Abstract
Most patients with pancreatic ductal adenocarcinoma (PDAC) are diagnosed with advanced disease and survive less than 12 months1. PDAC has been linked with obesity and glucose intolerance2-4, but whether changes in circulating metabolites are associated with early cancer progression is unknown. To better understand metabolic derangements associated with early disease, we profiled metabolites in prediagnostic plasma from pancreatic cancer cases and matched controls from four prospective cohort studies. We find that elevated plasma levels of branched chain amino acids (BCAAs) are associated with a greater than 2–fold increased risk of future pancreatic cancer diagnosis. This elevated risk was independent of known predisposing factors, with the strongest association observed among subjects with samples collected 2 to 5 years prior to diagnosis when occult disease is likely present. We show that plasma BCAAs are also elevated in mice with early stage pancreatic cancers driven by mutant Kras expression, and that breakdown of tissue protein accounts for the increase in plasma BCAAs that accompanies early stage disease. Together, these findings suggest that increased whole–body protein breakdown is an early event in development of PDAC.
PDAC is a leading cause of cancer–related death worldwide, and most patients have incurable disease at diagnosis1. The best–characterized predisposing factors, current tobacco use and a first–degree relative with PDAC, both impart an approximate 1.8–fold increased risk for the disease5,6. These risk factors, however, have thus far provided limited insight into the biology of early disease progression of sporadic tumors. Development and progression of PDAC is associated with altered systemic metabolism including obesity2, glucose intolerance3,4, and cancer-induced cachexia7. Nevertheless, no systematic examination of circulating metabolites has been performed to determine whether altered metabolism may indicate subclinical pancreatic cancer or inform our understanding of early disease progression when interventions might improve patient outcomes.
Prior efforts to identify changes in circulating metabolites related to cancer have employed a cross–sectional design, comparing cancer–free controls to cases with blood samples collected at diagnosis8-10. This approach is problematic for discovery of changes related to early cancer progression, as consequences of advanced disease are likely to impact circulating metabolite profiles. This is particularly true for patients with pancreatic cancer, who commonly have significant anorexia, weight loss, and pancreatic insufficiency at the time of diagnosis1. To investigate how altered metabolism might contribute to pancreatic malignancy, we profiled plasma metabolites in cases with PDAC and matched controls drawn from four prospective cohort studies with blood collected at least two years before cancer diagnosis (Supplementary Table 1). The median time between blood collection and PDAC diagnosis was 8.7 years.
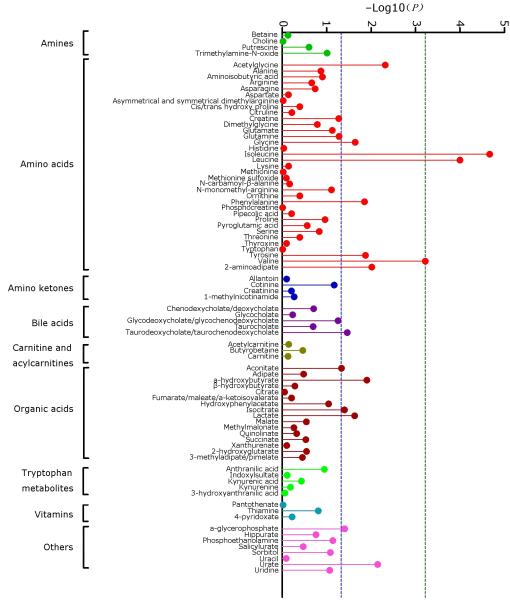
In conditional logistic regression models, 15 plasma metabolites were associated with future diagnosis of PDAC to P<0.05; 3 metabolites, the branched chain amino acids (BCAAs) isoleucine, leucine and valine were significant to P≤0.0006, the predefined significance threshold after correction for multiple–hypothesis testing (Fig. 1 and Supplementary Table 2). To evaluate the magnitude of risk for PDAC diagnosis, we divided participants into quintiles of increasing BCAA levels. Compared to the bottom quintile, subjects in the top quintile had at least a 2–fold increased risk of developing PDAC (Table 1, Supplementary Table 3 and Supplementary Fig. 1). As noted previously11, circulating levels of the three BCAAs were highly correlated (Supplementary Table 4), reflecting their common pathways of metabolism12 and leading to similar results for the sum total of BCAAs (Table 1 and Supplementary Table 3).
Fig. 1. Plasma metabolites and risk of future pancreatic cancer diagnosis.
P–values of the log–transformed, continuous metabolite levels from human plasma comparing pancreatic cancer cases and controls in conditional logistic regression models conditioned on matching factors and adjusted for age at blood draw (years, continuous), fasting time (<4, 4–8, 8–12, ≥12 hours, missing) and race/ethnicity (White, Black, other, missing). The dashed green line indicates the statistically significant P–value threshold after Bonferroni correction for multiple–hypothesis testing, P–trend ≤0.0006 (0.05/83). The dashed blue line indicates P– trend of 0.05.
Table 1.
Odds ratios for pancreatic cancer by prediagnostic plasma levels of branched chain amino acids
| Modela | Isoleucine | Leucine | Valine | Total BCAAsc | ||||
|---|---|---|---|---|---|---|---|---|
| Extreme Quintilesb | Per S.D. | Extreme Quintilesb | Per S.D. | Extreme Quintilesb | Per S.D. | Extreme Quintilesb | Per S.D. | |
| Base model | 2.11 (1.40-3.18) | 1.30 (1.15–1.48) | 2.08 (1.38–3.13) | 1.31 (1.14–1.50) | 2.00 (1.37–2.92) | 1.23 (1.09–1.39) | 2.13 (1.43–3.15) | 1.30 (1.14–1.48) |
| + BMI and physical activity | 2.05 (1.34–3.12) | 1.29 (1.14–1.48) | 2.01 (1.32–3.06) | 1.29 (1.12–1.49) | 1.94 (1.31–2.86) | 1.21 (1.07–1.38) | 2.06 (1.37–3.09) | 1.28 (1.12–1.47) |
| + BMI, physical activity, and reported diabetes at blood collection | 2.00 (1.31–3.05) | 1.28 (1.13–1.46) | 1.97 (1.29–2.99) | 1.28 (1.11–1.48) | 1.90 (1.28–2.81) | 1.20 (1.06–1.37) | 2.01 (1.34–3.03) | 1.27 (1.11–1.46) |
| + BMI, physical activity, reported diabetes, HbA1c, plasma insulin, proinsulin and C–peptide | 1.86 (1.13–3.03) | 1.24 (1.06–1.45) | 1.81 (1.11–2.96) | 1.24 (1.04–1.47) | 1.67 (1.06–2.63) | 1.14 (0.99–1.33) | 1.89 (1.17–3.06) | 1.22 (1.04–1.44) |
| Exclude subjects with reported diabetes or HbA1c ≥6.5% at blood collection | 2.12 (1.37–3.27) | 1.33 (1.16–1.52) | 2.16 (1.39–3.35) | 1.32 (1.14–1.54) | 1.91 (1.28–2.85) | 1.23 (1.08–1.41) | 2.19 (1.44–3.34) | 1.31 (1.14–1.51) |
| Exclude subjects with reported diabetes or HbA1c ≥6.5% at blood collection and those with reported diabetes after blood collection | 2.18 (1.39–3.43) | 1.32 (1.15–1.53) | 2.21 (1.40–3.49) | 1.33 (1.13–1.55) | 1.94 (1.27–2.96) | 1.22 (1.07–1.40) | 2.25 (1.45–3.49) | 1.31 (1.13–1.52) |
Odds ratio (95% confidence interval) from conditional logistic regression models conditioned on matching factors and adjusted for age at blood draw (years, continuous), fasting time (<4, 4–8, 8–12, ≥12 hours, missing) and race/ethnicity (White, Black, other, missing). Subsequent models also adjusted for indicated measure of energy balance, hyperglycemia or insulin resistance: body–mass index (kg/m2, continuous), physical activity (MET–hr/week, continuous), reported diabetes at blood collection (yes/no), Hemoglobin A1c (%, continuous), plasma insulin (μIU/ml, continuous), plasma proinsulin (pM, continuous), and plasma C–peptide (ng/ml, continuous).
