Abstract
This chapter describes a mass spectrometry-based strategy that facilitates the unambiguous identification and characterization of proteins modified by lipid peroxidation-derived 2-alkenals. The approach employs a biotinylated hydroxyl amine derivative as an aldehyde/keto reactive probe in conjunction with selective enrichment and tandem mass spectrometric analysis. Methodological details are given for model studies involving a distinct protein and 4-hydroxy-2-nonenal (HNE). The method was also evaluated for an exposure study of a cell culture system with HNE that yielded the major protein targets of HNE in human monocytic THP-1 cells. The application of the approach to complex biological systems is demonstrated for the identification and characterization of endogenous protein targets of aldehydic lipid peroxidation products present in cardiac mitochondria.
Introduction
Reactive oxygen species are constantly generated within cells, for instance, by environmental insults (e.g. UV light), metal-catalyzed reactions, as products of the inflammatory response in neutrophils and macrophages and as a byproduct of oxidative phosphorylation. The production of ROS and derived secondary products are largely counteracted by an intricate antioxidant system. The extent of imbalance between ROS production and removal by cellular antioxidant defenses determines the degree of oxidative stress (Beckman and Ames, 1998; Finkel and Holbrook, 2000).
Non-enzymatic peroxidation of polyunsaturated fatty acids (PUFA) present in membranes and lipoproteins results in the formation of reactive aldehydes (Esterbauer et al., 1991). For example, radical mediated hydrogen abstraction and oxidation of the prototypic ω-6 PUFA linoleic acid results in the formation of a set of unsaturated hydroperoxides: 13-hydroperoxy-octadecadienoic acid (13-HPODE) and 9-hydroperoxy-octadecadienoic acid (9-HPODE). Subsequent oxidative cleavage results in the formation of 2-alkenals that contain the ω-tail of the PUFA or retain the carboxyl terminus (Esterbauer et al., 1991; Sayre et al., 2006). Examples of α,β-unsaturated aldehydic lipoxidation products derived from linoleic acid are a) 4-oxo-2-noneal and 4-hydroxy-2-nonenal, and b) the carboxy terminating aldehydes 9,12-dioxo-10-dodecenoic acid (DODE) and 9-hydroxy-12-oxo-10-dodecenoic acid (HODA) (Sayre et al., 2006; Spiteller et al., 2001). The latter carboxylated alkenals remain usually esterified to the phospholipid but can be liberated by lipases (Spiteller, 2001).
Oxidative decomposition of ω-3 PUFAs leads to formation of 4-hydroxy-2(E)-hexenal (Catalá, 2009). Acrolein, 2-propenal, is commonly considered as a lipid peroxidation product, but, can also be formed by diverse metabolic routes, e.g. polyamine oxidation and myeloperoxidase-mediated oxidation of threonine. Acrolein is also a byproduct of cigarette smoking and burning of fossil fuels (Stevens and Maier, 2008). Other α,β-unsaturated carbonylic lipoxidation products include the cyclopentenone-containing isoprostanes. Radical-induced oxidation of arachidonic acid gives rise to cyclopentenone-A2- and J2-isoprostanes (Chen et al., 1999), while eicosapentaenoic acid (EPA) yields cyclopentenone-A3/J3-isoprostanes (Brooks et al., 2008).
There is increasing interest in the characterization of the modifications of proteins by electrophilic lipid peroxidation products. Elevated levels of oxidatively modified proteins have been linked to diverse cardiovascular diseases (Uchida, 2000), liver inflammation (Poli et al., 2008), renal failure (Helga et al., 2004) and neurodegenerative disorders (Butterfield and Sultana, 2008), as well as aging (Beckman and Ames, 1998; Montine et al., 2002). Because lipid peroxidation-derived aldehydes are electrophiles, one possible route in which LPO-derived aldehydes exerts their impact on cellular functions and cytotoxicity is the direct modification of proteins. An overview of the many possible structures of LPO-derived protein side-chain adducts is given in recent review by Sayre et al. (Sayre et al., 2006). Modification of proteins by reactive oxylipids has been linked to protein misfolding (Bieschke et al., 2006; Qin et al., 2007), protein dysfunction (Stewart et al., 2007), aberrant protein processing and degradation (Carbone et al., 2004; Farout et al., 2006; Powell et al., 2005) and modulation of diverse intracellular signaling pathways (Lee et al., 2009; West and Marnett, 2006).
This chapter describes a chemical labeling approach in combination with mass spectrometry to facilitate the unambiguous identification and characterization of proteins modified by lipid peroxidation-derived 2-alkenals. The approach is based on selective labeling of aldehyde/keto groups present in oxidatively modified proteins and the subsequent targeted analysis of the modified proteins by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Modification of proteins by aldehydic lipid peroxidation products
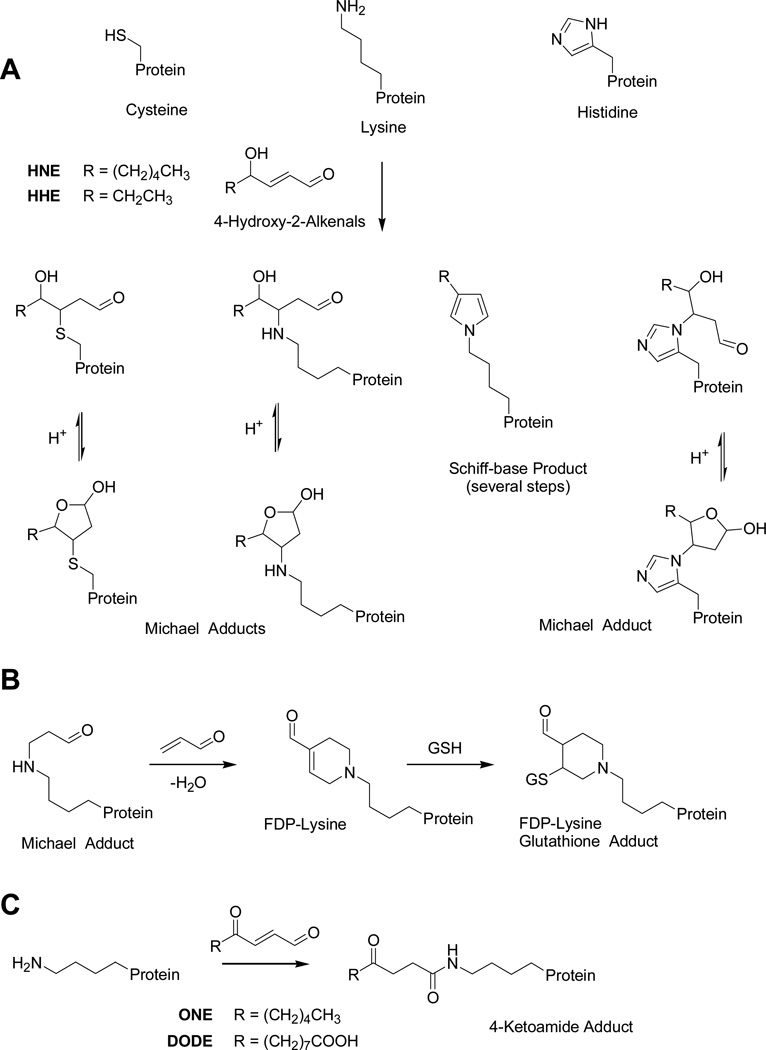
Protein adduct formation is predominately caused by 1,4-Michael-type addition of α,β-unsaturated aldehydes to nucleophilic side chains, e.g. the cysteine thiol, the histidine imidazol moiety and the ε-amino group of lysine residues (Berlett and Stadtman, 1997; Sayre et al., 2006). Representative Michael-type adducts of 4-hydroxy-2-alkenals are given in Figure 1A. In addition, in vitro studies with arginine-containing peptides have demonstrated that ONE can form adducts with the guanidinyl side chain (Doorn and Petersen, 2003; Oe et al., 2003b). Model studies at the amino acid level indicated that the Michael adduct reactivity of HNE and ONE declines in the following order: Cys >> His > Lys (> Arg for ONE) (Sayre et al., 2006). Uchida et al. reported that the modification of lysine residues by 2-enals resulted in the formation of Nε-(3-formyl-3,4-dehydropiperidino)lysine (FDP-lysine) derivatives (Furuhata et al., 2002; Ichihashi et al., 2001) (Figure 1B). FDP-lysine functions as electrophile in the reaction with glutathione (Furuhata et al., 2002) and retains a free aldehyde group which should make the FDP-lysine-containing protein amenable to the described chemical labeling approach. The Sayre group reported that the long-lived ONE-derived adduct of lysine residues is actually a 4-ketoamide adduct rather a Michael adduct (Zhu and Sayre, 2007). Because DODE and ONE share the same 4-oxo-2-enal functional element, adduct formation of DODE with the ε-amino group of lysine may also yield ketoamide adducts (Figure 1C). Ketoamide adducts retain the keto functionality and, therefore, our strategy to chemically tag the aldehyde/keto group in oxidatively modified proteins by utilizing an aldehyde/keto reactive hydroxylamine-functionalized biotin derivative should be applicable to these long-lived protein modifications. LPO-derived 4-hydroxy- and 4-oxo-2-alkenals Michael protein adducts can also further engage in protein cross linking resulting in the loss of the carbonyl functionality (Stewart et al., 2007; Zhang et al., 2003).
Fig. 1.
Possible modifications of proteins by aldehydic lipid peroxidation products. A) Michael-type addition and Schiff’s base formation of 2-alkenals involving nuclephilic side chains commonly found in proteins. B) Proposed formation and structure of FDP-lysine adducts. The Michael adduct of lysine reacts further with an second 2-enal (e.g. acrolein) molecule via Michael addition followed by an aldol condensation yielding the FDP-lysine adduct. FDP-lysine is an electrophile due to its 2-enal moiety and can readily react with nucleophiles, such as glutathione. C) 4-Ketoamide lysine adduct formation involving the ε-amino group of lysine and lipids with a 4-oxo-2-enal reactive moiety.
