Abstract
Many natural underwater adhesives harness hierarchically assembled amyloid nanostructures to achieve strong and robust interfacial adhesion under dynamic and turbulent environments. Despite recent advances, our understanding of the molecular design, self-assembly, and structure-function relationship of those natural amyloid fibers remains limited. Thus, designing biomimetic amyloid-based adhesives remains challenging. Here, we report strong and multi-functional underwater adhesives obtained from fusing mussel foot proteins (Mfps) of Mytilus galloprovincialis with CsgA proteins, the major subunit of Escherichia coli amyloid curli fibers. These hybrid molecular materials hierarchically self-assemble into higher-order structures, in which, according to molecular dynamics simulations, disordered adhesive Mfp domains are exposed on the exterior of amyloid cores formed by CsgA. Our fibers have an underwater adhesion energy approaching 20.9 mJ/m2, which is 1.5 times greater than the maximum of bio-inspired and bio-derived protein-based underwater adhesives reported thus far. Moreover, they outperform Mfps or curli fibers taken on their own at all pHs and exhibit better tolerance to auto-oxidation than Mfps at pH ≥7.0. This work establishes a platform for engineering multi-component self-assembling materials inspired by nature.
Strong underwater adhesives are needed for technological and biomedical applications in water or high-moisture settings1, 2. An emerging strategy for developing such advanced molecular materials is based on mimicking and improving upon naturally occurring underwater adhesives from marine organisms2–4. The versatility of 3,4-dihydroxyphenylalanine (Dopa) for cross-linking and coupling in natural underwater interfacial adhesion phenomena has promoted a wide range of biomimetic research focused on Dopa-containing or Dopa-analog-containing peptides5, 6, hydrogels7, polymer constructs3, 8, and recombinant Mfp variants9. In contrast, the rational design of biomimetic underwater adhesives through molecular self-assembly has lagged behind, even though the importance of hierarchical assembly of protein complexes into higher-order structures is increasingly recognized in natural underwater adhesive systems10, 11.
Several marine organisms, including barnacles, algae, and marine flatworms, exhibit remarkable moisture-resistant adhesion to a variety of substrata by utilizing functional amyloid nanostructures12,13. Amyloids are characterized by β-strands that are oriented perpendicularly to the fibril axis and connected through a dense hydrogen-bonding network, which leads to supramolecular β-sheets that usually extend continuously over thousands of molecular units14–16. Such fibrillar structures have intrinsic advantages for interfacial underwater adhesion. These advantages include tolerance to environmental deterioration, self-healing arising from self-polymerization, and large fiber surface areas10, 16–18, which appear to enhance adhesion by increasing contact area in the adhesive plaques of barnacles13. In addition, potential mechanical benefits of amyloid nanostructures include the cohesive strength associated with the generic amyloid intermolecular β-sheet structure and adhesive strength related to adhesive residues external to the amyloid core12, 16. Amyloid structures can therefore constitute the basis for a promising new generation of bio-inspired adhesives for a wide range of applications3, 12. Despite advances in both amyloid self-assembly14–16 and amyloid-enabled nanotechnology16, 19, 20, the rational design of biomimetic amyloid-based underwater adhesives remains challenging and has not been demonstrated experimentally, in part due to limited understanding of the underlying biological design principles.
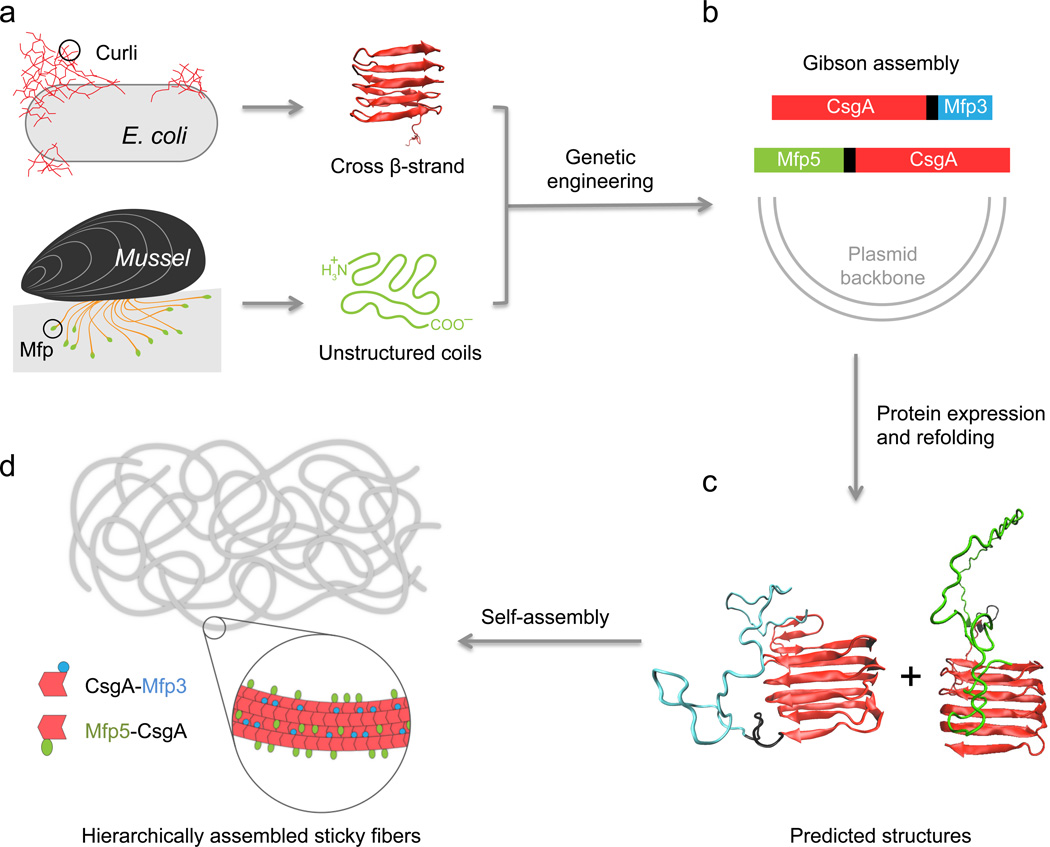
Here, we rationally designed a new generation of bio-inspired adhesives that combine two independent natural adhesion systems, Dopa-based adhesives and amyloid-based adhesives, using synthetic-biology techniques (Fig. 1). To achieve strong interfacial underwater adhesion, we selected Mfp3 and Mfp5 (representatives of Dopa-based mussel adhesives originating from M. galloprovincialis2) and CsgA (an amyloidogenic protein which constitutes the major subunit of adhesive curli fibers in E. coli18) (Fig. 1a and Supplementary Fig. 1) from an inventory of adhesive biomolecules and genes compiled over decades of molecular biology and functional genomics research. Two genetic fusion constructs, CsgA-Mfp3 and Mfp5-CsgA, were constructed using isothermal one-step Gibson DNA assembly (Fig. 1b and Supplementary Fig. 2–5). Because CsgA is amyloidogenic, we reasoned that both CsgA-Mfp3 and Mfp5-CsgA would self-assemble into fibrous bundles or films with adhesive properties19 by displaying the mussel adhesion domains on the surface of amyloid scaffolds. We further hypothesized that the co-assembly of the two fusion proteins together would lead to hierarchically assembled copolymer structures that could integrate synergistic features from the two different types of adhesive modules and potentially recapitulate the intermolecular interactions between Mfp3 and Mfp5 found in natural adhesion systems2 (Fig. 1c–d).
