Abstract
A simple and accurate test to detect early-stage breast cancer has not been developed. Previous studies indicate that the level of human endogenous retrovirus type K (group HERV-K(HML-2)) transcription may be increased in human breast tumors. We hypothesized that HERV-K(HML-2) reactivation can serve as a biomarker for early detection of breast cancer. Serum samples were collected from women without cancer (controls) and patients with ductal carcinoma in situ (DCIS) and invasive breast cancer. ELISA assays were employed to detect serum anti-HERV-K(HML-2) antibody titers. RNA was extracted from sera and analyzed by real-time RT-PCR to quantitate the level of HERV-K(HML-2) mRNA. We measured significantly higher serum mRNA and serum antibody titers against HERV-K(HML-2) proteins in women with DCIS and stage I disease than in women without cancer. At optimized cutoffs for the antibody titers, the assay produced an area under the ROC curve (AUC) of 0.89 (95% confidence interval 0.77 to 1.00) for DCIS and of 0.95 (95% confidence interval 0.89 to 1.00) for invasive breast cancer. These AUCs are comparable to those observed for mammograms. We also found that serum HERV-K(HML-2) mRNA tended to be higher in breast cancer patients with a primary tumor who later on developed the metastatic disease than in patients who did not develop cancer metastasis. Our results show that HERV-K(HML-2) antibodies and mRNA are already elevated in the blood at an early stage of breast cancer, and further increase in patients who are at risk of developing a metastatic disease.
Keywords: HERV-K, endogenous retroviruses, early detection, biomarkers, breast cancer, metastasis
Introduction
Breast cancer is the most common cancer in women in the US. Approximately 229,000 women in the US will be diagnosed with invasive breast cancer in 2012, and about 40,000 will die from the disease1. Although early detection and improved treatment have reduced the mortality rates of breast cancer, it is still the second leading cause of cancer death in American women.
The current classification scheme for breast cancer includes ductal carcinoma in situ (DCIS), which is considered breast cancer, stage 0. Although a diagnosis of DCIS increases the risk for subsequent development of invasive breast cancer, the overall 10-year survival rate is nevertheless greater than 98%2. After diagnosis of DCIS, the treatment regimen, which is usually successful, is surgery with or without radiation and hormone therapy. Around 4% of women treated with a combination of lumpectomy and radiation therapy develop ipsilateral invasive breast cancer by 90 months post-treatment3. Deaths from DCIS are attributed to factors such as poor diagnosis of invasive carcinoma at the time of the original diagnosis, or incomplete excision of DCIS, which leads to progression to invasive carcinoma. The incidence of DCIS is rising, likely due to increased breast cancer screening and awareness. DCIS incidence rates have risen from 1.87 per 100,000 women from 1973–1975 to 32.5 per 100,000 in 2004, and the rise has been greatest in those older than 50 years of age, where the rate increase from five per 100,000 in 1980 to 59–77 per 100,000 in 20044. DCIS now accounts for approximately 20% of all breast carcinomas detected at screening5, with an estimate of over 60,000 patients diagnosed with DCIS in the United States each year6.
One group of viruses that has emerged recently because of their association with cancer is the family of human endogenous retroviruses (HERVs), which are remnants of ancient germ line infections that have become part of our genome. It has been shown that HERV transcription is increased in several human cancers7, 8 and in breast cancer cell lines9, 10. We have found that expression of one HERV group, HERV-K(HML-2), is increased in breast cancer11–14. The HML-2 group includes the most recently active retroviruses, and a comprehensive analysis of HML-2 element distribution within the human genome has recently been reported15.
Some cancers have well-accepted biomarker-based screening techniques for early detection, such as human papillomavirus for cervical cancer and prostate-specific antigen for prostate cancer. Using these markers, a positive test for HPV types 16 and 18 indicates increased risk for cervical cancer, and elevated serum PSA levels indicates the presence of malignant prostate cells in the prostate. Analogous early detection tests for breast cancer are not available. We hypothesized that either antibodies against serum HERV-K(HML-2) or serum HERV-K(HML-2) mRNA, or the combination of both, could indicate the presence of early stage breast cancer and examined their association with DCIS and stage I breast cancer. Here, we report that HERV-K(HML-2) antibody titers and viral mRNA copy numbers are increased in the serum of women with the preneoplastic breast disease, DCIS, and early-stage invasive breast cancer.
Materials and Methods
Materials
Sera derived from early- and late-stage female breast cancer patients and from sex-matched controls were collected prior to definitive treatment, using a standardized protocol, and stored at −80°C until use. The age of patients and healthy controls was not matched. Sera used in the early detection studies were provided by the Laboratory of Human Carcinogenesis, National Cancer Institute, Bethesda, MD. They were collected between 1993 and 2003 from consented patients with a confirmed diagnosis of DCIS or invasive breast cancer. The sera used in the metastasis biomarker studies were provided by the MD Anderson Cancer Center Breast Cancer Serum Bank. All samples were analyzed retrospectively, and cases and controls were processed simultaneously. The human investigations were performed after approval by the MD Anderson Cancer Center Institutional Review Board. ELISA tests were developed by us using anti-HERV-K(HML-2) antibodies and HERV-K(HML-2) proteins generated from expression vectors in our laboratory11. HERV-K(HML-2) envelope (env) surface fusion proteins were used to immunize BALB/c mice. Hybridoma cells were derived from the splenocytes of immunized mice by fusion with a myeloma cell line using standard techniques. Monoclonal antibody (mAb) clones 6H5, 4D1, 4E11, 6E11, and 4E6 against HERV-K(HML-2) env surface protein were generated in our laboratory, and mAb 6H5 had the highest specificity and sensitivity toward HERV-K(HML-2)11.
ELISA assay
The serum antibody titers against HERV env proteins were determined by ELISA using purified recombinant HERV fusion proteins produced in our laboratory. ELISA assays were carried out as described previously16, 17 to detect anti-HERV antibody in human sera. Briefly, a 96-well ELISA plate was coated with HERV env fusion proteins (10 µg/ml) and blocked, and sera (1:100 to 1:2,700 serial dilutions with PBS) were added to the coated wells, followed by detection with HRP-conjugated anti-human IgG or IgM antibodies. Serum antibodies against HERV-K(HML-2) env surface (SU) protein as well as the Np9 and Rec splice variants of HERV-K(HML-2) env were determined. The HERV-K(HML-2) group comprises around 100 copies per human haploid genome15, and at least 6 proviral loci of this group may express full-length env proteins18. These env proteins are closely related and differ by only a few point mutations, and thus may not be discriminated by antibodies against HERV-K(HML-2) env.