Redox proteomics of protein targets of reactive lipid peroxidation products
Many studies have been reported that use a gel-based redox proteomics approach to identify protein targets using a combination of 2D gel electrophoresis, immuno staining or oxyblots, image analysis, in-gel digestion followed by mass spectrometry-based protein identifications in cultured cells, organelles and tissues (Butterfield and Sultana, 2008; Chung et al., 2009; Reed et al., 2009; Sultana et al., 2009). Although these studies commonly provide only putative protein identifications, they provide good starting points for in depths studies of distinct proteins in order to determine the site and the functional consequences of an aldehydic modification (Carbone et al., 2005; Eliuk et al., 2007; Roede et al., 2008).
Mass spectrometry-based approaches for the identification and characterization of protein adducts of aldehydic lipid peroxidation products
Gel-free mass spectrometry-based proteomics studies have emerged that attempt to characterize protein targets of aldehydic modifications in complex biological systems. These strategies focus on covalently tagging of oxidatively modified protein and their subsequent targeted analysis by tandem mass spectrometry (Table 1) (Chavez et al., 2006; Codreanu et al., 2009; Danni et al., 2007; Han et al., 2007). Alternative approaches utilize solid phase capture of protein-HNE adducts in conjunction with LC-MS/MS analyses for obtaining protein and modification site identifications (Roe et al., 2007). In addition, reactive HNE surrogate probes have been developed that allow biotin/streptavidin catch and photorelease of protein-HNE adducts employing ex vivo click chemistry (Kim et al., 2009; Vila et al., 2008).
Table 1.
Chemical Probes (in alphabetical order) used for MS/MS-based analyses of protein Adducts of LPO-derived Enals in complex biological systems
| Probe1 | Application | Ref. |
|---|---|---|
| ARP2 | Mitochondrial protein targets of LPO-derived alkenals | (Chavez et al., 2006); |
| Protein targets of HNE in THP-1 cells | (Chavez et al., 200x) | |
| Biotin hydrazide2 | Protein carbonyls in skeletal muscle mitochondria5 | (Danni et al., 2007) |
| Biotin-LC-hydrazide2 | Protein targets of HNE exposure in RKO cells | (Codreanu et al., 2009) |
| HICAT3 | Mitochondrial protein targets of HNE exposure; endogenous protein adducts | (Han et al., 2007) |
| Hydrazide-functionalized beads4 | Protein targets of HNE in yeast whole cell lysate after treatment with HNE | (Roe et al., 2007) |
| Model peptide-HNE adducts in mouse brain tryptic digest | (Rauniyar et al., 2008) |
ARP, aldehyde reactive probe (N’-Aminooxymethylcarbonylhydrazino D-biotin); Biotin-LC-hydrazide, biotinamidohexanoic acid hydrazide; HICAT, Hydrazide-functionalized isotope-coded affinity tag;
Commerically available;
Synthesis described by Han et al. (Han et al., 2007);
Preparation described by Roe et al. (Roe et al., 2007);
No specific carbonylation sites were reported.
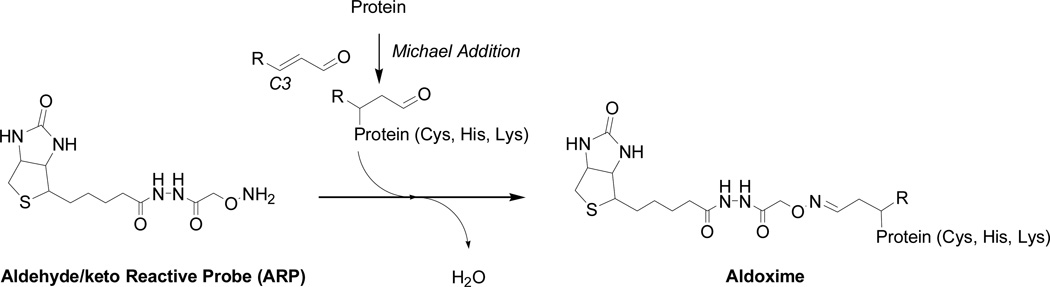
Our laboratory emphasizes chemical approaches that allow the specific tagging of oxidatively modified protein and their subsequent analysis by mass spectrometry techniques to obtain high content information on the target molecule and post-translational modification chemistry. In this context, we have explored the use of hydrazine-functionalized isotope-coded affinity probes (HICATs) (Han et al., 2007) and a biotinylated hydroxylamine derivative, N’-aminooxymethylcarbonylhydrazino D-biotin (aldehyde-reactive probe, ARP) for the derivatization, enrichment and mass spectrometric characterization of oxylipid-modified proteins (Chavez et al., 2006). The hydroxyl amine group of ARP forms with the aldehyde/keto groups present in lipoxidation-derived protein adducts aldoxime/ketoxime derivatives which are sufficiently stable for the subsequent analysis by LC-MS/MS (Figure 2). This contribution details a gel-free mass spectrometry-based strategy to unambiguously identify and characterize protein adducts of α,β-unsaturated aldehydes based on our recently introduced ARP labeling strategy (Figure 3). We demonstrate the utility of this method to determine mechanisms of oxidative protein insult at the molecular level in proteins, cellular and tissue samples.
Fig. 2.
Reaction of aldeyde/keto-containing peptide and protein adducts of lipoxidation products with N'-amino-oxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe (ARP).
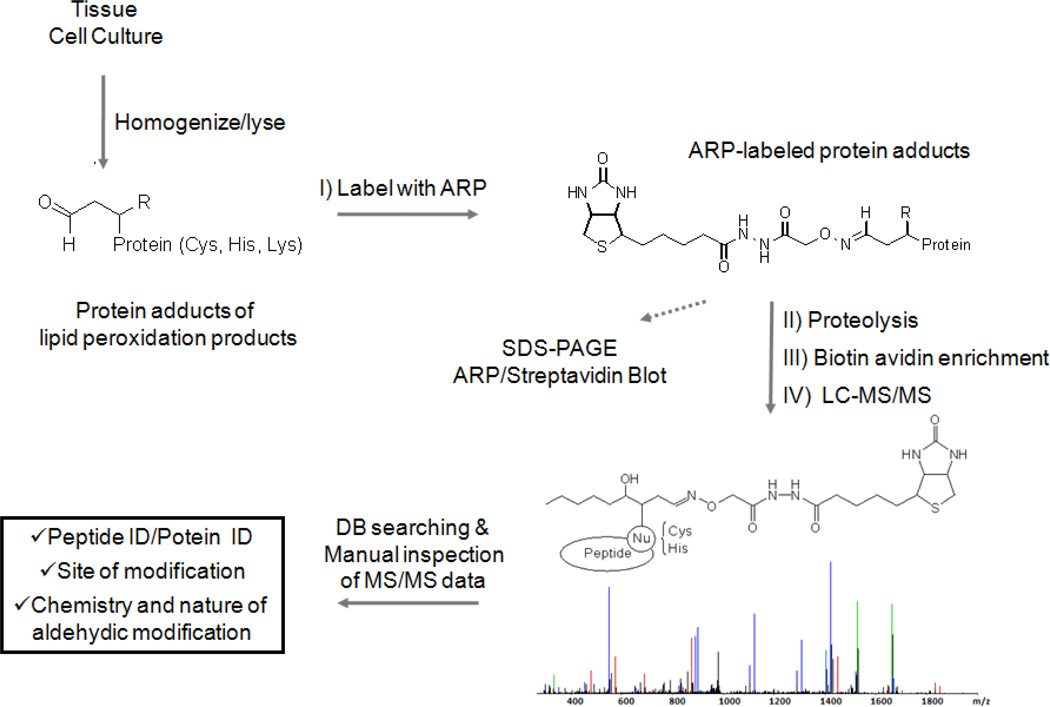
Fig. 3.
The ARP labeling strategy in conjunction with mass spectrometry-based identification and characterization of protein adducts of lipid peroxidation-derived aldehydes.