Figure 1. Combinatorial and modular genetic strategy for engineering self-assembling underwater adhesives.
a, Schematic illustration of two independent natural adhesive proteins: curli from E. coli and mussel foot proteins (Mfps) from mussels. Curli are adhesive amyloid fibers that are composed of the major functional subunit, CsgA. CsgA contains five stacked strand-loop-strand motifs mediated by conserved residues (Supplementary Fig.1) and can self-assemble into nanofibers via a rate-limiting nucleation step followed by fibril extension18. Mfp3 and Mfp5, major mussel adhesive foot proteins, have unstructured coil structures in solution32, 49 and are critical to the underwater interfacial adhesion of mussels2. b, Modular design of artificial adhesive materials is enabled by rationally fusing genes encoding the two natural adhesive elements shown in a). Two gene constructs (CsgA-Mfp3 and Mfp5-CsgA) were independently created using one-pot isothermal Gibson assembly and tagged with poly-histidine residues to enable purification. c, Schematic illustration of the predicted cross-β-strand structures for CsgA-Mfp3 and Mfp5-CsgA in solution. In both predicted structures, CsgA forms the amyloid core, with Mfp3 and Mfp5 extending from CsgA’s C-terminal or N-terminal, respectively, as unstructured coil structures. d, Because of the amyloidogenic domains, CsgA-Mfp3 and Mfp5-CsgA monomers can self-assemble into large bundles of fibrils or hierarchical networks of filaments. CsgA domains are key to fiber self-assembly by enabling fibril extension through self-polymerization and β-strand lamination by lateral stacking, with adhesive domains Mfp3 and Mfp5 exposed on the fibril surfaces. In vitro copolymerization of the CsgA-Mfp3 and Mfp5-CsgA monomers can lead to hierarchically co-assembled structures with two different adhesive domains displayed on amyloid scaffolds, potentially recapitulating intermolecular interactions between Mfp3 and Mfp5 molecules in natural mussel adhesion systems2. Enhanced underwater adhesion is expected to arise from the synergy between the high fiber surface area of curli and adhesive residues from the Mfp domains.
Molecular-Dynamics Simulations and Molecular Characterization
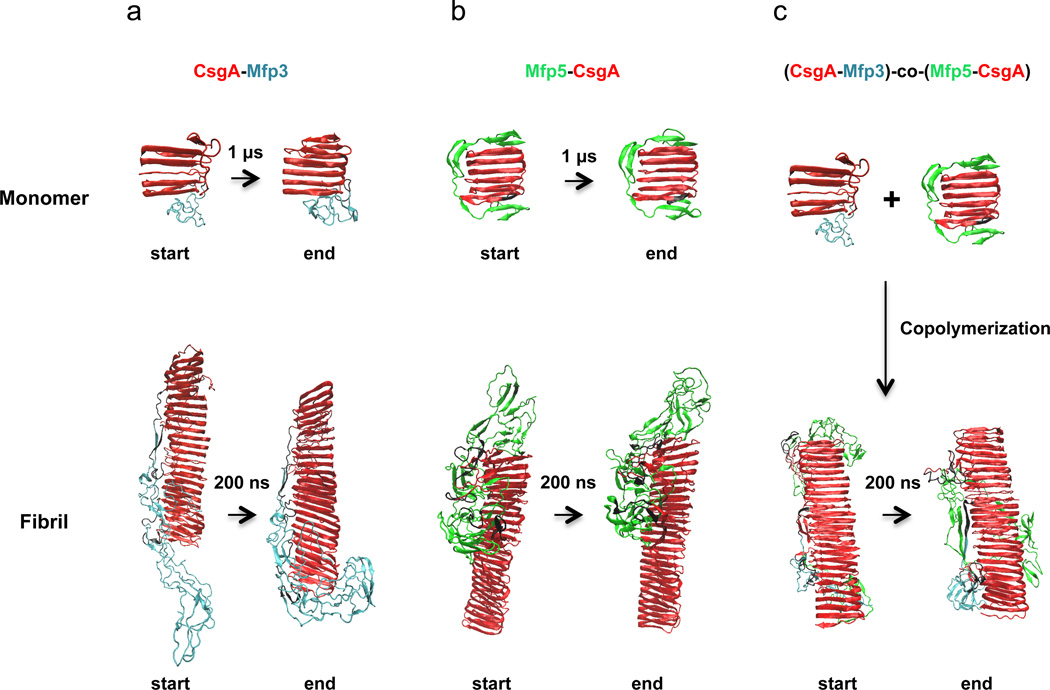
To investigate whether the presence of a disordered Mfp domain would affect the overall structure of CsgA amyloid cores, we built molecular-dynamics models representing both monomeric and fibrillar states of the CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer constructs (Fig. 2 and Supplementary Fig. 6). Simulations of the monomeric proteins (1 µs) and the fibrillar states (200 ns) indicated that the core amyloid structure does not significantly diverge from that of a prototypical amyloid structure when the disordered domains are present (Fig. 2). These results suggest that CsgA-Mfp3 alone, Mfp5-CsgA alone and copolymers of the two fusion proteins should form stable amyloid structures dominated by the CsgA domains, with the highly disordered Mfp domains displayed external to the amyloid core in all cases (Fig. 2a–c) and potentially interacting with each other in the copolymer construct (Fig. 2c).
Figure 2. Comparison of monomer, individual fibril and co-assembled fibril structures before and after molecular-dynamics simulations for modified CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer constructs.
a, The start and end simulated structures for the CsgA-Mfp3 monomer (top) and fibril (bottom). b, The start and end simulated structures for the Mfp5-CsgA monomer (top) and fibril (bottom). c, The start and end simulated structures for (CsgA-Mfp3)-co-(Mfp5-CsgA) fibril (bottom), which are assembled via copolymerization of CsgA-Mfp3 and Mfp5-CsgA (top). For both monomeric and fibrillar states of the constructs, the start and end structures display similar structures: the CsgA domains always dominate the well-ordered amyloid cores, while disordered Mfp5 or Mfp3 domains are external to the amyloid cores. The simulation times for the monomeric and the fibrillar structures were 1 µs and 200 ns, respectively.