Odds ratios (95% CI) for the comparison of the fifth quintile to the first quintile (referent) for the circulating branched chain amino acids
Sum of the concentrations of the three circulating branched chain amino acids, isoleucine, leucine and valine
Abbreviations: BCAAs, branched chain amino acids; BMI, body–mass index; CI, confidence interval; HbA1c, hemoglobin A1c; S.D., standard deviation
Circulating BCAAs are elevated in obese individuals and those with insulin resistance13. In study participants, plasma BCAAs were modestly correlated with markers of energy balance, obesity and glucose intolerance (Supplementary Table 4). To evaluate the independent effect of BCAAs on PDAC risk, we assessed models that included these markers and found that the associations of BCAAs with PDAC remained largely unchanged (Table 1). Circulating levels of BCAAs are also associated with future risk of diabetes11,14. Since type 2 diabetes is a predisposing factor for PDAC15, we questioned whether the intermediate development of diabetes was underlying the BCAA association. Exclusion of subjects with diabetes at blood collection did not change our results (Table 1), indicating that we had not identified a signature of prevalent diabetes associated with later PDAC diagnosis. To determine whether circulating BCAAs were identifying a population at risk for diabetes, who are then at elevated risk of PDAC, we excluded subjects who developed diabetes between the time of blood collection and cancer diagnosis and found the results unchanged (Table 1). These data suggest that the association of BCAAs with future PDAC diagnosis is not dependent on intermediate development of diabetes.
To examine the contribution of circulating BCAAs to risk stratification models for PDAC, we evaluated area under the curve (AUC) of receiver–operating–characteristic (ROC) curves16 and net reclassification improvement (NRI)17 with low–risk and high–risk categories. Compared to the base model, including circulating BCAAs led to a significant increase in AUC (Supplementary Table 5a and Supplementary Fig. 2), and a net 8.2% of cases moving to the high–risk category with a NRI of 5% (Supplementary Table 5b). Thus, in our population, inclusion of circulating BCAAs improved the ability of risk stratification models to identify future PDAC cases.
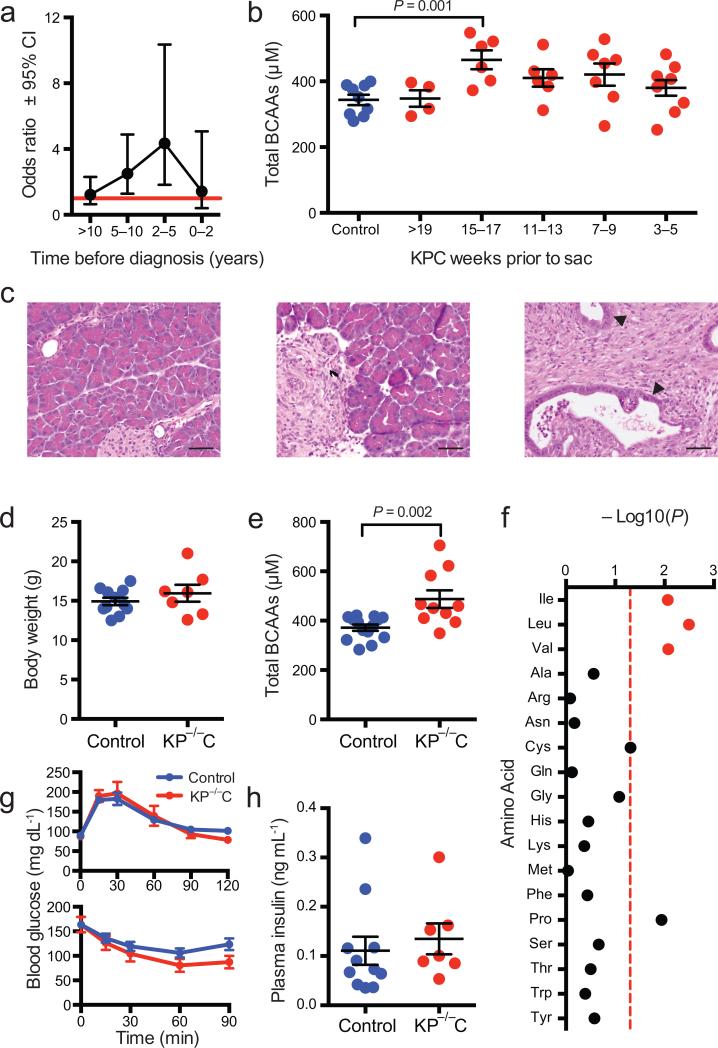
In stratified analyses, we noted no significant differences in the association of BCAAs with PDAC by cohort, sex, smoking status, BMI, or fasting status at blood collection (Supplementary Fig. 3, all P–interaction ≥0.14). To examine when circulating BCAAs were most associated with PDAC, we stratified cases and matched controls by time interval between blood collection and PDAC diagnosis (<2 years, 2–<5 years, 5–<10 years, ≥10 years). These analyses demonstrated particularly strong associations between 2 and 5 years prior to diagnosis (Fig. 2a and Supplementary Table 6).
Fig. 2. Plasma BCAAs are elevated during subclinical disease.
a, Graph of odds ratio (95% confidence interval) for future pancreatic cancer diagnosis among cohort cases and matched controls comparing highest versus lowest quintiles of circulating BCAAs stratified by time from blood collection to the case's cancer diagnosis. Odds ratio (95% confidence interval) from conditional logistic regression models conditioned on matching factors and adjusted for age at blood draw (years, continuous), fasting time (<4, 4–8, 8–12, ≥12 hours, missing) and race/ethnicity (White, Black, other, missing). Red horizontal line marks an odds ratio of 1.0. b. Graph of total plasma BCAAs in KPC mice over time and in littermate controls. Each control data point is an average for one mouse over the course of the study (Supplementary Fig. 3b) and values for KPC mice living longer than 19 weeks are averaged for the >19 weeks time point. For weeks 15-17, n = 6 KPC and n = 9 control, t-test, P=0.001. c, H&E staining of pancreatic tissue obtained from KP–/–C mice and littermate controls at 3-4 weeks of age. Tissue are shown from a control mouse with histologically normal pancreas (left); a KP–/–C mouse with areas of PDAC adjacent to areas of normal pancreas (middle); and a KP–/–C mouse with areas of PDAC and pancreatic intraepithelial neoplasia (closed arrow heads) (right); scale bar = 50μM. d, Mean (±SEM) body weights at 3-4 weeks of age for KP–/–C mice and littermate controls (n = 7 KP–/–C, n = 11 control mice). e, Mean (±SEM) total plasma branched chain amino acid levels from KP–/–C mice and littermate controls at 3-4 weeks of age (n = 10 KP–/–C, n = 14 control mice, t-test, P=0.002). f, P–values for comparison of circulating amino acid levels in KP–/–C mice and littermate controls at 3-4 weeks of age, cys = cystine (n = 10 KP–/–C, n = 14 control mice), The dashed red line indicates P–value of 0.05. g, top, glucose tolerance test in KP–/–C mice and littermate controls at the time of weaning (n = 7 KP–/–C, n = 11 control mice) and bottom, insulin tolerance test in KP–/–C mice and littermate controls at four weeks of age (n = 7 KP–/–C, n = 15 control mice). h, Mean (±SEM) fasting plasma insulin levels from KP–/–C mice and littermate controls at four weeks of age (n = 7 KP–/–C, n = 11 control mice).
Experimental studies indicate years elapse between formation of the initial malignant clone and cancer diagnosis18, suggesting that occult PDAC might be present when we observe the strongest associations with elevated BCAAs. We therefore hypothesized elevated circulating BCAAs were a marker of early PDAC. To test this possibility, we conducted a prospective serial blood sampling study utilizing LSL–KrasG12D/+;LSL–Trp53R172H/+;Pdx–1–Cre (KPC) mice which develop PDAC with variable latency19. KPC mice progress through all histological stages of disease from normal pancreata to invasive adenocarcinoma with a median survival of approximately 21 weeks19 (Supplementary Fig. 4a). KPC mice initially displayed similar BCAA levels to littermate controls, but developed significant elevations from 15–17 weeks prior to sacrifice (Fig. 2b and Supplementary Fig. 4b). These data suggest circulating BCAA elevations accompany early PDAC.
LSL–KrasG12D/+; Trp53flox/flox;Pdx–1–Cre (KP–/–C) mice develop PDAC with more consistent kinetics, displaying precursor lesions (PanINs) with limited invasive cancer by 3–4 weeks of age (Fig. 2c) and a median lifespan of 10-12 weeks20. At 3–4 weeks of age we observed no difference in weight or food consumption between KP–/–C mice and littermate controls suggesting animals with early PDAC had not yet developed overt constitutional symptoms (Fig. 2d and Supplementary Fig. 4c). Consistent with findings in patients and KPC mice, circulating BCAAs were higher in KP–/–C animals with subclinical PDAC when compared with controls (Fig. 2e), a pattern not observed for most other amino acids (Fig. 2f and Supplementary Fig. 4d-e).
We observed no significant differences in fasting blood glucose, response to glucose load, response to insulin challenge, or fasting insulin levels during intraperitoneal glucose and insulin tolerance tests in four–week–old KP–/–C and control mice (Fig. 2g–h and Supplementary 4f-i). These findings argue that BCAA elevations are not reflective of hyperglycemia or insulin resistance and instead represent an early consequence of subclinical PDAC.