Experimental strategy of using an aldehyde/keto reactive probe for the targeted analysis of protein adducts of aldehydic lipid peroxidation products
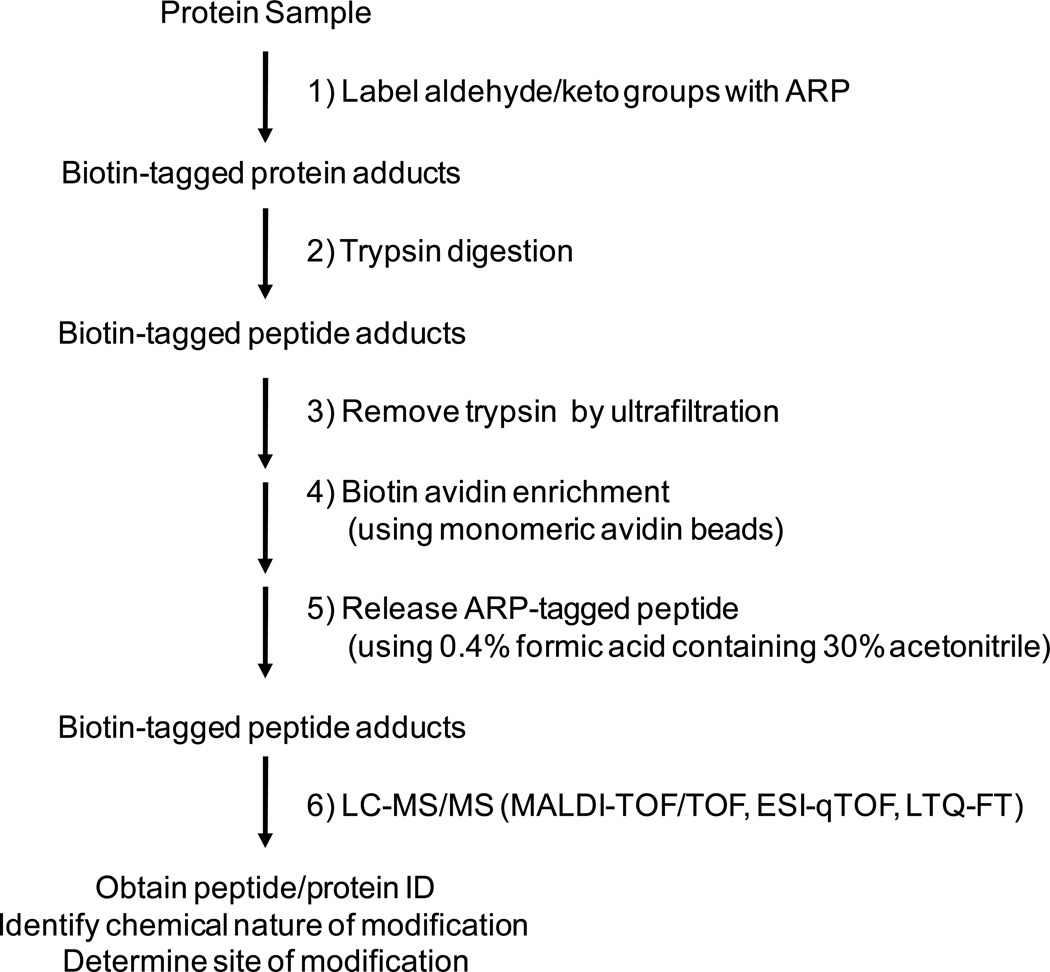
Only tandem mass spectrometric approaches allow for unambiguous assignments of protein adducts because these techniques enable the localization of the modification to a specific residue and provide mass assignments for the modifying entity. The targeted tandem mass spectrometry-based analysis of aldehydic protein adducts using an aldehyde/keto-specific probe involves four steps: I) chemical tagging of proteins that contains the aldehydic modification using the aldehyde/keto reactive probe; II) proteolysis, III) an enrichment step that involves capture of the chemically tagged aldehydic oxylipid adduct and subsequent release of the tagged peptide adduct and IV) LC-MS/MS analysis of the peptide adduct (Figure 3). This strategy allows the unambiguous assignment of the modified peptide including mass information on the nature of the α,β-unsaturated aldehyde and the localization of the aldehydic modification to a partial sequence, in many cases, to a distinct residue. Figure 4 outlines the experimental workflow for the ARP-labeling strategy. The ARP labeling strategy is applicable to distinct protein systems, cellular systems as well as to complex biological matrices to identify and characterize low-abundance protein adducts of lipoxidation-derived aldehydes. The targeted nature of the analyses should allow obtaining more complete data on the distribution of oxidative protein modifications caused by LPO-derived aldehydes in biological samples, a prerequisite for the successful conduction of proteome-wide studies for investigating and accessing protein-oxylipid adducts under condition of oxidative stress associated with certain diseases and aging.
Fig. 4.
Experimental flow chart for the ARP labeling strategy and subsequent mass spectrometry-based identification of aldehydic protein adducts.
ARP labeling of aldehydic protein adducts of 2-alkenals and tryptic proteolysis
The ARP labeling reaction of Michael-type protein adducts of 2-alkenals is carried out by using a final ARP (Dojindo Laboratories, Kumamoto, Japan) concentration of 2.5–5 mM in 10 mM sodium phosphate buffer, pH 7.4, for 1–2 hr at room temperature. Unreacted ARP is removed by using centrifugal ultrafiltration units (Biomax (Millipore) or Microcon (Amicon) units, both with 10-kDa MWCO) or gel filtration columns (Zeba desalting spin column, 7-kDa MWCO, Thermo/Pierce). In the case of protein model studies, ARP-labeled aldehydic protein adducts can be chromatographically isolated and subjected to mass analysis. In order to determine the site of modification, protein samples are subjected to digestion with trypsin (E:S 1:50, modified trypsin, Promega) overnight at 37 °C.
Enrichment of ARP-labeled peptide adducts using biotin avidin affinity chromatography
Peptides containing the ARP label are captured on monomeric avidin affinity columns (Ultralink monomeric avidin, Thermo/Pierce) as described (Chavez et al., 2006). Briefly, monomeric avidin beads are packed into a 0.5 ml Handee mini-spin column (Thermo/Pierce) and prepared for the capture/release protocol according to the manufacture’s instruction. Peptide digests are passed over the avidin beads whereby the ARP-labeled peptides are captured. Non-specific and non-labeled peptides are removed by extensive washing with PBS (20 mM NaH2PO4, 300 mM NaCl). ARP-labeled peptides are eluted from the affinity column by using 0.4 % formic acid containing 30 % acetonitrile and the resulting fractions are concentrated by vacuum centrifugation.
Tandem mass spectrometry for peptide identification and determining the site of adduction
For determining the peptide IDs and site of modifications of ARP-labeled adducts, reversed phase (C18) chromatography in combination with tandem mass spectrometry (LC-MS/MS) is used. So far, three different mass spectrometry systems were employed and evaluated for the identification and characterization of ARP-labeled peptide adducts: a) nanoLC separation of ARP-labeled peptides and subsequent tandem mass spectral (MS/MS) analysis using a MALDI time-of-flight/time-of-flight (TOF/TOF) instrument (4700 Proteomics Analyzer, Applied Biosystems); b) an Electrospray Ionization (ESI) quadrupole TOF (qTOF) mass spectrometer (Micromass/Waters) coupled to nanoAcquity UPLC system (Waters) and c) a capillary LC (Waters) coupled to a hybrid linear ion trap-FTICR (7-Tesla) mass spectrometer (LTQ-FT Ultra, Thermo Finnigan). Because the different instruments utilize different ionization types and collision-induced fragmentation techniques, tandem mass spectra of peptide adducts obtained on different LC-MS/MS systems are provided. Distinct mass spectral features are discussed in the respective figure legends. It is recommended that the LC-MS/MS analyses and data interpretation are performed in close consultation with mass spectrometry professionals.
4700 Proteomics Analyzer
This TOF/TOF instrument is equipped with a MALDI source utilizing a 200-Hz frequency-tripled Nd:YAG laser operating at a wavelength of 355 nm. Protein mass spectra are acquired in the linear mode, whereas peptide mass spectra are obtained in the reflector mode. For both modes, the accelerating voltage is set to 20 kV. For all peptide MS/MS analyses described in this chapter, the following instrument settings are used. The precursor ion is selected by operating the timed gate with an approximately 3–10 Da-width. A collision energy of 1 kV is used and the gas pressure (air) in the collision cell is set to 6 × 10−7 Torr. Fragment ions are accelerated with 15 kV into the reflector.
In order to prepare the fraction containing the ARP-labeled peptides for MALDI-MS/MS analysis, the peptides are loaded onto a trap cartridge, back-flushed onto a PepMap C18 column (150 × 75 µm inner diameter, mm), chromatographically separated and spotted to stainless steel MALDI target plate using a Dionex/LC Packings Ultimate nanoLC system coupled to a Probot™ target spotter. Gradient elutions have to be tailored to the respective peptide sample. For most peptide separations, a binary gradient system can be used that consists of solvent A, 0.1% aqueous trifluoroacetic acid (TFA) containing 5% acetonitrile, and solvent B, 80% aqueous acetonitrile containing 0.1 % TFA. Peptides are eluted using a linear gradient with a slope of 0.8%/min over 60 min. The eluate is continuously mixed with the MALDI matrix solution (2 mg/mL α-cyano-4-hydroxycinnamic acid in 50% aqueous acetonitrile containing 0.1% TFA) and spotted onto a 144-spot MALDI target. The spotting time is typically set to 20 seconds but depends on the complexity of the peptide sample.
ESI qTOF instrument
For the described peptide LC-MS/MS analyses, the electrospray source is operated in the positive mode with a spray voltage of 3.5 kV. The mass spectrometer is operated in data-dependent acquisition mode: a 0.6s a survey scan is followed by a 2.4-second period in which MS/MS analyses on the three most abundant precursor ions detected in the survey scan are acquired. A 60-second dynamic exclusion of previously selected ions is used. The collision energy for MS/MS (25 to 65 eV) is dynamically selected based on the charge state of the ion selected by the quadrupole analyzer. To obtain optimal mass measurement accuracy, lock spray mass correction is performed on the doubly charged ion of Glu1-fibrinopeptide ([M+2H]2+ 785.8426 Th) every 30 seconds.
For nanoLC-MS/MS analyses of peptides, the ESI-qTOF instrument is connected to a nanoAcquity UPLC system (Waters Milford, MA). Typically, ARP-labeled peptide adducts are fractionated on a BEH C18 column (100 µm i.d. × 200 mm, 1.7 µm; Waters, Milford, MA) using a linear 60 min-gradient of a binary solvent system consisting of solvent A (2% acetonitrile/ 0.1% formic acid) and solvent B (acetonitrile containing 0.1% formic acid). The solvent composition was changed with a rate of 0.8%/min.
LTQ-FT Ultra
For the described analyses, the instrument was equipped with a Michrom ADVANCE electrospray ionization source and coupled to a capillary HPLC (CapLC, Waters). Peptide mixtures are separated on C18 column (Agilent Zorbax 300SB-C18, 250 × 0.3 mm, 5 µm) using a flow rate of 4 µL/min with a binary solvent system consisting of solvent A, 0.1% formic acid, and solvent B, 0.1% formic acid in acetonitrile.