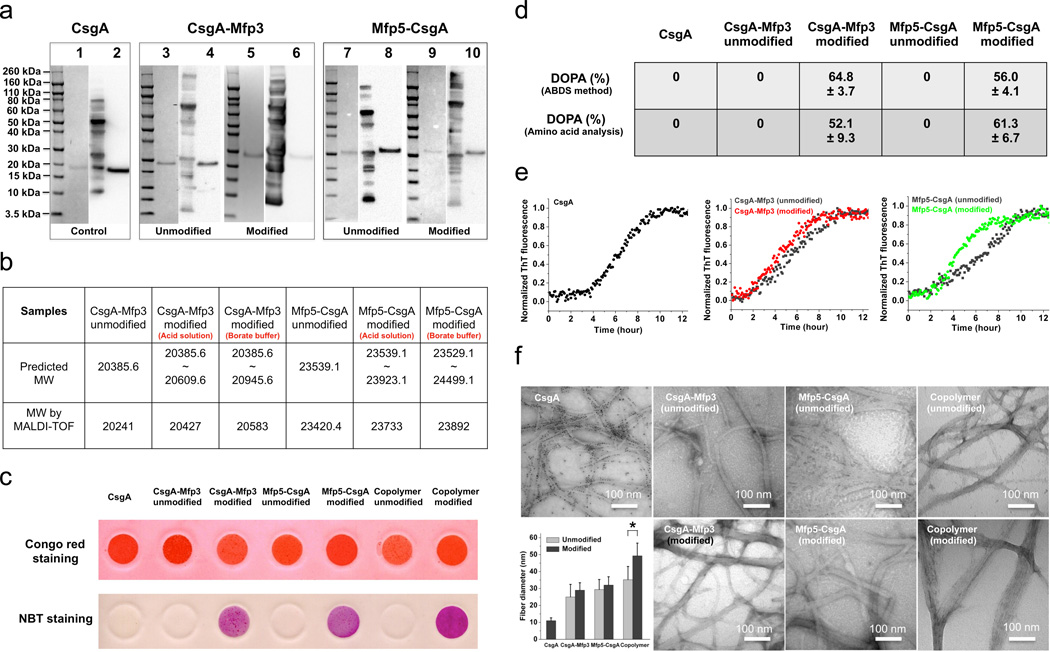
We expressed the adhesive proteins in E. coli, purified them, and exposed them to the enzyme tyrosinase to convert tyrosine residues to Dopa (Supplementary Fig. 7). Hereafter, “unmodified” and “modified” adhesive proteins refer to the proteins before and after tyrosinase conversion. CsgA-Mfp3 (unmodified and modified) and Mfp5-CsgA (unmodified and modified) migrated as single bands at ~28.5 kDa and ~32 kDa, respectively, under SDS-polyacrylamide gel electrophoresis (SDS-PAGE), in contrast with the single band at ~17 kDa for CsgA (Fig. 3a). No clear differences were detected between the band positions of the unmodified and modified versions of the same protein based on SDS-PAGE and Western blotting (Fig. 3a). However, more accurate molecular weight (MW) assessment by MALDI-TOF (matrix-assisted-laser-desorption-ionization-time-of-flight) showed higher MWs in the modified adhesive proteins (compared with their unmodified counterparts), which we attribute to the conversion of tyrosine residues into Dopa with tyrosinase modification (Fig. 3b and Supplementary Fig. 8). Based on MALDI-TOF spectra, we found that adhesive proteins collected from borate buffer had higher MWs than those collected from acid solutions, likely due to the formation of diol-borate bonds between Dopa and borate21, thus confirming successful modification of the adhesive proteins (Supplementary Fig. 8). In addition, Dopa residues were detected in mature hybrid amyloid fibers with the nitroblue tetrazolium (NBT) staining assay, in which only the modified samples turned purple due to redox-cycling of Dopa residues22 (Fig. 3c). Further quantitative analysis by acid-borate difference spectrum (ABDS) analysis21 revealed conversion percentages from tyrosine to Dopa of 64.8% and 56.0% for modified CsgA-Mfp3 and Mfp5-CsgA, respectively (Fig. 3d and Supplementary Fig. 10), in agreement with results obtained from amino acid analysis (Fig. 3d and Supplementary Fig. 12–13).
Figure 3. Purification, in vitro self-assembly and characterization of CsgA, CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) fibers.
a, Coomassie-stained SDS/PAGE gels and Western blots with anti-His antibodies confirm purification of the expressed proteins by cobalt-resin columns. Lanes 1, 3, 5, 7 and 9 represent data from SDS/PAGE, while lanes 2, 4, 6, 8 and 10 refer to corresponding data from Western blots. b, MALDI-TOF confirms the molecular weights of CsgA-Mfp3 (unmodified and modified) and Mfp5-CsgA (unmodified and modified), which are consistent with the predicted molecular weights. c, The Congo red (CR) assay confirms the amyloidogenic features of all fibers, while the Nitro Blue Tetrazolium (NBT) assay detects Dopa residues in modified fibers only. d, Quantitative analysis of Dopa residues in modified samples by acid-borate difference spectrum (ABDS) analysis and amino acid analysis (AAA) (N = 3). e, Thioflavin T (ThT) assay reveals the kinetics of amyloid formation for CsgA, CsgA-Mfp3 (unmodified and modified) and Mfp5-CsgA (unmodified and modified), respectively. f, TEM images of purified CsgA, CsgA-Mfp3 (unmodified and modified), Mfp5-CsgA (unmodified and modified) and (CsgA-Mfp3)-co-(Mfp5-CsgA) (unmodified and modified) solutions after three-day incubations at 4°C demonstrate the formation of self-assembled fibers in all cases. CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) fibers all had larger diameters than CsgA fibers based on measurements from TEM images. Significant differences were detected between unmodified and modified (CsgA-Mfp3)-co-(Mfp5-CsgA) fibers, while no significant differences were found among unmodified and modified fibers of the other two types (P < 0.05). The error bars represent standard deviations (SD) of the fiber diameters (N = 50). The copolymer refers to fiber structures formed via copolymerization of CsgA-Mfp3 and Mfp5-CsgA at a molar ratio 1:1 (15 µM:15 µM) in solution.
Our hybrid adhesive proteins formed hierarchically self-assembled structures (Fig. 1d). Immediately after elution from cobalt resin columns, solutions containing CsgA-Mfp3 (unmodified or modified) or Mfp5-CsgA (unmodified or modified) were clear with no evidence of aggregation. However, after about two hours of incubation at ambient conditions, the solutions became opaque and noticeably viscous. Transitions of soluble proteins to insoluble amyloid aggregates can be monitored using Thioflavin T (ThT), an amyloid-specific dye commonly used to assay amyloid formation23. The ThT fluorescence of all samples followed a sigmoidal curve with distinguishable lag, growth, and stationary phases (Fig. 3e). However, the polymerization lag phases for CsgA-Mfp3 (unmodified or modified) and Mfp5-CsgA (unmodified or modified) were typically shorter than that for CsgA, suggesting that the fusion of Mfp domains to CsgA accelerates amyloid formation. This observation is consistent with the fly-casting mechanism, which postulates that a relatively unstructured protein can have a greater “capture radius” and enhance the rate of intermolecular association24. In addition, partial conversion of tyrosine into Dopa in modified proteins shortened the time required to reach stationary phase, implying a higher fiber formation rate in modified proteins (Fig. 3e).
The formation of long fibers was confirmed for all proteins under Transmission Electron Microscopy (TEM), with no apparent differences in morphology and fiber length (Fig. 3f). However, the mean diameters of the three types of functionalized fibers in both unmodified and modified forms were 3–5-fold larger than that of CsgA (Fig. 3f). In particular, the mean diameter of modified (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer fibers reached as high as ~50 nm, which was noticeably larger than fibers composed of the unmodified copolymer and both unmodified and modified samples of CsgA-Mfp3 and Mfp5-CsgA. Note that all instances of the copolymer described hereafter were assembled from 1:1 molar ratios of CsgA-Mfp3:Mfp5-CsgA, unless otherwise noted. We believe that this increased diameter arises from additional intermolecular associations between the Mfp3 and Mfp5 domains in the copolymer construct. Circular dichroism studies (Supplementary Fig. 14) indicated that the fibers were generally rich in beta-sheet secondary structure in solution, in agreement with molecular-dynamics modeling (Fig. 2). Interestingly, all of the unmodified or modified proteins, either upon incubation of freshly made soluble proteins over existing nanofiber seeds or upon incubation at high solution concentrations, assembled into larger fiber bundles and even thick hierarchical films composed of fibrils (Supplementary Fig. 15–16).
