Summary
Contact-dependent growth inhibition (CDI) is a mode of bacterial competition orchestrated by the CdiB/CdiA family of two-partner secretion proteins. The CdiA effector extends from the surface of CDI+ inhibitor cells, binds to receptors on neighboring bacteria and delivers a toxin domain derived from its C-terminal region (CdiA-CT). Here, we show that CdiA-CT toxin translocation requires the proton-motive force (pmf) within target bacteria. The pmf is also critical for the translocation of colicin toxins, which exploit the energized Ton and Tol systems to cross the outer membrane. However, CdiA-CT translocation is clearly distinct from known colicin-import pathways because ΔtolA ΔtonB target cells are fully sensitive to CDI. Moreover, we provide evidence that CdiA-CT toxins can be transferred into the periplasm of de-energized target bacteria, indicating that transport across the outer membrane is independent of the pmf. Remarkably, CDI toxins transferred under de-energized conditions remain competent to enter the target-cell cytoplasm once the pmf is restored. Collectively, these results indicate that outer- and inner-membrane translocation steps can be uncoupled, and that the pmf is required for CDI toxin transport from the periplasm to the target-cell cytoplasm.
Introduction
Bacteria are often regarded as isolated unicellular organisms, but they clearly engage in cooperative and antagonistic relationships with other microorganisms. Contact-dependent growth inhibition (CDI) is one mechanism of intercellular competition that is common amongst Gram-negative pathogens (Aoki et al., 2010, Ruhe et al., 2013a). CDI is mediated by the CdiB/CdiA two-partner secretion proteins, which constitute a sub-family of type V secretion systems (Hayes et al., 2010, Jacob-Dubuisson et al., 2013). CdiB is an outer membrane β-barrel protein that uses periplasmic polypeptide-transport domains to recognize and secrete CdiA effectors. CdiA proteins are very large (180 – 650 kDa) and contain peptide repeats similar to those found in filamentous hemagglutinin (FHA) from Bordetella pertussis (Kajava et al., 2001). Like FHA, CdiA proteins are thought to adopt an elongated β-helical structure that allows the effector to reach receptors on the surface of susceptible bacteria. After binding its receptor, CdiA is presumably cleaved to release a C-terminal toxin domain (CdiA-CT), which is subsequently transported into the target bacterium (Aoki et al., 2010, Aoki et al., 2005). CDI+ bacteria protect themselves from auto-inhibition by producing CdiI immunity proteins. CdiI binds to the CdiA-CT toxin domain and neutralizes its activity (Aoki et al., 2010, Nikolakakis et al., 2012, Morse et al., 2012). CDI systems encode a variety of sequence-diverse CdiA-CT toxins. Analysis of CdiA-CT coding sequences from Escherichia coli strains reveals at least 18 distinct toxin types (Ruhe et al., 2013a). Each CdiA-CT sequence type is associated with a specific CdiI immunity protein, which together constitute a cognate pair. The different sequence types often correspond to unique toxin activities. For example, CdiA-CTEC93 from E. coli EC93 appears to be an ionophore toxin (Aoki et al., 2009), whereas CdiA-CTUPEC536 from uropathogenic E. coli 536 (UPEC 536) is a nuclease that degrades tRNA molecules (Aoki et al., 2010, Diner et al., 2012). Remarkably, cdiA-CT/cdiI gene pairs are modular and can be exchanged horizontally between bacteria (Poole et al., 2011, Arenas et al., 2013). These observations suggest that bacteria can abruptly change their toxin/immunity type through the acquisition and recombination of new cdiA-CT/cdiI modules. This hypothesis is supported by experimental work demonstrating that cdiA-CT/cdiI pairs can be fused to heterologous cdiA genes to produce functional chimeric CdiA proteins (Aoki et al., 2010, Poole et al., 2011, Morse et al., 2012, Nikolakakis et al., 2012, Webb et al., 2013). Thus, CDI systems encode a large network of toxin/immunity pairs that are deployed in the competition for environmental resources.
The mechanisms underlying CdiA-CT toxin translocation into target bacteria are poorly understood, though some insights have been gained through analysis of E. coli mutants that are resistant to the CDI system from Escherichia coli EC93 (Aoki et al., 2008). CdiAEC93 uses BamA as a receptor to bind target bacteria, and mutations that decrease BamA expression or alter its surface epitopes confer resistance to CDIEC93 (Aoki et al., 2008, Ruhe et al., 2013b). BamA is an essential outer-membrane protein that forms the core of the β-barrel assembly machine (BAM) complex. The BAM complex is required for the assembly of β-barrel proteins into the outer membrane of Gram-negative bacteria (Gentle et al., 2004, Voulhoux et al., 2003, Wu et al., 2005, Werner & Misra, 2005), but the biogenesis function of BamA does not appear to play a role in CDIEC93 (Aoki et al., 2008). Because BamA is itself a β-barrel protein (Noinaj et al., 2013), it may also serve as a conduit for toxin translocation across the target cell outer membrane. Additionally, E. coli acrB mutants are resistant to CDIEC93 (Aoki et al., 2008). AcrB is a trimeric inner-membrane protein that functions together with AcrA and TolC to form a multi-drug efflux pump (Ma et al., 1995, Nikaido & Zgurskaya, 2001). The AcrAB-TolC complex spans the entire cell envelope, providing a potential pathway for toxin transport. However, ΔacrA and ΔtolC mutants are not resistant to CDIEC93 (Aoki et al., 2008). Because CdiA-CTEC93 is hypothesized to be a membrane-bound ionophore, AcrB could facilitate insertion of the toxin into the cytoplasmic membrane of target bacteria. Regardless of its function, AcrB appears to be specifically required for CDIEC93 because E. coli ΔacrB mutants are sensitive to other CdiA-CT toxins (J.L.E. Willett, C.M.B. & C.S.H., unpublished data).
Initial work on the CDI phenomenon showed that continuous protein synthesis within CDI+ cells is required for the inhibition of target-cell growth (Aoki et al., 2005). This finding suggests that CdiA synthesis and CdiA-CT delivery are functionally linked, raising the possibility that active translation provides the energy for toxin translocation into target bacteria. Here, we reexamine those results and find that ongoing protein synthesis is only required to replenish CdiAEC93 on the surface of inhibitor cells. This result led us to explore what energy source, if any, is required for CDI toxin translocation. Using the colicin translocation paradigm, we hypothesized that the proton-motive force (pmf) across the inner membrane of target cells may be required for CdiA-CT toxin import. We find that dissipation of the target-cell pmf prevents the translocation of several different CdiA-CT toxins. Notably, CdiA-CT import does not require tolA nor tonB, indicating that the CDI translocation pathway is distinct from that of the group A and group B colicins, respectively (Braun et al., 1980, Jiang et al., 1997). Moreover, though colicins require the pmf for transport across the outer membrane, our results suggest that the pmf is harnessed to translocate CdiA-CT toxins across the inner membrane of target bacteria.
Results
Active protein synthesis in CDI+ inhibitor cells is not required for CDI
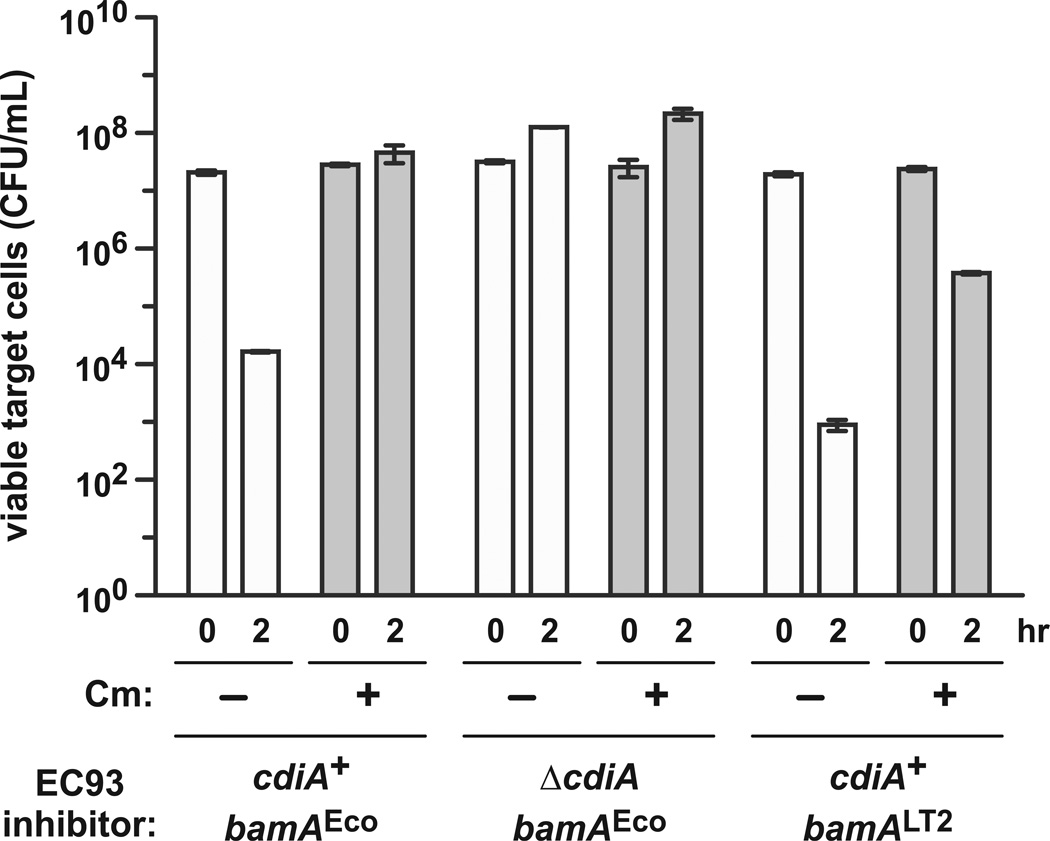
Previous work showed that chloramphenicol (Cm)-treated CDI+ cells are unable to inhibit target bacteria (Aoki et al., 2005). This finding suggests that CdiA synthesis is mechanistically linked to toxin delivery and that active secretion of CdiA may provide the energy to translocate toxins into target cells. We revisited these experiments to determine whether protein synthesis does in fact provide the driving force for CDI toxin delivery. We co-cultured wild-type E. coli isolate EC93 with E. coli MC4100 target cells in shaking broth for two hr, which resulted in a ~1,000-fold decrease in viable targets (Fig. 1). This inhibition is attributable to CDI, because the growth of target cells was unaffected during co-culture with E. coli EC93 ΔcdiA mutants (Fig. 1). We next examined the effect of protein synthesis inhibitors on CDI. We pre-treated E. coli EC93 cells with Cm for 20 min to block protein synthesis, then mixed the inhibitor cells with Cm-resistant E. coli MC4100 target cells in Cm-supplemented broth. Viable target cells increased ~1.6-fold under these conditions (Fig. 1), demonstrating that CDI is attenuated during translational arrest. However, we note that target cells grew ~8.5-fold when co-cultured with ΔcdiA mock inhibitors under the same Cm-treatment regimen (Fig. 1). Although this difference in target-cell growth is not statistically significant (p = 0.0934), it suggests that Cm-treated E. coli EC93 cells retain residual inhibition activity and that active protein synthesis may not be required for toxin delivery. Because CDI+ cells deliver toxins to one another (Webb et al., 2013), we reasoned that this exchange between inhibitors could deplete cell-surface CdiA during Cm-pretreatment. If true, then the inhibitor cells would lack deployable CdiA molecules when the target cells are introduced, thus explaining the decrease in CDI potency. To test this hypothesis, we replaced the bamAEco receptor gene in E. coli EC93 with bamALT2 from Salmonella Typhimurium LT2. CdiAEC93 does not recognize BamALT2 as a receptor (Ruhe et al., 2013b), so the resulting E. coli EC93 bamALT2 cells cannot deliver toxin to one another. We found that these cells were more potent inhibitors than wild-type E. coli EC93 (Fig. 1), presumably because CdiA-CTEC93 toxin is not “wasted” through delivery to immune sibling cells. We then tested whether Cm blocks the inhibition activity of E. coli EC93 bamALT2 cells. Though inhibition activity was clearly diminished by Cm treatment, viable target cells were reduced ~20-fold compared to mock competitions with E. coli EC93 ΔcdiA cells (Fig. 1). These results demonstrate that active protein synthesis is not required for CDI-mediated growth inhibition.