The electrospray ionization source voltage is set to 1.8 kV. The LTQ-FT mass spectrometer is operated in a data-dependent MS/MS acquisition mode. Full scan mass spectral data from 400–1800 m/z with the resolving power set to 100,000 at m/z 400 is obtained by using the ICR cell (AGC target 1 × 106 ions; maximum ion accumulation time, 1000 ms). MS/MS data of the five most abundant doubly or triply charged ions detected in the full scan survey MS experiment is acquired by using the linear ion trap. For precursor ion selection an isolation width of ± 2 Th is used. An AGC target value of 3 × 105 ions and a maximum ion accumulation time of 50 ms are used for MS/MS scans in the ion trap. The normalized collision energy is set to 35%, and three microscans are acquired per spectrum. Previously selected ions are excluded from further sequencing for 60 s.
Searching of MS/MS data against protein sequence databases for the identification of peptide sequences and peptide adducts
There are numerous software tools available to aid the interpretation of fragment ion spectra to obtain peptide identifications. Dependent on the instrument with which the fragment ion mass spectral data have been acquired, software tools will facilitate the transformation of the instrument-specific data files in peaklist files that are suitable for processing using the different bioinformatic software packages available.
The mass spectral data discussed in this contribution were processed using MASCOT (Matrix Sciences, Inc.) (David et al., 1999). Instrumentation-independent search parameters include choice of database (e.g. for the discussed data, SwissProt database limited to human or rodentia) and proteolytic processing (e.g. Trypsin/P was selected as the digesting enzyme allowing for the possibility of one missed cleavage site). The instrument-dependent search parameters include instrument type and the respective mass tolerances. For MALDI-ToF/ToF and ESI qTOF MS/MS data the following settings were used: precursor ion tolerance ± 100 ppm and fragment ion tolerance ± 0.1 Da. For data acquired on the LTQ-FT instrument, the precursor ion mass tolerance was set to 10 ppm and the fragment ion tolerance to 0.5 Da. Mascot allows inclusion of two types of peptide modifications: fixed modification and variable modifications. For the peptide adduct analyses discussed, the following variable modifications are critical: Met oxidation (147.04 Da, monoisotopic residue mass), ARP-acrolein modified Cys, His, and Lys (monoisotopic residue masses 472.16 Da, 506.21 Da, 497.24 Da, respectively) and ARP-HNE modified Cys, His, and Lys (monoisotopic residue masses 572.25 Da, 606.29 Da, and 597.2 Da, respectively).
Applications of the ARP labeling strategy
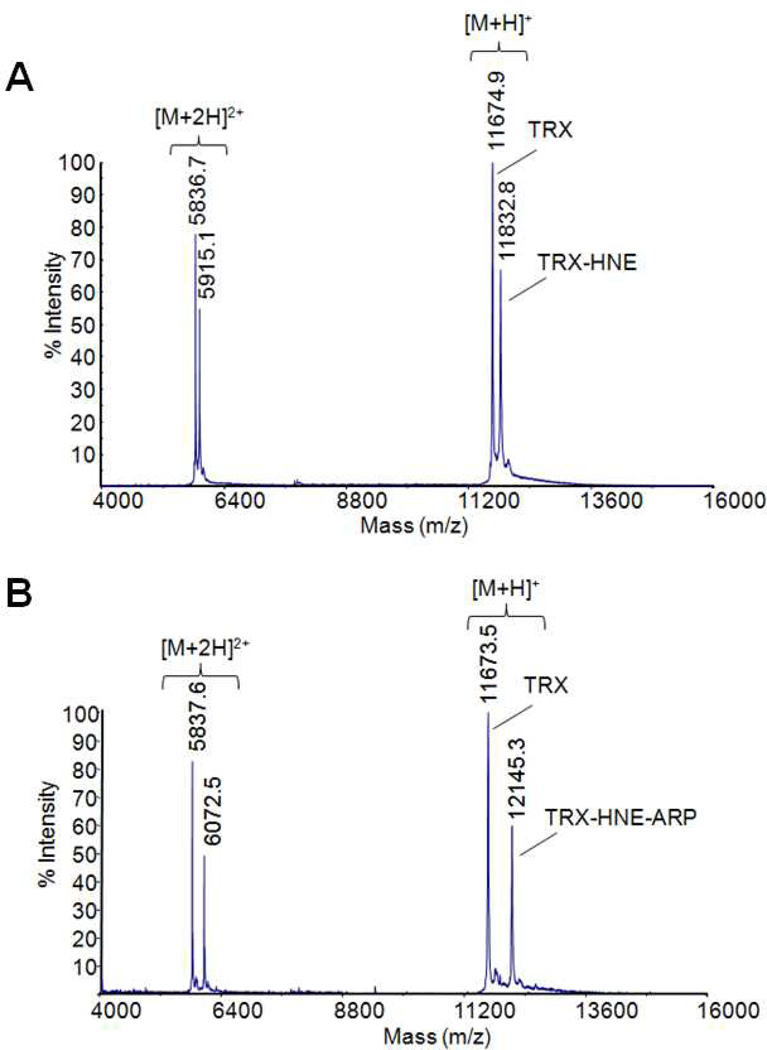
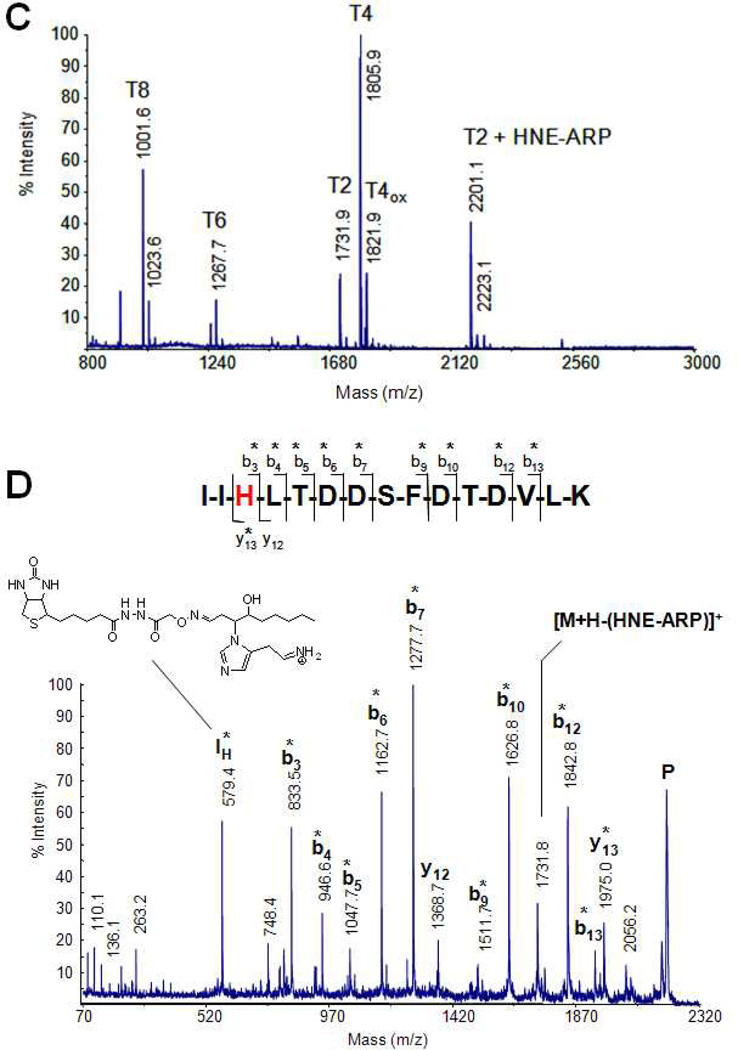
Figure 5 summarizes the typical analytical stages of the ARP method illustrated on the modification of a model protein by HNE and the subsequent characterization of the resulting product (Chavez et al., 2006). In Figure 5A, the MALDI mass spectrum of the model protein after modification with HNE is shown. The mass difference (Δm 156 Da) between the molecular ion of the protein and the protein adduct indicates Michael adduction by HNE. Mass spectral analysis of the reaction mixture after incubation with ARP reveals that the protein-HNE adduct was tagged by the aldehyde/keto-specific biotinylation probe, ARP. Aldoxime formation between the Michael addition product of HNE and ARP results in a mass difference of 313.1 Da between the HNE-modified protein and the ARP-labeled protein adduct (Figure 5B). Figure 5C depicts the MALDI mass fingerprint of the respective tryptic digest; the ARP-HNE-modified peptide T2 (m/z 2201.1) is shifted by 469.2 Da compared to the unmodified peptide T2 (m/z 1731.9). Subsequent tandem mass spectral analysis of the ARP-labeled T2 HNE-peptide adduct reveals modification of the His residue at position 3 of T2 based on the m/z-difference of 606.3 Da between the y12 and y13-ion (monoisotopic residue mass for His-HNE-ARP 606.29 (C27H42N8O6S)) (Figure 1D).
Fig. 5.