Intrinsic Fluorescence of Adhesive Fibers
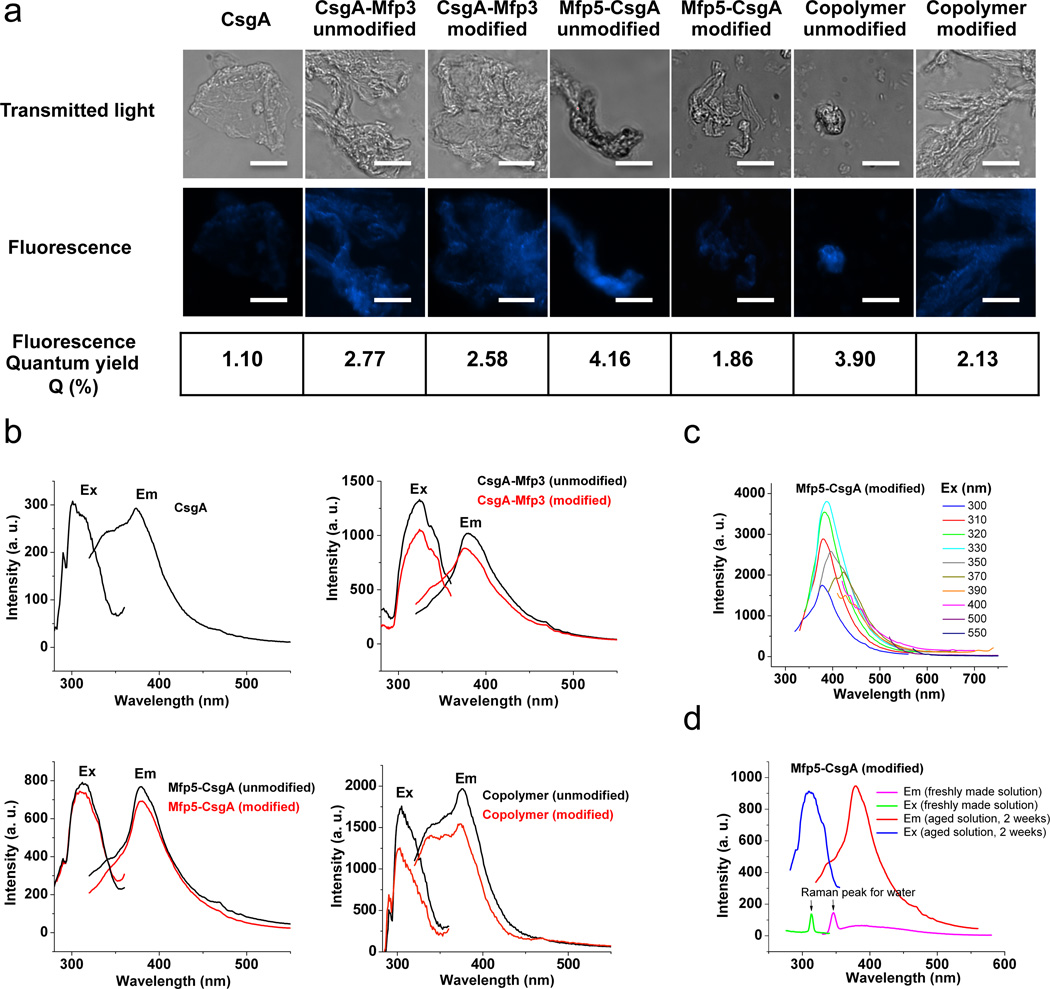
One remarkable feature of the adhesive fibers is that they all exhibited intrinsic fluorescence signatures in the visible spectrum range (Fig. 4a). Adhesive biomaterials that display intrinsic fluorescence may be useful for biological and imaging applications, similar to photoluminescent materials25. The unmodified adhesive fibers displayed higher fluorescence intensities than their modified counterparts when measured in bulk fiber bundles (Fig. 4a). These observations were in agreement with fluorescence quantum yields (Q (%)) determined by the Williams comparative method26 (Fig. 4a and Supplementary Fig. 17). Among the unmodified fibers, Mfp5-CsgA had the highest Q value (4.2), followed by 3.9 for the (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer and 2.8 for CsgA-Mfp3. However, for the modified fibers, the Q values followed an opposite trend, with 1.9, 2.1 and 2.6 for Mfp5-CsgA, copolymer and CsgA-Mfp3, respectively. Both unmodified and modified hybrid adhesive fibers had ~2–4-fold higher Q values than CsgA control fibers. Spectral analysis revealed that the excitation maxima for Mfp5-CsgA (unmodified or modified), CsgA-Mfp3 (unmodified or modified), copolymer (unmodified or modified) and CsgA fibers was near 318, 312, 315 and 306 nm, respectively, while the emission maxima was close to 378 nm for all constructs (Fig. 4b). Interestingly, when excited at different wavelengths, the adhesive fibers emitted fluorescence that could be detected with a wide range of filters with fluorescence microscopy, ranging from blue to red (Supplementary Fig. 18), in agreement with the spectral results (Fig. 4c). In addition, fluorescence was detected in aged solutions, but not in freshly made solutions, implying that fiber formation is a precondition for fluorescence (Fig. 4d).
Figure 4. Intrinsic fluorescence of CsgA, CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer fibers.
a, Comparison of intrinsic blue fluorescence of CsgA with both unmodified and modified samples of CsgA-Mfp3, Mfp5-CsgA and copolymer fibers, detected with fluorescence microscopy with an EBFP fluorescence filter (excitation wavelength 385 nm and emission wavelength 448 nm). Corresponding fluorescence quantum yields (ratios of photons absorbed to photons emitted) were determined following the standard Williams comparative method (Supplementary Fig. 17). Scale bar represents 20 µm. b, Excitation and emission spectra of CsgA, CsgA-Mfp3 (unmodified and modified), Mfp5-CsgA (unmodified and modified) and copolymer (unmodified and modified) fibers. Excitation spectra were measured with the emission wavelength fixed at 375 nm, while emission spectra were measured with the excitation wavelength fixed at 300 nm. c, Emission spectra of modified Mfp5-CsgA fibers measured at different excitation (exc) wavelengths. d, Excitation and emission spectra for freshly made Mfp5-CsgA solutions as well as Mfp5-CsgA solutions aged for 2 weeks. No fluorescence features were detected with freshly made Mfp5-CsgA. The two peaks in the curves for freshly made Mfp5-CsgA could be assigned to Raman peaks for water50. In contrast, significant fluorescence signals were detected in solution after Mfp5-CsgA solutions were aged for two weeks, during which fibers formed. The fluorescence signal of aged solutions almost covered the Raman peaks assigned to water. The copolymer refers to fiber structures that formed via copolymerization of CsgA-Mfp3 and Mfp5-CsgA at a molar ratio 1:1 (15 µM:15 µM) in solution.