Figure 1. CDI does not require active protein synthesis in the inhibitor cell.
Cm-sensitive E. coli EC93 inhibitor cells and Cm-resistant E. coli MC4100 target cells were co-cultured for 2 hr and viable target-cell counts determined as colony-forming units (CFU mL−1). Where indicated, inhibitors and targets were treated with Cm for 20 min prior to cell mixing. The cdiA and bamA genotypes for the E. coli EC93 strains are provided under the histogram. Data are presented as the mean ± SEM for two independent experiments.
CDI toxin delivery
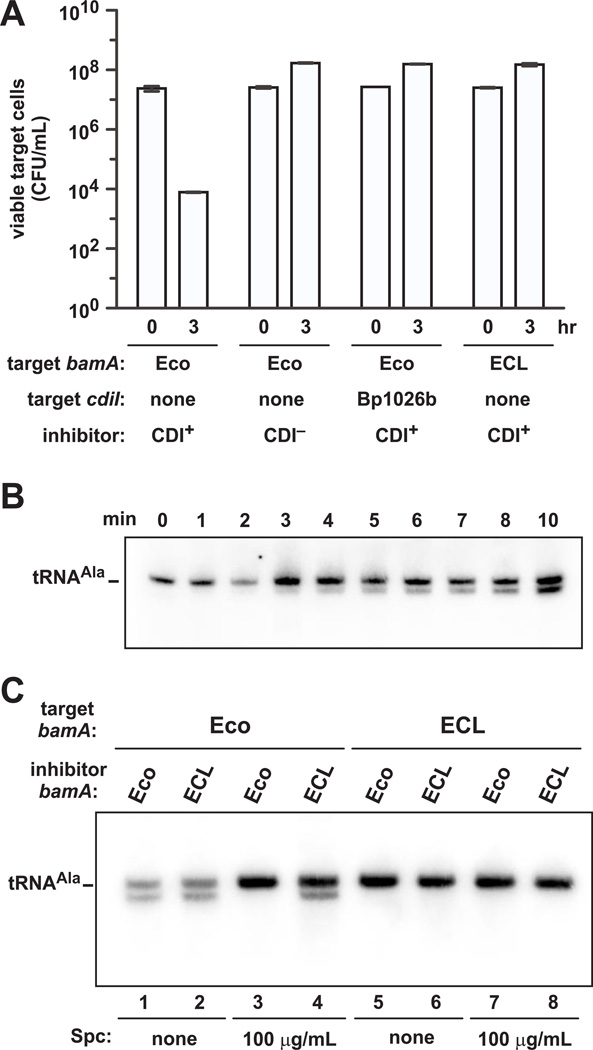
Although ongoing protein synthesis is not required for CDI, actively translating inhibitor cells are clearly more potent than non-translating inhibitors. Moreover, because inhibitors were used in 10-fold excess over target cells in the preceding experiments, it appears that a single delivered toxin domain may be insufficient to kill target bacteria. Therefore, we examined CdiA proteins that deploy RNase domains, reasoning that sub-lethal delivery of these toxins could be easily monitored by northern blot analysis. We chose the tRNase domain from CdiA-CTBp1026b of Burkholderia pseudomallei 1026b as a model because this toxin specifically cleaves tRNAAla molecules (Morse et al., 2012, Nikolakakis et al., 2012). We fused the CdiA-CTBp1026b tRNase domain onto CdiAEC93 and tested the chimeric effector in competition experiments. The CdiAEC93-CTBp1026b chimera was functional and reduced viable target-cell counts ~1,000-fold after three hr of co-culture (Fig. 2A). Furthermore, target bacteria were protected by a plasmid-borne copy of the cdiIBp1026b immunity gene; and cells expressing heterologous bamAECL (from Enterobacter cloacae ATCC 13047) were also resistant (Fig. 2A). Together, these results demonstrate that the CdiA-CTBp1026b tRNase domain inhibits growth and its delivery is dependent upon the BamAEco receptor. We then co-cultured CDIBp1026b inhibitors and target cells at a 1:1 ratio and isolated total RNA for northern blot analysis using a probe to E. coli tRNAUGCAla. Only full-length tRNAUGCAla was detected within the first two min of co-culture, but cleaved molecules accumulated progressively thereafter (Fig. 2B). Much of the tRNAUGCAla remained intact because the inhibitor cells produce CdiIBp1026, which prevents tRNA cleavage in the inhibitor-cell cytoplasm. We also note that although substantial tRNase activity was detected after 10 min of co-culture, there was no loss of target-cell viability at this time (data not shown), suggesting that target cells recover from transient exposure to CdiA-CTBp1026b toxin.
Figure 2. Detection of CdiA-CT toxin delivery into target cells.
A) Chimeric CdiAEC93-CTBp1026b inhibits growth of E. coli target cells. Inhibitor cells (E. coli EPI100 carrying pCH10415) were cultured with E. coli X90 target cells for three hr. Viable target cell counts were determined as CFU mL−1, and data presented as the mean ± SEM for two independent experiments. Target cells expressed the cdiIBp1026b immunity gene or E. cloacae bamA (bamAECL) where indicated. B) CdiA-CTBp1026b delivery during CDI. CDIBp1026b inhibitor cells (EPI100 pCH10415) were cultured with E. coli X90 target cells at a 1:1 ratio in shaking broth. Samples were removed at the indicated times for RNA isolation and northern blot analysis of tRNAUGCAla. C) Active protein synthesis is not required for delivery of the CdiA-CTBp1026b tRNase. CDIBp1026b inhibitor cells (EPI100 pCH10415) and E. coli X90 target cells were co-cultured for two hr, then total RNA was isolated for northern blot analysis of tRNAUGCAla. Where indicated, inhibitor and target cells were pre-treated with Spc for 6 hr, then mixed for 2 hr in media supplemented with Spc. Inhibitor and target cells expressed bamA from either E. coli or E. cloacae (ECL).
We next examined the efficiency of CdiA-CTBp1026 delivery when translation is blocked in inhibitor cells. Because the chimeric CDIBp1026b system resides on a cosmid that also encodes chloramphenicol acetyltransferase, we used spectinomycin (Spc) to arrest protein synthesis in all subsequent experiments. No tRNase activity was detected by northern blot when the inhibitor cells were pre-treated with Spc (Fig. 2C, compare lanes 1 and 3). According to our model, this result reflects the depletion of CdiA during antibiotic pre-treatment. Therefore, we repeated the experiment with bamAECL inhibitors, which cannot exchange toxin with one another. As predicted, cleaved tRNAUGCAla was detected in these latter co-cultures (Fig. 2C, lane 4), indicating that bamAECL inhibitor cells delivered CdiA-CTBp1026b toxin even after pre-treatment with Spc. We also performed control experiments with target cells that express bamAECL (Fig. 2C, lanes 5 – 8), demonstrating that tRNase delivery requires the appropriate CDI receptor. Taken together, these findings suggest that pre-assembled CdiA on the surface of inhibitor cells is competent to deliver toxin into target bacteria.
Uncoupling agents prevent CDI toxin translocation
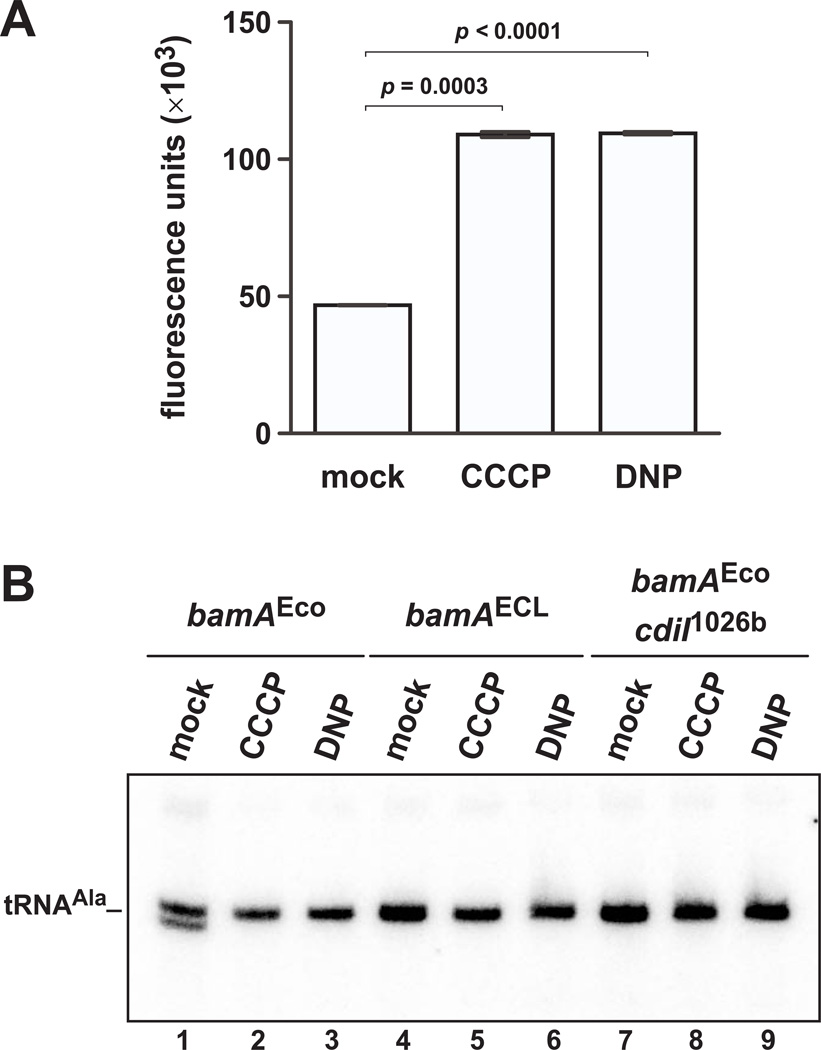
Having excluded co-translational secretion as a mechanism to energize CDI, we next asked whether the proton-motive force (pmf) plays a role in toxin translocation. The pmf powers a variety of cellular activities including ATP synthesis, flagellar rotation and multi-drug efflux (Maloney et al., 1974, Manson et al., 1977, Thanassi et al., 1997). Moreover, many colicin toxins require the pmf to translocate across the outer membrane of E. coli cells (Cascales et al., 2007). Therefore, we tested CDI toxin delivery in the presence of the uncoupling agents carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and dinitrophenol (DNP), which dissipate the pmf by transporting protons across the inner membrane. We first confirmed that the uncoupling agents dissipate the pmf using an ethidium bromide (EtBr) exclusion assay. EtBr permeates bacteria and is highly fluorescent when bound to intracellular nucleic acids. However, E. coli exports EtBr using pmf-dependent efflux pumps, and therefore whole-cell fluorescence is inversely related to the pmf. We pre-treated E. coli X90 cells with Spc for 20 min, then measured EtBr fluorescence in the presence of CCCP or DNP. As expected, whole-cell fluorescence was significantly increased when cells were treated with the uncoupling agents (Fig. 3A), consistent with disruption of the pmf. We then examined the effects of CCCP and DNP on CdiA-CTBp1026b toxin delivery to E. coli X90 target cells. Inhibitors and targets were each pre-treated with Spc for 20 min, then mixed at a 1:1 ratio in media supplemented with uncoupling agent. Toxin activity was observed in the mock-treated control, but no cleaved tRNAUGCAla was detected in either the CCCP- or DNP-treated co-cultures (Fig. 3B, lanes 2 and 3). Moreover, RNA from CCCP- and DNP-treated cultures resembled samples from target cells that lack the CDI receptor (Fig. 3B, lanes 4 – 6), and targets that express the cdiIBp1026b immunity gene (Fig. 3B, lanes 7 – 9). Together, these results indicate that uncoupling agents interfere with toxin activity in target cells, perhaps by blocking CdiA-CTBp1026b delivery.