Identification and characterization of a Michael-type protein adduct of HNE. A) MALDI mass spectrum of the model protein thioredoxin after modification with HNE. B) MALDI mass spectrum of the HNE-modified thioredoxin after labeling with ARP. The mass difference of ~472 Da between the molecular ion at m/z 11,673.5 and 12145.3 is indicative for the presence of the HNE-ARP oxime moiety (theoretical mass shift: Δm 469 Da). Michael adduct formation results in a mass increase of 156 Da and the subsequent derivatization with ARP under aldoxime formation yields an additional mass shift of 313 Da. C) Mass spectrum of the tryptic digest of mixture of TRX and ARP-labeled TRX-HNE adduct. The ARP-labeled HNE-modified peptide T2 is observed at m/z 2201.1 (i.e. 469 Da higher compared to the unmodified peptide T2 at m/z 1731.9. D) Tandem mass spectrum of the ARP-labeled HNE-modified peptide of thioredoxin encompassing the residue 4 to 18. The b-type fragment ions (b3 to b7, b9, b10, b12 and b13) are shifted by 469 Da to higher m/z values compared to the theoretical m/z values that would be expected for the unmodified peptide. The coherent mass shift of the bn ions indicates that the ARP-HNE moiety is present near the N-terminus of this peptide. The mass difference of 606.3 Da between the fragment ions y12 and y13 localize the APR-HNE moiety to the His residue at position 3 in this peptide. In addition, the intense ARP-HNE-modified His immonium ion at m/z 579.4 supports the assignment of the His residue as the site of modification by HNE. Mass spectral analyses were performed on a MALDI ToF/ToF instrument (4700 Applied Biosystems Proteome Analyzer) using α-cyano-4-hydroxycinnamic acid as matrix. Fragment ions marked with an asterisk (*) retained the ARP-HNE moiety during collision induced fragmentation.
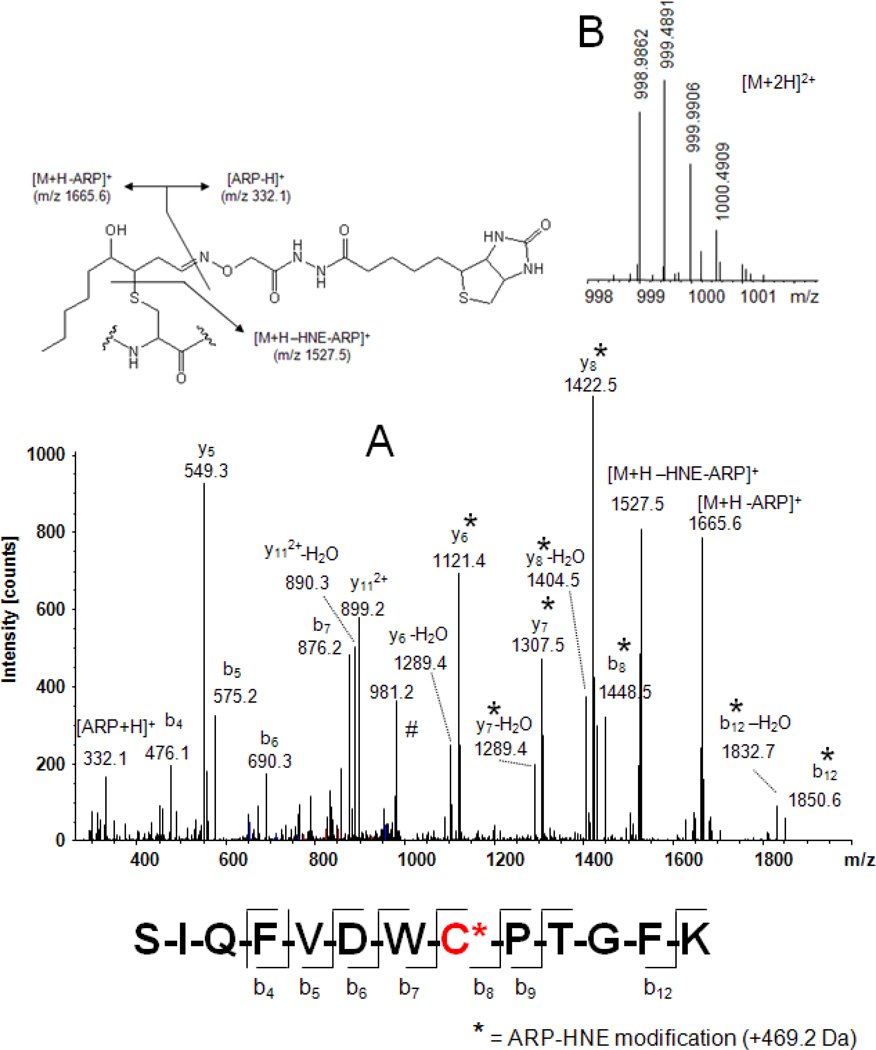
We recently reported the unambiguous identification of the major protein targets of HNE exposure in human monocytic THP-1 cells. Employing the ARP labeling strategy, 18 peptides were identified with ARP-HNE modification to distinct cysteine or histidine residues. The majority of the identified protein targets of HNE were cytoskeletal proteins, proteins involved in glycolysis, metabolic processes and RNA binding and regulation of translation (Chavez et al., 200x). The use of high resolution mass spectrometers that provide high mass accuracy for the characterization of complex peptide mixture becomes increasingly popular. The confidence in the identification of a peptide is increased by accurately measuring the mass of a peptide. This is of particular relevance for the ARP- labeling strategy because identification is based on the correct assignment of a single peptide adduct. Figure 6 features the type of data that are obtained using a hybrid linear quadrupole ion trap/FT ICR (LTQ-FT) mass spectrometer for the identification of HNE-modified peptides. Accurate determination of peptide masses was achieved using the FT-ICR MS, whereas MS/MS data were acquired using the linear ion trap.
Fig. 6.
Mass spectral analysis of the ARP-labeled HNE-modified peptide SIQFVDWC*PTGFK from tubulin α-1B using a LTQ-FT mass spectrometer. A) Tandem mass spectrum of the doubly protonated precursor ion which was used for collision-induced fragmentation in the linear ion trap of an LTQ-FT mass spectrometer. Fragment ions marked with an asterisk (*) retained the ARP-HNE moiety during collision-induced fragmentation and allowed the unambiguous assignment of the Cys residue as site of HNE adduction. B) FT-ICR full scan mass spectrum showing the doubly protonated [M+2H]2+ ion cluster region. Exact mass determination using the ICR cell of the instrument yielded for the monoisotopic ion an m/z value of 998.9862 Th which reflects a mass accuracy of −0.4 ppm (calculated m/z 998.9866). Having both analytical information, sequencing data and exact mass, enables the identification of the peptide as the partial sequence 340–352 of tubulin α-1B chain (TBA1B_Human; Swiss-Prot P68363) with Cys-347 modified by HNE with high confidence.
For instance, Figure 6 depicts the mass spectral data that led to the assignment of Cys-347 as one of the adduction sites of HNE in the tubulin-α1 protein. The tandem mass spectrum provides peptide assignment and localization of the HNE adduction site (Figure 6A). The unambiguous assignment of the adduction site is predominately derived from the m/z-difference between the bn-ions at m/z 876.2 9 (b7) and 1448.5 (b8) ion (monoisotopic residue mass for Cys-HNE-ARP 572.25 (C24H40N6O6S2)). In addition, the observation of a diagnostic ion at m/z 1527.5, which originates from fragmentation of the ARP tag, supports the assignment of the peptide adduct. A possible caveat of the ARP approach is that peptide assignments are usually based on single peptide identification. Therefore, it is desirable to conduct peptide mass measurements with the highest possible accuracy to enhance the confidence in the peptide assignment. The FT-ICR full scan spectrum of the doubly protonated ion, [M+2H]2+, of the ARP-labeled HNE-modified peptide SIQFVDWC*PTGFK from tubulin α-1B is depicted in Figure 6B. The monoisotopic [M+2H]2+ ion at m/z 998.9862 was determined with an accuracy of −0.4 ppm (monoisotopic m/zcalc 998.9866) ascertaining high confidence in the peptide assignment.