We also evaluated serum antibody titers using multiple antigen peptides (MAPs), which are branched peptides designed to replicate single epitopes to stimulate the immune system for vaccine discovery. The MAP system consists of a large number of the same or different synthetic peptides bound to the groups of a dendritic core molecule providing a high concentration of antigen in a low molecular volume. We produced and used HERV-K MAPs, which are protease and peptidase resistant, as substitutes for the HERV-K(HML-2) protein to coat the ELISA plate. HERV-K(HML-2) MAPs containing a poly-lysine core were produced by BiomerTechnology (Pleasanton, CA). The three MAPs produced, designated MAPs 73, MAPs 75–76, and MAPs 92–93, had sequences corresponding to three immunogenic peptides that are candidate T and B cell epitopes we recently identified in the HERV-K(HML-2) env SU protein sequence. MAPs 73 contained replicates of a single peptide, while MAPs 75–76 and MAPs 92–93 contained replicates of two overlapping peptide sequences. The MAPs peptide sequences are as follows: MAPs 73, SQTITCENCRLLSCI; MAPs 75–76, CRLLSCIDSTFNWQHRILL; MAPs 92–93, GLIAVTATAAVAGVALHSS.
RNA extraction and real-time RT-PCR
RNA was extracted from serum using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA), following the manufacturer's protocol. We used the forward, reverse and probe TaqMan real-time RT–PCR primers coding for both type 1 and type 2 HERV-K(HML-2) env transcript to amplify RNA extracted from serum. One-step real-time RT–PCR was performed using an ABI PRISM 7700 sequence detector. We quantitated the amount of target in patient samples using the standard curve method19. To rule out contamination with genomic DNA, a no reverse transcriptase control was run during the RT step. Known amounts of HERV-K(HML-2) cRNA molecules were used to generate an absolute standard curve, and the copy number of HERV-K(HML-2) mRNA was then determined by linear extrapolation of the CT values using the equation of the line obtained from the absolute HERV standard curve. A BLAST search revealed that the primers used for real-time RT-PCR analyses14 most closely matched the HERV-K HML-2_22q11.21 locus, which is also known as K101, K(C22) and ERVK-2415. Both primers also showed 100% identity with HERV-K HML-2_19p12b (K113), HERV-K HML-2_12q13.2, HERV-K HML-2_5q33.3 (K107/K10, K(C5), ERVK-10), HERV-K HML-2_3q27.2 (K50b, K117, ERVK-11), HERV-K HML-2_1q22 (K102, K(C1b), K50a, ERVK-7), and HERV-K HML-2_1p31.1 (K4, K116, ERVK-1)15.
Serum HERV-K(HML-2) mRNA levels and breast cancer distant metastasis
To evaluate HERV-K(HML-2) as a candidate biomarker for the risk to develop breast cancer metastasis, we identified 21 patients with stage II/III breast cancer who subsequently developed distant metastases within 3–4 years (MBC) and 21 patients with stage II/III breast cancer with no evidence of metastatic disease (NED) at 3–4 years. We evaluated expression of HERV-K(HML-2) gag mRNA by real-time RT-PCR in serum samples drawn at the time of diagnosis. The copy number was determined for the MBC and NED groups. We repeated this experiment in an additional set of 56 MBC patients and 57 NED patients to independently validate the initial results. In this repeat experiment, each patient with MBC was paired with an NED patient based on ER, PR and HER2 status. These studies were conducted in a double-blind fashion.
Statistical analysis
Continuous variables were summarized using mean (SD) and median (range). The Wilcoxon rank sum test was employed to compare the mean of OD between breast cancer versus normal controls for titers of anti-HERV antibodies and for serum mRNA levels. Receiver operating characteristic (ROC) curves were prepared and analyzed using GraphPad Prism 5.
