SUMMARY
The nuclear receptor peroxisome-proliferation activated receptor gamma (PPARγ), a transcriptional master regulator of glucose and lipid metabolism, inhibits the growth of several common cancers including lung cancer. In this study, we show that the mechanism by which activation of PPARγ inhibits proliferation of lung cancer cells is based on metabolic changes. We found that treatment with the PPARγ agonist pioglitazone triggers a metabolic switch that inhibits pyruvate oxidation and reduces glutathione levels. These PPARγ-induced metabolic changes result in a marked increase of reactive oxygen species (ROS) levels that lead to rapid hypophosphorylation of retinoblastoma protein (RB) and cell cycle arrest. The antiproliferative effect of PPARγ activation can be prevented by suppressing pyruvate dehydrogenase kinase 4 (PDK4) or β-oxidation of fatty acids in vitro and in vivo. Our proposed mechanism also suggests that metabolic changes can rapidly and directly inhibit cell cycle progression of cancer cells by altering ROS levels.
INTRODUCTION
A hallmark of cancer is the reprogramming of glucose, lipid and glutamine metabolism to provide energy and substrates for biosynthesis, which are required for proliferation and survival of transformed cells (DeBerardinis et al., 2008; Hanahan and Weinberg, 2011; Vander Heiden et al., 2009). A classical metabolic change in cancer is the increased consumption of glucose and inhibition of pyruvate oxidation, leading to high rates of lactate secretion even under normoxic conditions (“Warburg effect”) (Hsu and Sabatini, 2008; Koppenol et al., 2011; Warburg, 1956). More recently, glutamine has been identified as a critical nutrient for cancer cells to replenish TCA cycle metabolites for the biosynthesis of lipids and mitochondrial ATP production (DeBerardinis et al., 2007; Hensley et al., 2013), complementing the altered state of glucose metabolism.
Therapeutic strategies harnessing altered metabolic properties of cancer cells are currently focused on developing specific inhibitors against key metabolic enzymes, such as glutaminase (Le et al., 2012; Robinson et al., 2007; Thangavelu et al., 2012) and PKM2 (Christofk et al., 2008; Vander Heiden et al., 2010). In principle, the activation or inhibition of transcriptional master regulators of metabolic processes presents another promising strategy to target cancer metabolism and may achieve a more selective response. In this vein, our group recently found that PPARγ is expressed at high levels in a subset of non-small cell lung cancer cells (Jeong et al., 2012), where its activation with thiazolidinediones (TZDs) causes growth inhibition both in vitro and in vivo. Also, growth-suppressing effects of PPARγ and TZDs have been shown for a variety of other cancers (reviewed in (Koeffler, 2003; Peters et al., 2012)). Considering the known functions of PPARγ in orchestrating glucose and lipid metabolism (Kliewer and Willson, 1998; Willson et al., 2001), it is conceivable that the anti-tumorigenic effects of PPARγ may be mediated in part through the activation of a genetic program that affects metabolism of cancer cells. Therefore, understanding the molecular basis of the effects of PPARγ activation on lung cancer cells may also provide general mechanistic insights into the impact of metabolic changes on cancer cell growth. In the present study, we sought to elucidate the molecular actions of PPARγ in lung cancer cells by integrating the analysis of genomic functions of PPARγ with biochemical follow-up analyses and metabolic profiling of changes caused by PPARγ activation.
RESULTS
PPARγ regulates the expression of genes implicated in metabolism and proliferation in lung cancer cells
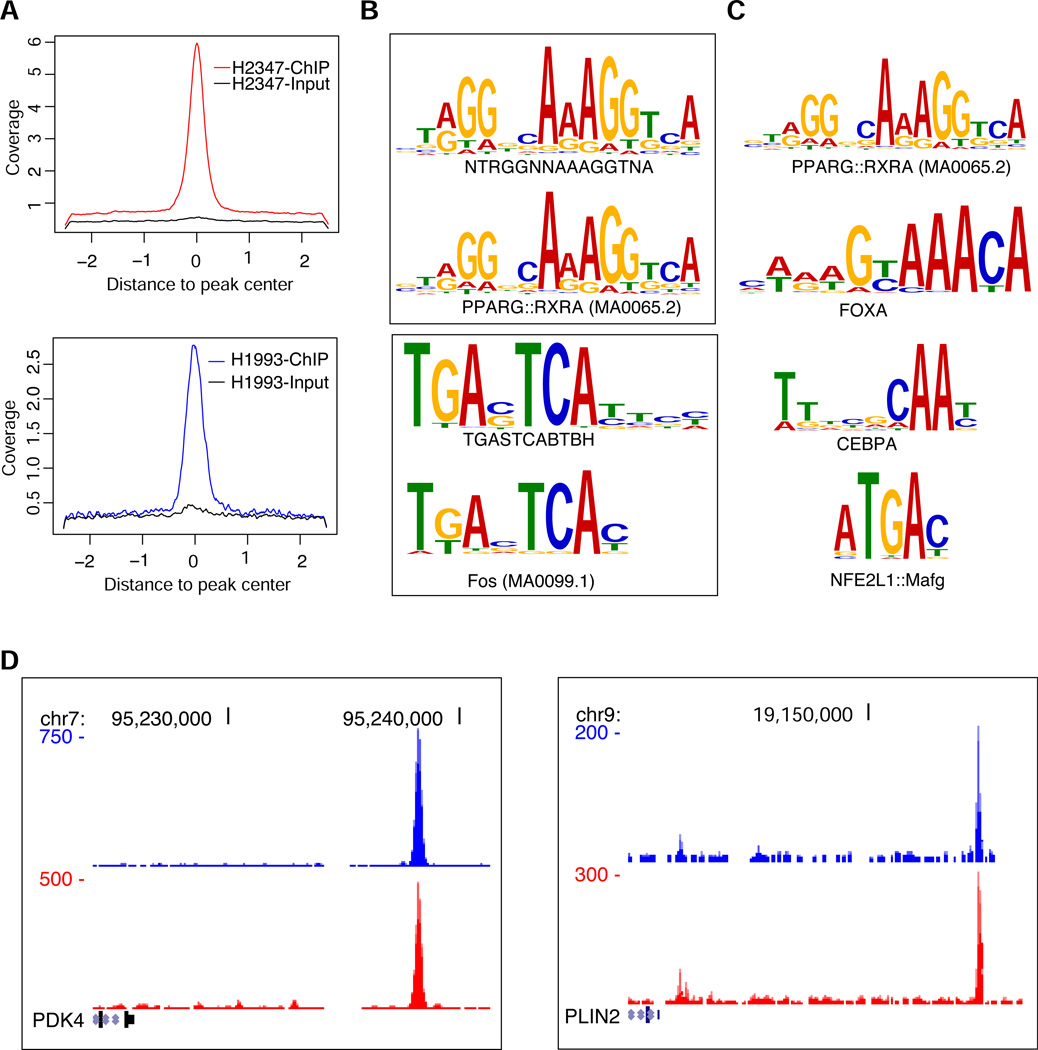
To characterize the genetic program regulated by PPARγ we analyzed the cistrome of this transcription factor and the effects of PPARγ activation by TZDs on transcription on a genome-wide scale. For the cistrome analysis, we used chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) with NCI-H2347 and NCI-H1993 lung adenocarcinoma cells, which express PPARγ at high levels and whose growth is inhibited by TZDs such as pioglitazone and troglitazone (Jeong et al., 2012). We found 2225 regions in NCI-H2347 cells and 1419 in NCI-H1993 cells with a significantly increased occupancy for PPARγ (Figure 1A, Table S1). Of these binding sites, 484 were shared by both cell lines. De novo motif analysis of the PPARγ binding regions identified a canonical PPARγ motif consisting of two direct hexanucleotide repeats with a one nucleotide spacer (DR1) as the most significant motif (Figure 1B, Table S2), which is similar to the motif elicited for PPARγ occupied regions in mouse 3T3-L1 cells (Lefterova et al., 2008; Nielsen et al., 2008). The second most significant motif identified by MEME is highly similar to the AP-1 (Fos) motif, which suggests that PPARγ target genes in lung adenocarcinoma cell lines may be co-regulated by AP-1 transcription factors. The canonical PPARγ motif was also the most highly and significantly enriched motif when we compared the frequency of known transcription factor motifs in PPARγ-bound regions with respect to their frequency distribution in the human genome (Figure 1C, Table S3). This analysis identified additional enriched motifs of other transcription factors such as Nrf2 (NFE2L1), Forkhead and CEBP family proteins, which are recruited to PPARγ binding regions (Lefterova et al., 2008). These findings demonstrate the presence of bona fide, direct binding sites of PPARγ in the genome of lung cancer cells. When we analyzed the putative PPARγ target genes (defined as genes whose transcription start sites are within 100 kB to PPARγ occupied loci) shared by both NCI-H2347 and NCI-H1993 cells, e.g. the established PPARγ target genes PDK4 and PLIN2 (Lefterova et al., 2008; Nielsen et al., 2008) (Figure 1D), for enrichment in functional annotation categories, we found biological processes that are relevant for known metabolic functions of PPARγ (Figure 2A). Importantly, we found a highly significant enrichment for genes implicated in several aspects of lipid metabolism among the predicted PPARγ targets.