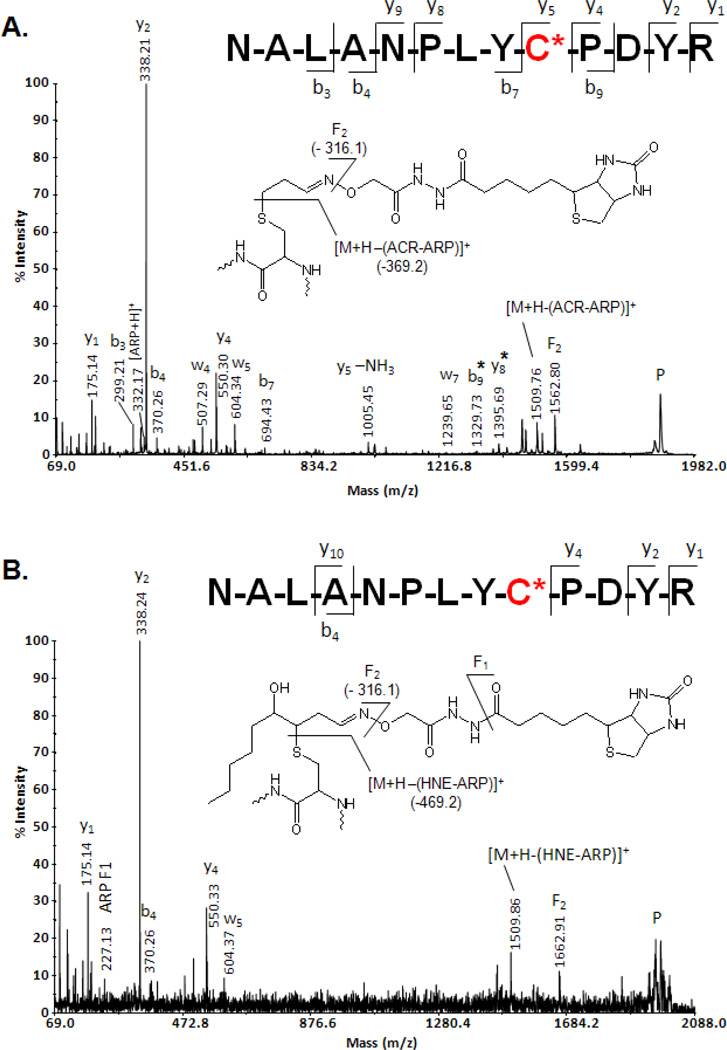
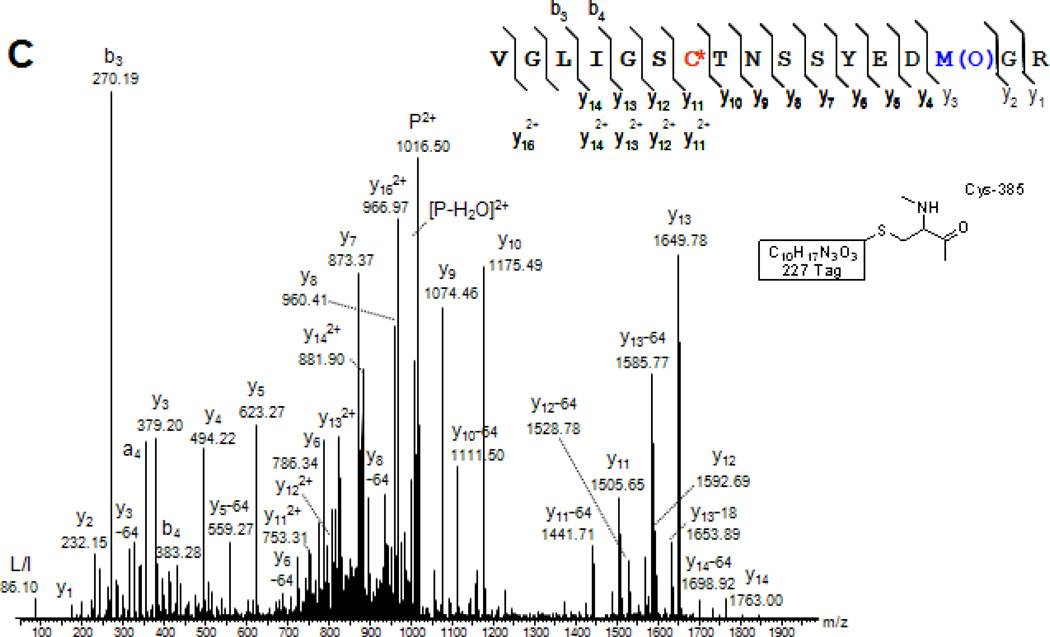
The applicability of the ARP labeling strategy to complex biological mixtures is illustrated on the identification of in vivo Michael-type protein adducts of LPO-derived aldehydes in mitochondria isolated from rat heart. Our studies repeatedly identified as target site of HNE the Cys-166 residue in long-chain-specific acyl-CoA dehydrogenase (ACADL-RAT; Swiss-Prot P15650) (Chavez et al., 2006). Several distinct adduction sites to constituents of the respiratory complexes were identified and characterized by tandem mass spectrometry. For example, the MALDI mass spectra depicted in Figure 7 provided the basis for the identification of a modification ‘hot spot’ of Complex III subunit 2 of the electron transport chain (UQCR2_RAT; Swiss-Prot P32551). Note, in this example, the peptide derived from the core 2 protein of Complex III was found to be susceptible to modification by different α,β–unsaturated aldehydes on the Cys-191 residue. This level of information describing site-specific protein modifications by endogenous lipid peroxidation products is exclusively achievable by tandem mass spectrometry-based strategies.
Fig. 7.
Tandem mass spectral identification and characterization of endogenous peptide adducts of lipid peroxidation products after applying the ARP derivatization and enrichment strategy to determine the protein targets of aldehydic lipid peroxidation products in rat cardiac mitochondria. A) MALDI-MS/MS spectrum of the ARP-labeled acrolein-modified peptide of the core protein 2 of the ubiquinol-cytochrome-c reductase complex (UQCR2_RAT; Swiss-Prot P32551) encompassing the residue 183 to 195 and B) the respective peptide with Cys-191 modified by HNE. Mitochondrial protein samples were treated with 5 mM ARP for 1 hr at room temperature. Subsequent to the removal of unreacted ARP using desalting spin column the protein sample was digested with trypsin. ARP-labeled peptides were enriched using a monomeric avidin column. The enriched peptide samples were subjected to reversed-phase (C18) nanoLC, automatically mixed with MALDI matrix, spotted onto a target plate and subjected to MALDI-MS/MS analyses (using a 4700 Applied Biosystems Proteomics Analyzer). Fragment ions marked with an asterisk (*) retained the ARP-HNE moiety during collision-induced fragmentation.
Darley-Usmar and colleagues demonstrated that the cyclopentenone-containing electrophile 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) modulates cell death pathways and induces apoptosis in liver mitochondria (Landar et al., 2006). Using a biotinylated 15d-PGJ2 derivative they were able to identify several putative mitochondrial targets of 15d-PGJ2, of which two seem to be relevant to triggering the permeability transition pore and leading to apoptosis: the adenine nucleotide translocase (ANT) and ATP-synthase. ANT shuffles ATP from the mitochondrial matrix to the cytosol and functions as one of the modulators of the pore opening process in which ANT thiols are proposed to serve as redox-sensitive sensors (Crompton, 1999; Crompton et al., 1999). We reported recently that ANT is a major target of acrolein adduction and identified the cysteine residue in position 256 as site of modification (Han et al., 2007). It seems plausible that cysteine modification by electrophilic lipids promotes activation of ANT (Dmitriev, 2007). Electrophile stress potentiates opening of the mitochondrial permeability transition pore (PTP) under conditions of elevated Ca2+ levels in the mitochondrial matrix (Brookes et al., 2004). Elevated Ca2+ levels result in upregulation of the oxidative phosphorylation machinery including the ATP synthase to meet the increased demands of ATP during apoptosis (Nicotera et al., 1998). Enhanced respiratory chain activity may lead to increased levels of ROS production and, concomitantly, lipid peroxidation. We identified several sites in subunits of ATP synthase that were modified by acrolein: Cys-294 in ATPase α-chain, Cys-78 in ATP synthase γ-chain, Cys-239 in ATP B-chain, Cys-41 in ATP synthase O subunit (OSCP) and Cys-100 in ATP synthase D chain. It seems possible that modifications of ATPase and ANT by α,β-unsaturated aldehydes may interfere with apoptotic cell death pathways. Further work is needed to determine the mechanisms by which α,β-unsaturated aldehydes and other electrophiles modulate mitochondrial processes and function.
Interpretation of MS/MS spectra of protein adducts and the use of diagnostic marker ions
Mass spectrometric studies of protein and peptide adducts of lipid aldehydes have been conducted using many different instrument types. Mass spectral analyses of proteins modified by diverse LPO-derived aldehydes indicate that Michael adducts of 4-hydroxy-2-alkenals on Cys and His seem to be sufficiently stable to a wide range of experimental conditions and can be readily observed without reductive stabilization or other derivatizations (Rauniyar et al., 2008; Roe et al., 2007; Roede et al., 2008; Shonsey et al., 2008; Williams et al., 2007). In contrast, the Michael adduct of HNE on Lys residue has been observed exclusively after reductive stabilization using sodium borohydride (NaBH4) (Lin et al., 2005). Studies of model proteins modified in vitro by α,β-unsaturated aldehydes indicate the potential of using proteolytic analyses in conjunction with mass spectrometry to identify and characterize crosslinks caused by LPO-derived aldehydes (Liu et al., 2003; Oe et al., 2003a).
Besides facilitating avidin affinity-based enrichment, the derivatization of protein adducts of LPO-derived aldehydes by a aldehdye/keto-specific probe has as additional advantage the introduction of an extra layer of confidence based on the specificity of the hydroxylamine chemistry for aldehyde/keto groups. ARP-derivatized protein and peptides adducts are well amenable to mass spectral measurements and the oxime bond is sufficiently stable for retaining the ARP-aldehyde moiety on the peptide fragment ions allowing the use of automated database search software for aiding in the analysis of tandem mass spectral data (Figures 5 to 8).
Fig. 8.
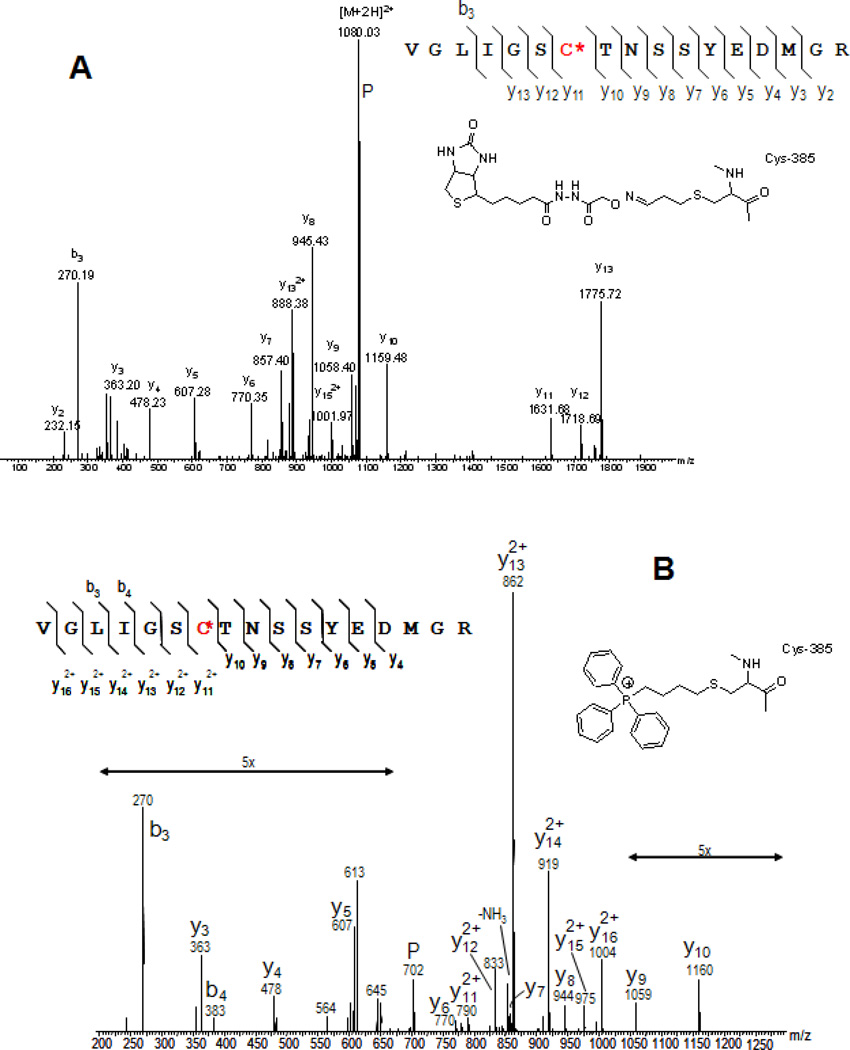
ESI-MS/MS spectral analysis of (A) the ARP-labeled acrolein-modified peptide of aconitase ([M+2H]2+, m/z 1079.47); (B) the respective peptide with Cys-385 alkylated with a sterically demanding triphenylphosphonium group ([M+2H]3+, m/z 702.3) and (C) tagged with the thiol-specific cleavable ICAT probe ([M+2H]2+, m/z 1016.50). Peptide identification and characterization of the respective modification was achieved by nanoLC-ESI-MSMS analysis using a Waters/Micromass Global quadrupole time-of-flight instrument operated in the data-dependent acquisition mode. P, denotes precursor ion.