Figure 3. Proton ionophore uncoupling agents prevent CDI toxin delivery.
A) E. coli X90 cells were treated with Spc and CCCP or DNP for 20 min. Cells were resuspended in 15 PBS supplemented with Spc, CCCP/DNP and EtBr, and whole-cell fluorescence was measured. Fluorescence is expressed in arbitrary units and data are presented as the mean ± SEM for two independent experiments. B) CDIBp1026b bamAECL inhibitors (CH9591 pCH10415) and E. coli X90 target cells were treated with Spc and CCCP/DNP for 20 min, then the cells were mixed for one hr. Total RNA was isolated for northern blot analysis of tRNAUGCAla. Target cells expressed cdiIBp1026b immunity and E. coli or E. cloacae (ECL) bamA as indicated above the blot.
CDI toxin translocation is independent of ATP levels
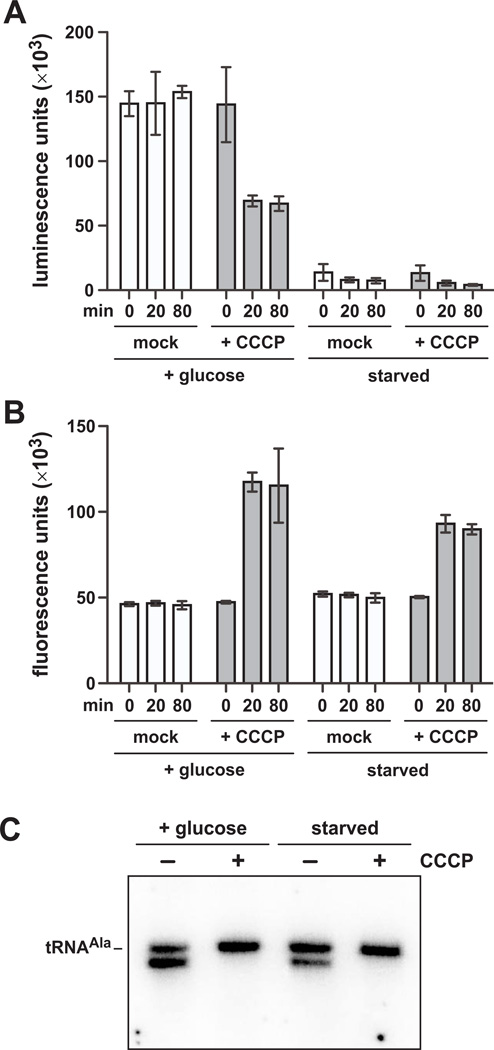
Uncoupling agents also reduce cellular ATP pools. Therefore, if toxin delivery is dependent on ATP hydrolysis, then CCCP/DNP may block CDI by depleting ATP rather than by dissipating the pmf. To address this possibility, we tested whether toxin delivery is directly correlated with ATP levels. We used X90 ΔatpA targets for these experiments because this mutant retains the pmf under starvation (low-ATP) conditions. We first examined the effects of CCCP on well-fed X90 ΔatpA cells in defined media containing glucose as a carbon source. We pre-treated cells with Spc for 20 min, then added CCCP for an additional 1 hr incubation. ATP levels and pmf were measured at 0, 20 and 80 min during the time course. Immediately after CCCP addition (at 20 min), ATP levels decreased to ~50% of untreated cells and remained essentially constant for the duration of the incubation (Fig. 4A). This decrease in ATP was mirrored by a loss of pmf as assessed by EtBr exclusion (Fig. 4B). To monitor toxin delivery under these conditions, we took mock- and CCCP-treated cells (from the 20 min time point) and mixed them at a 1:1 ratio with CDIBp1026b inhibitors. As expected, toxin activity was detected in the mock control, but not in the CCCP-treated co-cultures (Fig. 4C). We then repeated the experiment using starved X90 ΔatpA target cells, which showed a 20-fold reduction in ATP compared to glucose-fed bacteria but fully retained the pmf (Figs. 4A & 4B). Even though ATP was much reduced in these starved cells, CdiA-CTBp1026b toxin activity was readily detected when mixed with inhibitors (Fig. 4C). Treatment of the starved target cells with CCCP reduced ATP levels further and also blocked toxin delivery (Figs. 4A & 4C). Together, these data show that CdiA-CTBp1026b toxin can be translocated over a wide range of ATP levels, but its delivery is blocked when the pmf is dissipated.
Figure 4. CDI toxin delivery is not dependent upon ATP.
A) Glucose-fed and carbon-starved E. coli X90 ΔatpA cells (CH12179) were treated with Spc for 20 min, then CCCP was added (where indicated) at the 0 min time point. Relative ATP levels were determined as described in Methods. B) Cells from panel A were also assayed for EtBr exclusion to determine the status of the pmf. C) Cell aliquots were taken from the 20 min time point and mixed with CDIBp1026b bamAECL inhibitors (CH9591 pCH10415) in Spc-supplemented M9 medium for one hr. Total RNA was isolated for northern blot analysis of tRNAUGCAla.
The target-cell pmf is required for CDI toxin translocation
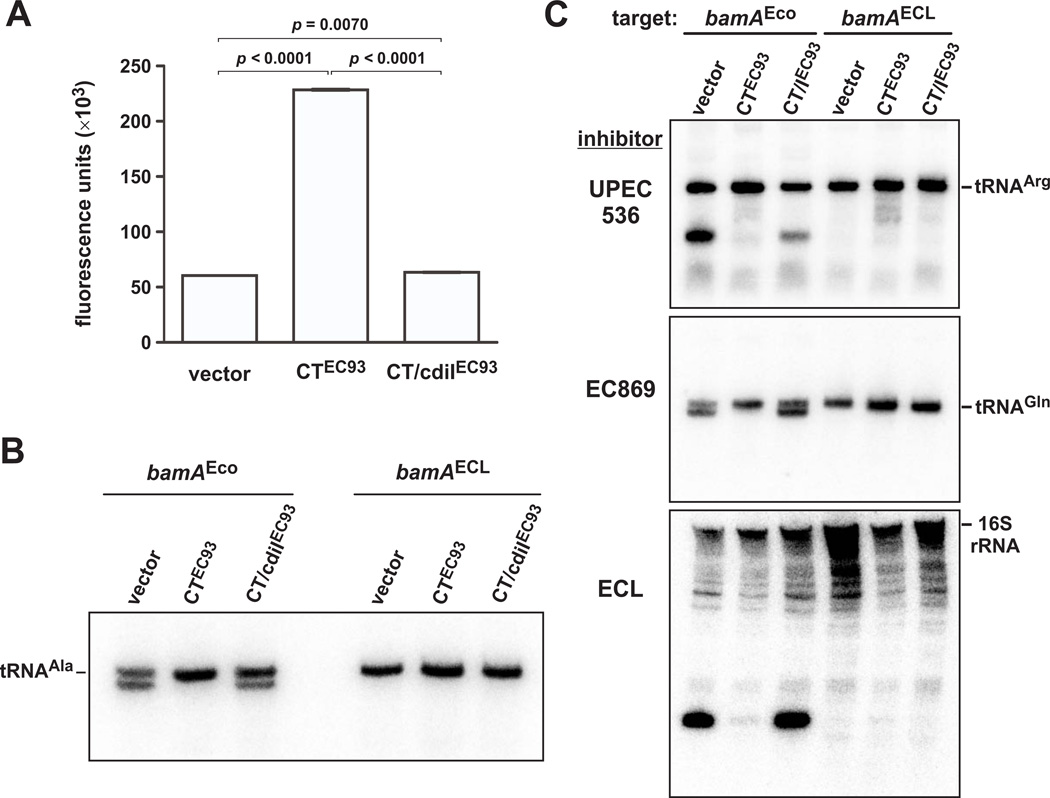
Because CCCP and DNP affect inhibitor cells as well as target cells, it is not clear whether the pmf is required in one or both cell populations. Based on the colicin paradigm, we hypothesized that the target-cell pmf is most critical for CDI toxin translocation. Therefore, we sought to dissipate the pmf specifically in target cells. Our previous work shows that the CdiA-CTEC93 ionophore rapidly dissipates the pmf when expressed inside E. coli (Aoki et al., 2009), thus providing a strategy to modulate the pmf in target cells. Notably, CdiA-CTEC93 expression is not immediately lethal, and inhibited cells can be revived hours later through expression of the cognate CdiIEC93 immunity protein (Aoki et al., 2009). We generated E. coli X90 target cells that harbor a cdiA-CTEC93 expression plasmid and confirmed that EtBr efflux was impaired when the cells expressed the ionophore (Fig. 5A). As a control, we also tested cells that co-express the cdiIEC93 immunity gene with cdiA-CTEC93 and found that EtBr efflux was unperturbed in these cells (Fig. 5A). We then used the cdiA-CTEC93 and cdiA-CT/cdiIEC93 expressing cells as targets in co-cultures with CDIBp1026b inhibitors and assessed toxin delivery by northern blot. No toxin activity was detected in cdiA-CTEC93-expressing target cells, but cleaved tRNAUGCAla accumulated when targets co-expressed cdiA-CTEC93 and cdiIEC93 (Fig. 5B). Taken together with the uncoupling agent experiments, these results suggest that the target-cell pmf is critical for CdiA-CTBp1026b delivery.