Figure 1. Characterization of the PPARγ cistrome in lung cancer cell lines.
(A) Density plots of ChIP-seq reads for PPARγ occupied regions in NCI-H2347 and NCI-H1993 cells. The x axis depicts 2 kb 5’ and 3’ to the predicted peak summits (0). The y axis depicts read coverage.
(B) de novo motif analysis for PPARγ occupied regions. Shared PPARγ occupied regions of NCI-H2347 and NCI-H1993 were analyzed with MEME. The two most significant motifs are shown. These motifs have the most significant similarity to the known PPARγ::RXRA or Fos/AP1 motif, respectively.
(C) Known ENCODE and JASPAR transcription factor motif enrichment in PPARγ bound regions. Enrichment scores and p values were calculated based on the distribution of each motif in the genome using 75,000 simulations.
(D) PPARγ binding regions near the transcription start sites of putative PPARγ target genes (PDK4 and PLIN2) as depicted in the UCSC Genome Browser. The x axis depicts genome coordinates, the y axis depicts ChIP-seq read density (red: NCI-H2347, blue NCI-H1993) normalized with respect to input reads.
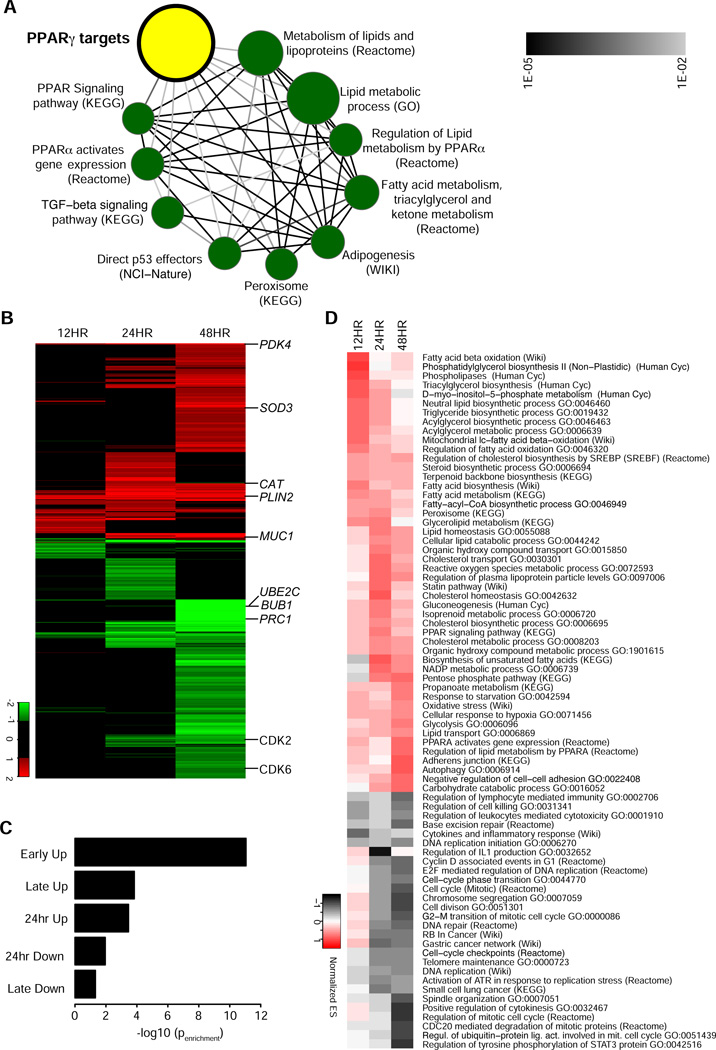
Figure 2. Genomic functions of PPARγ in lung cancer cell lines.
(A) Concept map of PPARγ targets predicted by ChIP-Seq. P values for enrichment between gene signature sets were calculated (hypergeometric test), and used to construct a network of significantly enriched signatures (FDR cut-off 1%). The node size corresponds to the number of genes in a signature set, the edge intensity correlates with the significance of enrichment between two signature sets.
(B) Time course analysis of gene expression for pioglitazone treatment. Heat map of log2 tranformed fold changes of 1781 genes with significantly differential gene expression (cut-off: fold change ≥ 2 and adjusted p value ≤ 0.05) after 12, 24 or 48 hours of pioglitazone treatment. Hierarchical clustering was performed using euclidean distance metric. Several genes are shown as examples for differentially expressed genes (PDK4, SOD3, CAT, PLIN2, MUC1, UBE2C, BUB 1, PRC1, CDK2, CDK6).
(C) Enrichment for PPARγ direct targets in different gene expression clusters. Clusters defined by k-means clustering and the significance of enrichment for PPARγ direct targets as predicted by ChIP-seq was calculated for each cluster (hypergeometric test).
(D) Gene set enrichment analysis for pioglitazone-regulated genes. Hierarchical clustering of normalized enrichment scores for gene sets that are significantly (FDR ≤ 0.05) overrepresented in pioglitazone regulated genes.