Collision-induced dissociation of multiply charged ARP-labeled peptide adducts yielded, beside the usual peptide-specific fragment ions of the bn and yn types, frequently non-peptide fragment ions that originated from the ARP tag, including the ions observed at m/z 227.1 (F1) and m/z 332.2 ([ARP+H]+). Additionally, in the ESI-MS/MS spectrum depicted in Figure 6 intense ion peaks at m/z 1665.6 and 1527.5 are visible indicating that [ARP-H]+ is expelled and the resulting ion, [M+H -ARP]+, undergoes loss of dehydrated HNE (−138 Da) resulting in a ion that appears at m/z 1527.5 ([M+H –(HNE-ARP)]+, i.e. charge reduction and a total loss of −462.Da is observed upon CID of the doubly protonated precursor ion (m/z 998.99). In contrast, fragment ions that indicate neutral loss of the ARP-lipid moiety from singly protonated precursor ions are visible in the MALDI MS/MS spectra (Figures 5D and 7). The observation of ions that relate to fragmentation involving the ARP-lipid aldehyde moiety may therefore serve as diagnostic marker ions in the interpretation of tandem mass spectra of peptide adducts of lipid aldehyde.
Noteworthy, Prokai and coworkers explore the high propensity of aldehydic protein adducts to undergo neutral loss fragmentation via retro Michael addition reaction for neutral-loss-dependent MS3 strategies to facilitate large-scale analyses of these protein modifications (Rauniyar et al., 2008).
Conclusion
This chapter summarizes our ongoing efforts to develop and apply chemical labeling approaches in combination with tandem mass spectrometry to determine protein targets of in vivo oxidative stress. Here, an experimental approach is described that enables the unambiguous identification and characterization of proteins modified by lipid peroxidation-derived 2-alkenals. This approach employs a biotinylated hydroxyl amine derivate as aldehyde/keto reactive probe in conjunction with selective enrichment and tandem mass spectrometric analysis. Model studies allow studying the chemistry of LPO-derived aldehydes and properties of the resulting peptide and protein adducts in mass spectrometry-based sequencing studies. Methodological details are given for an exposure study of a cell culture system with HNE that yielded the major protein targets of HNE in human monocytic THP-1 cells. The utility of the ARP-labeling approach is also demonstrated for the identification and characterization of endogenous mitochondrial protein targets of aldehydic lipid peroxidation products. The diverse applications indicate that the described method can possibly be equally well applied to the target analysis of other aldehydic addition products, such FDP-lysine and 4-keto-amide lysine residues.
Which factors direct the modification of proteins by LPO-derived aldehydes under physiological conditions? A few reports have appeared that started the discussion to what extent do protein structural parameters govern the sensitivity of distinct sites towards modification by LPO-derived aldehydes and other toxic electrophiles. A scenario is emerging in which the dielectric constant, surface accessibility and the modulation of the nucleophilicity and basicity of a certain residue by its microenvironment may govern the propensity of a distinct site to modifications by electrophiles (Roe et al., 2007; Sayre et al., 2006). In this context, various alkylating reagents have been employed to determine preferred target sites of modifications (Marley et al., 2005; Shin et al., 2007; Wong and Liebler, 2008). We explored the use of a sterically demanding butyltriphenylphosphonium group as a chemical tool for the identification of mitochondrial protein thiols that exhibit a large degree of surface accessibility (Marley et al., 2005). In this study, Cys-385 of aconitase was readily alkylated by (4-iodo)butyltriphenylphosphonium iodide (IBTP). The same cysteine residue was also found to be susceptible to modification by another alkylation reagent and has been identified to be modified by acrolein in rat cardiac mitochondria (Figure 8). The same cysteine residue was also described as being susceptible to oxidative modification by peroxynitrite (Han et al., 2005). The compilation of reactivity parameters for distinct sites that show high reactivity towards electrophilic agents may provide the foundation for future attempts to predict protein susceptibility to these modifications.
To conclude, the ARP labeling method is a versatile analytical strategy for the identification and characterization of carbonyl-containing protein adducts of lipid peroxidation products. We further anticipate that our approach may also find some applicability in the area of oxidative stress insults by metal-catalyzed oxidations (Maisonneuve et al., 2009; Mirzaei and Regnier, 2005; Mirzaei and Regnier, 2007).
Additional experimental methods (not related to ARP derivatization of aldehydic protein adducts)
Derivatization of protein thiols using iodobutyl triphenylphosphonium (IBTP)
Frozen mitochondria (isolated according to Sue et al. (Suh et al., 2003)) were suspended in 10 mM potassium phosphate buffer (pH 8.2) containing 250 mM sucrose. Surface accessible protein thiols were alkylated with 1 mM IBTP for 30 min at 37 °C. For this purpose, a 33 mM stock solution of IBTP prepared in 50 mM potassium phosphate buffer containing 30% acetonitrile was prepared. The final IBTP concentration in the reaction mixture was approximately 1 mM. After several freeze-thaw cycles to disrupt mitochondrial membranes, the soluble mitochondrial proteins were recovered after centrifugal removal of the insoluble membrane fraction. Tryptic digests were submitted to LC-ESI MS/MS analysis using Micromass/Water qToF instrument.
Labeling of protein thiols using a cleavable ICAT probe
Soluble protein lysate was prepared by shaking isolated rat cardiac mitochondria (ref) in the presence of 1% Triton X-100 (v/v) for 1 hr on ice. ICAT labeling of protein thiols was performed under non-denaturing conditions but otherwise following the procedure outlined in Applied Biosystems’ cICAT labeling kit. Briefly, cICAT labeling was performed for 2 hrs at 37 °C in the dark. The protein sample was incubated overnight at 37 °C with trypsin (E:S ratio 1:40). Proteolytic digests were cleaned up using a strong cation-exchange (SCX) spin column. cICAT-labeled peptides were enriched using ultraLink-immobilized monomeric avidin column. After release of cICAT-lableled peptides using 0.4 % formic acid/30 % acetonitrile, the eluate was lyophilized and the biotin moiety was removed by using cleaving reagent A and B from the Applied Biosystems kit. Prior to LC-ESI qToF MS/MS analysis the lyophilized sample was dissolved in 0.1% aqueous TFA containing 1% acetonitrile.
Acknowledgement
We are grateful to Brian Arbogast and Michael Hare for technical assistance. This work was supported by grants from the NIH (AG025372, HL081721, ES000210) and the Oregon Medical Research Foundation. The Mass Spectrometry Facility of the Environmental Health Sciences Center at Oregon State University is supported in part by NIH/NIEHS grant P30ES000210.