Results
Detection of anti-HERV antibodies in breast cancer and DCIS patient sera
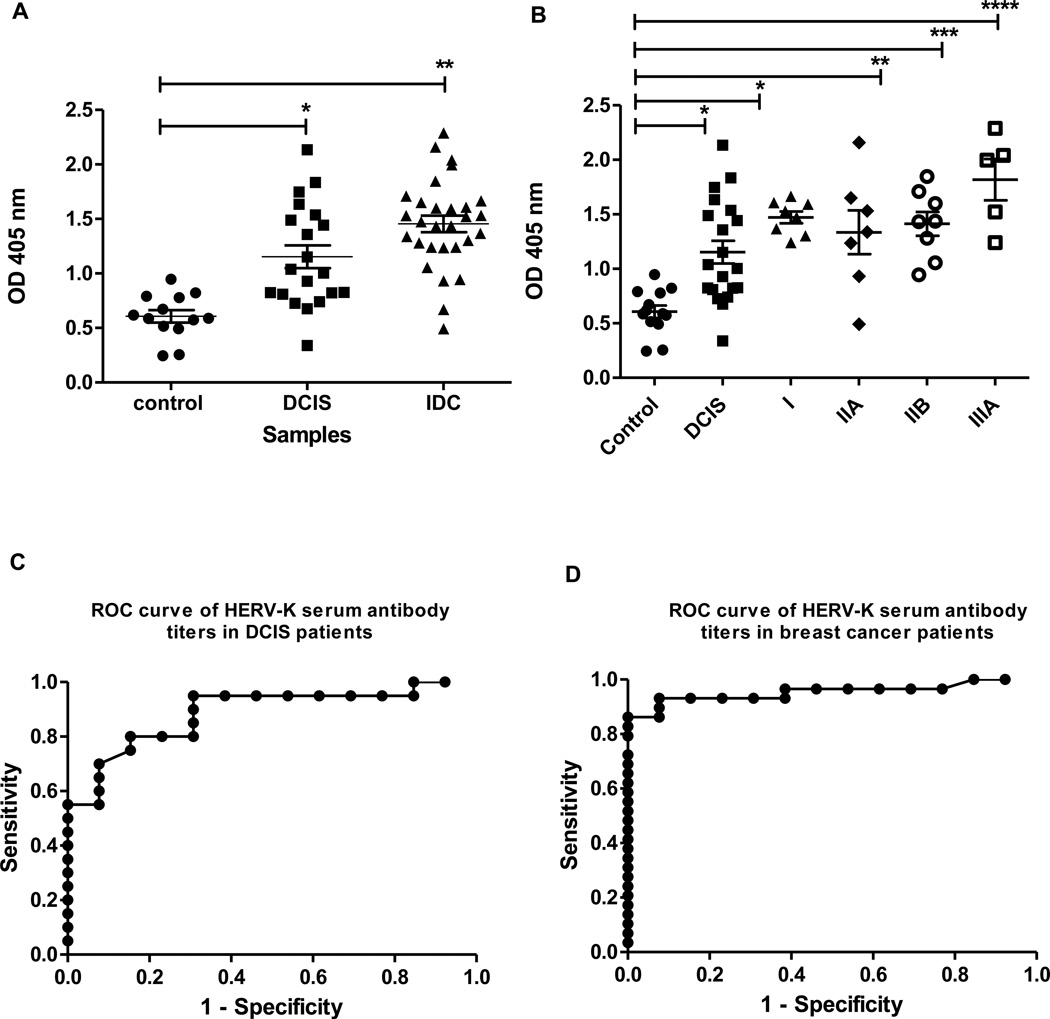
We conducted assays of serum anti-HERV-K(HML-2) env antibody titers in women with ductal carcinoma in situ (DCIS) and invasive breast cancer (Fig. 1A). Patients with both DCIS and invasive ductal carcinoma (IDC) presented with significantly elevated antibody titers when compared with the control group. When the breast cancer patients were further stratified according to TNM disease stage (Fig. 1B), significantly elevated antibody titers were detected in women presenting with early-stage breast cancer, defined as DCIS or stage I disease, indicating that the serum anti-HERV-K(HML-2) antibody titer is a candidate biomarker for the early-stage disease. Receiver operating characteristic curves to distinguish the cancer groups from the controls were used to assess the diagnostic accuracy of our antibody titer assays. Control sera were compared to DCIS (Fig. 1C) and IDC (Fig. 1D) sera. For IDC, the area under the ROC curve (AUC) was 0.95 and the 95% confidence interval was 0.89 to 1.00. For DCIS, the AUC was 0.89 and the 95% confidence interval was 0.77 to 1.00. For the breast cancer curve, the optimum cut-off value was 1.00 OD405 units at which we achieved an 86.2% sensitivity and 100% specificity. For the DCIS curve, the optimum cut-off value was 0.80 OD405 units at which we achieved an 80.0% sensitivity and 84.6% specificity. These AUCs compare favorably with the diagnostic performance of mammograms20. When the irrelevant protein glutathione S-transferase (GST) was coated on the wells of ELISA plates, the OD values averaged 10.4% of the mean value of controls in Figure 1A. Thus, our serum antibody assay may effectively distinguish between a control and a breast cancer patient using an antibody titer as cutoffs that is not generally reached in healthy women.
Figure 1.
Serum antibody titers in patients at various stages of breast cancer. A, Anti-HERV-K serum antibody titers in ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC). Sera from patients were diluted 1:100 and assayed for the presence of serum anti-HERV-K antibodies by ELISA. The antigen used for detection was the HERV-K env surface fusion protein. DCIS and IDC were significantly different from controls by the Wilcoxon rank-sum test (*, p = 0.0002; **, p < 0.0001). N=20 for DCIS, N=29 for IDC and N=13 for control. B, Anti-HERV-K serum antibody titers, analyzed by breast cancer stage. The differences between stages of breast cancer and controls were significant by the Wilcoxon rank-sum test (*, p = 0.0002; **, p = 0.079; ***, p = 0.0003; ****, p = 0.0016). N=20 for DCIS, N=8 for stage I, N=7 for stage IIA, N=8 for stage IIB, N=5 for stage IIIA and N=13 for control. Results in A and B are presented as individual data points for each subject. C, ROC curve of HERV-K serum antibody titers in DCIS patients. D, ROC curve of HERV-K serum antibody titers in breast cancer patients.
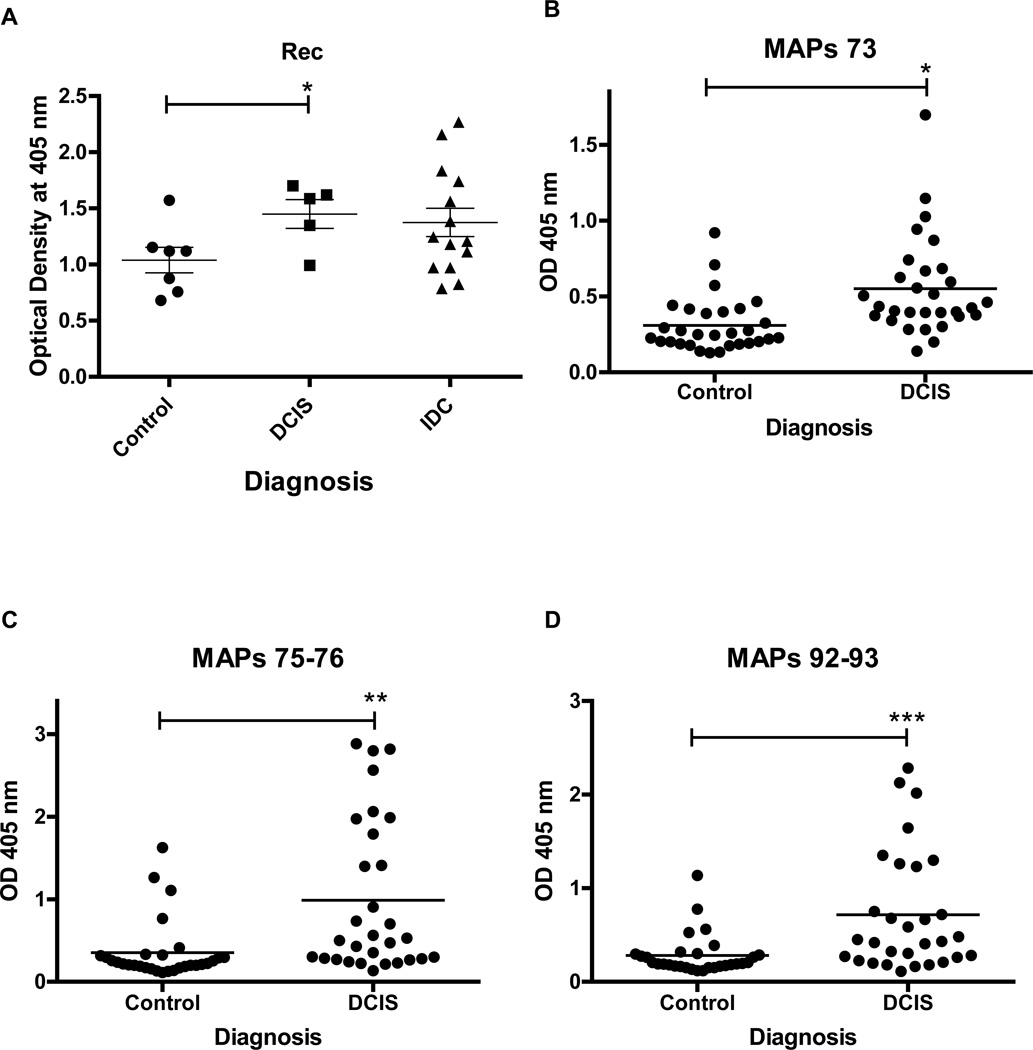
Next, we examined antibody titers against HERV-K(HML-2) Rec protein, which can induce carcinoma in situ, and HERV-K(HML-2) Np9 protein, which is overproduced in many tumors including breast cancers21. Both Np9 and Rec interact with the promyelocytic leukemia zinc finger (PLZF) tumor suppressor, a transcriptional repressor and chromatin remodeler that plays an important role in cancer22. Our ELISA results show that women with DCIS have increased serum antibody levels against Rec, compared with the women who did not have cancer (Fig. 2A). Our results suggest that the presence of anti-Rec serum antibodies is as good a marker for the presence of DCIS as an anti-HERV env antibody. Np9 is a