We next combined the analysis of the PPARγ cistrome with the analysis of the transcriptional effects of PPARγ activation by TZDs. For that purpose, we performed a time course analysis of gene expression, where we profiled the effects of pioglitazone treatment on the transcriptome of NCI-H2347 cells after 12, 24 and 48 hours of treatment (Figure 2B, Table S4). We identified 1781 genes whose expression was either significantly upregulated or downregulated at any of the three time-points (adjusted p ≤ 0.05; fold change ≥ 2). We used k-means clustering to group genes into five clusters with similar expression profiles. For the obtained clusters we analyzed PPARγ binding site enrichment (Figure 2C). We found that genes in the ‘Early Up’ cluster whose expression is upregulated after 12 hours are most significantly enriched for PPARγ target genes predicted by ChIP-seq, and thus likely present putative direct targets of PPARγ. Genes whose expression is upregulated after 24 or 48 hours were less significantly enriched among genes with PPARγ binding sites, and genes whose expression was decreased showed the least enrichment among PPARγ direct targets suggesting that these genes are not directly regulated by PPARγ. Next, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) to elucidate the gene programs regulated by PPARγ. Among the significantly enriched gene sets (Figure 2D) we found that genes implicated in lipid metabolism such a fatty acid beta oxidation were among the early upregulated genes (e.g., PDK4 and PLIN2), while genes implicated in cell cycle progression (e.g. UBE2C, BUB1, PRC1, CDK2, CDK6) were enriched among the genes whose expression is downregulated late (after 48 hours), and which are not enriched for PPARγ target genes. Furthermore, we found genes implicated in oxidative stress response (e.g. SOD3, CAT, MUC1) among the genes that were upregulated at later times points. Collectively, these findings suggest that PPARγ activation causes the direct and early upregulation of genes controlling lipid metabolism.
PPARγ activation inhibits cell proliferation by an ROS dependent-mechanism
We hypothesize that the increased expression of these genes causes a change in cancer cell metabolism, which in turn causes oxidative stress and ultimately leads to cell cycle arrest in G1. Hence, there was a reduction in transcript levels for genes that are expressed in the S and G2/M phases of the cell cycle. To test this hypothesis, we analyzed the effects of pioglitazone treatment on cell cycle progression and the levels of reactive oxygen species in the lung cancer cell lines. Using DNA content analysis by FACS we found that pioglitazone treatment caused G1 arrest in both NCI-H2347 and NCI-H1993 cells (Figures 3A and 3B) but not in NCI-H1299 and NCI-H1395 cells (Figures S1A and S1B) that are essentially PPARγ-negative (Jeong et al., 2012). The pioglitazone-induced cell cycle arrest coincided with a marked reduction in the levels of phosphorylated RB in the NCI-H2347 and NCI-H1993 cells within 24 hours of treatment (Figure 3C), but not in the PPARγ-negative NCI-H1299 and NCI-H1395 cells (Figure S1C). We found that pioglitazone-induced RB hypophosphorylation was PPARγ-dependent as PPARγ knockdown with two independent siRNAs prevented a reduction of phosphorylated RB by pioglitazone in NCI-H2347 cells (Figure S1D). Also, we observed RB hypophosphorylation when we treated NCI-H2347 and NCI-H1993 cells with an alternative TZD, troglitazone (Figure S1E). Pioglitazone treatment of NCI-H2347 cells resulted in decreased BrdU incorporation in a time-dependent manner (Figure S1F), but did not cause apoptosis as PARP or Caspase-3 cleavage (Figure S1G) were not increased. Thus, pioglitazone treatment appears to inhibit cell proliferation in an RB-dependent manner without inducing apoptosis. We found that pioglitazone treatment of the PPARγ-expressing breast cancer cell lines HCC-1954 and HCC-1187 (but not of the PPARγ-negative breast cancer cell lines T-47D and HCC-1143) also resulted in RB hypophosphorylation (Figure S1H), which suggests that our findings are applicable to other major cancer types.
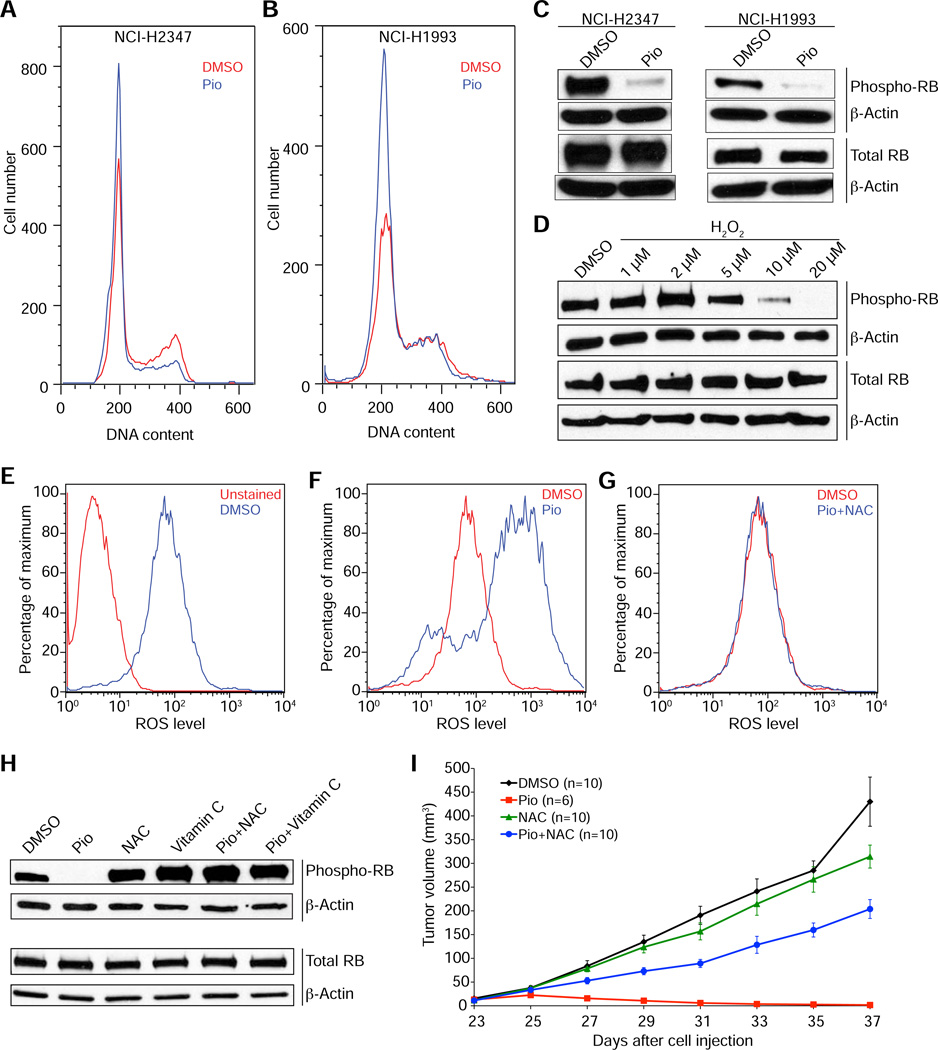
Figure 3. PPARγ activation inhibits cell proliferation by an ROS dependent-mechanism.
(A and B) DNA content analysis of NCI-H2347 (A) and NCI-H1993 (B) cells after 24 hours of treatment with with pioglitazone (Pio) or vehicle (DMSO) indicates a G1 arrest upon PPARγ activation.
(C) Pioglitazone treatment causes a decrease in the levels of phosphorylated RB in both NCI-H2347 and NCI-H1993 cells within 24 hours.
(D) Hydrogen peroxide treatment causes a decrease in the levels of phosphorylated RB in NCI-H2347 within 24 hours.