Figure 5. The target-cell pmf is required for CDI toxin delivery.
A) Expression of cdiA-CTEC93 and cdiA-CTEC93/cdiIEC93 was induced in E. coli X90 with L-arabinose for one hr, and then the cells were treated with Spc for 20 min. Treated cells were resuspended in 15 PBS supplemented with Spc and EtBr and whole-cell fluorescence was measured. B) CDIBp1026b bamAECL inhibitors (CH9591 pCH10415) were treated with Spc for 20 min prior to mixing with target cells. Target E. coli X90 expressing cdiA-CTEC93 or cdiA-CTEC93/cdiIEC93 were prepared as described in panel A before mixing with inhibitor cells in media supplemented with Spc. After 1 hr, total RNA was isolated for northern blot analysis of tRNAUGCAla. Target cells expressed bamA from E. coli or E. cloacae (ECL) as indicated. C) Target-cell pmf is required for other CDI toxins. Inhibitor and target cells were treated in the same manner described in panel B, but with EPI100 bamAECL (CH9591) inhibitor cells that deploy CdiA-CTUPEC536 (pCH10540), CdiA-CTEC869 (pCH10525), or CdiA-CTECL (pCH10445). Total RNA was isolated for northern blot analysis of tRNAICGArg, tRNACUGGln and 16S rRNA, respectively.
CDI toxins have diverse primary sequences, therefore we asked whether other CdiA-CTs also require the pmf. We chose toxins from UPEC 536, enterohemorrhagic E. coli EC869 and Enterobacter cloacae ATCC 13047, which are not related in primary sequence (Fig. S1) and have distinct RNase activities. CdiA-CTUPEC536 is a tRNA anticodon nuclease (Aoki et al., 2010, Diner et al., 2012), CdiA-CTEC869 cleaves near the 3′-end of tRNAGln, and CdiA-CTECL cleaves a 3′-fragment from 16S ribosomal RNA (rRNA) (Beck et al., 2014). We fused each cdiA-CT/cdiI coding sequence to cdiAEC93 to generate chimeric CDI systems and tested for toxin activities in co-cultures. The resulting chimeras were functional in toxin delivery based on the appearance of cleaved substrate RNA in co-cultures (Fig. 4C). Additionally, RNase activity was not detected with target cells that express bamAECL (Fig. 5C), indicating that toxin delivery requires the BamAEco receptor. As with CdiA-CTBp1026b delivery, we did not detect toxin activity when the target-cell pmf was disrupted through internal cdiA-CTEC93 expression (Fig. 5C). However, cleaved RNAs were apparent when the target cells co-expressed cdiA-CTEC93 and cdiIEC93 (Fig. 5C). These observations suggest that the target-cell pmf is required for the delivery of several different CDI toxins.
Target-cell pmf is not required for cell-cell adhesion during CDI
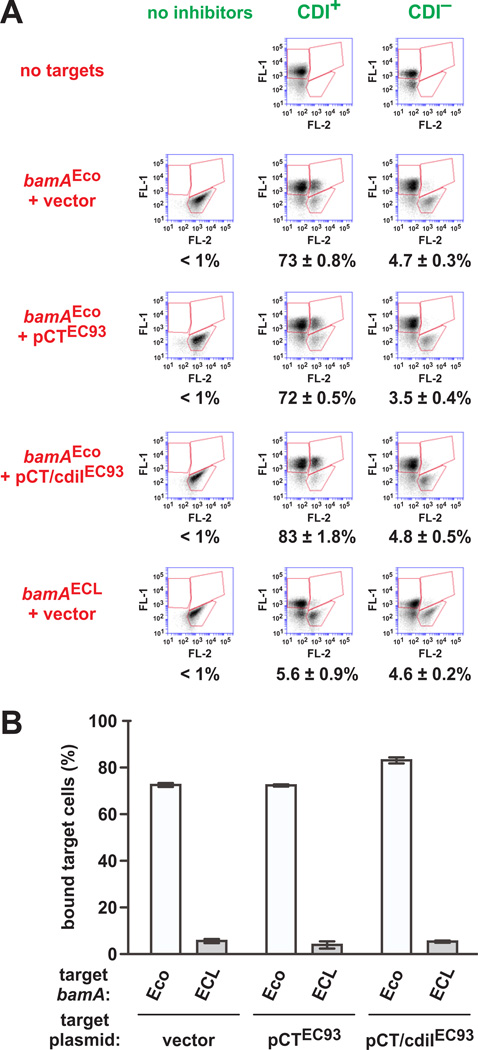
Abrupt changes in the pmf can evoke an envelope-stress response in bacteria (Darwin, 2013). In principle, this response could alter the target-cell envelope and interfere with cell-cell binding during CDI. Therefore, we used a previously established flow-cytometry assay (Aoki et al., 2005, Ruhe et al., 2013b) to determine whether cell-cell binding is disrupted when the target-cell pmf is dissipated. GFP-labeled inhibitor cells bind to DsRed-labeled E. coli X90 targets to form stable aggregates with both green and red fluorescence (Fig. 6A). Under these conditions, approximately 73% of target bacteria were bound to CDI+ inhibitor cells (Figs. 6A & 6B). By contrast, CDI− mock inhibitors did not aggregate with target cells (Fig. 6A), indicating that CdiAEC93 is required for cell-cell binding. Moreover, CDI+ inhibitors did not adhere to targets expressing bamAECL (Figs. 6A & 6B), indicating that cell-cell adhesion reflects specific binding interactions between CdiAEC93 and E. coli BamA. We then expressed cdiA-CTEC93 in target cells to dissipate the pmf and found that these cells aggregated with inhibitors to the same extent as those with an intact pmf (Figs. 6A & 6B). Target cells that express both cdiA-CTEC93 and the cdiIEC93 immunity gene also aggregated with inhibitor cells at a comparable level (~80%) (Figs. 6A & 6B). Together, these experiments demonstrate that dissipation of the target-cell pmf has no discernable effect on adhesion to CDI+ inhibitor cells.
Figure 6. The pmf is not required for adhesion of CDI+ inhibitors to target cells.
A) GFP-labeled EPI100 inhibitors (CDI+) or mock inhibitors (CDI−) and DsRed-labeled EPI100 target strains were treated with Spc for 20 min, then mixed at a 5:1 ratio for analysis by flow cytometry. Prior to Spc-treatment, cdiA-CTEC93 or cdiA-CTEC93/cdiIEC93 expression was induced with L-arabinose for one hr. Target cells also expressed bamA from E. coli (Eco) or E. cloacae (ECL) as indicated. FL-1 and FL-2 represent green and red fluorescence, respectively. The upper-left bin corresponds to GFP+ inhibitors, the lower-right bin to DsRed+ targets, and the upper-right bin to GFP+ DsRed+ cell aggregates. The percentage of target-cell events associated with green fluorescence is provided under the raw data. B) Inhibitor-target cell binding was quantified as the percentage of target cells bound to inhibitors. Data in both panels are expressed as the mean ± SEM for two independent experiments.
CDI toxin translocation is independent of TolA and TonB
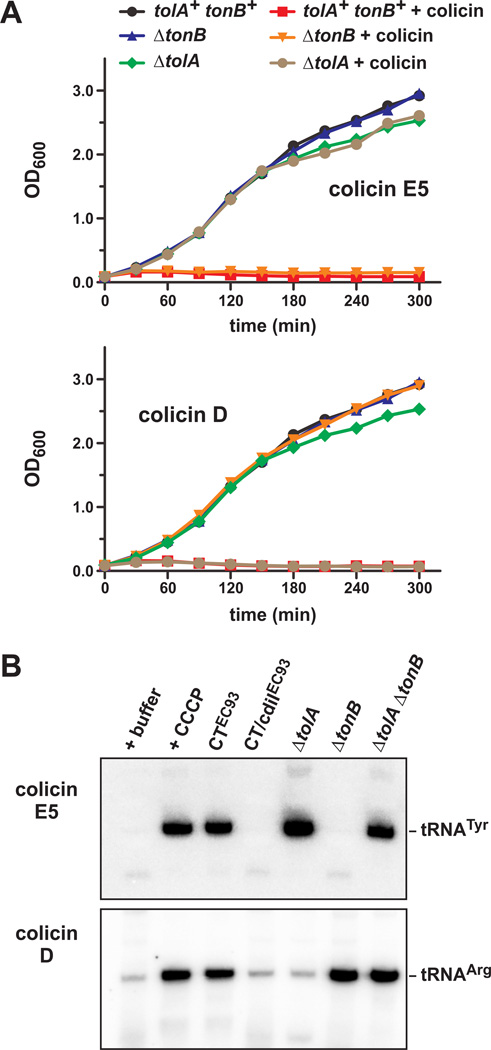
Given that cell-cell adhesion is maintained when the target-cell pmf is disrupted, we next asked whether CDI toxins use the pmf to translocate across the outer membrane similar to colicins. Group B colicin import requires the Ton system, which transduces energy from the pmf to the outer membrane (Braun et al., 1980, Cascales et al., 2007). Tol proteins are also energized by the pmf and may function in the same manner to translocate group A colicins across the outer membrane (Cascales et al., 2007, Vankemmelbeke et al., 2009). We generated E. coli X90 ΔtolA and ΔtonB mutants and tested them for resistance to colicin E5 (group A) and colicin D (group B). As expected, ΔtolA cells were resistant to colicin E5 but killed by colicin D; and conversely, ΔtonB cells were resistant to colicin D but killed by colicin E5 (Fig. 7A). We also isolated RNA from the colicin-treated cells and examined nuclease activity by northern analysis. We found that tRNAGUATyr was degraded in colicin E5-treated cells and tRNAICGArg was degraded in colicin D-treated cells (Fig. 7B), consistent with the known activities of these bacteriocins (Ogawa et al., 1999, Tomita et al., 2000). However, colicin D activity was not detected in ΔtonB mutants and colicin E5 activity was not detected in ΔtolA cells (Fig. 7B). Further, we also confirmed that the pmf is required for colicin translocation. Colicin nuclease activity was not detected in cells treated with CCCP nor in cells that express cdiA-CTEC93 (Fig. 7B). These results are in accord with the previously established roles of TolA, TonB and the pmf in colicin-mediated cell killing.
Figure 7. Colicin import is blocked by cdiA-CTEC93 expression.