small protein, and because of its small size it may not be as antigenic as the HERV env and Rec proteins in patients with DCIS. However, Np9 antibodies were present in the serum of DCIS patients at levels that approached being significantly higher than controls (P~0.07 for 1:100 dilutions) (data not shown). These findings support our hypothesis that mRNA and proteins derived from HERV-K(HML-2) such as Rec and Np9 are candidate serum-based biomarkers for early-stage breast cancer. We also used HERV-K(HML-2) MAPs (see methods), which are protease and peptidase resistant, as substitutes for the HERV-K(HML-2) protein to coat the ELISA plate. We evaluated MAPs in a larger sample set than Rec and Np9 because the MAPs synthetic peptides would be easier to produce and, because they are much smaller fragments than full-length proteins, should give more reproducible results than an expressed protein having multiple epitopes. The MAPs-based assays were able to discriminate between DCIS and controls (Fig. 2B–2D) as well as the HERV-K(HML-2) protein-based assays, suggesting that synthetic peptides can be used instead of HERV-K(HML-2) proteins for detecting serum antibodies in ELISA assays. A MAPs peptide lacking an HERV-K epitope gave an OD value in ELISA assays that was not different from controls.
Figure 2.
Serum antibody titers of HERV-K Rec and multiple antigen peptides (MAPs) in breast cancer. A, Anti-Rec serum antibody titers in ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC). The difference between controls and DCIS is significant by the Wilcoxon rank-sum test (*, p = 0.048). N=5 for DCIS, N=14 for IDC and N=7 for control. B-D, Anti-HERV-K antibody titers in sera from patients with DCIS or women without cancer (Control), using MAPs. Figures represent three MAPs, designated MAPs 73, MAPs 75–76, and MAPs 92–93, that had sequences corresponding to three immunogenic peptides we recently identified in the HERV-K(HML-2) env SU protein sequence. The differences between controls and DCIS were significant by the Wilcoxon rank-sum test, with *, p = 0.0001; **, p < 0.0001; and ***, p = 0.0002 for MAPs 73, MAPs 75–76 and MAPs 92–93, respectively. N=30 for both DCIS and control for each of the MAPs.
Moreover, the presence of anti-HERV-K(HML-2) env protein antibodies in the serum of breast cancer patients indicates that there was a humoral response to HERV-K(HML-2). More importantly, our results suggest that this assay might be useful for rapidly screening women for breast cancer.
Detection of HERV-K(HML-2) mRNA in DCIS tissue and sera
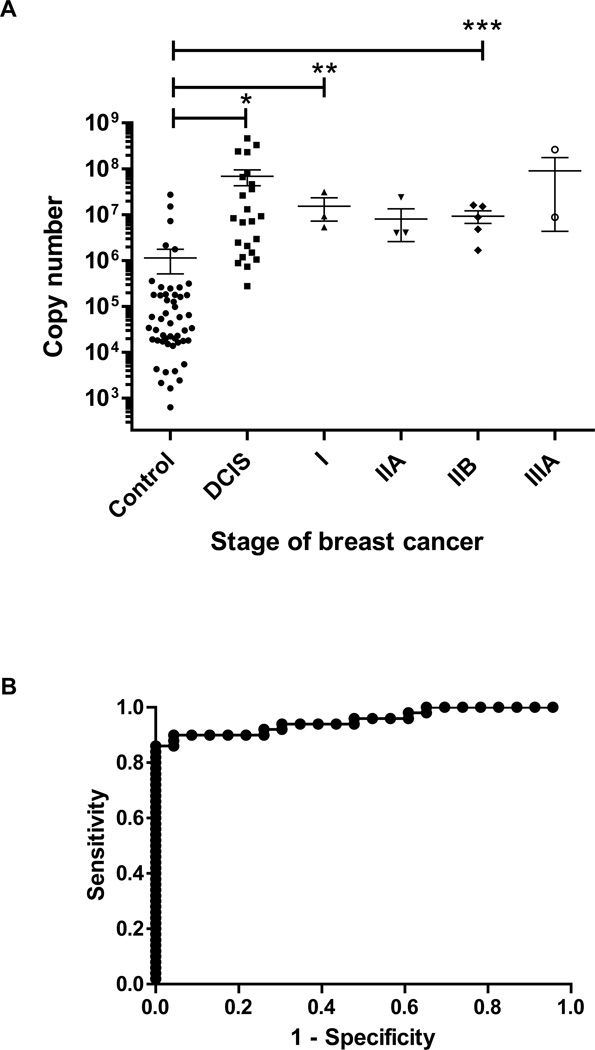
Our laboratory previously reported that the expression of HERV-K(HML-2) env mRNA was elevated in approximately 70–80% of breast cancer tissues compared with normal breast tissues11. Based on the tissue viral mRNA data, we tested whether the increased expression of HERV-K(HML-2) mRNA in breast tissue would also be reflected in the serum of patients with preneoplastic breast lesions such as DCIS. We purified viral RNA from serum of women with DCIS and IDC, or women without the disease, using a QIAamp Viral RNA Mini Kit (Qiagen, Inc.). We developed a real-time RT-PCR assay for HERV-K(HML-2) viral RNA in serum, and analyzed sera for viral HERV-K(HML-2) mRNA titers. This assay is analogous to serum HIV and high-risk HPV viral load tests, which seek to determine the degree of cervical neoplasia. Our assays show an excellent separation between women without cancer and women with DCIS and IDC (Fig. 3A), with a ROC curve AUC of 0.95 and a 95% confidence interval of 0.91 to 0.997 for DCIS (Fig. 3B). The optimum cut-off value to separate controls from DCIS cases was a copy number of 2.64 × 106, with 90.0% sensitivity and 95.7% specificity.