Collectively, we suggest that electron delocalization via hydrogen bonds within ordered β-sheet structures27 and the high percentage of aromatic side-chains may both contribute to the intrinsic fluorescence signatures of our hybrid adhesive fibers. The higher fluorescence intensity of unmodified Mfp5-CsgA may arise from its higher fluorescent aromatic residue content (12.5%) compared with CsgA-Mfp3 (10.8%) and CsgA (5.8%), respectively. The decrease in fluorescence intensity within modified fibers versus their unmodified counterparts might be associated with partial conversation of tyrosine residues to Dopa, as the fluorescence quantum yield of Dopa residue (0.09)28 is lower than that of tyrosine (0.14)29 in acid-neutral solutions and is further substantially reduced in conjugate base forms (e.g., with deprotonation of the phenolic hydroxyl group)28. Decreased fluorescence in modified fibers might also be related to fluorescence quenching due to auto-oxidation or potential cross-linking30.
Underwater Adhesion Performance of Adhesive Fibers
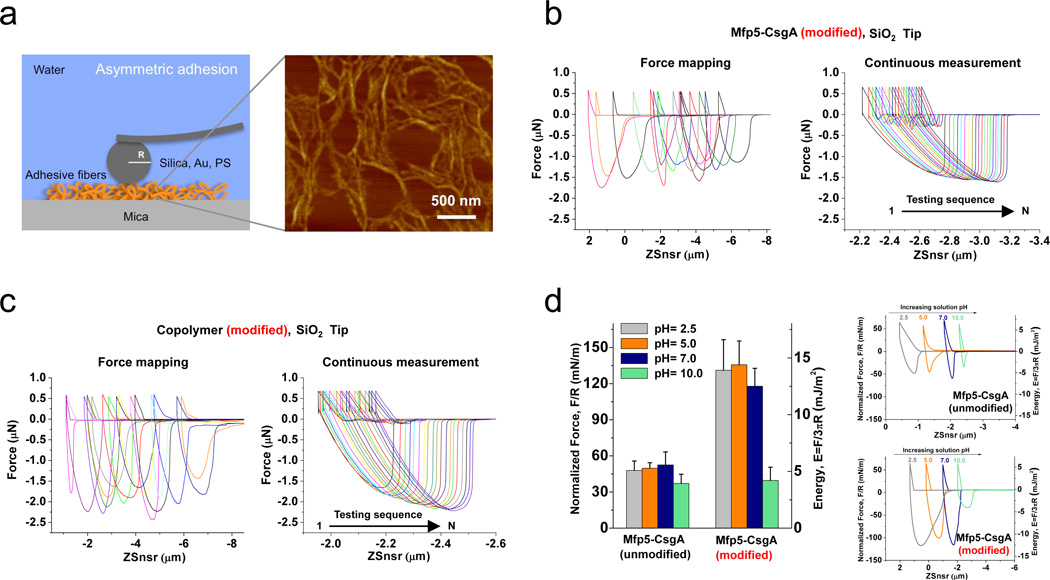
To assess the underwater adhesion performance of our adhesive fibers, we used the atomic force microscopy (AFM) colloidal probe technique31 (Fig. 5a). This technique essentially measures the asymmetric adhesion of fibers, which pre-bind firmly to mica and bind to the AFM tip surface temporarily during measurement. AFM tips used in this study included silica, gold and polystyrene (PS) surfaces, which were chosen as representative inorganic, metal and polymeric surfaces. Proteins were deposited and adsorbed on clean mica surfaces under buffered aqueous conditions with a wide range of pH values. Fibrous structures were found covering the mica surfaces after one hour of adsorption (Fig. 5a and Supplementary Fig. 19). To comprehensively understand the adhesive behavior of our fibers, we performed both force mapping mode and continuous measurement mode studies, which were consistent with each other (Fig. 5b and 5c). Thus, we concluded that our fibers exhibited repeatable adhesion properties (rather than just a single adhesion event) and that the adhesion characteristics were indeed dependent on the specific adhesive fibers studied (Fig. 5b, 5c and 6).
Figure 5. Adhesion force measurements and adhesion stability of hybrid adhesive fibers determined by the atomic force microscopy (AFM) colloidal probe technique.
a, Schematic illustration of a spherulitic particle AFM probe (R = 10 µm for silica and gold tips, R = 12.5 µm for polystyrene tips) used to measure asymmetric adhesion of nanofibers that were deposited on smooth mica surfaces in the presence of buffered aqueous solutions. The graph on the right is a representative AFM image showing modified Mfp5-CsgA fibers one hour after deposition on a mica surface. b, Representative adhesion force curves collected through force mapping mode and continuous measurement mode for modified Mfp5-CsgA samples measured with silica tips. The force mapping mode enables statistical analyses of fiber adhesion based on the random measurement of spots containing nanofibers on the mica surfaces with a scanning range of 20–100 µm. The continuous measurement mode includes 20 continuous measurements on the same specific fiber spot, thus enabling assessment of the cyclic behaviour of contacts between nanofibers and probes. c, Representative adhesion force curves collected through force mapping mode and continuous measurement mode for modified (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer (1:1 ratio) measured with silica tips. d, Adhesion stability of Mfp5-CsgA (unmodified and modified) fibers measured with silica tips in the presence of buffered aqueous solutions of varied pH values (left). (N = 64) The graphs on the right show representative adhesion curves for both unmodified and modified samples of Mfp5-CsgA. Note: the adhesion force curves are plotted as force-displacement curves. The x axis, Zsnsr (Z sensor), stands for the displacement between the sample surface and the resting position of the cantilever (rather than the actual distance between the sample surface and the AFM tip).
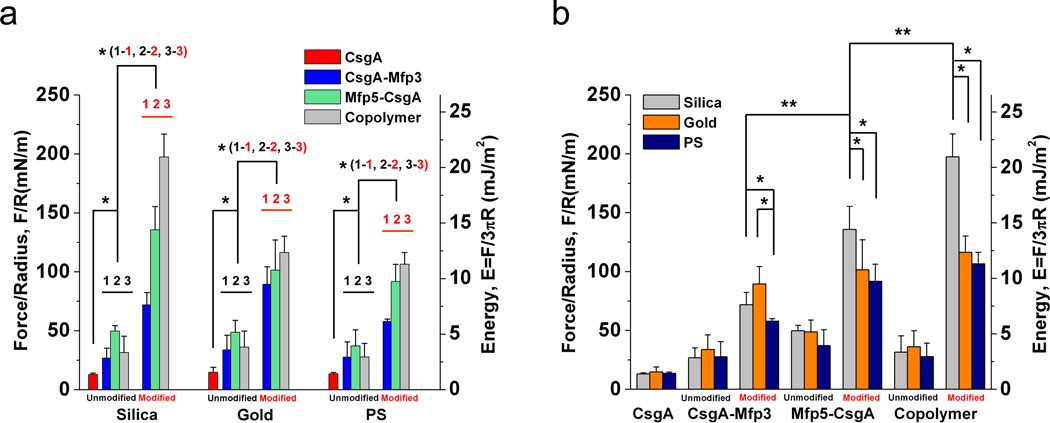
Figure 6. Comparison of the adhesion performances of different functionalized adhesive fibers with different AFM tips.
a, Comparison of adhesion forces (normalized force (Force/Radius) and adhesion energies (Ead = F/3πR)) for CsgA (control) with unmodified and modified versions of functionalized adhesive fibers measured with the use of silica, gold and PS tips, respectively. b, Effects of AFM tips with varied surface energies on the adhesion properties of all adhesive fibers, including CsgA and unmodified and modified versions of CsgA-Mfp3, Mfp5-CsgA and (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer (1:1 ratio). * P < 0.05, ** P < 0.01. The error bars in the figure represent standard deviations (SD). The number of measurements for each statistical analysis was 64 (4 groups × 16 random spots/group). All data collected in this figure were measured under force mapping mode in phosphate buffer at pH=5.0. Figure 6a and 6b contain the same data plotted in two different formats to enable convenient comparisons. In a), the asterisk (*) for CsgA versus unmodified fibers indicates three independent tests between CsgA and the three different fibers, while 1-1, 2-2 and 3-3 refer to comparisons between unmodified and modified samples for the same type of fiber.
