h1 and Btg1, as well as the pro-apoptotic gene Bim. Our results highlight how MYC maintains tumorigenesis through the regulation of miR-17-92-dependent epigenetic and survival programs.
Oncogene-induced tumorigenesis is generally suppressed through intrinsic barriers, such as apoptosis and senescence (Braig et al., 2005; Lowe et al., 2004; Nardella et al., 2011). The inactivation of a driver oncogene can restore these tumor suppressor mechanisms, even in a tumor that is genetically complex (Karlsson et al., 2003), thereby eliciting the phenomenon of oncogene addiction (Felsher, 2008; Weinstein, 2002). Hence, our results suggest that MYC via miR-17-92 maintains autonomous proliferation, self-renewal, and survival. Correspondingly, MYC inactivation induces a loss of tumor hallmark features as a consequence of restoration of proliferative arrest, apoptosis, and senescence (Figure 7).
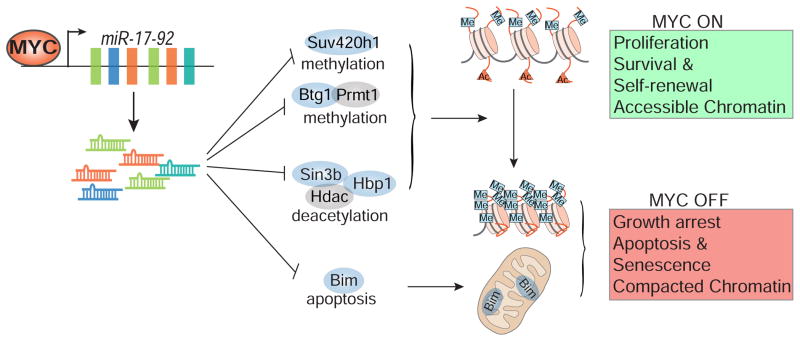
Figure 7. A Model illustrating MYC, through miR-17-92 and its target genes, controls a chromatin regulatory and survival switch that is required to sustain a neoplastic state.
MYC through miR-17-92 suppresses specific genes to maintain autonomous proliferation, self-renewal, and survival. Correspondingly, MYC inactivation induces a loss of neoplastic features as a consequence of restoration of proliferative arrest, apoptosis, and senescence.
We have identified specific miR-17-92 target genes that are essential for the reversal of neoplasia upon MYC inactivation. These genes drive proliferation arrest, senescence, and apoptosis by regulating chromatin modification and apoptosis. Sin3b interacts with Hbp1 and recruits HDACs to silence proliferation-related genes and mediate cell cycle exit and senescence (David et al., 2008; Grandinetti et al., 2009; Swanson et al., 2004). Suv420h1 is a histone methyltransferase that dimethylates and trimethylates H4K20 (Fraga et al., 2005; Greer and Shi, 2012). H4K20me3 is known to direct chromatin compaction and is a marker of heterochromatin formation and senescence (Greer and Shi, 2012; Lu et al., 2008; Nelson, 2012). Loss of H4K20me3 is a common feature in human cancer (Fraga et al., 2005). Btg1 is a tumor suppressor that is frequently lost in acute lymphoblastic leukemia (Lundin et al., 2012; Waanders et al., 2012). It is also a biomarker of chemotherapy-induced cellular senescence (Roninson, 2003). Finally, Bim is a major tumor suppressor in MYC-induced lymphomagenesis (Egle et al., 2004). Bim is also frequently lost in human B-cell lymphomas and its loss can cause chemoresistance in patients (Richter-Larrea et al., 2010). Thus, these miR-17-92 targets are some of the critical players in the maintenance of the neoplastic state by MYC.