(E-G) Pioglitazone treatment causes an increase in ROS levels in NCI-H2347 cells.
FACS with DCFDA was performed after 12 hour treatment with vehicle (DMSO) (E) or pioglitazone (F) or pioglitazone and NAC (G). Note that the increase in ROS levels was completely blocked by co-treatment with NAC.
(H) NAC and Vitamin C prevent the reduction in phosphorylated RB levels by pioglitazone in NCI-H2347 cells after 24 hours of treatment.
(I) Tumor growth inhibition by pioglitazone in vivo is ROS-dependent. Mice xenografted with NCI-H2347 cells were treated upon appearance of tumors with pioglitazone, DMSO (vehicle), NAC, or a combination of pioglitazone and NAC (i.p.) every other day beginning on day 23 after cell injection. Error bars represent s.e.m.
Next, we tested if treatment of NCI-H2347 cells with H2O2 caused RB hypophosphorylation, and found that low micromolar concentrations of H2O2 (>5 µM) caused a dose-dependent decrease in the levels of phosphorylated RB (Figure 3D). To analyze the impact of PPARγ activation on ROS levels, we used the fluorescent ROS probe DCFDA for FACS (Figure 3E) and found that pioglitazone markedly increased ROS levels in PPARγ-positive NCI-H2347 (Figure 3F) and NCI-H1993 cells (Figures S2A and S2B) but not in the PPARγ-negative NCI-H1299 and NCI-H1395 cells (Figures S2C-F). Likewise, pioglitazone markedly increased ROS levels in the PPARγ-positive HCC-1954 and HCC-1187 breast cancer cells (Figures S2G-J) but not in the PPARγ-negative T-47D and HCC-1143 cells (Figures S2K-N). The increase in ROS levels upon pioglitazone treatment could be rescued both by treatment with the antioxidant N-Acetylcysteine (NAC) (Figure 3G) or PPARγ knockdown by siRNAs (Figures S2O-R). Furthermore, we found ROS levels increased when we treated NCI-H2347 and NCI-H1993 cells with troglitazone (Figures S2S-V).
The above findings suggested that PPARγ activation causes an increase in both ROS levels and RB hypophosphorylation. To corroborate this hypothesis we performed two experiments. First, we tested if antioxidant treatment could prevent RB hypophosphorylation after pioglitazone treatment in PPARγ-expressing cells. We found that NCI-H2347 cells co-treated with NAC or with another antioxidant, vitamin C, showed no decrease in phosphorylated RB levels upon pioglitazone treatment (Figure 3H). Likewise, NAC prevented RB hypophosphorylation by pioglitazone in HCC-1954 and HCC-1187 breast cancer cells (Figure S1I). Second, we tested if increased ROS levels were required for the growth inhibition observed for TZDs in vivo (Jeong et al., 2012). For this purpose we tested the effects of NAC on pioglitazone-mediated inhibition of NCI-H2347 tumor xenograft growth. We found that NAC co-treatment significantly mitigated the antitumorigenic effects of pioglitazone (p=1e-0.4) (Figure 3I).
To pinpoint the mechanistic link between increased ROS levels and RB hypophosphorylation, we tested if active dephosphorylation of RB by the RB-dephosphorylating enzyme protein phosphatase 2A (PP2A) (Alberts et al., 1993; Avni et al., 2003) was required for the effect of pioglitazone on RB phosphorylation. We found that low nanomolar concentrations of okadaic acid, which achieves selective and effective inhibition of PP2A prevented the increase in dephosphorylated RB after pioglitazone treatment (Figures S1J) suggesting that increased ROS levels activate PP2A catalytic activity.
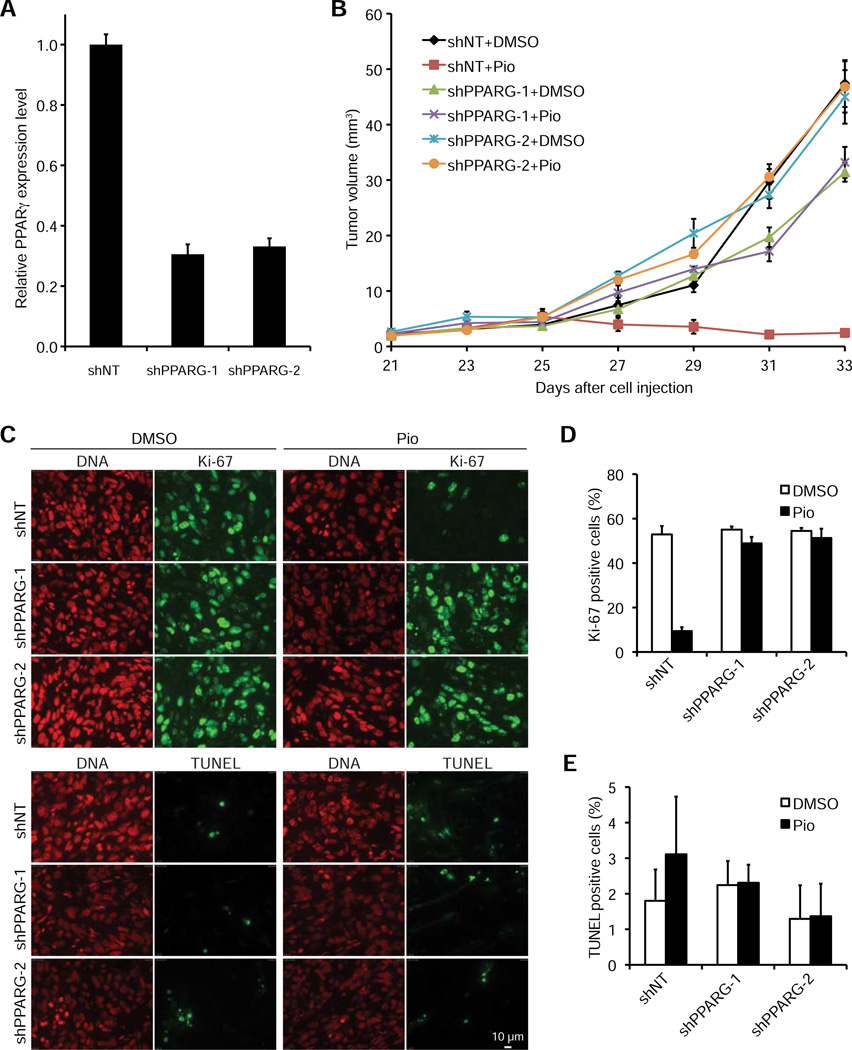
To further validate the specificity and characterize the effects of TZDs on the growth of PPARγ-expressing tumors, we generated NCI-H2347 cell lines that stably express two independent shRNAs targeting PPARγ (shPPARG-1 and shPPARG-2, achieving ~70% PPARγ knockdown, Figure 4A) and a non-targeting shRNA (shNT) as negative control. We found that the growth of NCI-H2347 tumors with stable PPARγ knockdown was not inhibited by pioglitazone treatment in vivo (Figure 4B). We further characterized the underlying cause of pioglitazone-mediated inhibition of tumor growth by analyses of xenograft tumor cell proliferation (Ki-67 staining) and apoptosis (TUNEL staining) (Figure 4C). We found that pioglitazone treatment dramatically reduced the percentage of Ki-67 positive (i.e. proliferating cells) in the shNT (p=0.0045) but not in the shPPARG-1 and shPPARG-2 tumors (Figure 4D) (p>0.05) and had no statistically significant effect on the percentage of apoptotic cells in either tumor (Figure 4E) (p>0.05). Collectively, these findings suggest that PPARγ causes metabolic changes that result in increased levels of ROS, which ultimately lead to active RB hypophosphorylation and thus inhibition of cancer cell proliferation.