Figure 3.
Serum mRNA levels in breast cancer patients. A, HERV-K mRNA levels in sera of patients with DCIS and stage I, IIA, IIB or IIIA breast cancer. Patients with DCIS (*, p < 0.001), stage I (**, p = 0.0075) breast cancer, and stage IIB (***, p = 0.0011) breast cancer had significantly higher serum mRNA levels than controls by the Wilcoxon rank-sum test. N=23 for DCIS, N=3 for stage I, N=4 for stage IIA, N=5 for stage IIB N=3 for stage IIIA and N=50 for control. B, ROC curve of serum HERV-K copy number (assessed by quantitative RT-PCR) in DCIS patients.
The Spearman correlation coefficient was used to verify the correlation between mRNA expression and anti-HERV-K env antibodies in selected breast cancer patient serum samples from Figures 1–3. Correlation analysis of OD and HERV-K copy number in serum samples from 3 controls, 1 stage I, 1 stage IIA, 1 stage IIB and 8 DCIS patients showed a correlation coefficient of 0.79 (p=0.0007), suggesting a strong positive correlation.
Serum samples that express env mRNA also express gag mRNA, and we plan in future studies to evaluate the potential of gag as a biomarker of early breast cancer.
Serum HERV-K(HML-2) as a biomarker of breast cancer metastasis
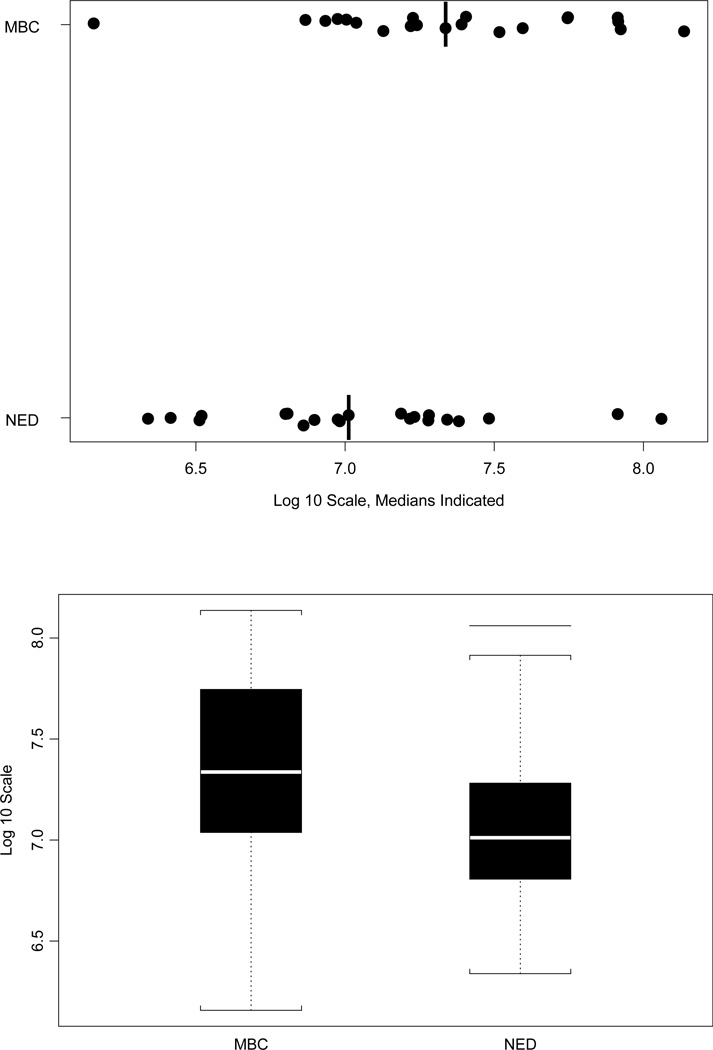
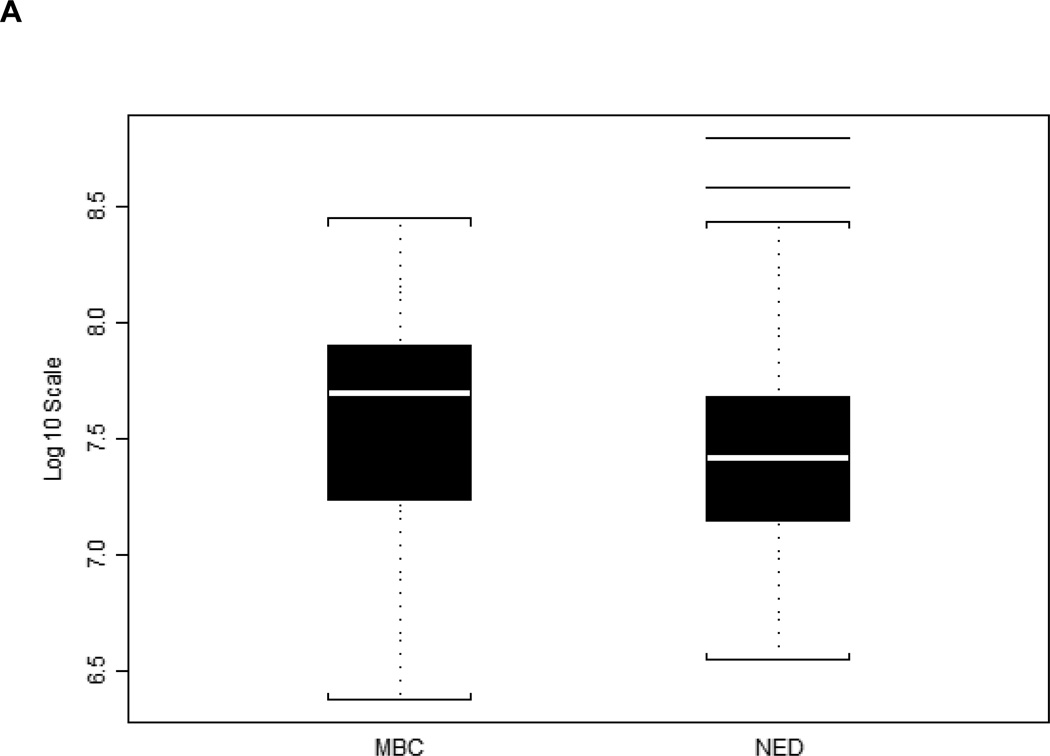
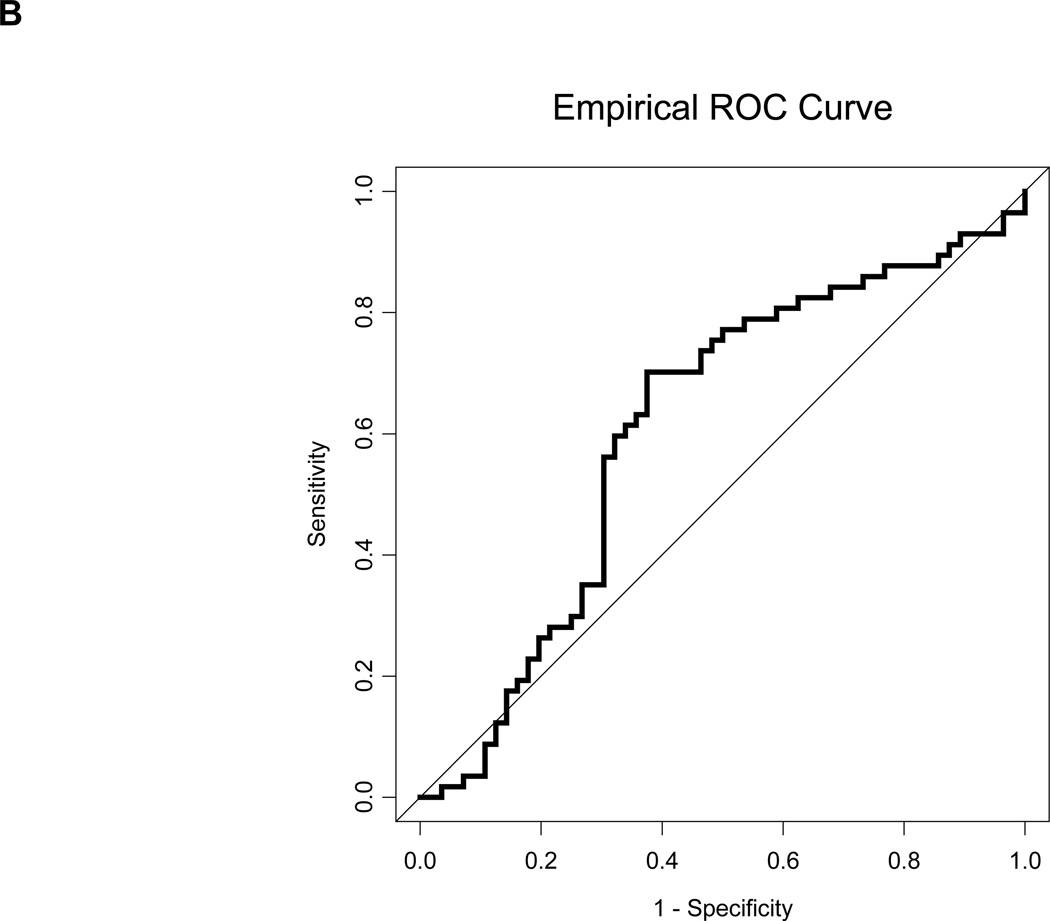
A biomarker that predicts the likelihood of metastatic breast cancer would aid in treatment decisions for women in the early stages of breast cancer. We initially compared 21 metastatic breast cancer (MBC) patients and 21 no evidence of disease (NED) patients, and found that the patients who developed metastatic disease had higher HERV-K(HML-2) gag copy number values at baseline (Fig. 4). This difference was significant by the Wilcoxon rank-sum test, with p = 0.036. We sought to confirm this initial finding with a larger set of 56 MBC patients and 57 NED patients. In this independent validation, each patient with MBC was paired with an NED patient based on ER, PR and HER2 status. Our results verified the findings with the initial group of patients, showing that MBC patients had higher (p = 0.041, Wilcoxon rank-sum test) gag mRNA expression at baseline (Fig. 5A) and an increased AUC (Fig. 5B).
Figure 4.