Recently it has been shown that MYC can function as a transcriptional amplifier of the already expressed genes within the cells without specificity (Lin et al., 2012; Nie et al., 2012). It is a remarkable finding and explains one aspect of MYC function that is consistent with prior studies (Dang, 2012; Guccione et al., 2006; Guney and Sedivy, 2006; Varlakhanova and Knoepfler, 2009). However, whether the transcriptional amplifier mechanism is required for MYC to maintain the neoplastic state is not known. Moreover, MYC’s function as an amplifier is likely only one of its many functions and does not account for the ability of MYC to suppress gene expression, nor does it provide an explanation for gene-specific effects on expression (Walz et al., 2013). Our results highlight an additional mechanism by which MYC controls several essential features of a neoplastic state (Figure 7). MYC, through miR-17-92, controls a general ON and OFF switch of chromatin state and thereby regulates the decision between survival versus death and self-renewal versus senescence. Our findings are also complementary to MYC’s role in transcriptional amplification. High levels of MYC can keep the chromatin transcriptionally accessible and allow for transcriptional amplification. When MYC is turned off, many genes are downregulated by the amplifier mechanism and this likely reduces the ability of tumor cells to grow and proliferate. However, the suppression of miR-17-92 upon MYC inactivation allows the induction of many genes, including chromatin modifiers and apoptosis regulators involved in apoptosis and senescence (Figure 7). Hence, MYC inactivation leads to a change in the neoplastic state.
MYC has been shown before to modulate global euchromatin structure that may contribute to self-renewal and pluripotency, but the specific mechanism has been elusive (Knoepfler et al., 2006; Varlakhanova and Knoepfler, 2009). Our finding that MYC, through miR-17-92, regulates the heterochromatin formation may provide an explanation. Hence, MYC suppression in cancer cells results in irreversible changes in gene expression and the permanent loss of a neoplastic phenotype (Jain et al., 2002). We infer that MYC’s ability to sustain autonomous proliferation, self-renewal, and survival is mediated through a miR-17-92-dependent chromatin regulatory and survival switch. The shut off of this epigenetic switch contributes to the mechanism of MYC-associated oncogene addiction.
EXPERIMENTAL PROCEDURES
Cell Lines, DNA Constructs, and Viruses
Conditional lymphoma and leukemia cell lines were derived from Eμ-tTA/tet-O-MYC mice. MYC inactivation was achieved with doxycycline treatment. The miR-17-92 was cloned into pMSCV retroviral vectors. Virus production and infection of tumor cells was performed as previously reported (Wu et al., 2007). Construction of the 3’UTR luciferase reporters and the shRNAs can be found in Supplemental Experimental Procedures.
Tumor Transplantation
All animal experiments were approved by Stanford’s Administrative Panel on Laboratory Animal Care (APLAC) and in accordance with national guidelines. The conditional MYC lymphoma cell line was transplanted into host mice and allowed to grow to 1.5cm diameter before MYC inactivation with doxycycline. Tumor diameters were measured with a caliper. Tumor volume (V) was calculated as: V = ab2 / 2, where a indicates length (mm) and b indicates breadth (mm). Further details can be found in Supplemental Experimental Procedures.
Flow cytometry, microRNA quantification, Western Blot, Immunohistochemistry, SA-β-gal Staining, and Chromatin Immunoprecipitation
The microRNAs were quantified with TaqMan microRNA assay kits (Applied Biosystems). Western blotting, immunofluorescence, SA-β-gal staining, and chromatin immunoprecipitation were performed as described (van Oevelen et al., 2010; Wu et al., 2007). Details can be found in Supplemental Experimental Procedures.
Microarray Analysis
Control and miR-17-92-expressing cell lines were used for the microarray analysis. MYC ON and MYC OFF samples were collected at 0 hour and 48 hours after MYC inactivation. Details of the microarray analysis can be found in Supplemental Experimental Procedures.
Multiple Knockdown with shRNAs
The individual knockdown of miR-17-92 target genes was accomplished using LMP miR-30-based shRNAs (OpenBiosystems). For multiple knockdown, the shRNAs were cloned into vectors with different drug selection markers to allow for simultaneous knockdown of multiple genes in the same cell. Further details can be found in Supplemental Experimental Procedures.
Supplementary Material
Significance.
The MYC oncogene is frequently overexpressed in human cancers. MYC overexpression coordinates the expression of thousands of genes that could potentially contribute to its neoplastic properties. However, it was unclear which specific genes are responsible for MYC to maintain a neoplastic state. Here we show that MYC maintains a neoplastic state through the regulation of the microRNA cluster miR-17-92, which controls specific chromatin regulatory and survival programs. MYC inactivation, through the downregulation of miR-17-92, induces a loss of neoplastic features as a consequence of restoration of proliferative arrest, apoptosis, and senescence. Our results highlight how MYC maintains tumorigenesis through the regulation of miR-17-92-dependent epigenetic and survival programs and provide a mechanistic explanation of the phenomenon of oncogene addiction.