We examined whether other malignancies induced by the same genetic lesions caused elevated plasma BCAAs. Cre–recombinase introduction into lung or muscle of mice with the Kras and Trp53 alleles from the KP–/–C model leads to non–small cell lung cancer and sarcoma, respectively21-23. Neither model displayed the BCAA alterations seen with early PDAC (Supplementary Fig. 5). Re-implantation of cell lines derived from the KP–/–C model into immunocompetent syngenic hosts also failed to cause BCAA elevations (Supplementary Fig. 6). These data argue BCAA elevations are associated with early stage autochthonous tumors arising in the pancreas and are not a general feature of Kras–driven cancer. They also suggest re–implanting cells from end–stage disease does not model the early disease state that produces BCAA elevations.
Chronic pancreatitis is a risk factor for PDAC24, and pancreatic inflammation can promote PDAC development and progression in mice25,26. Therefore, we examined whether BCAA elevations might be a cause or consequence of pancreatic inflammation in early disease. Mild, chronic pancreatitis in the absence of tumorigenesis failed to recapitulate BCAA elevations (Supplementary Fig. 7a-h), and prolonged increases in plasma BCAAs from dietary intervention did not cause pancreatic inflammation or pancreatitis (Supplementary Fig. 7i-o). Further studies are needed to understand the relationship between BCAAs and more severe pancreatitis.
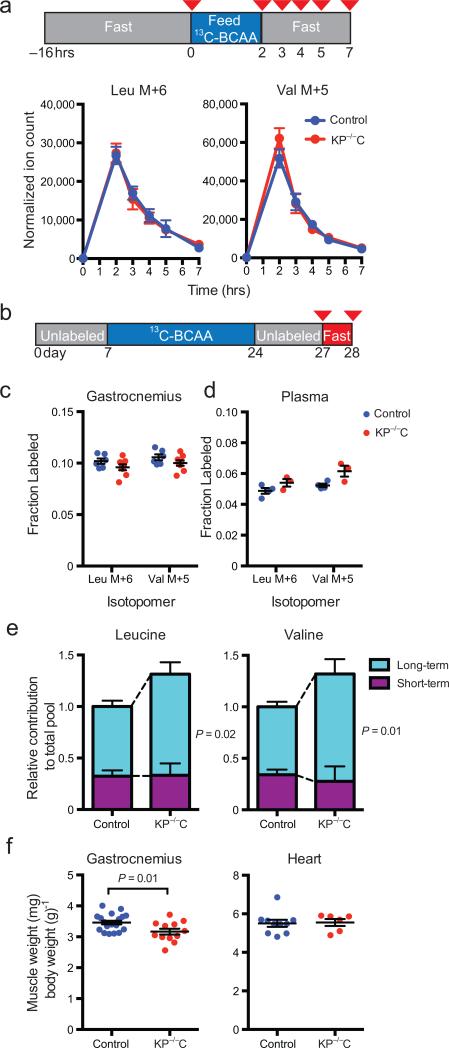
Unlike other amino acids, the liver does not regulate plasma BCAAs levels27,28; instead, levels are determined by dietary uptake, tissue utilization, and breakdown of muscle and other body protein stores27,29. Therefore, plasma BCAAs may originate from short–term pools related to dietary uptake and disposal or long–term pools related to breakdown of tissue proteins. To interrogate the short–term pool, we fed four–week–old KP–/–C and control mice an amino acid defined diet, in which 20% of leucine and valine were 13C–labeled. KP–/–C and control mice consumed similar amounts of food when exposed to labeled diet for two hours, (Supplementary Fig. 8a) and we observed no difference in appearance and disappearance of plasma 13C–label (Fig. 3a and Supplementary Fig. 8b), arguing that gut uptake and peripheral disposal are similar in mice with or without PDAC.
Fig. 3. BCAA elevations are derived from a long–term pool of amino acids.
a, Plasma levels (mean ± SEM) of 13C–labeled leucine and valine normalized to food intake over time following a two–hour exposure to diets containing 13C–labeled leucine and valine. The time points correspond to the red arrowheads in the diagram. b, Diagram of experiment using labeled diets to investigate contributions to plasma BCAA levels from long–term pools. Two cohorts of mice were used for these experiments, one sacrificed in the fed state and a second sacrificed in the fasted state at the time points indicated by the red arrowheads. c, Fractional labeling of total amino acids in protein hydrolysate of gastrocnemius muscle from fasted KP–/–C mice and control littermates (n = 8 KP–/–C, n = 6 control). d, Fractional labeling of plasma amino acids in KP–/–C and control mice in the fed state (n = 3 KP–/–C, n = 4 control). e, The calculated contribution of the short– and long–term BCAA pools to the BCAAs present in plasma . f, Mean (±SEM) gastrocnemius weight (left panel, t-test, P=0.01), a predominantly fast–twitch muscle, and heart weight (right panel) normalized to body weight (n = 6 KP–/–C, n = 10 control).
To determine the contribution of long–term BCAAs pools to plasma levels, we exposed mice to labeled diet during a period of rapid growth in early life, and then switched them to unlabeled diet for three days to chase label from the short–term pool (Fig. 3b). Despite similar peripheral tissue protein labeling (Fig. 3c and Supplementary Fig. 8c), the fraction of labeled BCAAs in plasma was elevated in four–week–old KP–/–C mice relative to littermate controls (Fig. 3d). Furthermore, by comparing plasma label under fed conditions, which is determined by both labeled long–term and unlabeled short–term pools, and fasted conditions, in which only labeled long–term pools contribute, we calculated that increased liberation of BCAAs from long–term body stores was solely responsible for BCAA elevations in KP–/–C mice (Fig. 3d–e and Supplementary Fig. 8d). These data suggest that an early consequence of PDAC is enhanced breakdown of tissue proteins leading to plasma BCAA elevations. Consistent with this hypothesis, KP–/–C mice with early PDAC have smaller fast–twitch muscles with sparing of slow–twitch and cardiac muscle (Fig. 3f and Supplementary Fig. 9). Notably, muscle atrophy associated with prolonged fasting and late–stage malignancy exhibits a similar pattern30-32.
Increased muscle catabolism represents one aspect of cancer–associated cachexia, a wasting syndrome frequently affecting patients with advanced PDAC and contributing to worse outcomes33-36. Our findings, however, suggest that protein breakdown begins much earlier than previously appreciated and pre–dates onset of clinical cachexia. Inflammatory cytokines produced by immune and/or tumor cells have been implicated in cachexia31,37 and the low disease burden at the time of BCAA elevations suggests hormonal factors may be involved here as well. Liberation of tissue amino acids could support the elevated amino acid requirements of pancreatic cancer cells38,39, with BCAAs and/or other amino acids derived from tissue breakdown contributing to disease progression. Because hepatic metabolism maintains relatively constant plasma levels of all amino acids except BCAAs27,29,40, increased liberation of tissue amino acids are expected raise BCAA concentrations in blood. The association between elevated BCAAs and other metabolic disease states11,13,14,41 suggests that plasma BCAAs could be a general marker of increased protein turnover, and elevated BCAAs could contribute to the peri-diagnostic hyperglycemia commonly found in patients with PDAC42.
In participants from four large prospective cohorts, circulating BCAAs were associated with future development of PDAC. We observed similar BCAA elevations in two mouse models of PDAC, and demonstrated these elevations result from breakdown of peripheral protein stores. These findings provide new insight into how early disease impacts whole-body metabolism and suggest that muscle protein loss occurs much earlier in disease progression than previously appreciated.
ONLINE METHODS
HUMAN STUDIES
Study Population
Our study population included pancreatic cancer cases and controls from four prospective cohort studies: Health Professionals Follow–Up Study (HPFS), Nurses’ Health Study (NHS), Physicians’ Health Study I (PHS), and Women's Health Initiative–Observational Study (WHI–OS). HPFS was initiated in 1986 when 51,529 U.S. men 40–75 years working in health professions completed a mailed biennial questionnaire. NHS was established in 1976 when 121,700 female nurses aged 30–55 years completed a mailed biennial questionnaire. PHS is a completed trial initiated in 1982 of aspirin and β–carotene among 22,071 male physicians, aged 40–84 years. After trial completion, study participants were followed as an observational cohort. WHI–OS consists of 93,676 postmenopausal women, aged 50–79 years, enrolled from 1994–1998 at 40 U.S. clinical centers. Participants completed a baseline clinic visit and annual mailed questionnaires. The study was approved by Human Research Committee at Brigham and Women's Hospital (Boston, MA) and participants provided consent.