Figure 4. The antitumorigenic effect of pioglitazone in vivo is caused by inhibition of cell proliferation and is PPARγ-dependent.
(A) PPARγ mRNA expression in NCI-H2347 cells stably expressing shNT (negative control), shPPARG-1 or shPPARG-2. PPARγ transcript levels were determined by quantitative RT-PCR using B2M as a calibrator (n=3, error bars represent s.d.).
(B) Mice xenografted with NCI-H2347 shNT, shPPARG-1 and shPPARG-2 cells were treated upon appearance of tumors with DMSO (vehicle) or pioglitazone every other day beginning on day 25 after cell injection. Error bars represent s.e.m. N = 10 for all treatment groups. Growth of the shNT tumors was inhibited by pioglitazone treatment (p<1e-5), but no such effect was observed for the shPPARG-1 and shPPARG-2 tumors (p>0.05).
(C-E) Tumors were obtained at the end of the treatment course and stained for Ki-67, TUNEL, and DNA (propidium iodide) (Scale bar is 10 µm) (C). For quantification of Ki-67 positivity (D) and TUNEL positivity (E) two microscopic fields containing at least 100 cells each were analyzed for each treatment group. Error bars represent s.d.
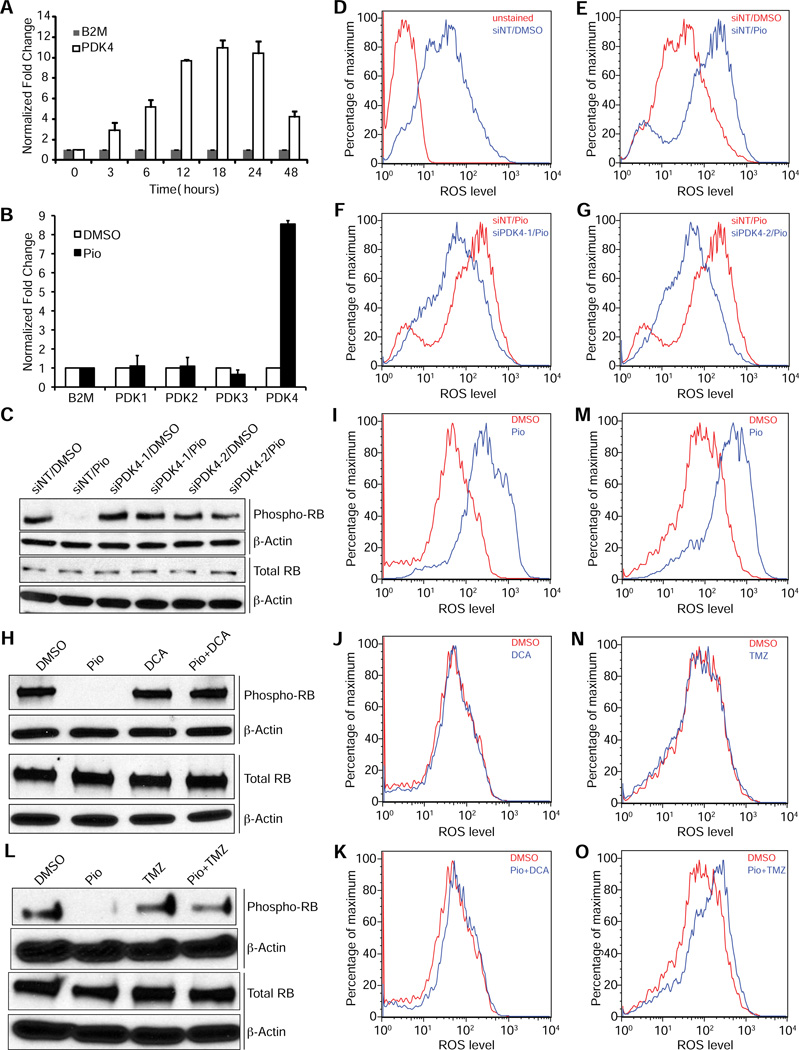
PDK4 and β-oxidation are required for the effects of pioglitazone on ROS levels and RB phosphorylation
In order to identify metabolic regulators responsible for the antitumorigenic effects of PPARγ activation we analyzed the functions of genes whose expression was upregulated early by pioglitazone in our gene expression time course analysis. Among these genes we found pyruvate dehydrogenase kinase 4 (PDK4) as the most highly upregulated direct PPARγ target gene. Using quantitative RT-PCR analysis we found a 3.1-fold increase in PDK4 transcript levels in NCI-H2347 cells after three hours of pioglitazone treatment, which further increased to more than 10-fold higher levels after 12 hours (Figure 5A). PDK4 is a regulatory kinase that phosphorylates and thereby inhibits pyruvate dehydrogenase. Thus, PDK4 inhibits pyruvate oxidation, and was shown to mediate the metabolic switch from glucose oxidation to fatty acid oxidation in normal tissues (Andrews et al., 1998; Holness et al., 2000; Hue and Taegtmeyer, 2009; Strand et al., 2012). Interestingly, upregulation of PDK4 expression was previously shown to inhibit cell proliferation of MCF-10A mammary epithelial cells (Grassian et al., 2011). Importantly, the same study found that PDK4 expression is markedly downregulated in many common solid tumors including lung and breast cancer, suggesting that PDK4 may cause metabolic changes that inhibit tumor cell proliferation. We did not observe increased expression of any of the other three pyruvate dehydrogenase kinase genes (PDK1, PDK2, and PDK3) upon pioglitazone treatment (Figure 5B). In order to test the role of PDK4 in the induction of oxidative stress and RB hypophosphorylation by TZDs we analyzed the effects of PDK4 knockdown by siRNAs or inhibition with the pyruvate dehydrogenase kinase inhibitor dichloroacetate (DCA) on ROS levels and RB phosphorylation. We found that PDK4 knockdown (Figure S3A) reversed the effects of pioglitazone on RB phosphorylation (Figure 5C) and ROS levels (Figures 3D-G) in NCI-H2347 and HCI-H1993 cells (Figures S3B and S3E-H), as did inhibition by DCA (Figures 5H-K and Figures S3C and S3I-L). Next, we tested whether fatty acid oxidation was required for the increased ROS levels and RB hypophosphorylation by TZDs. For that purpose we used trimetazidine, an inhibitor of acetyl-coenzyme A acyltransferase 2 (ACAA2), which catalyzes the terminal step of the β-oxidation cycle. This compound prevented RB hypophosphorylation by pioglitazone (Figure 5L, Figure S3D) and the increase of ROS levels (Figures 5M-O, Figures S3M-P) by pioglitazone in NCI-H2347 and NCI-H1993 cells. We tested whether pioglitazone treatment affected mitochondrial superoxide generation directly using the superoxide fluorescent FACS probe MitoSOX. We observed no effect of pioglitazone on mitochondrial super oxide generation using antimycin A and rotenone as positive controls (Figures S3Q-T). We note that inhibition of NADPH oxidase, which is one major source of ROS by apocynin prevented the increase in ROS levels by pioglitazone, but also caused a slight reduction of ROS levels without pioglitazone treatment (Figures S3U-X).