The stability of adhesion was assessed on unmodified and modified Mfp5-CsgA fibers under aqueous conditions with different pH values (pH = 2.5, 5.0, 7.0 and 10.0) (Fig. 5d). Both unmodified and modified Mfp5-CsgA fibers maintained adhesion under acidic and neutral conditions (pH 2.5–7.0). Surprisingly, both modified and unmodified Mfp5-CsgA fibers still exhibited considerable levels of adhesion even under basic conditions (pH = 10.0). Similar trends were observed for the other fiber types (data not shown). These data contrast with previous findings that reported soluble and non-fibrous Mytilus edulis foot protein-5 (Mefp5) had a 3-fold lower adhesion energy32 at pH = 5.5 versus pH = 2.6 as well as studies showing that adhesion by Mfp3 and Mfp5 is almost completely abolished when exposed to slightly basic conditions (pH ≥ 7.5)32, 33. These results suggest that our adhesive fibers generally exhibit better tolerance to auto-oxidation compared with previous reports on Mfps alone, possibly arising from the protective effects of hydrophobic aromatic residues and the general hydrophobic features of amyloid fibers34, 35. In particular, partial conversion (~50%–70%) of tyrosine to Dopa in our adhesive proteins may resemble the so-called Mfp3 “slow” variant, which is comprised of both Dopa and more hydrophobic tyrosine residues and was recently reported to preserve significant adhesion even at pH 7.0 due to hydrophobicity compensation for Dopa oxidation33.
We further compared the adhesive behaviours of different functionalized adhesive fibers (Fig. 6). For the same protein, modified fibers always displayed higher adhesion (at least a 2–3-fold increase) compared with unmodified fibers (Fig. 6a), independent of the AFM tips (silica, gold and PS) used. These data suggest that Dopa residues in the self-assembled nanofibers contribute significantly to underwater adhesion. However, unmodified fibers also displayed higher adhesion than the CsgA control, implying that features of Mfp domains other than Dopa can participate in underwater adhesion (Fig. 6a). Further comparison of different modified adhesive fibers measured under the same conditions with the same type of tips indicated that adhesion performance had the following general trend, independent of the AFM tips used: (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer > Mfp5-CsgA > CsgA-Mfp3 > CsgA. Among the three tips, the most obvious trend was observed with silica. Testing with silica tips showed very high underwater adhesion for modified Mfp5-CsgA and copolymer fibers (Fig. 5b, 5c and 6a). Specifically, modified Mfp5-CsgA fibers had a normalized adhesion force (Force/Radius, F/R) and adhesion energy (Ead) of 136 mN/m and 14.4 mJ/m2, respectively. Of note, the adhesion energy of Mfp5-CsgA fibers was 3-fold greater than the reported adhesion energy for recombinant Mfp5 alone under the same conditions9, and was 3-fold higher than Mefp5 (Mytilus edulis foot protein 5), the most adhesive mussel protein reported so far, at pH ~5.032.
Interestingly, the (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer (1:1 ratio) had significantly higher adhesion than Mfp5-CsgA and CsgA-Mfp3 when measured with silica tips (P < 0.01), Specifically, the copolymer fibers had F/R = 197.5 mN/m and Ead = 20.9 mJ/m2. These values were ~2–3-fold higher than modified CsgA-Mfp3 measured under the same conditions (Fig. 6a). Moreover, copolymers with a CsgA-Mfp3:Mfp5-CsgA monomer ratio of 1:1 displayed higher adhesion than copolymers assembled from monomer ratios of 3:7 or 7:3 (Supplementary Fig. 23). To our knowledge, (CsgA-Mfp3)-co-(Mfp5-CsgA) copolymer (1:1 ratio) fibers exhibited the strongest underwater adhesion among all known bio-derived and bio-inspired protein-based underwater adhesives reported to date2, 5, 32, 36. We hypothesize that amyloid fiber structures enable large surface areas for contact, with multiple disordered Mfp domains on fiber surfaces interacting to achieve enhanced ultra-strong underwater adhesion. This hypothesis is supported by the larger mean fiber diameters of copolymer-based fibers compared with fibers assembled from the individual proteins, as noted earlier (Fig. 3f).
We compared how different AFM tips with varied surface energies affect the adhesive behaviours of the same type of proteins (Fig. 6b). Among all of the adhesive proteins studied, significant adhesion differences between tips were only found in tyrosinase-modified samples of CsgA-Mfp3, Mfp5-CsgA and copolymer (1:1 ratio) (Fig. 6b and Supplementary Fig. 25–27). Specifically, for modified CsgA-Mfp3, the general trend of adhesion was silica ≈ gold > PS (P < 0.05). For modified Mfp5-CsgA and copolymer (1:1 ratio), the trend was silica > gold ≈ PS (P < 0.05). Both CsgA-Mfp3 and Mfp5-CsgA have basic pIs (~9.0–9.4) and are therefore positively charged at acidic and neutral conditions and can effectively bind to negatively charged SiO2 surfaces. Hydrogen bonding (between hydroxyl or amine groups on proteins and oxygen atoms on SiO2), bidentate H-bonding by Dopa, or hydrogen bonds coupled with coordinate bonds between Dopa and silica could contribute to strong adhesion of fibers with SiO2 surfaces36–38. In contrast, the adhesion of fibers to PS and gold surfaces could be mainly due to hydrophobic interactions, with additional cation-π interactions or π- π interactions likely associated with PS surfaces36, 38. Previous studies in which adhesion was measured with a surface force apparatus revealed that non-fibrous Mfp3 and Mfp5 molecules exhibited the highest adhesion to PS among four substrates (silica, PS, mica and PMMA) with short contact times and had the same level of adhesion to silica and PS with longer contacts36. In contrast, we found that stronger adhesion was found with silica tips and tyrosinase-modified fibers. These observations suggest that, other than providing high surface area for contact, the amyloid domains of our adhesive fibers may modulate how the Mfp domains interact with the substrates and achieve different adhesion levels.
Outlook
We have demonstrated a modular genetic strategy for the design of bio-inspired hybrid fibers for underwater adhesives. Our strategy combines the properties of amyloids and Dopa-containing mussel-foot proteins, two natural adhesives used as building blocks. The resulting fibers have hierarchical structures and multi-functional properties, such as strong wet bonding strength, material robustness, enhanced stability, and intrinsic fluorescence. In particular, the underwater adhesion energy reached 20.9 mJ/m2, which is 1.5-fold greater than the maximum of all bio-derived and bio-inspired protein-based underwater adhesives reported thus far. We envision that engineering complex self-assembly of multiple biomolecular building blocks is a promising strategy to enhance the properties of bio-inspired and bio-mimetic materials. As demonstrated in our work, this strategy can be particularly successful for amyloid-based materials, which have attractive properties16, 19 and have been discovered in several biological settings39.