We included incident pancreatic adenocarcinoma cases diagnosed after blood collection through 2010 with available plasma and no prior history of cancer. Cases were identified by self– report or follow–up of deaths. Deaths were ascertained from next–of–kin, postal service, or National Death Index; this method captures >98% of deaths43. Medical records were reviewed by physicians blinded to exposure data to confirm pancreatic cancer diagnoses. Similar to prior studies in these cohorts4,44-46 and based on a predefined analysis plan, we included only cases diagnosed ≥2 years after blood collection, as the weight loss and insulin resistance which develop due to pancreatic cancer manifest in the 2 years before diagnosis42,47. For each case, we randomly selected two controls, matching on cohort (also matches on sex), year of birth (+/–5 years), smoking status (never, past, current, missing), fasting status (<8 hours, ≥8 hours), and month/year of blood collection (+/–3 months in HPFS, +/– 3 months in NHS, +/–6 months in PHS, and exact matching in WHI). Controls were alive without cancer at the case's diagnosis date and provided a blood sample. Covariate data were obtained from baseline questionnaires in PHS and WHI and questionnaires prior to blood collection in HPFS and NHS, as described previously4,45. In HPFS, NHS, and PHS, cancer stage among cases was directly classified based on medical record review as: (1) local disease amenable to surgical resection; (2) locally advanced disease that is unresectable, but without distant metastases, or (3) distant metastatic disease. In WHI, medical records were coded using Surveillance Epidemiology End Results summary staging, which classifies tumors as localized, regional, or distant. These stages were then classified in the same manner as in HPFS, NHS and PHS, as local, locally advanced and metastatic disease, respectively.
The initial dataset included 454 cases and 908 controls. Seven controls had insufficient plasma for metabolite profiling. One case and control were excluded due to missing data for >10% of metabolites.
Plasma Samples
Blood samples in EDTA tubes were collected from 18,225 men in HPFS from 1993–1995, 14,916 men in PHS from 1982–1984, and 93,676 women in WHI–OS from 1994– 1998, and in Heparin tubes for 32,826 women in NHS from 1989–1990. Comparisons of participant characteristics in the blood collection cohort and the full cohort are provided for each study in Supplementary Table 7. Blood samples in HPFS and NHS were collected by participants, mailed overnight on cold–packs, and then spun to collect and store plasma (delayed processing), while PHS and WHI participants’ whole blood was separately immediately into plasma and stored. An overview of procedures for collection and storage of samples from each cohort is provided below and summarized in Supplementary Table 8.
Health Professionals Follow–up Study
Upon arrival at the blood lab, vials were centrifuged in order to separate the various component parts. Cryo storage tubes were labeled with the appropriate study members ID number and the separated blood components were pipetted into them. This process produced 5 tubes of plasma, 2 tubes of white blood cells, and 1 tube of red blood cells for each cohort member. The tubes were then stored in liquid nitrogen freezers. A bulk tank, holding up to 3,000 gallons of LN2 automatically feeds each individual freezer whenever the freezer's sensors indicate that coolant is required.
Nurses’ Health Study
Blood samples were separated into components (plasma, white blood cells and red blood cells) and pipetted into 8 cryotubes with 5 tubes of plasma, 2 tubes of white blood cells and 1 tube of red blood cells. Samples were immediately frozen in vapor phase liquid nitrogen freezers. The NHS Blood Lab stores all biologic samples associated with the Blood Study in–house in a large liquid–nitrogen (LN2) freezer farm. The cryotubes are stored in the vapor phase of LN2 freezers; the highest freezer temperature is –130°C near the top of the freezer and the lowest temperature is –196°C at the bottom near the liquid nitrogen. All freezers are alarmed and monitored continuously either by NHS laboratory staff or a central security desk (nights and weekends).
Physicians Health Study
Blood collection kits were sent to all participants with instructions to have blood drawn into the EDTA tubes that were provided. Two tubes were centrifuged for plasma, and a third tube was for whole blood. The specimens were received in the laboratory on chill packs within 24 hours of being drawn. Upon receipt, the samples were refrigerated and re–aliquotted into nine 1.2 mL tubes (three whole blood and six plasma), all frozen at –82°C.
Women's Health Initiative
Blood samples were collected on all WHI–OS participants at a baseline clinic visit in the fasting state. Blood samples were maintained at 4°C for up to one hour until plasma or serum was separated from cells. Centrifuged aliquots were put into –70°C freezers within two hours of collection. Samples were shipped frozen by overnight delivery to a central facility and kept within –70°C freezers.
Plasma samples were grouped based on cohort, so that all cases and controls from a single cohort study underwent metabolite profiling as a batch. Sample triplets (pancreatic cancer case, matched control #1, and matched control #2) were distributed randomly within the batch, and the order of the case and two matched controls within each triplet was also randomly designated. Therefore, the case and its two controls were always run in the same batch and were always directly adjacent to each other in the analytic run, thereby limiting variability in platform performance across matched case–control triplets.
For participants from all four cohorts, plasma samples were thawed once to aliquot them from large volume vials into the smaller volumes needed for shipment to the Broad Institute of the Massachusetts Institute of Technology and Harvard University (Cambridge, MA). The samples were refrozen at the Broad Institute and then thawed a second time to perform metabolite profiling. Therefore, for all cases and controls, plasma samples had been thawed twice at the time of metabolite profiling.
We previously measured hemoglobin A1c (HbA1c) in 389 cases and 757 controls in the laboratory of Dr. Nader Rifai (Children's Hospital, Boston, MA) using reagents from Roche Diagnostics (Indianapolis, IN). We measured plasma insulin in 386 cases and 743 controls, plasma proinsulin in 388 cases and 746 controls, and plasma C–peptide in 408 cases and 785 controls using reagents from Diagnostic Systems Laboratory (Webster, TX) and Millipore Corporation (Billerica, MA). Randomly inserted samples from quality control (QC) plasma pools had mean intra–assay coefficients of variance (CVs) of 2.0% for HbA1c, 5.4% for insulin, 3.1% for proinsulin, and 4.9% for C–peptide4.
Metabolite Profiling
Profiles of endogenous, polar metabolites were obtained using liquid chromatography–tandem mass spectrometry (LC–MS) at the Broad Institute of the Massachusetts Institute of Technology and Harvard University (Cambridge, MA). The LC–MS methods were designed to enable broad measurement of metabolic markers and intermediates, including metabolites from central metabolism and amino acid metabolism, using low plasma sample volumes48. LC–MS parameters for targeted analyses, including chromatographic retention times and mass spectrometry multiple reaction monitoring settings (declustering potentials, collision energies, and lens voltages), were determined using over 300 commercially available reference compounds. A subset of 133 polar metabolites were measurable in plasma using a combination of two distinct hydrophilic interaction liquid chromatography (HILIC) methods, one operated under acid mobile phase conditions with positive ion mode MS detection and the other under basic elution conditions with negative ion mode MS detection.
The acidic HILIC method using positive ionization mode MS analyses was similar to the method described in Wang et al11. Briefly, the LC–MS system consisted of a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX) coupled to an 1100 Series pump (Agilent) and an HTS PAL autosampler (Leap Technologies). Plasma samples (10μL) were extracted using nine volumes of 74.9:24.9:0.2 (v/v/v) acetonitrile/methanol/formic acid containing stable isotope– labeled internal standards (valine–d8, Isotec; and phenylalanine–d8, Cambridge Isotope Laboratories). The samples were centrifuged (10 min, 9,000 x g, 4°C) and the supernatants (10 μL) were injected onto an Atlantis HILIC column (150 x 2.1 mm, 3 μm particle size; Waters Inc.). The column was eluted isocratically at a flow rate of 250 μL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. The ion spray voltage was 4.5 kV and the source temperature was 450°C.
A second method using basic HILIC separation and negative ionization mode MS detection was established on an LC–MS system consisting of an ACQUITY UPLC (Waters Inc.) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (AB SCIEX). Plasma samples (30μL) were extracted using 120 μL of 80% methanol (VWR) containing the internal standards inosine–15N4, thymine–d4, and glycocholate–d4 (Cambridge Isotope Laboratories). The samples were centrifuged (10 min, 9,000 x g, 4°C) and the supernatants were injected directly onto a Luna NH2 column (150 x 2.0 mm, 5 μm particle size; Phenomenex) that was eluted at a flow rate of 400 μL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide (Sigma–Aldrich) in water (VWR)) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol (VWR)) followed by a 10 min linear gradient to 100% mobile phase A. The ion spray voltage was –4.5 kV and the source temperature was 500°C.
Raw data were processed using MultiQuant 1.2 software (AB SCIEX) for automated LC– MS peak integration. All chromatographic peaks were manually reviewed for quality of integration and compared against a known standard for each metabolite to confirm compound identities. Internal standard peak areas were monitored for quality control, to assess system performance over time, and to identify any outlier samples requiring re–analysis. A pooled plasma reference sample was also analyzed after sets of 20 study samples as an additional quality control measure of analytical performance and to serve as reference for scaling raw LC–MS peak areas across sample batches. Metabolites with a signal to noise ratio <10 were considered unquantifiable. Metabolite signals were analyzed in relation to pancreatic cancer risk as LC–MS peak areas, which are proportional to metabolite concentration and appropriate for metabolite clustering and correlative analyses.