Figure 5. PDK4 and β-oxidation are required for the effects of pioglitazone on ROS levels and RB phosphorylation.
(A and B) PDK4 mRNA expression rapidly increases upon pioglitazone treatment (A), but expression of PDK1, PDK2 and PDK3 mRNA is not increased after 24 hours of pioglitazone treatment (B). PDK1, PDK2, PDK3, and PDK4 transcript levels in NCI-H2347 cells were determined by quantitative RT-PCR using B2M as a calibrator for a pioglitazone treatment time course (A) or after 24 hours of treatment (B) (n=3, error bars represent s.d.).
(C-G) PDK4 knockdown by siRNA prevents the decrease in phosphorylated RB (C) and the increase in ROS levels (D-G) upon pioglitazone treatment. NCI-H2347 cells were transfected with two siRNAs targeting PDK4 or a non-targeting siRNA (siNT), after 48 hours the cells were treated with vehicle or pioglitazone for 24 (C) or 12 hours (D-G). Then the cells were analyzed by immunoblotting (C) or stained with DCFDA and assayed by FACS (D-G).
(H-K) Co-treatment with the PDK inhibitor DCA prevents the decrease in phosphorylated RB levels (H) and the increase in ROS levels (I-K) upon pioglitazone treatment. NCI-H2347 cells were treated with vehicle, pioglitazone, DCA, or pioglitazone and DCA for 24 (H) or 12 hours (I-K). Then the cells were analyzed by immunoblotting (H) or stained with DCFDA and assayed by FACS (I-K).
(L-O) Co-treatment with the β-oxidation inhibitor trimetazidine prevents the decrease in phosphorylated RB levels (L) and the increase in ROS levels (M-O) upon pioglitazone treatment. NCI-H2347 cells were treated with vehicle, pioglitazone, trimetazidine, or pioglitazone and trimetazidine for 24 (L) or 12 hours (M-O). Then the cells were analyzed by immunoblot (L) or stained with DCFDA and assayed by FACS (M-O).
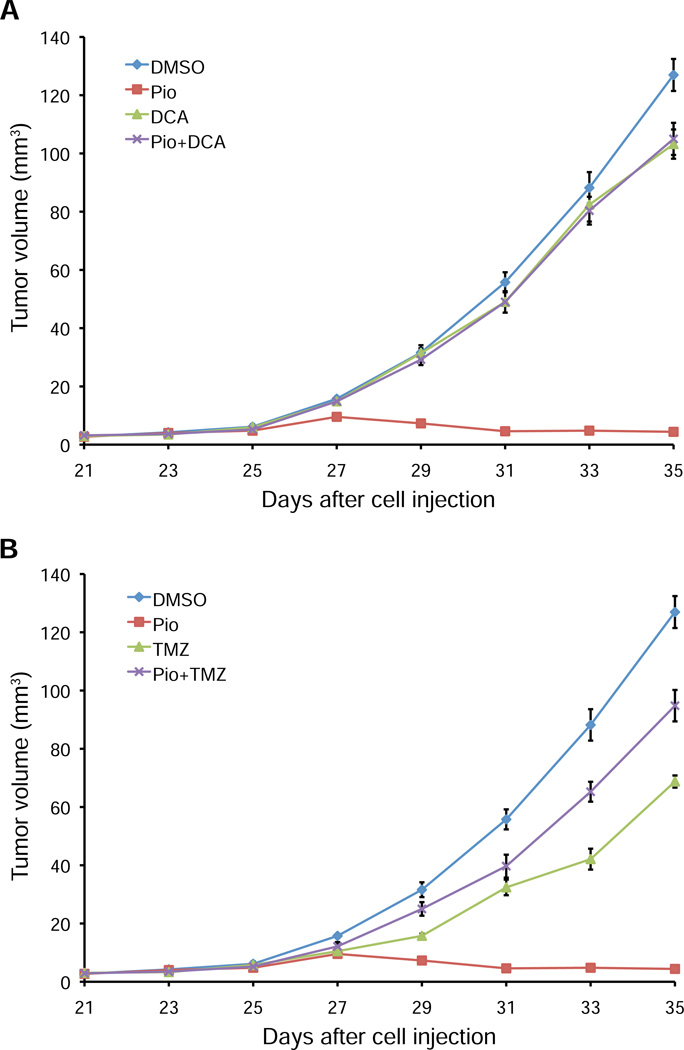
To test the relevance of PDK4 and fatty acid oxidation on tumor growth inhibition by PPARγ activation in vivo, we analyzed the effects of DCA (Figure 6A) and trimetazidine (Figure 6B) in NCI-H2347 xenograft experiments. We found that both compounds suppressed the growth-inhibiting effects of pioglitazone, which indicates a critical role of PDK4 and fatty acid oxidation in the antitumorigenic effects of PPARγ activation. Collectively, these findings suggest that the upregulation of PDK4, a key regulator of glucose and fatty acid utilization is necessary for the increase in ROS levels that ultimately leads to RB hypophosphorylation and tumor growth arrest.
Figure 6. Tumor growth inhibition by pioglitazone in vivo requires PDK activity and fatty acid oxidation.
(A and B) Mice xenografted with NCI-H2347 cells were treated upon appearance of tumors with DMSO (vehicle), pioglitazone, DCA (A), trimetazidine (B), a combination of pioglitazone and DCA (A), or pioglitazone and trimetazidine (B) (i.p.) every other day beginning on day 25 after cell injection. Error bars represent s.e.m. N = 10 for all treatment groups. Both DCA and trimetazidine suppress the growth-inhibiting effects of pioglitazone (p<1e-5).