A) E. coli X90 cells were treated with colicin E5 (group A) or colicin D (group B) and growth monitored by measuring the optical density at 600 nm (OD600). Strains carried ΔtolA or ΔtonB deletion mutations where indicated. B) E. coli X90 cells were treated with Spc and CCCP for 20 min prior to the addition of colicins E5 and D. Expression of cdiA-CTEC93 and cdiA-CTEC93/cdiIEC93 was induced in E. coli X90 cells for one hr, then treated with Spc for 20 min prior to the addition of colicins. All cells were colicin-treated for 1 hr and RNA isolated for northern blot analysis of tRNAGUATyr (cleaved by colicin E5) and tRNAICGArg (cleaved by colicin D).
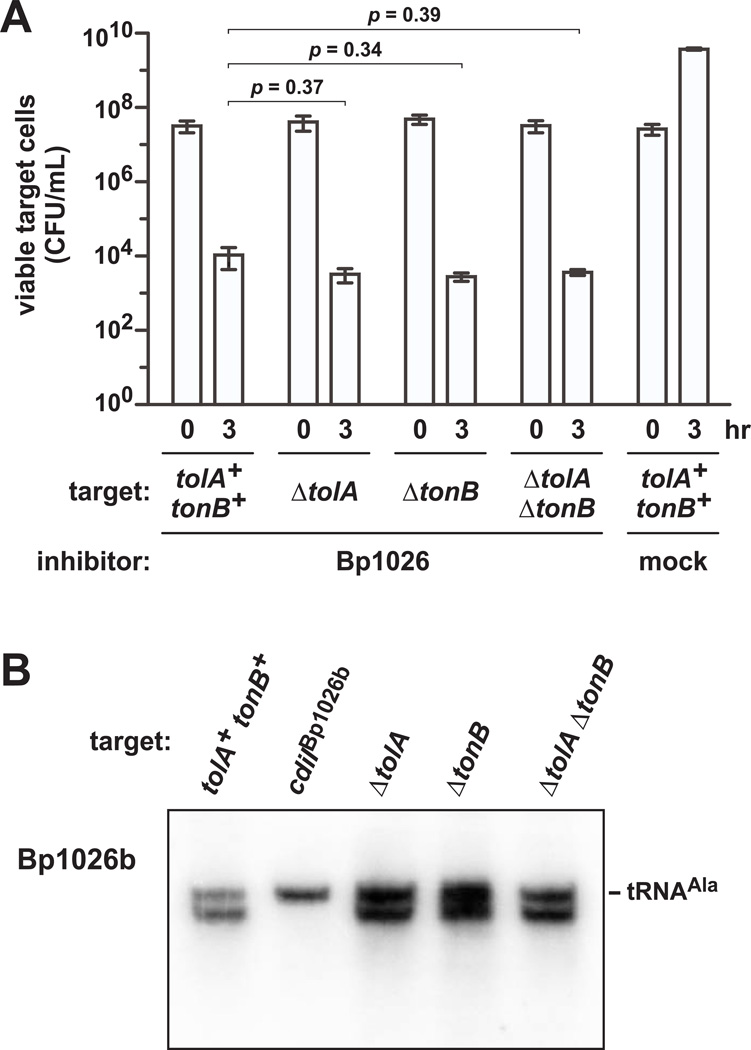
We next tested X90 ΔtolA and ΔtonB target cells in competition co-cultures with CDIBp1026b inhibitor cells and found that each mutant was inhibited to the same extent as wild-type tolA+ tonB+ targets (Fig. 8A). We also examined the ΔtolA ΔtonB double mutant to uncover possible redundancy between the import pathways, but found that these targets were also fully sensitive to CDIBp1026b (Fig. 8A). In accord with the observed growth inhibition, northern blot analysis detected cleaved tRNAUGCAla in each competition (Fig. 8B). Thus, TolA and TonB are not required for CdiA-CTBp1026b toxin delivery. We extended this analysis to test eight additional CDI toxins (Fig. S1). These toxins have ionophore activity (CdiA-CTEC93), tRNase activity (CdiA-CTUPEC536 and CdiA-CTEC869), 16S rRNase activity (CdiA-CTECL), and DNase activity (CdiA-CTDd3937 and CdiA-CTo11EC869). We also tested inhibitors that deploy CdiA-CTo5EC869 and CdiA-CTo10EC869 (Fig. S1), for which the toxin activities are unknown. These competition experiments revealed that ΔtolA ΔtonB cells are fully sensitive to each of the tested CDI toxins (Fig. S2). Taken together, these results demonstrate that the target-cell pmf is harnessed for CDI toxin import using a TolA- and TonB-independent pathway.
Figure 8. CDI toxin delivery is independent of TolA and TonB.
A) E. coli X90 ΔtolA and ΔtonB target cells were co-cultured for three hr with CDIBp1026b inhibitors (EPI100 pCH10415). Viable target cell counts were determined as CFU mL−1, and the data expressed as the mean ± SEM for two independent experiments. Two-tailed p-values from unpaired t tests are reported. B) CDIBp1026b bamAECL inhibitors (CH9591 pCH10415) and E. coli X90 targets were treated with Spc for 20 min, then mixed together for 1 hr in media supplemented with Spc. Total RNA was isolated and analyzed by northern blot using a probe to tRNAUGCAla. Where indicated, the target cells expressed the cdiIBp1026 immunity gene.
The pmf is required to transport CDI toxins across the inner membrane of target cells
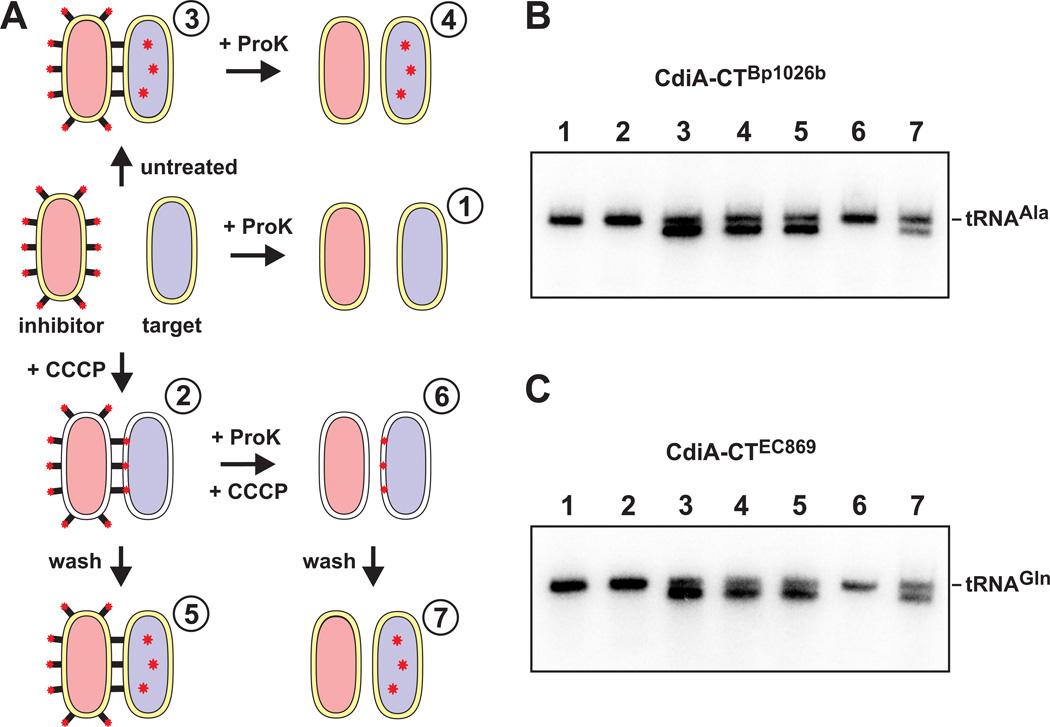
Because CDI is independent of Tol- and Ton-mediated import pathways, we considered the possibility that another uncharacterized mechanism may energize toxin translocation. To test whether the pmf is required for transport across the outer membrane, we used extracellular proteinase K treatment to monitor toxin delivery. We first established that pre-treatment of inhibitor and target cells with proteinase K blocks toxin delivery (Fig. 9A, condition 1 & Fig. 9B, lane 1), presumably through the degradation of surface-exposed CdiA and/or BamAEco. Consistent with this hypothesis, we found that pre-formed inhibitor-target cell aggregates were readily disrupted by proteinase K treatment (Fig. S3). Together, these results indicate that CdiA-BamAEco interactions are sensitive to extracellular proteinase K. Next, we asked whether proteinase K interferes with the activity of delivered toxin. We mixed CDIBp1026b bamAECL inhibitors with target cells in Spc-supplemented media for 20 min (Fig. 9A, condition 3), then added proteinase K to disaggregate the cells (Fig. 9A, condition 4). Northern analysis showed that toxin delivery was essentially complete (Fig. 9B, compare lanes 3 and 4), indicating that delivered toxin is protected from the protease. We then tested whether CCCP blocks the transfer of toxin to a protease-insensitive compartment. We mixed the same inhibitor and target cells in Spc/CCCP-supplemented medium for 20 min and re-confirmed that toxin delivery was blocked under these conditions (Fig. 9A, condition 2 & Fig. 9B, lane 2). However, CCCP does not irreversibly block toxin delivery, because the treated cells showed tRNase activity when re-suspended in Spc-supplemented medium that lacks CCCP (Fig. 9A, condition 5 & Fig. 9B, lane 5). This latter result demonstrates that toxin delivery can resume once the pmf is reestablished. We next treated the Spc/CCCP cell suspension with proteinase K to disrupt cell-cell interactions (Fig. 9A, condition 6), then restored the pmf by re-suspending the cells in Spc-supplemented medium without CCCP (Fig. 9A, condition 7). Remarkably, we detected tRNase activity in the CCCP/proteinase K-treated cells after the pmf was restored (Fig. 9B, compare lanes 6 and 7). Because CCCP prevents toxin delivery into the target-cell cytoplasm, these results suggest that CdiA-CTBp1026b resides within another protease-insensitive compartment, presumably the target-cell periplasm, during these treatments. In this model, periplasmic toxin would continue along its translocation pathway into the cytoplasm once the pmf is restored. To determine whether other CDI toxins are also transferred to the periplasm of de-energized target bacteria, we performed the same series of experiments using CDIEC869 inhibitors, which deploy a different tRNase toxin (see Fig. S1) that specifically cleaves tRNAGln. As expected, toxin activity was not detectable when inhibitors and targets were pre-treated with proteinase K (Fig. 9C, lane 1) or CCCP (Fig. 9C, lane 2). Moreover, removal of CCCP again led to detectable toxin activity in CCCP/proteinase K-treated cell suspensions (Fig. 9C, lane 7). Together, these results suggest that toxin transfer to the periplasm of de-energized target cells is a general feature of CDI.
Figure 9. Transport of CdiA-CT toxin across target-cell outer membranes does not require the pmf.