Of the 133 metabolites measured (Supplementary Fig. 10), 83 were included in the analyses of our nested pancreatic cancer case–control population (Supplementary Table 9). In pilot work49, we determined that 32 metabolites had poor reproducibility in samples with delayed processing, so these metabolites were excluded as they could not be reliably measured in two of the participating cohorts. In the current study, three Heparin plasma pools (57 total QC samples) and three EDTA plasma pools (128 total QC samples) were randomly interspersed among participant samples as blinded QC samples. We calculated mean CVs for each metabolite across QC plasma pools and set an a priori threshold of ≤25% for satisfactory reproducibility. Based on this criterion, 13 metabolites with mean CV>25% were excluded from our analyses. Five metabolites had undetectable levels for >10% of participants and were also excluded. We evaluated plasma from ten volunteers with plasma collected simultaneously in Heparin and EDTA tubes. For the branched chain amino acids, Spearman correlation coefficients between Heparin and EDTA samples were 0.85 for isoleucine, 0.88 for leucine, and 0.95 for valine.
For metabolites meeting the threshold for statistical significance after multiple– hypothesis correction (isoleucine, leucine and valine), LC–MS peak areas were converted to absolute concentrations using stable isotope–labeled standards. Briefly, external calibration curves of MS response were determined using solutions of isotope–labeled 13C6, 15N–leucine, 13C6, 15N–isoleucine (Cambridge Isotope Laboratories), and d8–valine (Isotec). A 1 μg/μL solution of each standard was prepared in water. 20 μL of each stock solution were added to 180 μL of reference pooled plasma and the resulting solution was then serially diluted using pooled plasma to generate a calibration curve. For multiple reaction monitoring MS analyses, the bond cleavage products and collision energy and declustering potential settings were the same as those used for the endogenous metabolites: natural leucine 132/86, CE=18 and DP=50; 13C6, 15N– leucine 134/87, CE=18 and DP=50; natural isoleucine 132/86, CE=18 and DP=50; 13C6, 15N– isoleucine 139/92, CE=18 and DP=50; natural valine 118/72, CE=18 and DP=25, and d8–valine 126/80, CE=18 and DP=25. Three separate plasma samples were prepared at each concentration and were analyzed using the acidic HILIC LC–MS method described above. The median concentrations of endogenous isoleucine, leucine and valine in the reference pooled plasma were calculated and the concentration of each metabolite in each study sample was determined from the response ratio relative to the nearest reference pooled plasma sample in the analysis queue.
Statistical Analysis
To compare baseline characteristics, we used conditional logistic regression conditioned on the matching factors and including the covariate of interest. Partial Spearman correlation coefficients were calculated for metabolites and covariates. Metabolites were log– transformed to improve normality and included as continuous variables in conditional logistic regression models conditioned on matching factors and adjusted for age at blood draw (years, continuous), fasting time (<4, 4–8, 8–12, ≥12 hours, missing) and race/ethnicity (White, Black, other, missing). Using a conservative Bonferroni correction for multiple–hypothesis testing50, metabolites with P≤0.0006 (0.05/83) were considered statistically significant.
To provide estimates of effect magnitude, significant metabolites were examined in conditional logistic regression models after categorization into quintiles based on log– transformed metabolite levels in controls. Separate quintiles were generated for fasting (≥8 hours since last meal) and non–fasting (<8 hours since last meal) participants, given the possible effect of fasting time on metabolite levels. Quintiles were generated from the population of selected controls, which may not exactly reflect the characteristics of the full cohort population. Odds ratios (OR) and 95% confidence intervals (CI) were also calculated per standard deviation (SD) change in log–transformed metabolite levels. To control for possible confounding, we evaluated regression models adjusted for body–mass index (BMI), physical activity, history of diabetes mellitus, HbA1c, plasma insulin, plasma proinsulin, and plasma C–peptide. We also evaluated regression models after excluding subjects with diabetes by self–report or HbA1c≥6.5% at blood collection (prevalent diabetes). We further evaluated models that excluded subjects who developed diabetes after blood collection but >2 years prior to cancer diagnosis (incident diabetes not thought to be recent onset from pancreatic cancer)42,47.
Metabolite values were considered missing when an LC–MS peak was below the limit of detection. In the primary analysis, any case or control with missing data for a metabolite was excluded from the analysis of that metabolite. However, we also conducted sensitivity analyses, in which participants with missing values were assigned the lower limit of detection or half of the lower limit of detection, and our results were unchanged.
We assessed heterogeneity of metabolite associations with pancreatic cancer risk across cohorts using Cochran's Q–statistic51. We examined associations in predefined subgroups by sex, smoking status, BMI, and fasting status. Statistical interactions were assessed by entering into models the main effect terms and cross–product terms of metabolites and stratification variables, evaluating likelihood ratio tests. We also examined associations by time between blood collection and the case's cancer diagnosis. In these time–based analyses, one stratum included 40 pancreatic cancer cases with blood collected within 2 years of diagnosis and their matched controls. These cases and controls were not part of the primary analysis population, but were included in the stratified analyses by time to more fully delineate the association of metabolites with pancreatic development by time before diagnosis. Associations were also examined for circulating BCAAs with previously explored risk factors for pancreatic cancer in our cohorts (Supplementary Table 10) and with cancer stage at diagnosis (Supplementary Table 11).
Since association of a marker with disease does not indicate the suitability of the marker to serve as a screening test for the disease, we examined two approaches to quantify the value of metabolites in a multi–factor risk discrimination tool for pancreatic cancer. Discrimination quantifies the ability of one or more disease markers to separate cases (individuals with the disease) from controls (individuals without the disease). We investigated the discrimination of risk models for predicting pancreatic cancer diagnosis in the ten years after measurement of circulating BCAAs, i.e. the 10–year risk of pancreatic cancer.
In the first approach, we investigated receiver–operating–characteristic (ROC) curve analysis and calculated of the area under the ROC curve (AUC), also known as the concordance (C) statistic16,52. The base model included age at blood collection (continuous), cohort (HPFS, NHS, PHS, WHI; which also accounts for sex), race/ethnicity (white, black, other/missing), smoking status (never, past, current, missing) and fasting time (<4, 4–8, 8–12, ≥12 hours, missing). Three subsequent models mirrored the base model, but additionally included: (1) body–mass index, physical activity, and history of diabetes, (2) circulating BCAAs, or (3) body– mass index, physical activity, history of diabetes, and circulating BCAAs. Each point on the ROC curve shows the effect of a rule for turning a risk estimate into a prediction of the development of an event. The y axis of the ROC curve is the true positive rate or sensitivity (i.e., the proportion of individuals with pancreatic cancer who were correctly predicted to have the disease). The x axis shows the false positive rate, which is the complement of specificity (i.e., the proportion of individuals without pancreatic cancer who were incorrectly predicted to have pancreatic cancer). The area under the ROC curve, the AUC, measures how well the model discriminates between case subjects and control subjects. An ROC curve that corresponds to a random classification of case subjects and control subjects is a straight line with an AUC of 50%. An ROC curve that corresponds to perfect classification has an AUC of 100%. The improvement in AUC for a model containing a new marker is defined as the difference in AUCs calculated using a model with and without the new marker of interest53.
For context, the Breast Cancer Risk Assessment Tool, commonly referred to as the Gail Model54,55, estimates a woman's risk for breast cancer using clinically available information including current age, age at menarche, age of first live birth, number of first degree relatives with breast cancer, number of previous breast biopsies, breast biopsies that show atypical hyperplasia, and race/ethnicity. The Gail Model is used to counsel women on appropriate screening tests for breast cancer56, the use of tamoxifen as a chemopreventative agent57, and for determining sample size calculations in randomized clinical trials of prevention strategies58. Several studies have evaluated the discrimination of the Gail model using ROC curve analysis and calculated the AUC to be 0.58 to 0.6352,59-61. Follow–up studies have described an AUC of 0.62 to 0.66 when breast density is added as an additional predictor in the original Gail Model59,60,62.
Although ROC curves are commonly used, they have a number of limitations and may underestimate the ability of a new marker to contribute to risk prediction when added to previously defined predictors63-66. Another approach to evaluating model discrimination is to evaluate the ability of a new marker to shift an individual's risk up or down between pre–defined risk categories. This is known as the prediction increment of a marker and has been codified in an approach known as net reclassification improvement (NRI)17. The NRI (sometimes referred to as the net reclassification index) constructs reclassification tables separately for participants with and without events, and quantifies the correct movement between categories of risk; namely, to higher risk categories for participants with events and to lower risk categories for those without events. Furthermore, incorrect movement in categories of risk (downwards for events and upwards for non–events) reduces the net correct reclassification of individuals within the study population.
The NRI calculation is represented by the following formula:
Upward movement (up) is defined as a change into a higher risk category based on the new model and downward movement (down) as a change into a lower risk category based on the new model, where P indicates probability and D denotes the event indicator (1, event; 0, non–event).