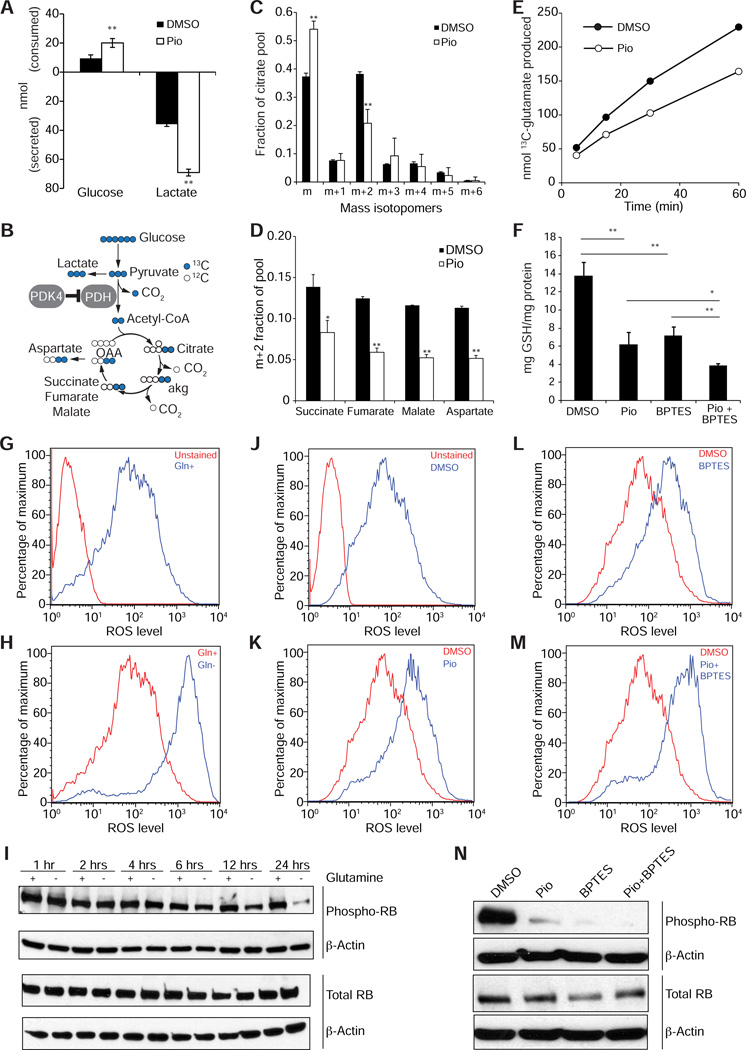
PPARγ activation alters glucose and glutamine metabolism in NCI-H2347 cells to culminate in suppressed glutathione abundance
To pinpoint the metabolic effects of PPARγ activation in cancer cells, we applied a suite of assays designed to interrogate key nodes of metabolic activity. Consistent with the known function of PDK4 in limiting pyruvate oxidation, examination of tissue culture medium revealed enhanced glucose consumption and lactate secretion in pioglitazone-treated cells (Figure 7A). Furthermore, culture with [U-13C]glucose (Figure 7B) revealed a suppressed production of citrate molecules (Figure 7C) and other TCA cycle metabolites (Figure 7D) containing two glucose-derived carbons (m+2), indicative of reduced PDH contribution to the acetyl-CoA pool. We next tested whether metabolism of glutamine, the predominant source of glutamate, was altered by pioglitazone. For this purpose, we pre-treated cancer cells with pioglitazone for 6 or 12 hours prior to adding [U-13C] glutamine to the medium, and followed the time course of labeling in the glutamate pool. Pioglitazone reduced the conversion rate of glutamine to glutamate (Figure 7E). Since glutamate is required for the synthesis of glutathione, a central component of cellular ROS detoxification, we tested whether pioglitazone affected the levels of glutathione. We found that pioglitazone treatment resulted in a marked decrease of both reduced and total glutathione (GSH) levels (Figure 7F, Figures S4A and S4B), which mirrored the effects of glutaminase inhibition by BPTES, consistent with an overall reduction in glutathione biosynthesis, as would be predicted to follow a suppression of glutamate synthesis. Furthermore, we demonstrate that pioglitazone markedly reduced the flux from [U-13C] glutamine in the medium to intracellular GSH within 12 hours of treatment, and also mirrored the effects of glutaminase inhibition by BPTES (Figure S4C). Also, we found that inhibition of fatty acid oxidation with trimetazidine prevented the reduction of glutamate synthesis from glutamine (Figure S4D). When considered together with our other observations, these results suggest that the metabolic changes induced by PPARγ activation impair glutaminolysis, reducing levels of glutamate and glutathione. According to this model, the reduction in glutathione levels diminishes the capacity of the cancer cells to detoxify reactive oxygen species. To corroborate this hypothesis, we tested if glutaminase inhibition or glutamine withdrawal inhibition would have similar effects in NCI-H2347 cells as pioglitazone treament. The glutaminase inhibitor BPTES also suppressed both reduced and total levels of GSH, and combining pioglitazone with BPTES enhanced these effects (Figure 7F). Also, we found that glutamine withdrawal resulted in a marked increase of ROS levels within twelve hours (Figures 7G and 7H), and led to a time-dependent decrease of phosphorylated RB levels (Figure 7I). Likewise, glutaminase inhibition with BPTES caused upregulation of ROS levels (Figures 7J-M), which was suppressed by NAC (Figures S4E-H), led to RB hypophosphorylation (Figure 7N), which was prevented by co-treatment with NAC (Figure S4I), and resulted in a marked inhibition of the growth of xenograft tumors in vivo (Figure S4J). Thus, glutaminase inhibition phenocopies essentially all effects of pioglitazone treatment, which supports our mechanistic model for the effects of PPARγ activation on cancer cell metabolism and cell proliferation.
Figure 7. PPARγ activation alters glucose and glutamine metabolism in NCI-H2347 cells to culminate in suppressed glutathione abundance.
(A) Glucose consumption and lactate secretion in vehicle and pioglitazone (Pio)-treated NCI-H2347 cells. Data are the average and s.d. of three independent cultures.
(B) Isotope tracing diagram illustrating the supply of carbon from glucose to the TCA cycle. Blue circles are 13C originating from [U-13C]glucose. Open circles are unlabeled carbon (12C). Abbreviations: PDH, pyruvate dehydrogenase; PDK4, pyruvate dehydrogenase kinase-4; aKG, α-ketoglutarate, OAA, oxaloacetate.
(C and D) Mass isotopomer distribution for citrate in vehicle and pioglitazone-treated cells cultured with [U-13C]glucose (C). Isotopomers are shown as mass (m) plus additional mass units from 13C. m+2 fraction of additional metabolites related to the TCA cycle (D). Data are the average and s.d. of three independent cultures.
(E) Time-dependent production of [U-13C]glutamate in control and pioglitazone-treated cells cultured with [U-13C]glutamine. Each time point is derived from an independent culture.
(F) Glutathione (GSH) abundance in control cells and in cells treated with pioglitazone, BPTES, or pioglitazone and BPTES. Data are the average and s.d. of three independent cultures. *, p<0.05; **, p<0.005.
(G and H) Glutamine depletion causes a rapid increase in ROS levels. NCI-H2347 cells were grown for 12 hours in the presence (Gln+) (G) or absence (Gln−) (H) of glutamine in the medium, and stained with DCFDA and assayed by FACS.
(I) Glutamine depletion causes a decrease in phosphorylated RB levels. NCI-H2347 cells were grown for 1, 2, 4, 6, 12 or 24 hours in the presence (+) or absence (−) of glutamine in the medium.
(J-M) Glutaminase inhibition by BPTES causes an increase in ROS levels. NCI-H2347 cells were treated for 12 hours with vehicle (J), pioglitazone (K), BPTES (L), or BPTES and pioglitazone (M), and stained with DCFDA and assayed by FACS.
(N) Glutaminase inhibition with BPTES causes a decrease in phosphorylated RB levels. NCI-H2347 cells were treated with vehicle, pioglitazone, BPTES, or pioglitazone and BPTES for 24 hours.
DISCUSSION
Tumor-suppressive functions have been proposed for PPARγ and its agonists in a variety of common cancers such as breast, colorectal and lung cancer (Koeffler, 2003; Peters et al., 2012). For lung cancer, activation of PPARγ with TZDs was shown to be effective in inhibiting tumor growth of PPARγ-expressing human cancer cells in vitro and in vivo (Jeong et al., 2012). Importantly, combination of TZDs with platinum-based chemotherapy in mouse models of lung adenocarcinoma (Girnun et al., 2008) was shown to be highly effective in reducing tumor growth, and upregulation of PPARγ expression (and its target genes) has been linked to the antitumorigenic effects of Myc oncogene ablation in a lung adenocarcinoma mouse model (Soucek et al., 2013). In this vein, we found that high expression of PPARγ in early stage lung adenocarcinomas predicts longer overall and/or progression-free survival in independent patient cohorts (Figures S4A-C). Collectively, these observations suggest an antitumorigenic role for PPARγ in lung adenocarcinoma.