REFERENCES
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol. Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Bieschke J, et al. Small molecule oxidation products trigger disease-associated protein misfolding. Acc. Chem. Res. 2006;39:611–619. doi: 10.1021/ar0500766. [DOI] [PubMed] [Google Scholar]
- Brookes PS, et al. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- Brooks JD, et al. Formation of Highly Reactive Cyclopentenone Isoprostane Compounds (A3/J3-Isoprostanes) in Vivo from Eicosapentaenoic Acid. J. Biol. Chem. 2008;283:12043–12055. doi: 10.1074/jbc.M800122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Sultana R. Redox proteomics: understanding oxidative stress in the progression of age-related neurodegenerative disorders. Expert Rev. Proteomics. 2008;5:157–160. doi: 10.1586/14789450.5.2.157. [DOI] [PubMed] [Google Scholar]
- Carbone DL, et al. Cysteine Modification by Lipid Peroxidation Products Inhibits Protein Disulfide Isomerase. Chem. Res. Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- Carbone DL, et al. 4-Hydroxynonenal regulates 26S proteasomal degradation of alcohol dehydrogenase. Free Radic. Biol. Med. 2004;37:1430–1439. doi: 10.1016/j.freeradbiomed.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Catalá A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Chavez J, et al. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: Protein carbonylation is dimished by ascorbic acid. Chem. Res. Toxicol. doi: 10.1021/tx9002462. 200x. in prep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez J, et al. New role for an old probe: affinity labeling of oxylipid protein conjugates by N'-aminooxymethylcarbonylhydrazino d-biotin. Anal. Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- Chen Y, et al. Formation of reactive cyclopentenone compounds in vivo as products of the isoprostane pathway. J. Biol. Chem. 1999;274:10863–10868. doi: 10.1074/jbc.274.16.10863. [DOI] [PubMed] [Google Scholar]
- Chung W-G, et al. Hop proanthocyanidins induce apoptosis, protein carbonylation, and cytoskeleton disorganization in human colorectal adenocarcinoma cells via reactive oxygen species. Food Chem. Toxicol. 2009;47:827–836. doi: 10.1016/j.fct.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codreanu SG, et al. Global Analysis of Protein Damage by the Lipid Electrophile 4-Hydroxy-2-nonenal. Mol. Cell. Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 1999;341(Pt 2):233–249. [PMC free article] [PubMed] [Google Scholar]
- Crompton M, et al. The mitochondrial permeability transition pore. Biochem. Soc. Symp. 1999;66:167–179. doi: 10.1042/bss0660167. [DOI] [PubMed] [Google Scholar]
- Danni LM, et al. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- David NP, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Dmitriev LF. The involvement of lipid radical cycles and the adenine nucleotide translocator in neurodegenerative diseases. J. Alzheimers Dis. 2007;11:183–190. doi: 10.3233/jad-2007-11206. [DOI] [PubMed] [Google Scholar]
- Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chemico-Biological Interactions. 2003;143–144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- Eliuk SM, et al. Active Site Modifications of the Brain Isoform of Creatine Kinase by 4-Hydroxy-2-nonenal Correlate with Reduced Enzyme Activity: Mapping of Modified Sites by Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry. Chem. Res. Toxicol. 2007;20:1260–1268. doi: 10.1021/tx7000948. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, et al. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Farout L, et al. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch. Biochem. Biophys. 2006;453:135–142. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Furuhata A, et al. Thiolation of Protein-bound Carcinogenic Aldehyde. J. Biol. Chem. 2002;277:27919–27926. doi: 10.1074/jbc.M202794200. [DOI] [PubMed] [Google Scholar]
- Han B. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal. Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- Han D, et al. Sites and Mechanisms of Aconitase Inactivation by Peroxynitrite: Modulation by Citrate and Glutathione. Biochemistry. 2005;44:11986–11996. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- Helga S, et al. Genomic damage in end-stage renal failure: Potential involvement of advanced glycation end products and carbonyl stress. Seminars in nephrology. 2004;24:474–478. doi: 10.1016/j.semnephrol.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Ichihashi K, et al. Endogenous Formation of Protein Adducts with Carcinogenic Aldehydes. J. Biol. Chem. 2001;276:23903–23913. doi: 10.1074/jbc.M101947200. [DOI] [PubMed] [Google Scholar]
- Kim H-YH, et al. An Azido-Biotin Reagent for Use in the Isolation of Protein Adducts of Lipid-derived Electrophiles by Streptavidin Catch and Photorelease. Mol. Cell. Proteomics. 2009;8:2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landar A, et al. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1777–H1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- Lee HP, et al. The essential role of ERK in 4-oxo-2-nonenal-mediated cytotoxicity in SH-SY5Y human neuroblastoma cells. J. Neurochem. 2009;108:1434–1441. doi: 10.1111/j.1471-4159.2009.05883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, et al. 4-Oxo-2-nonenal Is Both More Neurotoxic and More Protein Reactive than 4-Hydroxy-2-nonenal. Chem. Res. Toxicol. 2005;18:1219–1231. doi: 10.1021/tx050080q. [DOI] [PubMed] [Google Scholar]
- Liu Z, et al. Mass Spectroscopic Characterization of Protein Modification by 4-Hydroxy-2-(E)-nonenal and 4-Oxo-2-(E)-nonenal. Chem. Res. Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, et al. Rules governing selective protein carbonylation. PLoS One. 2009;4:e7269. doi: 10.1371/journal.pone.0007269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley K, et al. Mass Tagging Approach for Mitochondrial Thiol Proteins. Journal of Proteome Res. 2005;4:1403–1412. doi: 10.1021/pr050078k. [DOI] [PubMed] [Google Scholar]
- Mirzaei H, Regnier F. Enrichment of carbonylated peptides using Girard P reagent and strong cation exchange chromatography. Anal. Chem. 2005;78:770–778. doi: 10.1021/ac0514220. [DOI] [PubMed] [Google Scholar]
- Mirzaei H, Regnier F. Identification of yeast oxidized proteins: Chromatographic top-down approach for identification of carbonylated, fragmented and cross-linked proteins in yeast. J. Chromatogr. A. 2007;1141:22–31. doi: 10.1016/j.chroma.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Montine TJ, et al. Lipid peroxidation in aging brain and Alzheimer's disease. Free Radic. Biol. Med. 2002;33:620–626. doi: 10.1016/s0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- Nicotera P, et al. Intracellular ATP, a switch in the decision between apoptosis and necrosis. Toxicology letters. 1998;102–103:139–142. doi: 10.1016/s0378-4274(98)00298-7. [DOI] [PubMed] [Google Scholar]
- Oe T, et al. A Novel Lipid Hydroperoxide-derived Cyclic Covalent Modification to Histone H4. J. Biol. Chem. 2003a;278:42098–42105. doi: 10.1074/jbc.M308167200. [DOI] [PubMed] [Google Scholar]
- Oe T, et al. A Novel Lipid Hydroperoxide-Derived Modification to Arginine. Chem. Res. Toxicol. 2003b;16:1598–1605. doi: 10.1021/tx034178l. [DOI] [PubMed] [Google Scholar]
- Poli G, et al. 4-Hydroxynonenal-protein adducts: A reliable biomarker of lipid oxidation in liver diseases. Mol. Aspects Med. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Powell SR, et al. Aggregates of oxidized proteins (lipofuscin) induce apoptosis through proteasome inhibition and dysregulation of proapoptotic proteins. Free Radic. Biol. Med. 2005;38:1093–1101. doi: 10.1016/j.freeradbiomed.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Qin Z, et al. Effect of 4-Hydroxy-2-nonenal Modification on α-Synuclein Aggregation. J. Biol. Chem. 2007;282:5862–5870. doi: 10.1074/jbc.M608126200. [DOI] [PubMed] [Google Scholar]
- Rauniyar N, et al. Characterization of 4-Hydroxy-2-nonenal-Modified Peptides by Liquid Chromatography-Tandem Mass Spectrometry Using Data-Dependent Acquisition: Neutral Loss-Driven MS3 versus Neutral Loss-Driven Electron Capture Dissociation. Anal. Chem. 2008;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed TT, et al. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Research. 2009;1274:66–76. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Roe MR, et al. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal. Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- Roede JR, et al. In vitro and in silico characterization of peroxiredoxin-6 modified by 4-hydroxynonenal and 4-oxononenal. Chem. Res. Toxicol. 2008;21:2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- Sayre LM, et al. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- Shin N-Y, et al. Protein Targets of Reactive Electrophiles in Human Liver Microsomes. Chem. Res. Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonsey EM, et al. Inactivation of human liver bile acid CoA:amino acid N-acyltransferase by the electrophilic lipid, 4-hydroxynonenal. J. Lipid Res. 2008;49:282–294. doi: 10.1194/jlr.M700208-JLR200. [DOI] [PubMed] [Google Scholar]
- Spiteller G. Lipid peroxidation in aging and age-dependent diseases. Experimental Gerontology. 2001;36:1425–1457. doi: 10.1016/s0531-5565(01)00131-0. [DOI] [PubMed] [Google Scholar]
- Spiteller P, et al. Aldehydic lipid peroxidation products derived from linoleic acid. Biochim. Biophys. Acta. 2001;1531:188–208. doi: 10.1016/s1388-1981(01)00100-7. [DOI] [PubMed] [Google Scholar]
- Stevens JF, Maier CS. Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BJ, et al. Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem. Res. Toxicol. 2007;20:1111–1119. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- Suh JH, et al. Two subpopulations of mitochondria in the aging rat heart display heterogenous levels of oxidative stress. Free Radic. Biol. Med. 2003;35:1064–1072. doi: 10.1016/s0891-5849(03)00468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, et al. Proteomics identification of oxidatively modified proteins in brain. Proteomics. 2009:291–301. doi: 10.1007/978-1-60761-157-8_16. [DOI] [PubMed] [Google Scholar]
- Uchida K. Role of reactive aldehyde in cardiovascular diseases. Free Radic. Biol. Med. 2000;28:1685–1696. doi: 10.1016/s0891-5849(00)00226-4. [DOI] [PubMed] [Google Scholar]
- Vila A, et al. Identification of Protein Targets of 4-Hydroxynonenal Using Click Chemistry for ex Vivo Biotinylation of Azido and Alkynyl Derivatives. Chem. Res. Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Marnett LJ. Endogenous Reactive Intermediates as Modulators of Cell Signaling and Cell Death. Chem. Res. Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- Williams MV, et al. Covalent Adducts Arising from the Decomposition Products of Lipid Hydroperoxides in the Presence of Cytochrome c. Chem. Res. Toxicol. 2007;20:767–775. doi: 10.1021/tx600289r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Liebler DC. Mitochondrial Protein Targets of Thiol-Reactive Electrophiles. Chem. Res. Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-H, et al. Model Studies on Protein Side Chain Modification by 4-Oxo-2-nonenal. Chem. Res. Toxicol. 2003;16:512–523. doi: 10.1021/tx020105a. [DOI] [PubMed] [Google Scholar]
- Zhu X, Sayre LM. Long-Lived 4-Oxo-2-enal-Derived Apparent Lysine Michael Adducts Are Actually the Isomeric 4-Ketoamides. Chem. Res. Toxicol. 2007;20:165–170. doi: 10.1021/tx600295j. [DOI] [PubMed] [Google Scholar]