Using the NRI, we evaluated the ability of the prediction model including circulating BCAAs to appropriately reclassify individuals into risk groups compared to the base model. The base model was calculated using conditional logistic regression conditioned on matching factors and adjusted for race/ethnicity, body–mass index, physical activity and history of diabetes. The subsequent model included the covariates in the base model with the addition of circulating metabolites. As in prior studies67-69, we defined the high–risk group as those individuals with risk for pancreatic cancer at least 2–fold greater than an individual with average risk.
For context, the Emerging Risk Factors Collaboration70 has examined the integration of novel risk factors into risk prediction models for cardiovascular disease. In these studies, additional potential risk predictors were added to a model of known risk predictors for cardiovascular disease, including age, sex, smoking status, blood pressure, history of diabetes, and cholesterol. The net reclassification improvement was then calculated for three 10–year risk categories for cardiovascular disease. C–reactive protein is a marker of systemic inflammation, and elevated CRP has been associated with an increased risk for cardiovascular events in numerous studies71-73. Circulating CRP is currently used to inform decisions in the clinic regarding screening and risk reduction strategies74,75 and to design clinical trials testing novel treatments to reduce cardiovascular events76,77 In an analysis of nearly 250,000 individuals78, the addition of CRP to know cardiovascular disease risk factors was associated with a statistically significant improvement in the area under the ROC curve and a NRI of 1.52% for 10–year risk of cardiovascular disease. In contrast, additional analyses demonstrated a <1% improvement in the NRI for body–mass index, waist circumference, waist–to–hip ratio, plasma fibrinogen, and circulating apolipoproteins78-80, such that the clinical utility of these additional predictors remains unclear75.
All analyses were performed with SAS 9.2 statistical package. All P–values were two– sided.
MOUSE STUDIES
All studies were approved by the MIT committee on animal care (IACUC).
Experimental Mice
All experimental groups were assigned based on genotype. All animals were numbered and experiments conducted blinded. After data collection, genotypes were revealed and animals assigned to groups for analysis.
KPC
Experimental KPC mice were male mice on a mixed background, heterozygous for the conditional lox–stop–lox KrasG12D allele, heterozygous for the conditional lox–stop–lox Trp53R172H allele and expressed Cre–recombinase under control of the Pdx–1–promoter (Tg(Ipf1-cre)1Tuv)19. Littermate controls lacked either the LSL–KrasG12D allele, the Cre allele, or both. Control mice were sacrificed at the same time as their tumor–bearing littermates.
KP–/–C
Experimental KP–/–C mice were male mice on a mixed background, heterozygous for the conditional lox–stop–lox KrasG12D allele, homozygous for loxP sites flanking exons 2–10 of Trp53 and expressed Cre–recombinase under control of the Pdx–1– promoter (Tg(Ipf1-cre)1Tuv)20. Littermate control mice lacked either the Cre–recombinase allele, LSL–KrasG12D allele, or both. Inbred C57B6/J male mice containing the same alleles were also examined where indicated, and cancer cell lines derived from these mice were used for syngenic re–implantation studies.
Non–small cell lung cancer
Six–month–old male mice on a pure 129 background, heterozygous for the conditional lox–stop–lox KrasG12D allele, homozygous for loxP sites flanking exons 2–10 of Trp53 were administered 2.5x107 pfu of Cre–expressing adenovirus as previously described21,22. High–titer adenovirus was obtained from the Gene Transfer Vector Core (University of Iowa).
Hindlimb sarcoma
Four–week–old male mice on a mixed background, heterozygous for the conditional lox–stop–lox KrasG12D allele and homozygous for loxP sites flanking exons 2–10 of Trp53 were administered 2.5x108 pfu of Cre–expressing adenovirus as previously described23. High–titer adenovirus was obtained from the Gene Transfer Vector Core (University of Iowa).
Re–implantation, Pancreatitis and BCAA diet studies
Male C57B6/J mice aged 4–6 weeks at the start of the study were used for these experiments.
Diets
Standard Chow Diet: RMH 3000 (Prolab).
Amino Acid Defined Diet
1x BCAA (TD.110839) and 2x BCAA (TD.110843) were designed in consultation with and subsequently obtained from Harlan Teklad.
Labeled Amino Acid Defined Diet
20% 13C–leucine and 20% 13C–valine labeled diets were based on diet TD.110839 and produced by Cambridge Isotopes and Harlan Teklad.
Plasma for Metabolomics
Plasma was collected for each experiment at the time points indicated. Mice were anesthetized under 2% isofluorane–oxygen mixture and retro–orbitally bled approximately 4.5 hours after the onset of the light cycle. Blood was immediately placed in EDTA–pretreated tubes and centrifuged to separate plasma. Plasma was aliquoted and frozen at –80°C for further analysis. Fasting blood samples were harvested in the same manner first thing in the morning after a 16–hour overnight fast.
Food Consumption
Mice were housed individually for 48 hours and remaining food pellets weighed at zero, 24 and 48 hours. A two–day average was then calculated for each mouse. Body weight was determined on the second day. To assess consumption of BCAA defined diets, mice were housed individually and fed diets for 5 days. Food was weighed after 2 days of feeding and again on day 5, and the average consumption per 24hrs over the 72hr period was calculated.
Blood glucose, plasma insulin, glucose tolerance test, and insulin tolerance test
Four–week– old KP–/–C mice were fasted overnight and blood glucose measured using a One Touch Ultra handheld glucometer. 25μL of plasma from the same mice was harvested in heparinized tubes, aliquoted, and frozen at –80°C for further analysis. Plasma insulin levels were determined using an ultrasensitive mouse insulin ELISA kit (Crystal Chem, #90080). After measuring fasting parameters, a glucose tolerance test was performed in accordance with published protocols81. Briefly, conscious mice received an intraperitoneal injection of 2g/kg glucose at time zero. Blood glucose was subsequently measured at 15, 30, 60, 90 and 120 minutes post–injection as described above. For insulin tolerance test, four–week–old KP–/–C mice were fasted for 6 hours during daytime hours. Following initial blood glucose measurement, conscious mice received an intraperitoneal injection of 0.75 IU/kg recombinant human insulin (Novolin, Novo Nordisk). Blood glucose was subsequently measured at 15, 30, 60 and 90 minutes post–injection as described above.
KP–/–C Cell Lines and Re–implantation Studies
End–stage tumors were dissected from C57B6/J KP–/–C mice and mechanically chopped prior to trypsin disaggregation, with tumor cells then propagated in DMEM with 10% FBS, 2mM glutamine and penicillin/streptomycin. Cell lines were negative for mycoplasma. For subcutaneous re–implantation studies, recipient mice were anesthetized with inhaled 2% isofluorane–oxygen mixture, low passage cell lines (passage 5 for each line) were resuspended at 2.5 x 105 cells/100μL in sterile PBS, and100μL of either PBS (control) or cell suspension was injected in the flank of syngeneic mice. For orthotopic re–implantation studies, recipient mice were anesthetized with inhaled 2% isofluorane–oxygen mixture, a vertical incision made in the abdomen at the left mid–calvicular line, the spleen mobilized, and 25μL of either PBS or PBS containing 1.0 x 105 cells (passage 3 for each line) was injected into the tail of the pancreas.
Caerulein–induced Chronic Pancreatitis
Mice were treated with either USP–grade saline or 5μg caerulein (Sigma) via intraperitoneal injection daily, 5 days per week for 10 weeks as previously described25. Blood was obtained and the mice sacrificed on the final day. Tissues were fixed in 10% formalin for subsequent histological analysis.
Plasma Markers of Pancreatitis
Plasma amylase and lipase measurements were performed by IDEXX BioResearch Laboratory (North Grafton, MA).
BCAA Diet Studies
Mice were fed either 1x or 2x BCAA diets for 10 weeks. Blood was obtained and mice sacrificed on the final day of the experiment. Tissues were fixed in 10% formalin for subsequent histological analysis.
Studies to determine source of BCAA elevations
Acute uptake and disposal
Following a 16– hour overnight fast, mice were fed 20% 13C–leucine and valine containing diet for two hours before removal of food, and food consumption during this period quantified as described above. At the time points indicated by the red arrowheads in Figure 3a, 10–25μL of plasma was harvested from the tail vein of conscious mice in a heparinized tube and centrifuged to separate plasma. Plasma was aliquoted and frozen at –80°C for subsequent GC–MS analysis. Total ion counts from GC–MS analysis of leucine and valine were then normalized to norvaline internal standard and multiplied by fractional labeling to determine the amount of label present. This number for each animal was then normalized to that animal's food intake to control for inter– animal variation in labeled food consumption.