Our approach builds upon decades of molecular biology and functional genomics research, which have resulted in a diverse library of functional biomolecules that are ripe for incorporation into new bio-inspired molecular materials using synthetic-biology tools. These strategies have already enabled the rational and high-throughput assembly of biological components40; have been used to produce living functional materials41, self-assembling nanostructures42, muscle-mimetic biomaterials43; and have been used to design biological devices with predictable, useful and novel functions44, 45. Recent advances in in vitro46 and cellular expression systems47 that can incorporate unnatural amino acids may open up new capabilities in materials design as well.
Methods Summary
Gene construction and sequencing
Mussel foot protein-3 or -5 (mfp3 or mfp5) gene sequences were synthesized by Integrated DNA Technologies (Coralville, IA). Recombinant genes combining csgA and mfp3 or mfp5 with appended C-terminal poly-histidine tags were constructed using isothermal Gibson assembly and cloned into pET-11d expression vectors. Plasmid construct sequences were confirmed by restriction digest and sequencing was performed by Genewiz (Cambridge, MA). The primers used for PCR and the parts that constitute the plasmids are described in Supplementary Table 1. Plasmid maps are described in Supplementary Figure 2. Sequencing results for csgA, csgA-mfp3 and mfp5-csgA are presented in Supplementary Figures 3, 4 and 5, respectively.
Molecular Dynamics Simulations
The initial model for the CsgA moiety was constructed by threading the CsgA amino acid sequence onto a structure of an amyloid β42 amyloid fibril using Modeller (PDB ID 2BEG)48. Molecular dynamics simulations were run for a total of 1µs and 200 ns for the monomeric proteins and fibrils, respectively. Further details on model construction are available in Supplementary Section 2.
Expression, Purification and Characterization of Adhesive Proteins
Detailed information about protein expression, purification and in vitro post-translational modification are described in Supplementary Section 5. Purified proteins were assayed with SDS/PAGE, Western blotting, matrix-assisted laser desorption ionization (MALDI) - time-of-flight (TOF) mass spectrometry and amino acid analysis (AAA). The general amyloid features of all adhesive fibers were detected with a Congo Red (CR) assay. Dopa residues in modified proteins were qualitatively detected by Nitro Blue Tetrazolium (NBT) staining and quantitatively analysed with acid-borate difference spectrum (ABDS) analysis, supported by amino acid analysis (AAA). Kinetics of amyloid fiber formation was assessed with a Thioflavin T (ThT) assay. The specific experimental protocols are described in detail in Supplementary Sections 5–10 and Supplementary Figures 7–14
Morphology Characterization
The morphology of fibers was assessed with Transmission Electron Microscopy (TEM) and Atomic Force Microscopy (AFM) imaging. Bright-field TEM images were collected on a FEI Tecnai G2 F20 S/TEM operated at an accelerating voltage of 200 kV after staining the samples with uranyl-acetate or gold nanoparticles. Tapping mode (TM) AFM was performed on an Asylum MFP-3D AFM (Asylum Research, Santa Barbara, CA, USA) using Veecoprobes Sb-doped Si cantilevers (ρ = 0.01–0.025 Ω-cm, k = 40 N/m, ν~ 300 kHz).
Fluorescence microscopy, fluorescence spectra and fluorescence quantum yields
Fluorescence images were recorded on a Nikon Ti-E PFS fluorescence microscopy coupled with an Olympus IX70 inverted microscope frame (Olympus UK, Essex, UK). Ultraviolet-Visible (UV-Vis) absorption spectra were recorded with a CARY-6000i spectrophotometer. Fluorescence emission and excitation spectra were measured using a FluoroLog 3 spectrometer manufactured by HORIBA Jobin Yvon. The fluorescence quantum yields (Q (%)) of adhesive fibers were assessed with the Williams comparative method26 using a standard blue fluorophore (Coumarin 102) with a known quantum yield of Q = 76.4% in ethanol. Detailed information about determination of fluorescence quantum yields are described in Supplementary Section 14 and Supplementary Figure 17.
Adhesion force measurements by AFM
AFM force measurements were made using an Asylum MFP-3D AFM (Asylum Research, Santa Barbara, CA, USA) mounted on top of an Olympus IX51 inverted optical microscope for visualizing and manually positioning regions to be probed. Force measurements (force-distance curves) were made at a rate of 0.3 to 2.0 Hz, using Si3N4 or Si cantilevers modified with a glass sphere (with a radius of 10 µm), a glass sphere coated with gold layers (with a radius of 10 µm), or a polystyrene sphere (with a radius of 12.5 µm) (Novascan), with calibrated spring constants between 0.6 and 14 N/m. Both force mapping mode and continuous measurement mode methods were used in this study. All adhesion plots shown in the figures were based on data collected from force mapping mode, which were confirmed with continuous measurement mode. More detailed information about adhesion force measurements are described in Supplementary Section 15 and Supplementary Figure 21–35.
Statistics
Data are presented as mean ± SD (standard deviation). SD was calculated based on at least 3 replicates (for force measurements, this refers to at least 4×16 = 64 force curves for each type of proteins). A Student’s t test was used to compare data sets and a P value less than 0.01 and 0.05 was considered statistically significant between two marked groups or samples.
Supplementary Material
Acknowledgments
We thank Alan Schwartzman for help in applying AFM colloid nanoparticle technology for measuring adhesive forces and Hadi Tavakoli Nia (Ortiz group, MIT) for initial discussions regarding this technology. We acknowledge help from the NERCE Biomolecule Production Laboratory (Harvard University) for producing part of the cell pellets for protein purification. We thank the Whitehead Institute, and the Biopolymers Laboratory in the David H. Koch Institute for Integrated Cancer Research and the Institute for Soldier Nanotechnologies for access to characterization equipment. This research was primarily supported by the Office of Naval Research. This work was also supported in part by the MRSEC Program of the National Science Foundation under award number DMR-0819762. T.K.L. also acknowledges support from the NIH New Innovator Award (1DP2OD008435). The molecular dynamics modeling used the computer cluster and corresponding materials based upon work supported by the National Science Foundation under award number 0821391.
Footnotes
Author Contributions. T.K.L. directed the research. C.Z. conceived the technical details and designed the experiments. C.Z. performed or participated in all the experiments. J.D. performed experiments in protein expression and purification. Z.D. assisted in collecting and analyzing the fluorescence emission and excitation spectra. A.C. constructed the genes. C.M.S. and T.G. designed the simulations. T.G. performed the simulations. C.Z. and T.K.L. wrote the manuscript with help from all the authors. All the authors revised the manuscript.
Additional Information. Supplementary information accompanies the paper at www.nature.com/naturenanotechnology. Preprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/. Correspondence and requests for materials should be addressed to T. K. L.
Competing financial interests
The authors declare competing financial interests. T.K.L. and C.Z. have filed a patent disclosure with the MIT Technology Licensing Office on this work.