A) CDIBp1026b bamAECL inhibitors (CH9591 pCH10415) and E. coli X90 target cells were treated with Spc for 20 min, then mixed under six different experimental conditions. Spc was maintain at 100 µg mL−1 throughout all treatments. Condition 1, inhibitors and targets were treated with proteinase K (ProK) prior to cell mixing. Condition 2, inhibitors and targets were mixed in media supplemented with CCCP. Condition 3, inhibitors and targets were mixed for 20 min (untreated). Condition 4, inhibitors and targets were mixed for 20 min, then treated with proteinase K. Condition 5, CCCP-treated cells were washed to restore the pmf. Condition 6, CCCP-treated cells were incubated with proteinase K. Condition 7, CCCP-treated cells were incubated with proteinase K and then washed to restore the pmf. B) Total RNA was isolated from each treatment and tRNAUGCAla analyzed by northern blot. Numbered lanes correspond to the conditions outlined in panel A. C) The experiments were repeated with CDIEC869 bamAECL inhibitors (CH9591 pCH10525) and tRNAGln examined by northern blot to monitor CdiA-CTEC869 activity in target cells. Numbered lanes correspond to the experimental conditions outlined in panel A.
Discussion
The experiments presented here shed light on two fundamental features of CDI. First, it is now clear that ongoing protein synthesis in CDI+ inhibitor cells is not required for toxin delivery. Previous observations suggested that CDI+ inhibitor cells must actively synthesize CdiA to block the growth of target cells (Aoki et al., 2005). Our new data indicate that protein synthesis is only required to replenish effector molecules on the inhibitor-cell surface. Because inhibitor cells express the receptor for CdiA, they freely exchange toxins with one another in shaking broth cultures. After protein synthesis is blocked, toxin-exchange eventually depletes CdiA from the inhibitor-cell surface such that little to no effector remains when target bacteria are introduced to the culture. By contrast, inhibitors lacking the appropriate CDI receptor retain cell-surface CdiA and are able to deliver toxins after protein synthesis is arrested. Thus, once assembled onto the cell surface, CdiA is poised to deliver its toxin domain without further activity on the part of the inhibitor cell. These observations are consistent with a “porcupine” model of CdiA presentation. This model postulates that fully synthesized CdiA filaments (or “quills”) extend from the inhibitor-cell surface to bind receptors on target bacteria. However, we note that active protein synthesis within CDI+ inhibitors may often be required to irreversibly inhibit target bacteria. For example, Fig. 2B shows that a significant proportion of tRNAAla is cleaved after 10 min of co-culture. Because inhibitor and target cells were mixed at a 1:1 ratio, presumably half of the RNA is from inhibitors (which are immune to the toxin) and the other half from targets. Therefore, most of the tRNAAla in the target-cell population is cleaved, indicating that the majority of target cells have received at least one toxin domain after 10 min. However, there is no loss in target-cell viability at this time (data not shown). Together, these data indicate that the delivery of one, or even multiple, toxin domains is insufficient to kill the target cell. This is in sharp contrast to colicins, for which a single delivered molecule is thought to be lethal (Cascales et al., 2007). The second insight is that the target-cell pmf is critical for CDI toxin translocation. The pmf is required for delivery of several different CdiA-CTs, suggesting that energized toxin transport is a general feature of CDI. These results suggest that cells growing exclusively through fermentation should be resistant to CDI. This has implications for when and where CDI is effective. CDI was discovered in E. coli isolate EC93, which was the predominant intestinal E. coli strain present in a rat colony (Aoki et al., 2005). The dominance of E. coli EC93 suggests that CDI provides a significant fitness advantage in this environment. Although the mammalian large intestine is largely anaerobic, E. coli and other enterobacteria are capable of anaerobic respiration using terminal electron acceptors other than oxygen. Thus, most metabolically active enterobacteria are predicted to be susceptible to CDI, provided they express the appropriate surface receptor.
Though colicins and CdiA proteins are not related, they share a number of functional similarities. Both protein families carry variable C-terminal toxin domains and exploit outer-membrane proteins (OMP) as target-cell receptors. Because colicins and CdiA-CT toxins are typically ionophores and nucleases, they must be translocated across the target-cell envelope to reach the inner membrane and cytosol, respectively. Colicins use two general strategies to enter cells. Group A colicins bind to β-barrel OMP receptors, then use a “fishing pole” mechanism to recruit a trimeric porin (usually OmpF) that serves as a translocation conduit (Yamashita et al., 2008, Jakes & Cramer, 2012, Housden et al., 2013, Housden et al., 2010). Translocation also requires Tol proteins, which are thought to directly bind the colicin and facilitate its transit to the cytoplasmic membrane. Group B colicins require only one OMP, which probably serves as both receptor and translocator (Jakes & Finkelstein, 2010, Jakes & Cramer, 2012). Although group B receptors are β-barrel proteins, the central pore is occluded by the N-terminal “plug” domain (Buchanan et al., 1999). There is currently a controversy as to whether the plug domain undergoes conformational changes during group B colicin transport (Devanathan & Postle, 2007, Smallwood et al., 2009). By contrast, CDI toxin translocation is relatively unexplored. Exhaustive genetic selections for CDI-resistant mutants led to the identification of E. coli BamA as the receptor for CdiAEC93 (Aoki et al., 2008), but no other OMPs have been implicated in the CDI pathway. Given that most OMPs are not essential for cell viability, these observations suggest that BamA is the only OMP required for CDI and that it may function as both receptor and translocator. The lumen of the BamA β-barrel is occluded by extracellular loops eL6 and eL4 (Noinaj et al., 2013). Therefore, these loops would have to be displaced to open the barrel aperture for toxin translocation. Intriguingly, eL6 undergoes conformational changes during the BAM activity cycle (Leonard-Rivera & Misra, 2012, Rigel et al., 2013), raising the possibility that CdiA exploits these movements to deliver its toxin. However, BamAEco lacking the POTRA-3 domain supports CDI toxin delivery, even though it is defective in OMP biogenesis (Aoki et al., 2008). Another possible mechanism entails CDI-gated opening of BamA. CdiAEC93 appears to bind directly to BamAEco loops eL6 and eL7 (Ruhe et al., 2013b), and perhaps this interaction triggers conformational changes that allow for CdiA-CT toxin passage.
Our data indicate that colicins and CDI toxins enter target bacteria through distinct pathways. The translocation of group A and B colicins across the outer membrane requires the Tol and Ton systems, respectively. It is generally accepted that energy from the pmf powers group B colicin transport across the outer membrane. Because the pmf exists across the inner membrane, the energy must be transduced to the outer membrane via the TonB-ExbB-ExbD complex. Group A colicins may be translocated in a similar fashion, but there are conflicting reports concerning the energetics of their outer-membrane transport. Early work showed that group A colicins E2 and E3 are rapidly internalized by energized cells, but can be removed from the surface of CCCP-treated bacteria with trypsin (Jetten & Jetten, 1975, Reynolds & Reeves, 1963, Reynolds & Reeves, 1969). These data suggest that the pmf is required translocate surface-bound colicin into the periplasm. However, subsequent studies concluded that TolA need not be energized to transport group A colicins across the outer membrane (Bourdineaud et al., 1990, Goemaere et al., 2007). More recently, Kleanthous and colleagues found that the pmf is critical for the dissociation of the colicin E9-immunity complex during translocation (Vankemmelbeke et al., 2009), arguing that energy is indeed required for this translocation step. We find that ΔtolA ΔtonB mutants are fully sensitive to CDI, indicating that Tol and Ton are not required for CDI toxin translocation. Moreover, our results strongly suggest that the pmf plays no role in CDI toxin translocation across the outer membrane. CDI toxins are transferred to a proteinase K-insensitive compartment when inhibitor and target cells are incubated together in the presence of CCCP. This is in contrast to many colicins, which bind to surface receptors but cannot be internalized in the presence of uncoupling agents (Reynolds & Reeves, 1969, Braun et al., 1980). The simplest explanation for these results is that CdiA-CT toxins are transferred to the target-cell periplasm independent of the pmf. Remarkably, the transferred CDI toxins are competent to resume translocation into the target-cell cytoplasm once the pmf is restored. Thus, CDI toxins can apparently dwell in the periplasm for several minutes prior to transport into the cytoplasm. These findings indicate that CdiA-CT translocation can be separated into two independent membrane-crossing events.