Long–term pool contribution of BCAA to plasma
Mice were exposed to 20% 13C–leucine and valine labeled diets from 7 days to age to 24 days of age followed by three days of unlabeled diet (according to the protocol depicted in Figure 3b). Two cohorts of mice were used in this study. One cohort of mice was sacrificed on day 27 in the fed state and a second cohort of mice was sacrificed on day 28 after a 16–hour overnight fast (the points indicated by the red arrowheads in Figure 3b). At sacrifice, anesthetized mice were terminally bled and tissues harvested within 5 minutes, snap frozen in liquid nitrogen using Biosqueezer (BioSpec Products), and stored at –80°C for subsequent GC–MS analysis. Plasma was aliquoted and frozen at –80°C for subsequent GC–MS analysis.
Total contributions from short and long–term pools were calculated according to the following equations:
Raw data and calculations are summarized in Supplementary Table 12.
Tissue and body weights
For measurement of tissue weights, mice were weighed prior to sacrifice, then gastrocnemius, tibialis anterior, soleus and heart were subsequently dissected and weighed. Muscle weights for each individual mouse were normalized to the body weight of that mouse.
LC–MS Plasma Amino Acid Measurements
Plasma amino acids were measured by LC–MS at the Koch Institute of the Massachusetts Institute of Technology (Cambridge, MA) using similar methods used for assessment of metabolites in human plasma. Raw data were analyzed as peak area tops using the open–access MAVEN software tool82.
GC–MS Assessment of13C–leucine and valine Labeling
Plasma polar metabolites were extracted in ice–cold 4:1 methanol:water with norvaline internal standard (5μL plasma in 200μL extraction solution). Extracts were clarified by centrifugation and the supernatant evaporated under nitrogen and frozen at –80°C for subsequent derivitization. Dried polar metabolites were dissolved in 20μL of 2% methoxyamine hydrochloride in pyridine (Thermo) and held at 37°C for 1.5hr. After dissolution and reaction, tert–butyldimethylsilyl derivatization was initiated by adding 25μL N–methyl–N–(tert–butyldimethylsilyl)trifluoroacetamide + 1% tert– butyldimethylchlorosilane (Sigma) and incubating at 37°C for 1hr. The acid hydrolysis protocol was adapted from Antoniewicz and colleagues83. Briefly, acid hydrolysis of tissue proteins was performed on snap frozen tissues by boiling 1–5mg tissue in 1mL 18% hydrochloric acid overnight at 100°C. 50μL supernatant was evaporated under nitrogen and frozen at –80°C for subsequent derivitization. Dried hydrolysates were re–dissolved in pyridine (10μL/1mg tissue) prior to tert–butyldimethylsilyl derivatization, which was initiated by adding N–methyl–N–(tert– butyldimethylsilyl)trifluoroacetamide + 1% tert–butyldimethylchlorosilane (12.5μL/1mg tissue, Sigma) and incubating at 37°C for 1h.
GC/MS analysis was performed using an Agilent 7890 GC equipped with a 30m DB– 35MS capillary column connected to an Agilent 5975B MS operating under electron impact ionization at 70eV. One microlitre of sample was injected in splitless mode at 270°C, using helium as the carrier gas at a flow rate of 1mlmin−1. For measurement of amino acids, the GC oven temperature was held at 100°C for 3min and increased to 300°C at 3.5°Cmin−1. The MS source and quadrupole were held at 230°C and 150°C, respectively, and the detector was run in scanning mode, recording ion abundance in the range of 100–605 m/z. MIDs were determined by integrating the appropriate ion fragments83 listed in Supplementary Table 13 and corrected for natural isotope abundance using an algorithm adapted from Ferandez and colleagues84.
Statistics
Appropriate statistical tests were performed where required. Two–sided unpaired student's t–tests were performed for all statistical analyses unless otherwise specified using Mircosoft Excel for Mac:2011 (Microsoft) or GraphPad Prism 6 (GraphPad Software). Two– sided repeated–measures ANOVA was performed to compare mean plasma glucose levels in the glucose tolerance test and insulin tolerance tests85, using SAS 9.2 statistical package.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge A. Deik and K. Bullock of the Broad Institute for assistance with LC–MS sample analyses, and the Tang Histology facility in the Koch Institute Swanson Biotechnology Center for assistance processing mouse tissues. Cambridge Isotope Laboratories supplied 13C-BCAA diets for mouse labeling studies. The authors would like to thank C. Newgard and A. Goldberg for their thoughtful discussions regarding this manuscript. The authors also would like to thank the participants and staff of the HPFS, NHS, PHS, and WHI for their contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. NHS and HPFS are supported by NIH grants P01 CA87969, P01 CA55075, P50 CA127003, R01 CA124908, R01 CA49449, 1UM1 CA167552. PHS is supported by NIH grants CA 97193, CA 34944, CA 40360, HL 26490, and HL 34595. The WHI program is funded by the NIH through contracts N01WH22110, 24152, 32100–2, 32105–6, 32108–9, 32111–13, 32115, 32118–32119, 32122, 42107–26, 42129–32, and 44221. We acknowledge additional support from F30 CA183474 to JRM; from a Nestle Research Center award to the Broad Institute; from R01 DK081572 to TJW and REG; from the Robert T. and Judith B. Hale Fund for Pancreatic Cancer, Perry S. Levy Fund for Gastrointestinal Cancer Research, Pappas Family Research Fund for Pancreatic Cancer to CSF; from the Burroughs Wellcome Fund, Damon Runyon Cancer Research Foundation, the Smith Family, and the Stern Family to MGVH; and from NIH/NCI K07 CA140790, American Society of Clinical Oncology Conquer Cancer Foundation, Howard Hughes Medical Institute, and Promises for Purple to BMW. MGVH additionally acknowledges support from 5–P30–CA14051–39, and major support for this project was provided by the Howard Hughes Medical Institute to BMW and the Lustgarten Foundation to CSF, MGVH and BMW.
Footnotes
AUTHOR CONTRIBUTIONS
All authors participated in the analysis and interpretation of data. CW, CBC, PK, CY, YB, MKT and BMW performed the statistical analyses of the human data. CBC, MKT, SST, TJW, REG and BMW evaluated the platform for analysis of plasma metabolites in large cohort studies. CBC, JRM, AS and KP performed metabolite profiling experiments. PK, YB, MKT, SST, SO, MJS, ELG, ZRQ, DAR, JM, HDS, JMG, BBC, SL JWW, JEM, MNP and CSF assisted in data acquisition, management, and interpretation from the four cohort studies. JRM and MET conducted all mouse experiments with assistance from BPF, SMD, TP, AY TLD and ACK. CW, CBC, JRM, CSF, MGVH and BMW designed the study and drafted the manuscript with input from all authors.
The authors report no financial conflict of interest.