Serum HERV-K10 gag mRNA expression data for 42 matched samples. Dot plot and boxplots depicting HERV-K gag mRNA mean copy number values for 21 patients with distant metastases (MBC) and 21 patients without distant metastases (NED). This difference is significant by the Wilcoxon rank-sum test, with p = 0.036.
Figure 5.
Serum HERV-K10 gag mRNA expression data of a repeat analysis of 113 additional matched samples. A, Boxplots depicting HERV-K gag mRNA mean copy number values for 56 patients with distant metastases (MBC) and 57 patients without distant metastases (NED). The MBC patients again had higher HERV-K mRNA expression on average, and the difference was again significant by the Wilcoxon rank-sum test, with p = 0.041. B, ROC curve for HERV-K gag for the repeat analysis. The AUC = 0.611 +/− 0.053.
Discussion
Our laboratory demonstrated that the HERV-K(HML-2) env gene is expressed in breast cancer11–14. In these studies we showed elevated expression of HERV-K(HML-2) env mRNA in preneoplastic breast lesions, and of HERV-K(HML-2) protein in surgical specimens of breast ductal epithelial tumors in humans. HERV-K(HML-2) was immunogenic in human breast cancer, leading to elevated titers of anti-HERV-K(HML-2) antibodies relative to normal donors. Most cancer tissues expressed HERV-K(HML-2) protein, with the majority of those displaying moderate or strong expression. In contrast, 95% of normal tissues exhibited no expression of HERV-K(HML-2) protein, whereas 29% of tissues with various benign breast diseases weakly expressed HERV-K(HML-2) protein.
Anti-HERV-K(HML-2) antibodies may have an important diagnostic value and suggest chemotherapy success23. We determined the correlation between anti-HERV-K(HML-2) antibody titers and the type of proliferative lesion, reasoning that the presence and amount of anti-HERV-K(HML-2) antibodies might have an important diagnostic value for breast cancer. Antibodies recognizing synthetic HERV-K(HML-2) proteins were detected at a very low frequency in the sera of healthy donors7. Quantitation of circulating antibodies to HERV and HERV mRNA in breast cancer patients in early stage disease is potentially of great importance because it could serve as a screening tool to detect breast cancers before they reach an advanced stage.