Acknowledgments
We thank members of the Felsher Laboratory for generously providing their suggestions and thoughtful discussions. This work was supported by the NIH R01CA170378 (D.W.F), U54CA149145 (D.W.F. and D.L.D.), U54CA143907 (D.W.F. and Y.L.), 1F32CA177139 and 5T32A107290 (S.C.C.), the Leukemia and Lymphoma Society Translational Research grant R6223-07 (D.W.F.), and a King Abdullah University of Science and Technology research grant (D.L.D.).
Footnotes
ACCESSION NUMBERS
The GEO accession number for the microarray analysis reported in this paper is GSE57507.
AUTHOR CONTRIBUTIONS
Y.L., P.S.C., and D.W.F designed research; Y.L. and P.S.C. performed experiments; Y.L. and D.L.D. analyzed the microarray data; S.C.C. contributed to data analysis; Y.L. and D.W.F. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berthet C, Guehenneux F, Revol V, Samarut C, Lukaszewicz A, Dehay C, Dumontet C, Magaud JP, Rouault JP. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells. 2002;7:29–39. doi: 10.1046/j.1356-9597.2001.00497.x. [DOI] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Bui TV, Mendell JT. Myc: Maestro of MicroRNAs. Genes & cancer. 2010;1:568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- Choi PS, van Riggelen J, Gentles AJ, Bachireddy P, Rakhra K, Adam SJ, Plevritis SK, Felsher DW. Lymphomas that recur after MYC suppression continue to exhibit oncogene addiction. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17432–17437. doi: 10.1073/pnas.1107303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nature medicine. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nature medicine. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW. Oncogene addiction versus oncogene amnesia: perhaps more than just a bad habit? Cancer Res. 2008;68:3081–3086. doi: 10.1158/0008-5472.CAN-07-5832. discussion 3086. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandinetti KB, Jelinic P, DiMauro T, Pellegrino J, Fernandez Rodriguez R, Finnerty PM, Ruoff R, Bardeesy N, Logan SK, David G. Sin3B expression is required for cellular senescence and is up-regulated upon oncogenic stress. Cancer Res. 2009;69:6430–6437. doi: 10.1158/0008-5472.CAN-09-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature cell biology. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guney I, Sedivy JM. Cellular senescence, epigenetic switches and c-Myc. Cell Cycle. 2006;5:2319–2323. doi: 10.4161/cc.5.20.3348. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner CS, Zhang P, Van Etten RA, Tenen DG. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nature genetics. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Giuriato S, Tang F, Fung-Weier J, Levan G, Felsher DW. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 2003;101:2797–2803. doi: 10.1182/blood-2002-10-3091. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. The EMBO journal. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR–targeted therapy and beyond. Mod Pathol. 2008;21(Suppl 2):S16–22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. The Journal of biological chemistry. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Hjorth L, Behrendtz M, Nordgren A, Palmqvist L, Andersen MK, Biloglav A, Forestier E, Paulsson K, Johansson B. High frequency of BTG1 deletions in acute lymphoblastic leukemia in children with down syndrome. Genes, chromosomes & cancer. 2012;51:196–206. doi: 10.1002/gcc.20944. [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nature reviews Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- Nelson DM. Chromatin modification contributes to senescence associated proliferation arrest. University of Glasgow; 2012. [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c–Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, Calasanz MJ, Panizo C, Richter JA, Hernandez JM, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, Chapman R, Weidle UH, Eick D. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- van Oevelen C, Bowman C, Pellegrino J, Asp P, Cheng J, Parisi F, Micsinai M, Kluger Y, Chu A, Blais A, et al. The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol Cell Biol. 2010;30:5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Knoepfler PS. Acting locally and globally: Myc's ever-expanding roles on chromatin. Cancer Res. 2009;69:7487–7490. doi: 10.1158/0008-5472.CAN-08-4832. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waanders E, Scheijen B, van der Meer LT, van Reijmersdal SV, van Emst L, Kroeze Y, Sonneveld E, Hoogerbrugge PM, van Kessel AG, van Leeuwen FN, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS genetics. 2012;8:e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S, Eilers M, Eisenman RN, Dang CV. Unlocking the mysterious mechanisms of Myc. Nature medicine. 2013;19:26–27. doi: 10.1038/nm.3060. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature immunology. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.