References
- 1.Dolgin E. The sticking point. Nature Medicine. 2013;19:124–125. doi: 10.1038/nm0213-124. [DOI] [PubMed] [Google Scholar]
- 2.Lee BP, Messersmith PB, Israelachvili JN, Waite JH. Annual Review of Materials Research. 41 doi: 10.1146/annurev-matsci-062910-100429. 99-1322011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brubaker CE, Messersmith PB. The Present and Future of Biologically Inspired Adhesive Interfaces and Materials. Langmuir. 2012;28:2200–2205. doi: 10.1021/la300044v. [DOI] [PubMed] [Google Scholar]
- 4.Stewart RJ, Ransom TC, Hlady V. Natural Underwater Adhesives. Journal of Polymer Science Part B-Polymer Physics. 2011;49:757–771. doi: 10.1002/polb.22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart RJ. Protein-based underwater adhesives and the prospects for their biotechnological production. Applied Microbiology and Biotechnology. 2011;89:27–33. doi: 10.1007/s00253-010-2913-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin M, Yuan Y, Liu CS, Wang J. Development of mussel adhesive polypeptide mimics coating for in-situ inducing re-endothelialization of intravascular stent devices. Biomaterials. 2009;30:2764–2773. doi: 10.1016/j.biomaterials.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker CE, Kissler H, Wang LJ, Kaufman DB, Messersmith PB. Biological performance of mussel-inspired adhesive in extrahepatic islet transplantation. Biomaterials. 2010;31:420–427. doi: 10.1016/j.biomaterials.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matos-Perez CR, White JD, Wilker JJ. Polymer Composition and Substrate Influences on the Adhesive Bonding of a Biomimetic, Cross-Linking Polymer. Journal of the American Chemical Society. 2012;134:9498–9505. doi: 10.1021/ja303369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang DS, Yoo HJ, Jun JH, Moon WK, Cha HJ. Expression of functional recombinant mussel adhesive protein Mgfp-5 in Escherichia coli. Applied and Environmental Microbiology. 2004;70:3352–3359. doi: 10.1128/AEM.70.6.3352-3359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamino K, Nakano M, Kanai S. Significance of the conformation of building blocks in curing of barnacle underwater adhesive. Febs Journal. 2012;279:1750–1760. doi: 10.1111/j.1742-4658.2012.08552.x. [DOI] [PubMed] [Google Scholar]
- 11.Kamino K. Underwater adhesive of marine organisms as the vital link between biological science and material science. Marine Biotechnology. 2008;10:111–121. doi: 10.1007/s10126-007-9076-3. [DOI] [PubMed] [Google Scholar]
- 12.Anika S, Mostaert SPJ. In: The Functional Fold: Amyloid Structures in Nature. Mostaert SJA, editor. Pan Stanford: 2012. pp. 131–146. [Google Scholar]
- 13.Barlow DE, et al. Characterization of the Adhesive Plaque of the Barnacle Balanus amphitrite: Amyloid-Like Nanofibrils Are a Major Component. Langmuir. 2010;26:6549–6556. doi: 10.1021/la9041309. [DOI] [PubMed] [Google Scholar]
- 14.Wasmer C, et al. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 15.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 16.Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nature Nanotechnology. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 17.Knowles TP, et al. >Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 18.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles TPJ, Oppenheim TW, Buell AK, Chirgadze DY, Welland ME. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nature Nanotechnology. 2010;5:204–207. doi: 10.1038/nnano.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li CX, Adamcik J, Mezzenga R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nature Nanotechnology. 2012;7:421–427. doi: 10.1038/nnano.2012.62. [DOI] [PubMed] [Google Scholar]
- 21.Waite JH, Benedict CV. Assay of dihydroxyphenylalanine (dopa) in invertebrate structural proteins. Methods in enzymology. 1983;107:397–413. doi: 10.1016/0076-6879(84)07028-2. [DOI] [PubMed] [Google Scholar]
- 22.Paz M, Flückiger R, Boak A, Kagan H, Gallop PM. Specific detection of quinoproteins by redox-cycling staining. Journal of Biological Chemistry. 1991;266:689–692. [PubMed] [Google Scholar]
- 23.Wang X, Zhou Y, Ren J-J, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, et al. Development of aliphatic biodegradable photoluminescent polymers. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10086–10091. doi: 10.1073/pnas.0900004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams ATR, Winfield SA, Miller JN. Relative fluorescence quantum yields using a computer-controlled luminescence spectrometer. Analyst. 1983;108:1067–1071. [Google Scholar]
- 27.del Mercato LL, et al. Charge transport and intrinsic fluorescence in amyloid-like fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18019–18024. doi: 10.1073/pnas.0702843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith GJ. The fluorescence of dihydroxyphenylalanine: the effects of protonation-deprotonation. Coloration Technology. 1999;115:346–349. [Google Scholar]
- 29.Chen RF. Fluorescence quantum yields of tryptophan and tyrosine. Analytical Letters. 1967;1:35–42. [Google Scholar]
- 30.Al-Hilaly YK, et al. A central role for dityrosine crosslinking of Amyloid-β in Alzheimer’s disease. Acta neuropathologica communications. 2013;1:83. doi: 10.1186/2051-5960-1-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leite F, Herrmann P. Application of atomic force spectroscopy (AFS) to studies of adhesion phenomena: a review. Journal of adhesion science and technology. 2005;19:365–405. [Google Scholar]
- 32.Danner EW, Kan YJ, Hammer MU, Israelachvili JN, Waite JH. Adhesion of Mussel Foot Protein Mefp-5 to Mica: An Underwater Superglue. Biochemistry. 2012;51:6511–6518. doi: 10.1021/bi3002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei W, Yu J, Broomell C, Israelachvili JN, Waite JH. Hydrophobic enhancement of dopa-mediated adhesion in a mussel foot protein. Journal of the American Chemical Society. 2012;135:377–383. doi: 10.1021/ja309590f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C, Lim JY, Fuller GG, Cegelski L. Quantitative Analysis of Amyloid-Integrated Biofilms Formed by Uropathogenic Escherichia coli at the Air-Liquid Interface. Biophysical journal. 2012;103:464–471. doi: 10.1016/j.bpj.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulter-Thorsen R, Taran E, Gentle I, Gobius K, Dykes G. CsgA production by Escherichia coli O157: H7 alters attachment to abiotic surfaces in some growth environments. Applied and environmental microbiology. 2011;77:7339–7344. doi: 10.1128/AEM.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Q, et al. Adhesion of mussel foot proteins to different substrate surfaces. Journal of The Royal Society Interface. 2013;10:20120759. doi: 10.1098/rsif.2012.0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, et al. Adaptive hydrophobic and hydrophilic interactions of mussel foot proteins with organic thin films. Proceedings of the National Academy of Sciences. 2013;110:15680–15685. doi: 10.1073/pnas.1315015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Qin M, Li Y, Cao Y, Wang W. Single molecule evidences for the adaptive binding of DOPA to different wet surfaces. Langmuir. 2014;30:4358–4366. doi: 10.1021/la501189n. [DOI] [PubMed] [Google Scholar]
- 39.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid - from bacteria to humans. Trends in Biochemical Sciences. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nature Reviews Genetics. 2012;13:21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nature materials. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair JC, Davies KM, Venien-Bryan C, Noble MEM. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nature Nanotechnology. 2011;6:558–562. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 43.Lv S, et al. Designed biomaterials to mimic the mechanical properties of muscles. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 44.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 45.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 46.Hong SH, et al. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific non-standard amino acid incorporation. ACS synthetic biology. 2014;3:398–409. doi: 10.1021/sb400140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eswar N, et al. Comparative protein structure modeling using Modeller. Current protocols in bioinformatics. 2006:5.6. 1–5.6. 30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang DS, Waite JH. Three intrinsically unstructured mussel adhesive proteins, mfp-1, mfp-2, and mfp-3: Analysis by circular dichroism. Protein Science. 2012;21:1689–1695. doi: 10.1002/pro.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakowicz JR. Principles of fluorescence spectroscopy. Springer; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.