Several serum biomarkers of breast cancer have recently been proposed25–27. Our studies demonstrate that HERV-K(HML-2) is a biomarker of breast cancer that is expressed at very high levels in breast cancer tissue and very low levels in normal breast tissue. Many breast cancer antigens being tested in clinical trials are overexpressed tissue differentiation antigens. In contrast to self-antigens, viral antigens may be better suited to trigger stronger antitumor T-cell responses due to their foreign nature. The amount of HERV-K(HML-2) viral mRNA was greater in the tissue and serum of breast cancer or DCIS patients than in the serum of controls without cancer. HERV-K(HML-2) viral mRNA and antibodies against HERV-K(HML-2) were found in the sera of patients with early breast cancer (DCIS) in titers that were greater than controls. HERV-K(HML-2) serum mRNA and antibody titers showed greater sensitivity and specificity than other recently proposed serum biomarkers of breast cancer28–30. The increased sensitivity and specificity of this biomarker may be due to the very low expression of HERV-K(HML-2) in normal breast tissue relative to breast cancer tissue11, which has also been reported in another study31. Although the expression of biomarkers is increased in breast cancer, in many cases the biomarker protein is also expressed in normal breast tissue, and that appears not to be the case for HERV-K(HML-2). It would be of interest to determine whether HERV-K(HML-2) mRNAs and antibodies can be detected in prediagnostic breast cancer sera, as was recently reported for several known serum breast cancer antigens32.
HERV-K(HML-2) serum antibodies and mRNA levels may also be useful to screen for breast cancer in women who cannot or will not be screened by mammography, and in younger women. There are few options for early detection of breast cancer, especially in young women, and HERV-K(HML-2) screening may be an additional option for early detection in women at increased risk for breast cancer, including women with a family history of breast cancer, women previously diagnosed with atypical hyperplasia, or women whose first child was born after age 30. In addition, women with genetic mutations that predispose them to increased breast cancer risk (for example, BRCA1 and BRCA2) should also benefit from HERV-K(HML-2) screening. It remains to be determined whether serum copy numbers of HERV-K(HML-2) antibody or viral mRNA are elevated in women with recurrent breast cancer. If this turns out to be the case, then treatments that lower serum HERV-K(HML-2), such as therapeutic antibodies we have recently developed in our laboratory13 might prove effective in shrinking the tumor. It is important to note that the presence of HERV-K(HML-2) transcripts in the serum does not necessarily indicate that anti-HERV-K(HML-2) antibodies will be produced, since not all HERV-K(HML-2) transcripts are capable of encoding proteins.
Prognostic and predictive biomarkers for breast cancer include the estrogen and progesterone receptors, biomarkers of proliferation such as Ki-67, and the HER2 gene. There are a number of less well established prognostic and predictive molecular factors, including pS2, PCNA, mitosin, EGFR, IGF, P53, BCL-2, cyclins, uPA, PAI, VEGF, PD-ECGF and FGF33. HERV-K(HML-2) screening might also be used as a predictor of metastasis at the time the patient seeks treatment for breast cancer. Our results provide evidence that HERV-K(HML-2) gag mRNA copy number in sera collected at the time of presentation may be a useful biomarker of breast cancer metastasis. We found that HERV-K(HML-2) gag mRNA expression is higher at baseline in those patients whose breast cancer subsequently metastasized, in comparison to breast cancer patients whose breast cancer did not metastasize. It remains to be seen whether other endogenous retroviral serum mRNAs in addition to HERV-K(HML-2) gag are also biomarkers of breast cancer metastasis. In addition, HERV-K(HML-2) might be combined with other breast tumor markers such as HER2/neu to increase the sensitivity and specificity of breast cancer detection and/or prognosis. Since our results suggest that gag mRNA serves as a predictor of metastasis we plan in future studies to determine whether env mRNA is also a predictor of metastasis. Likewise, although we have not tested whether gag mRNA differs in DCIS vs. breast cancer, we also plan to focus on this question in future studies.
Our results suggest that endogenous retroviral sequences, which are harbored in the genome of all humans, are reawakened during the early stages of breast cancer. Our results also suggest that HERV-K(HML-2) levels in early-stage breast cancer may predict metastasis. These retroviruses have not been previously proposed as biomarkers for early detection and diagnosis of breast cancer, and are conceptually different from other breast cancer biomarkers, which have included hormones, growth factors, components of cell signalling pathways, methylated genes, cytokines, oncoproteins and others. In contrast to many other breast cancer biomarkers, our biomarker is present at high levels in breast cancer patient sera, it begins to appear in early breast cancer, and it is present in very small amounts in normal breast sera. The analysis of serum for HERV-K(HML-2) antibodies and RNA may enable clinicians to more easily identify women with non-invasive early breast cancer and treat these women at an early stage before the breast cancer can spread to other tissues.
Acknowledgments
Grant support: Avon Foundation grant 07-2007-070 01, Department of Defense grant DAMD1700-1-0123, Susan G. Komen Breast Cancer Foundation (Susan G. Komen for the Cure) grants BCTR0402892 and BCTR000007888, the National Institute of Environmental Health Sciences Center grant ES007784 and the Breast Cancer Research Foundation. We acknowledge the Breast Cancer Management System and the Nellie B. Connally Breast Cancer Research Fund for support of the MD Anderson Cancer Center Breast Cancer Serum Bank.
Abbreviations used
- AUC
area under the ROC curve
- DCIS
ductal carcinoma in situ
- Env
envelope
- ER
estrogen receptor
- HERV-K
Human endogenous retrovirus type K
- IDC
invasive ductal carcinoma
- mAb
monoclonal antibody
- MAPs
multiple antigen peptides
- MBC
metastatic breast cancer
- NED
no evidence of disease
- ORC
open reading frame
- PLZF
promyelocytic leukemia zinc finger
- PR
progesterone receptor
- ROC
receiver operating characteristic
- SD
standard deviation
- SU
surface
Footnotes
Novelty of the data presented: The significant and novel findings of our work are that human endogenous retrovirus antibodies and mRNA are already elevated in the blood at an early stage of breast cancer, and further increase in patients who are at risk of developing a metastatic disease. Our results suggest that the sensitive and specific serum assays we have described may usher in a new era of viral biomarkers for detection of early cancer and prediction of cancer metastasis.