Because outer membrane translocation occurs in the presence of CCCP, we hypothesize that the pmf is required for CDI toxins to cross the target-cell inner membrane. This same general mechanism may be shared with some nuclease colicins. The C-terminal nuclease domains of colicins E3 and E9 associate with anionic membrane lipids and form molten-globule-like structures (Mosbahi et al., 2004, Mosbahi et al., 2006). Kleanthous and colleagues have proposed that the molten-globule state facilitates insertion into membranes and is important for entry into the cytoplasm. The pmf could promote molten-globule formation by lowering the pH of the periplasm. In at least one instance, the nuclease domain forms channels in lipid bilayers, suggesting that it mediates its own passage across the inner membrane (Mosbahi et al., 2002). In other instances, inner membrane transport may be dependent upon FtsH. FtsH is a membrane-bound, ATP-dependent protease that functions primarily in the turnover of integral membrane proteins. Mutant alleles of ftsH (tolZ) confer resistance to some colicins (Matsuzawa et al., 1984, Qu et al., 1996), and de Zamaroczy and colleagues have proposed that FtsH releases the C-terminal toxin domains of colicins through its protease activity (Chauleau et al., 2011). Kleanthous and colleagues have speculated that colicin molten globules may be recognized as misfolded membrane proteins and pulled into the cytoplasm by FtsH (Walker et al., 2007). How the toxins avoid degradation to small peptides during this process is not clear. Intriguingly, FtsH activity is stimulated by the pmf (Akiyama, 2002), suggesting another mechanistic link between toxin import and the energized inner membrane. We have found that E. coli ftsH mutants are resistant to a small subfamily of CDI systems, but are still susceptible to most of the CDI toxins tested here (J.L.E. Willett & C.S.H., unpublished data), indicating that FtsH is not a general mediator of toxin import. Further, AcrB is an inner-membrane protein required for inhibition by E. coli EC93 (Aoki et al., 2008), but it plays no role in other CDI pathways. Together, these observations suggest that multiple import pathways are exploited during CDI. Elucidation of the mechanisms underlying this transport could inform new strategies to deliver small molecules and peptides into Gram-negative bacteria.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains used in this study are shown in Table 1. Bacteria were grown at 37 °C in LB medium or on LB-agar unless otherwise noted. Media were supplemented with antibiotics at the following concentrations: ampicillin (Amp), 150 µg mL−1; kanamycin (Kan), 50 µg mL−1; chloramphenicol (Cm), 66 µg mL−1; and spectinomycin (Spc), 100 µg mL−1. The bamALT2 gene from Salmonella Typhimurium LT2 was introduced into the bamA locus of E. coli EC93 using Red-mediated recombination. Regions upstream and downstream of bamAEco were amplified from EC93 genomic DNA using primer pairs bamA(KO)-Kpn/bamA(KO)-Xho and bamA(KO)-Bam/bamA(KO)-Sac, respectively (all oligonucleotide sequences are given in Table S1). The products were sequentially ligated to pKAN (Hayes & Sauer, 2003) to generate plasmid pKAN-BamA(KO). Next, bamALT2 was amplified with primers Salty-bamA-for-Xho/Salty-bamA-rev-Eco and ligated to pKAN-BamA(KO) using XhoI/EcoRI restriction sites to create pKAN-Salty-BamA(KI). The large SacI/KpnI restriction fragment from plasmid pKAN-Salty-BamA(KI) was electroporated into E. coli EC93 cells expressing the bacteriophage λ Red proteins from plasmid pSIM6 (Datta et al., 2006). Kan-resistant transformants were selected and replacement of the endogenous bamA gene was confirmed by PCR and DNA sequencing. The introduced Kan-resistance cassette was excised using FLP recombinase as described previously (Datsenko & Wanner, 2000). The tolA::kan and tonB::kan mutations were obtained from the Keio collection (Baba et al., 2006) and transduced into strain X90 using bacteriophage P1vir.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| X90 | F′ lacIq lac′ pro′/ara Δ(lac-pro) nal1 argE(amb) rifr thi-1, RifR | (Beckwith & Signer, 1966) |
| EPI100 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacXcZΔM15 ΔlacX recA1 endA1 araD139 Δ(ara, leu) 7697 galU galK ⌊− rpsL nupG, StrR | Epicentre |
| DY378 | W3110 λcI857 Δ(cro-bioA) | (Thomason et al., 2007) |
| EC93 | Escherichia coli isolate from rat feces | (Aoki et al., 2005) |
| CH276 | MC4100 sspB::cat, StrR CmR | (Levchenko et al., 2000) |
| CH2016 | X90 (DE3) Δrna ΔslyD::kan, RifR KanR | (Garza-Sánchez et al., 2006) |
| CH6479 | X90 ΔtolA::kan, KanR | This study |
| CH6480 | X90 ΔtonB::kan, KanR | This study |
| CH9223 | X90 ΔtonB | This study |
| CH9350 | X90 ΔbamA::cat pZS21-bamA+, CmR KanR | (Ruhe et al., 2013b) |
| CH9354 | X90 ΔtonB ΔtolA::kan, KanR | This study |
| CH9371 | X90 ΔbamA::cat pZS21-bamAECL, CmR KanR | (Ruhe et al., 2013b) |
| CH9591 | EPI100 ΔbamA::cat pZS21-bamAECL, CmR KanR | (Ruhe et al., 2013b) |
| CH9604 | EPI100 ΔbamA::cat pZS21-bamA+, CmR KanR | (Ruhe et al., 2013b) |
| CH10053 | EC93 bamALT2 ::kan, KanR | This study |
| CH10094 | EC93 ΔcdiA | This study |
| CH12179 | X90 ΔbamA::cat ΔatpA::kan pZS21amp-bamA+, CmR KanR AmpR | This study |
| Plasmids | ||
| pTrc99a | IPTG-inducible expression plasmid, AmpR | GE Healthcare |
| pCH450 | pACYC184 derivative with E. coli araBAD promoter for arabinose-inducible expression, TetR | (Hayes & Sauer, 2003) |
| pKAN | pBluescript with FRT-flanked kanamycin-resistance cassette ligated into SmaI restriction site, AmpR KanR | (Hayes & Sauer, 2003) |
| pKAN-BamA(KO) | pKAN containing regions upstream and downstream of bamAEco, AmpR KanR | This study |
| pKAN-Salty-BamA(KI) | Integration construct for replacing bamAEco with bamALT2, AmpR KanR | This study |
| pDsRedExpress2 | Constitutive expression of DsRed, AmpR | Clontech |
| pTrc-cdiIBp1026b | IPTG-inducible expression of cdiIBp1026b immunity gene, AmpR | (Nikolakakis et al., 2012) |
| pCP20 | Heat-inducible expression of FLP recombinase, CmR AmpR | (Cherepanov & Wackernagel, 1995) |
| pCH450-cdiA-CTEC93 | Expresses cdiA-CTEC93 toxin encoding residues Val2905 – Lys3132 of CdiAEC93, TetR | (Ruhe et al., 2013b) |
| pCH450-cdiA-CT/cdiIEC93 | Expresses cdiA-CTEC93 toxin and cdiIEC93 immunity gene, TetR | (Ruhe et al., 2013b) |
| pZS21-bamA+ | pZS21 derivative that expresses bamAEco, KanR | (Kim et al., 2007) |
| pZS21-bamAECL | Expresses bamAECL from Enterobacter cloacae ATCC 13047, KanR | (Ruhe et al., 2013b) |
| pET21::colD-imD | Over-production of colicin D and ImD-His6 proteins, AmpR | (Garza-Sánchez et al., 2008) |
| pET21::colE5-imE5 | Over-production of colicin E5 and ImE5-His6 proteins, AmpR | This study |
| pDAL660Δ1-39 | Constitutive expression of cdiAEC93 and cdiIEC93 genes, CmR AmpR | (Aoki et al., 2005) |
| pCH9305 | Constitutive expression of chimeric cdiAEC93 CTo11EC869 and cdiIo11EC869 genes, CmR | (Morse et al., 2012) |
| pCH10163 | Cosmid pCdiA-CT/pheS* that carries a kan-pheS* cassette in place of the E. coli EC93 cdiA-CT/cdiI coding sequence. Used for allelic exchange and counter-selection. CmR KanR | (Morse et al., 2012) |
| pCH10165 | Constitutive expression of chimeric cdiAEC93-CTo5EC869 and cdiIo5EC869 genes, CmR | This study |
| pCH10166 | Constitutive expression of chimeric cdiAEC93-CTo10EC869 and cdiIo10EC869 genes, CmR | This study |
| pCH10415 | Constitutive expression of chimeric cdiAEC93-CTBp1026b and cdiIBp1026b genes, CmR | This study |
| pCH10445 | Constitutive expression of chimeric cdiAEC93-CTECL and cdiIECL genes, CmR | (Beck et al., 2014) |
| pCH10525 | Constitutive expression of chimeric cdiAEC93-CTEC869 and cdiIEC869 genes, CmR | This study |
| pCH10540 | Constitutive expression of chimeric cdiAEC93-CTUPEC536 and cdiIUPEC536 genes, CmR | This study |
| pCH11009 | Constitutive expression of chimeric cdiAEC93-CTDd3937 and cdiIDd3937 genes, CmR | (Webb et al., 2013) |
Abbreviations: AmpR, ampicillin-resistant; CmR, chloramphenicol-resistant; KanR, kanamycin-resistance; RifR, rifampicin-resistant; TetR, tetracycline-resistant
CDI competitions were performed in LB medium with shaking. Cells were first grown to mid-log phase, then mixed at a 10:1 ratio of inhibitor to target cells. Protein synthesis in inhibitor cells was blocked with freshly prepared Cm (100 µg mL−1) or Spc for 20 min prior to and during co-culture with target cells. Antibiotics at these concentrations were sufficient to completely arrest inhibitor-cell growth. Target cell viability was determined by enumerating colony forming units per mL (CFU mL−1) on LB agar supplemented with the appropriate antibiotic.
Plasmids
Plasmids used in this study are listed in Table 1. Chimeric CDI systems were constructed by allelic exchange of the counter-selectable pheS* marker from cosmid pCH10163 as described (Morse et al., 2012, Webb et al., 2013, Beck et al., 2014). Briefly, each cdiA-CT/cdiI pair was amplified by PCR, then joined to upstream and downstream homology fragments from the cdiAEC93 locus using overlapping end-PCR (OE-PCR). Details for all chimeric cdiA constructs are provided in Supplementary Information. The final OE-PCR fusion product (100 ng) was electroporated together with plasmid pCH10163 (300 ng) into E. coli strain DY378 cells (Thomason et al., 2007) and recombinants selected on yeast extract glucose-agar supplemented with 33 µg/mL chloramphenicol and 10 mM D/L-p-chlorophenylalanine. The colE5-imE5 coding sequences were amplified from plasmid ColE5–099 using primers colE5-Nco-for and imE5-Xho-rev. The resulting product was digested with NcoI/XhoI and ligated to pFG21b to generate plasmid pET21::colE5-immE5.
Ethidium bromide efflux
E. coli X90 cells were grown to mid-log phase at 37 °C in LB medium, then treated with 100 µg mL−1 Spc for 20 min prior to the addition of CCCP (100 µM) or DNP (1 mM). Cells were harvested by centrifugation and resuspended in 15 phosphate buffered saline (PBS) supplemented with CCCP or DNP, Spc and 25 µM EtBr. After incubation at room temperature for 15 min, cell fluorescence was measured using a PerkinElmer Victor3V 1420 multi-label counter at 531 nm/595 nm excitation/emission.
ATP assays
The effect of ATP concentration on CDI toxin delivery was assessed using E. coli X90 ΔatpA target cells. Cells were grown to late-log phase in M9 medium supplemented with 0.2% glucose, harvested by centrifugation, then re-suspended in pre-warmed Spc-supplemented M9 medium with or without glucose. The cells were incubated at 37 °C for an additional five hr (to ensure nutrient depletion in the starved cultures), then treated with CCCP for 20 min prior to mixing with inhibitor cells. Inhibitor cells (CH9591 pCH10415) were grown to mid-log phase in M9 medium supplemented with 0.2% glucose, then washed twice in pre-warmed, Spc-supplemented M9 medium. Inhibitors were resuspended in M9 medium supplemented with Spc for 20 min at 37 °C, then CCCP was added 20 min prior to mixing with target cells. Inhibitors and targets were mixed at a 1:1 ratio for 1 hr, then cells were harvested for the isolation of total RNA. Target cell ATP levels were measured using the BacTiter-Glo™ (Promega) reagent using the manufacturer's instructions. Relative ATP levels were measured immediately prior to CCCP addition (defined as 0 min), and after 20 and 80 min of incubation with CCCP. The 20 and 80 min time points correspond to the beginning and end of the co-incubation with inhibitor cells.
Toxin delivery assays
Inhibitor cells were grown to mid-log phase, then treated with 100 µg mL−1 Spc for 6 hr (for Fig. 2C) or 20 min (for all other experiments) prior to mixing at a 1:1 ratio with E. coli X90 target cells in pre-warmed, Spc-supplemented LB medium. To assess the role of the pmf, Spc-treated inhibitor and target cells were incubated with 100 µM CCCP or 1 mM DNP for 20 min prior to mixing at a 1:1 ratio. Target cells carrying pCH450-cdiA-CTEC93 or pCH450-cdiA-CT/cdiIEC93 were induced with 0.2% L-arabinose for one hr prior to the addition of Spc. All co-cultures were incubated for one hr with shaking in medium supplemented with Spc and CCCP or DNP (where indicated). Cultures were poured into an equal volume of ice-cold methanol, cells collected by centrifugation and the cell pellet frozen at –80 °C. Total RNA was isolated with guanidine isothiocyanate-phenol and five µg of each sample analyzed by northern blot as described (Hayes & Sauer, 2003, Garza-Sánchez et al., 2006). Oligonucleotides Ala1B probe, Arg2 probe, Gln2 probe and 16S 3′-probe (Table S1) were radiolabeled and used as probes to detect tRNAUGCAla, tRNAICGArg, tRNACUGGln and 16S rRNA, respectively.