REFERENCES
- 1.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaud DS, et al. Physical activity, obesity, height, and the risk of pancreatic cancer. JAMA. 2001;286:921–929. doi: 10.1001/jama.286.8.921. [DOI] [PubMed] [Google Scholar]
- 3.Stolzenberg-Solomon RZ, et al. Insulin, glucose, insulin resistance, and pancreatic cancer in male smokers. JAMA. 2005;294:2872–2878. doi: 10.1001/jama.294.22.2872. [DOI] [PubMed] [Google Scholar]
- 4.Wolpin BM, et al. Hyperglycemia, Insulin Resistance, Impaired Pancreatic beta-Cell Function, and Risk of Pancreatic Cancer. J Natl Cancer Inst. 2013;105:1027–1035. doi: 10.1093/jnci/djt123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch SM, et al. Cigarette smoking and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Am J Epidemiol. 2009;170:403–413. doi: 10.1093/aje/kwp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51:14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan BH, Birdsell LA, Martin L, Baracos VE, Fearon KC. Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Cancer Res. 2009;15:6973–6979. doi: 10.1158/1078-0432.CCR-09-1525. [DOI] [PubMed] [Google Scholar]
- 8.Claudino WM, Goncalves PH, di Leo A, Philip PA, Sarkar FH. Metabolomics in cancer: a bench-to-bedside intersection. Critical reviews in oncology/hematology. 2012;84:1–7. doi: 10.1016/j.critrevonc.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 10.Kobayashi T, et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:571–579. doi: 10.1158/1055-9965.EPI-12-1033. [DOI] [PubMed] [Google Scholar]
- 11.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosnan JT, Brosnan ME. Branched-chain amino acids: enzyme and substrate regulation. J Nutr. 2006;136:207S–211S. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- 13.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floegel A, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huxley R, Ansary-Moghaddam A, Berrington de Gonzalez A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–2083. doi: 10.1038/sj.bjc.6602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 17.Pencina MJ, D'Agostino RB, Sr., D'Agostino RB, Jr., Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207-112. [DOI] [PubMed] [Google Scholar]
- 18.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 22.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirsch DG, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 24.Duell EJ, et al. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann Oncol. 2012;23:2964–2970. doi: 10.1093/annonc/mds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosnan JT. Interorgan amino acid transport and its regulation. J Nutr. 2003;133:2068S–2072S. doi: 10.1093/jn/133.6.2068S. [DOI] [PubMed] [Google Scholar]
- 28.Matthews DE, Marano MA, Campbell RG. Splanchnic bed utilization of leucine and phenylalanine in humans. The American journal of physiology. 1993;264:E109–118. doi: 10.1152/ajpendo.1993.264.1.E109. [DOI] [PubMed] [Google Scholar]
- 29.Cynober LA. Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition. 2002;18:761–766. doi: 10.1016/s0899-9007(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 30.Acharyya S, et al. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest. 2004;114:370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell metabolism. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 33.Lundholm K, Bylund AC, Holm J, Schersten T. Skeletal muscle metabolism in patients with malignant tumor. Eur J Cancer. 1976;12:465–473. doi: 10.1016/0014-2964(76)90036-0. [DOI] [PubMed] [Google Scholar]
- 34.Heymsfield SB, McManus CB. Tissue components of weight loss in cancer patients. A new method of study and preliminary observations. Cancer. 1985;55:238–249. doi: 10.1002/1097-0142(19850101)55:1+<238::aid-cncr2820551306>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 35.Dewys WD, et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am J Med. 1980;69:491–497. doi: 10.1016/s0149-2918(05)80001-3. [DOI] [PubMed] [Google Scholar]
- 36.Wigmore SJ, Plester CE, Richardson RA, Fearon KC. Changes in nutritional status associated with unresectable pancreatic cancer. Br J Cancer. 1997;75:106–109. doi: 10.1038/bjc.1997.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsoli M, Robertson G. Cancer cachexia: malignant inflammation, tumorkines, and metabolic mayhem. Trends in endocrinology and metabolism: TEM. 2013;24:174–183. doi: 10.1016/j.tem.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013 doi: 10.1038/nature12138. Epub ahead of Print May 15, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annual review of nutrition. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 41.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88–95. doi: 10.1016/S1470-2045(08)70337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rich-Edwards JW, Corsano KA, Stampfer MJ. Test of the National Death Index and Equifax Nationwide Death Search. Am J Epidemiol. 1994;140:1016–1019. doi: 10.1093/oxfordjournals.aje.a117191. [DOI] [PubMed] [Google Scholar]
- 44.Michaud DS, et al. Prediagnostic plasma C-peptide and pancreatic cancer risk in men and women. Cancer Epidemiol Biomarkers Prev. 2007;16:2101–2109. doi: 10.1158/1055-9965.EPI-07-0182. [DOI] [PubMed] [Google Scholar]
- 45.Wolpin BM, et al. Plasma 25-hydroxyvitamin D and risk of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:82–91. doi: 10.1158/1055-9965.EPI-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao Y, et al. A prospective study of plasma adiponectin and pancreatic cancer risk in five US cohorts. J Natl Cancer Inst. 2013;105:95–103. doi: 10.1093/jnci/djs474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pannala R, et al. Temporal association of changes in fasting blood glucose and body mass index with diagnosis of pancreatic cancer. Am J Gastroenterol. 2009;104:2318–2325. doi: 10.1038/ajg.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts LD, Souza AL, Gerszten RE, Clish CB. Targeted metabolomics. Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb3002s98. Chapter 30, Unit 30 32 31-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Townsend MK, et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 52.Wacholder S, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 54.Gail MH, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 55.Gail MH, Benichou J. Validation studies on a model for breast cancer risk. J Natl Cancer Inst. 1994;86:573–575. doi: 10.1093/jnci/86.8.573. [DOI] [PubMed] [Google Scholar]
- 56.Saslow D, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57:75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 57.Gail MH, et al. Weighing the risks and benefits of tamoxifen treatment for preventing breast cancer. J Natl Cancer Inst. 1999;91:1829–1846. doi: 10.1093/jnci/91.21.1829. [DOI] [PubMed] [Google Scholar]
- 58.Costantino JP, et al. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst. 1999;91:1541–1548. doi: 10.1093/jnci/91.18.1541. [DOI] [PubMed] [Google Scholar]
- 59.Tice JA, et al. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148:337–347. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barlow WE, et al. Prospective breast cancer risk prediction model for women undergoing screening mammography. J Natl Cancer Inst. 2006;98:1204–1214. doi: 10.1093/jnci/djj331. [DOI] [PubMed] [Google Scholar]
- 61.Rockhill B, Spiegelman D, Byrne C, Hunter DJ, Colditz GA. Validation of the Gail et al. model of breast cancer risk prediction and implications for chemoprevention. J Natl Cancer Inst. 2001;93:358–366. doi: 10.1093/jnci/93.5.358. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, et al. Projecting absolute invasive breast cancer risk in white women with a model that includes mammographic density. J Natl Cancer Inst. 2006;98:1215–1226. doi: 10.1093/jnci/djj332. [DOI] [PubMed] [Google Scholar]
- 63.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 64.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 65.Cook NR, Buring JE, Ridker PM. The effect of including C-reactive protein in cardiovascular risk prediction models for women. Ann Intern Med. 2006;145:21–29. doi: 10.7326/0003-4819-145-1-200607040-00128. [DOI] [PubMed] [Google Scholar]
- 66.Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. Jama. 2007;297:611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 67.Roberts NJ, et al. The predictive capacity of personal genome sequencing. Sci Transl Med. 2012;4:133ra158. doi: 10.1126/scitranslmed.3003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Ravesteyn NT, et al. Tipping the balance of benefits and harms to favor screening mammography starting at age 40 years: a comparative modeling study of risk. Ann Intern Med. 2012;156:609–617. doi: 10.1059/0003-4819-156-9-201205010-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatterjee N, et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–405. 405e401–403. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danesh J, et al. The Emerging Risk Factors Collaboration: analysis of individual data on lipid, inflammatory and other markers in over 1.1 million participants in 104 prospective studies of cardiovascular diseases. European journal of epidemiology. 2007;22:839–869. doi: 10.1007/s10654-007-9165-7. [DOI] [PubMed] [Google Scholar]
- 71.Ridker PM, Hennekens CH, Buring JE, Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 72.Ridker PM. Clinical application of C-reactive protein for cardiovascular disease detection and prevention. Circulation. 2003;107:363–369. doi: 10.1161/01.cir.0000053730.47739.3c. [DOI] [PubMed] [Google Scholar]
- 73.Pradhan AD, et al. Inflammatory biomarkers, hormone replacement therapy, and incident coronary heart disease: prospective analysis from the Women's Health Initiative observational study. Jama. 2002;288:980–987. doi: 10.1001/jama.288.8.980. [DOI] [PubMed] [Google Scholar]
- 74.Ridker PM, et al. C-reactive protein levels and outcomes after statin therapy. N Engl J Med. 2005;352:20–28. doi: 10.1056/NEJMoa042378. [DOI] [PubMed] [Google Scholar]
- 75.Greenland P, et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2010;56:e50–103. doi: 10.1016/j.jacc.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 76.Ridker PM, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359:2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 77.Ridker PM, et al. Reduction in C-reactive protein and LDL cholesterol and cardiovascular event rates after initiation of rosuvastatin: a prospective study of the JUPITER trial. Lancet. 2009;373:1175–1182. doi: 10.1016/S0140-6736(09)60447-5. [DOI] [PubMed] [Google Scholar]
- 78.Kaptoge S, et al. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367:1310–1320. doi: 10.1056/NEJMoa1107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wormser D, et al. Separate and combined associations of body-mass index and abdominal adiposity with cardiovascular disease: collaborative analysis of 58 prospective studies. Lancet. 2011;377:1085–1095. doi: 10.1016/S0140-6736(11)60105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Di Angelantonio E, et al. Lipid-related markers and cardiovascular disease prediction. JAMA. 2012;307:2499–2506. doi: 10.1001/jama.2012.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ayala JE, et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Disease models & mechanisms. 2010;3:525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis ... [et al.] Chapter 14, Unit14 11. 2012. [DOI] [PMC free article] [PubMed]
- 83.Antoniewicz MR, et al. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metabolic engineering. 2007;9:277–292. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. Journal of mass spectrometry : JMS. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 85.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab. 2008;295:E1323–1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- 86.Fuchs CS, et al. A prospective study of cigarette smoking and the risk of pancreatic cancer. Arch Intern Med. 1996;156:2255–2260. [PubMed] [Google Scholar]
- 87.Schernhammer ES, et al. A prospective study of aspirin use and the risk of pancreatic cancer in women. J Natl Cancer Inst. 2004;96:22–28. doi: 10.1093/jnci/djh001. [DOI] [PubMed] [Google Scholar]
- 88.Wolpin BM, et al. Circulating insulin-like growth factor binding protein-1 and the risk of pancreatic cancer. Cancer Res. 2007;67:7923–7928. doi: 10.1158/0008-5472.CAN-07-0373. [DOI] [PubMed] [Google Scholar]
- 89.Wolpin BM, et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst. 2009;101:424–431. doi: 10.1093/jnci/djp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.