There are no potential conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer Statistics, 2012. Ca-Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Viale G. Histopathology of primary breast cancer 2005. Breast. 2005;14:487–492. doi: 10.1016/j.breast.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, Dignam J, Wolmark N, Mamounas E, Costantino J, Poller W, Fisher ER, Wickerham DL, Deutsch M, Margolese R, Dimitrov N, Kavanah M. Lumpectomy and radiation therapy for the treatment of intraductal breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-17. J Clin Oncol. 1998;16:441–452. doi: 10.1200/JCO.1998.16.2.441. [DOI] [PubMed] [Google Scholar]
- 4.Virnig BA, Wang SY, Shamilyan T, Kane RL, Tuttle TM. Ductal carcinoma in situ: risk factors and impact of screening. J Natl Cancer Inst Monogr. 2010;2010:113–116. doi: 10.1093/jncimonographs/lgq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuerer HM, Lari SA, Arun BK, Hu CY, Brewster A, Mittendorf EA, Albarracin CT, Babiera GV, Caudle AS, Wagner JL, Litton JK, Bedrosian I, et al. Biologic features and prognosis of ductal carcinoma in situ are not adversely impacted by initial large body mass. Breast Cancer Res Treat. 2012 doi: 10.1007/s10549-012-1999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lari SA, Kuerer HM. Biological Markers in DCIS and Risk of Breast Recurrence: A Systematic Review. J Cancer. 2011;2:232–261. doi: 10.7150/jca.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Research. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 8.Ryan FP. Human endogenous retroviruses in health and disease: a symbiotic perspective. J Roy Soc Med. 2004;97:560–565. doi: 10.1258/jrsm.97.12.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifarth W, Skladny H, Kriegschneider F, Reichert A, Hehlmann R, Leibmosch C. Retrovirus-Like Particles Released from the Human Breast-Cancer Cell-Line T47-D Display Type-B-Related and Type-C-Related Endogenous Retroviral Sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etkind PR, Lumb K, Du J, Racevskis J. Type 1 HERV-K genome is spliced into subgenomic transcripts in the human breast tumor cell line T47D. Virology. 1997;234:304–308. doi: 10.1006/viro.1997.8670. [DOI] [PubMed] [Google Scholar]
- 11.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Research. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- 13.Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin BN, Frerich K, Garza JG, Shen JJ, Lin K, Yan P, Glynn SA, Dorsey TH, et al. Immunotherapeutic Potential of Anti-Human Endogenous Retrovirus-K Envelope Protein Antibodies in Targeting Breast Tumors. J Natl Cancer I. 2012;104:189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang-Johanning F, Frost AR, Jian BX, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang-Johanning F, Gillespie GY, Grim J, Rancourt C, Alvarez RD, Siegal GP, Curiel DT. Intracellular expression of a single-chain antibody directed against human papillomavirus type 16 E7 oncoprotein achieves targeted antineoplastic effects. Cancer Res. 1998;58:1893–1900. [PubMed] [Google Scholar]
- 17.Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 18.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang-Johanning F, Lu DW, Wang YY, Johnson MR, Johanning GL. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer. 2002;94:2199–2210. doi: 10.1002/cncr.10439. [DOI] [PubMed] [Google Scholar]
- 20.Chlebowski RT, Anderson G, Manson JE, Pettinger M, Yasmeen S, Lane D, Langer RD, Hubbell FA, McTiernan A, Hendrix S, Schenken R, Stefanick ML. Estrogen alone in postmenopausal women and breast cancer detection by means of mammography and breast biopsy. J Clin Oncol. 2010;28:2690–2697. doi: 10.1200/JCO.2009.24.8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res. 2002;8:1800–1807. [PubMed] [Google Scholar]
- 22.Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol. 2007;81:5607–5616. doi: 10.1128/JVI.02771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleiman A, Senyuta N, Tryakin A, Sauter M, Karseladze A, Tjulandin S, Gurtsevitch V, Mueller-Lantzsch N. Herv-K(HML-2) Gag/Env antibodies as indicator for therapy effect in patients with germ cell tumors. Int J Cancer. 2004;110:459–461. doi: 10.1002/ijc.11649. [DOI] [PubMed] [Google Scholar]
- 24.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Ali SM, Carney WP, Esteva FJ, Fornier M, Harris L, Kostler WJ, Lotz JP, Luftner D, Pichon MF, Lipton A. Serum HER-2/neu and relative resistance to trastuzumab-based therapy in patients with metastatic breast cancer. Cancer. 2008;113:1294–1301. doi: 10.1002/cncr.23689. [DOI] [PubMed] [Google Scholar]
- 26.Baek JM, Jin QR, Ensor J, Boulbes DR, Esteva FJ. Serum CD44 levels and overall survival in patients with HER2-positive breast cancer. Breast Cancer Res Tr. 2011;130:1029–1036. doi: 10.1007/s10549-011-1691-z. [DOI] [PubMed] [Google Scholar]
- 27.Shen YF, Tolic N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, Moore RJ, Purvine SO, Esteva FJ, Smith RD. Blood Peptidome-Degradome Profile of Breast Cancer. Plos One. 2010;5 doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padler-Karavani V, Hurtado-Ziola N, Pu MY, Yu H, Huang SS, Muthana S, Chokhawala HA, Cao HZ, Secrest P, Friedmann-Morvinski D, Singer O, Ghaderi D, et al. Human Xeno-Autoantibodies against a Non-Human Sialic Acid Serve as Novel Serum Biomarkers and Immunotherapeutics in Cancer. Cancer Research. 2011;71:3352–3363. doi: 10.1158/0008-5472.CAN-10-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson KS, Sibani S, Wallstrom G, Qiu J, Mendoza EA, Raphael J, Hainsworth E, Montor WR, Wong J, Park JG, Lokko N, Logvinenko T, et al. Protein Microarray Signature of Autoantibody Biomarkers for the Early Detection of Breast Cancer. Journal of Proteome Research. 2011;10:85–96. doi: 10.1021/pr100686b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, Barnes A, Robertson J. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–873. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- 31.Pichon JP, Bonnaud B, Cleuziat P, Mallet F. Multiplex degenerate PCR coupled with an oligo sorbent array for human endogenous retrovirus expression profiling. Nucleic Acids Research. 2006;34 doi: 10.1093/nar/gkl086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H, Ladd J, Feng Z, Wu M, Goodell V, Pitteri SJ, Li CI, Prentice R, Hanash SM, Disis ML. Evaluation of Known Oncoantibodies, HER2, p53, and Cyclin B1, in Prediagnostic Breast Cancer Sera. Cancer Prev Res (Phila) 2012 doi: 10.1158/1940-6207.CAPR-11-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteva FJ, Sahin AA, Cristofanilli M, Arun B, Hortobagyi GN. Molecular prognostic factors for breast cancer metastasis and survival. Semin Radiat Oncol. 2002;12:319–328. doi: 10.1053/srao.2002.35251. [DOI] [PubMed] [Google Scholar]