For the proteinase K protection assays, inhibitor and target strains were grown individually to mid-log phase, then treated with Spc for 20 min to arrest translation. Inhibitors and targets were treated with 100 µg mL−1 proteinase K (condition 1), 100 µM CCCP (condition 2) or buffer (condition 3) for 20 min prior to cell mixing. After mixing for 20 min, proteinase K (100 µg mL−1) was added to the buffer- and CCCP-treated co-cultures followed by an additional 20 min incubation. CCCP was removed through centrifugation and re-suspension of cells in pre-warmed, Spc-supplemented LB medium. All co-cultures were harvested into ice-cold methanol for RNA extraction as described above.
Colicin D and colicin E5 were purified as complexes with their His6-tagged cognate immunity proteins as described (Garza-Sánchez et al., 2008). E. coli X90 strains were grown to mid-log phase, then diluted to OD600 = 0.05 in fresh pre-warmed LB medium. Purified colicin/immunity complexes were added to the cultures at a final concentration of 10 nM and cell growth monitored. After one hr of incubation with colicin, the cultures were harvested into an equal volume of ice-cold methanol and cells collected by centrifugation. Total RNA was isolated and analyzed by northern blot using radiolabeled oligonucleotides Tyr2 probe and Arg2 probe as probes to detect tRNAGUATyr and tRNAICGArg, respectively.
Cell-cell adhesion assay
CDI-dependent cell-cell adhesion was measured as described previously (Ruhe et al., 2013b). DsRed-labeled EPI100 target cells were grown to mid-log phase at 37 °C in tryptone broth supplemented with Amp. E. coli X90 target cells were mixed with GFP-labeled E. coli EPI100 (CDI− mock inhibitors) or EPI100 carrying pDAL660Δ1–39 (CDI+ inhibitors) at a 1:5 ratio in tryptone broth and incubated 15 min at 30 °C. The cell suspensions were diluted into 15PBS and analyzed on an Accuri C6 flow cytometer using FL1 (533/30 nm, GFP) and FL2 (585/40 nm, DS-Red) filters (Becton Dickinson). The percentage of target cells bound to inhibitors was estimated by quantifying events with red and green fluorescence divided by total red-fluorescent events.
Supplementary Material
Acknowledgments
We thank Haruhiko Masaki for generously providing plasmid ColE5-099, and Stephanie Aoki, Grant Gucinski and Julia Willett for generating chimeric CDI system plasmids. Z.C.R. was supported by the Tri-Counties Blood Bank Postdoctoral Fellowship. This work was supported by grant GM102318 (D.A.L & C.S.H.) from the National Institutes of Health.
References
- Akiyama Y. Proton-motive force stimulates the proteolytic activity of FtsH, a membrane-bound ATP-dependent protease in Escherichia coli . Proc Natl Acad Sci U S A. 2002;99:8066–8071. doi: 10.1073/pnas.122616899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, Roodenbeke CTde, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, Low DA. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli . Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli . J Bacteriol. 2009;191:1777–1786. doi: 10.1128/JB.01437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J, Schipper K, van Ulsen P, Van der Ende A, Tommassen J. Domain exchange at the 3’ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics. 2013;14:622. doi: 10.1186/1471-2164-14-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, Hayes CS. CdiA from Enterobacter cloacae Delivers a Toxic Ribosomal RNase into Target Bacteria. Structure. 2014;22:707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith JR, Signer ER. Transposition of the lac region of Escherichia coli. I. Inversion of the lac operon and transduction of lac by phi80. J Mol Biol. 1966;19:254–265. doi: 10.1016/s0022-2836(66)80003-7. [DOI] [PubMed] [Google Scholar]
- Bourdineaud JP, Boulanger P, Lazdunski C, Letellier L. In vivo properties of colicin A: channel activity is voltage dependent but translocation may be voltage independent. Proc Natl Acad Sci U S A. 1990;87:1037–1041. doi: 10.1073/pnas.87.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Frenz J, Hantke K, Schaller K. Penetration of colicin M into cells of Escherichia coli . J Bacteriol. 1980;142:162–168. doi: 10.1128/jb.142.1.162-168.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli . Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauleau M, Mora L, Serba J, Zamaroczy Mde. FtsH-dependent processing of RNase colicins D and E3 means that only the cytotoxic domains are imported into the cytoplasm. J Biol Chem. 2011;286:29397–29407. doi: 10.1074/jbc.M111.242354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Darwin AJ. Stress relief during host infection: the phage shock protein response supports bacterial virulence in various ways. PLoS Pathog. 2013;9:e1003388. doi: 10.1371/journal.ppat.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Devanathan S, Postle K. Studies on colicin B translocation: FepA is gated by TonB. Mol Microbiol. 2007;65:441–453. doi: 10.1111/j.1365-2958.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26:515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemaere EL, Cascales E, Lloubes R. Mutational analyses define helix organization and key residues of a bacterial membrane energy-transducing complex. J Mol Biol. 2007;366:1424–1436. doi: 10.1016/j.jmb.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- Housden NG, Hopper JT, Lukoyanova N, Rodriguez-Larrea D, Wojdyla JA, Klein A, Kaminska R, Bayley H, Saibil HR, Robinson CV, Kleanthous C. Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science. 2013;340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden NG, Wojdyla JA, Korczynska J, Grishkovskaya I, Kirkpatrick N, Brzozowski AM, Kleanthous C. Directed epitope delivery across the Escherichia coli outer-membrane through the porin OmpF. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21412–21417. doi: 10.1073/pnas.1010780107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Guerin J, Baelen S, Clantin B. Two-partner secretion: as simple as it sounds? Res Microbiol. 2013;164:583–595. doi: 10.1016/j.resmic.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Jakes KS, Cramer WA. Border crossings: colicins and transporters. Annu Rev Genet. 2012;46:209–231. doi: 10.1146/annurev-genet-110711-155427. [DOI] [PubMed] [Google Scholar]
- Jakes KS, Finkelstein A. The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol. 2010;75:567–578. doi: 10.1111/j.1365-2958.2009.06966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Jetten ME. Energy requirement for the initiation of colicin action in Escherichia coli . Biochim Biophys Acta. 1975;387:12–22. doi: 10.1016/0005-2728(75)90048-1. [DOI] [PubMed] [Google Scholar]
- Jiang X, Payne MA, Cao Z, Foster SB, Feix JB, Newton SM, Klebba PE. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Leonard-Rivera M, Misra R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of beta-barrel outer membrane proteins, including that of BamA itself. J Bacteriol. 2012;194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli . Mol. Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- Maloney PC, Kashket ER, Wilson TH. A protonmotive force drives ATP synthesis in bacteria. Proc Natl Acad Sci U S A. 1974;71:3896–3900. doi: 10.1073/pnas.71.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson MD, Tedesco P, Berg HC, Harold FM, Drift CVan der. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa H, Ushiyama S, Koyama Y, Ohta T. Escherichia coli K-12 tolZ mutants tolerant to colicins E2, E3, D, Ia, and Ib: defect in generation of the electrochemical proton gradient. J Bacteriol. 1984;160:733–739. doi: 10.1128/jb.160.2.733-739.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbahi K, Lemaitre C, Keeble AH, Mobasheri H, Morel B, James R, Moore GR, Lea EJ, Kleanthous C. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat Struct Biol. 2002;9:476–484. doi: 10.1038/nsb797. [DOI] [PubMed] [Google Scholar]
- Mosbahi K, Walker D, James R, Moore GR, Kleanthous C. Global structural rearrangement of the cell penetrating ribonuclease colicin E3 on interaction with phospholipid membranes. Protein Sci. 2006;15:620–627. doi: 10.1110/ps.051890306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbahi K, Walker D, Lea E, Moore GR, James R, Kleanthous C. Destabilization of the colicin E9 Endonuclease domain by interaction with negatively charged phospholipids: implications for colicin translocation into bacteria. J Biol Chem. 2004;279:22145–22151. doi: 10.1074/jbc.M400402200. [DOI] [PubMed] [Google Scholar]
- Nikaido H, Zgurskaya HI. AcrAB and related multidrug efflux pumps of Escherichia coli . J. Mol. Microbiol. Biotechnol. 2001;3:215–218. [PubMed] [Google Scholar]
- Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, Tuanyok A, Keim PS, Peacock S, Hayes CS, Low DA. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature. 2013;501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, Roodenbeke Ct’Kint de, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu JN, Makino SI, Adachi H, Koyama Y, Akiyama Y, Ito K, Tomoyasu T, Ogura T, Matsuzawa H. The tolZ gene of Escherichia coli is identified as the ftsH gene. J Bacteriol. 1996;178:3457–3461. doi: 10.1128/jb.178.12.3457-3461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BL, Reeves PR. Some observations on the mode of action of colicin F. Biochem Biophys Res Commun. 1963;11:140–145. doi: 10.1016/0006-291x(63)90081-0. [DOI] [PubMed] [Google Scholar]
- Reynolds BL, Reeves PR. Kinetics of adsorption of colicin CA42-E2 and reversal of its bactericidal activity. J Bacteriol. 1969;100:301–309. doi: 10.1128/jb.100.1.301-309.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli . Proc Natl Acad Sci U S A. 2013;110:5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013a;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio. 2013b;4 doi: 10.1128/mBio.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood CR, Marco AG, Xiao Q, Trinh V, Newton SM, Klebba PE. Fluoresceination of FepA during colicin B killing: effects of temperature, toxin and TonB. Mol Microbiol. 2009;72:1171–1180. doi: 10.1111/j.1365-2958.2009.06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli . J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. Recombineering: genetic engineering in bacteria using homologous recombination. Current protocols in molecular biology / edited by Frederick M. Ausubel …[et al. 2007 doi: 10.1002/0471142727.mb0116s78. Chapter 1: Unit 1 16. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci U S A. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankemmelbeke M, Zhang Y, Moore GR, Kleanthous C, Penfold CN, James R. Energy-dependent immunity protein release during tol-dependent nuclease colicin translocation. J Biol Chem. 2009;284:18932–18941. doi: 10.1074/jbc.M806149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Walker D, Mosbahi K, Vankemmelbeke M, James R, Kleanthous C. The role of electrostatics in colicin nuclease domain translocation into bacterial cells. J Biol Chem. 2007;282:31389–31397. doi: 10.1074/jbc.M705883200. [DOI] [PubMed] [Google Scholar]
- Webb JS, Nikolakakis KC, Willett JL, Aoki SK, Hayes CS, Low DA. Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS ONE. 2013;8:e57609. doi: 10.1371/journal.pone.0057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli . Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli . Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 2008;27:2171–2180. doi: 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.