Several mechanisms have been proposed for growth inhibition of PPARγ and TZDs (Koeffler, 2003; Peters et al., 2012), but despite the established role of PPARγ as a key regulator of glucose and fatty acid metabolism in normal tissue, specific changes in cancer metabolism have not been proposed previously as a key mechanism for the anitumorigenic actions of PPARγ. Our findings show that PPARγ activation causes changes to glucose, fatty acid and glutamine metabolism that ultimately result in ROS-mediated RB hypophosphorylation, which can be prevented either by suppressing the PPARγ target PDK4 or by inhibiting β-oxidation. Thus we propose a mechanism by which PPARγ-induced metabolic changes elicit direct effects on cell cycle progression. Importantly, our study demonstrates an essential role of ROS in this metabolic regulation of cell proliferation and thus highlights a key role of ROS in the integration of metabolism and proliferation. Furthermore, we establish a proof of principle that this pathway may be therapeutically targeted with FDA-approved antidiabetic drugs such as pioglitazone and troglitazone. An altered redox state has long been observed in cancer cells, making transformed cells vulnerable to further increases in ROS levels and highly dependent on ROS detoxification systems (Cairns et al., 2011; Trachootham et al., 2009). In this context, our findings suggest that modulating glucose, lipid and glutamine metabolism with nuclear receptor ligands, alone or in combination with existing therapies, may provide an avenue for the development of novel cancer treatment strategies.
EXPERIMENTAL PROCEDURES
Cell Culture
NCI-H2347 and NCI-H1993 Cells were cultured in RPMI with 5% FBS and incubated at 37 degree at 5% CO2. For all experiments, cells were seeded one day prior to treatment.
Chromatin immunoprecipitation sequencing (ChIP-seq)
We generated PPARγ-LAP BAC transgenic NCI-H2347 and NCI-H1993 cell lines using the BAC-transgenesis approach to perform ChIP as previously described (Hua et al., 2009; Kittler et al., 2013; Poser et al., 2008). Barcoded libraries of ChIP and input DNA were generated with the SOLiD Fragment Library Barcoding Kit (Applied Biosystems), and 35-nt single-end reads were generated with the SOLiD4 HQ system (Applied Biosystems). PPARγ-bound regions were identified as genomic regions with a significant read enrichment and binding peak profile in the PPARγ reads over the input reads by using the Model-based Analysis of ChIP-Seq (MACS) (Poser et al., 2008) software tool (v.1.4.2) with 1% FDR. De novo motif discovery analysis for shared PPARγ-bound regions was performed with the Multiple EM for Motif Elicitation (MEME) software tool (Bailey et al., 2006). We analyzed the enrichment of known transcription factor motifs by determining the frequency of known motifs in PPARγ-bound regions and for 75,000 random sets of the same sample size by using Motif Scanner (Aerts et al., 2003). We performed pathway enrichment analysis on shared PPARG target genes in NCI-H2347 and NCI-H1993 cells. Gene sets used for this analysis were obtained from Merico et. al. (Merico et al., 2010). Enrichment p-values were calculated using hypergeometric test and significantly enriched pathways (FDR ≤ 1%) were used to construct an enrichment map.
Quantitative PCR and microarray analysis
Total DNA-free RNA was prepared and used for quantitative real time PCR using SYBR® Green. Quantification was performed with the the comparative CT method (ΔΔCt). Microarray analysis was performed with the Illumina Human HT12v4.0 Expression Beadchip (Illumina, Inc.). Linear models for microarray data (LIMMA) (Smyth, 2004) was used to identify genes that showed significant expression changes between the pioglitazone treated and vehicle samples for each time point (cut-off: adjusted p < 0.05 and log2 fold change of ≥1 and ≤-1). Clustering analysis was performed as hierarchical clustering with euclidean distance metric and “agglomerative average” method, or centroid based clustering (k-means clustering). To determine the enrichment of gene sets at the top or bottom of a ranked list of differentially regulated genes upon pioglitazone treatment, we performed gene set enrichment analysis (GSEA) using the time course expression data and a priori defined gene sets.
Cell cycle and ROS level analysis
DNA content analysis was performed by FACS of ~10,000 propidium-iodide stained cells. Intracellular ROS and mitochondrial superoxide was detected by FACS of ~10,000 cells with oxidation-sensitive fluorescent probes (DCFDA and MitoSOX).
Xenograft experiments
NCI-H2347 cells (2×106/site, 2 sites/animal) were injected subcutaneously into both flank regions of Female athymic NOD SCID mice. When tumors became visible treatment (i.p.) was started with various compounds or DMSO (vehicle), every other day for a total of four to five injections. Tumor volume was calculated as (width) X (width) X (length)/2 where length is greater than width.
Immunohistochemistry
Tumors harvested from anesthetized mice were processed for immunhistochemistry by standard procedures. Mouse anti-sera and subsequent biotin/streptavidin-fluorescein detection of bound primary was used for Ki-67 immunolabeling. Terminal deoxynucleotidyltransferase-mediated UTP end label (TUNEL) staining for apoptotic cells was done according to the protocol supplied with DeadEnd Fluorometric TUNEL System (Promega). Apoptotic cells were labeled with fluorescein, and the sections were counterstained with propidium iodide. Image acquisition was performed with a DM5500B fluorescent microscope (Leica).
Metabolic assays
For the analysis of glucose metabolism, NCI-H2347 cells were incubated with DMSO or Pioglitazone for 6 hours, medium was then replaced with medium containing 25mM [U-13C]glucose. After 6 hours of labeling, medium was removed for glucose and lactate analysis as previously described (Yang et al., 2009). For analysis by mass spectrometry, the cells were processed as previously described (Cheng et al., 2011). To quantify the production of 13C-glutamate from 13C-glutamine, cells were cultured in medium containing 2mM [U-13C]glutamine, and lysed at several time points over 60 minutes. Glutathione quantitation was performed using ultra high performance liquid chromatography. For measuring flux of extracellular glutamine to GSH, cells were cultured in medium containing 2mM [U-13C]glutamine, and lysed after 6, 12 or 24 hours. The measurement of GSH was performed using a liquid chromatography-tandem mass spectrometry (LC/MS/MS) approach.
Data deposition
ChIP-seq and microarray data has been deposited with NCBI Gene Expression Omnibus (GEO) (accession number GSE59736).
Statistical analysis
We tested the statistical significance between tumor volumes by calculating P values for 10,000 permutations with the compareGrowthCurves function from the Statistical Modeling package, statmod, available from the R Project (http://www.r-project.org). For other experiments statistical significance was determined by a two-tailed unpaired Student’s t test. P values < 0.05 were considered statistically significant.
Supplementary Material
PPARγ activates an antiproliferative gene regulatory program in lung adenocarcinoma
PPARγ agonists induce a metabolic switch that reduces glutathione levels
Reduced glutathione levels increase ROS levels, which mediate cell cycle arrest
ACKNOWLEDGEMENTS
We would like to thank David Mangelsdorf and John Minna, and all other colleagues of the Multi-Investigator Research Awards RP101251 and RP120732 of the Cancer Prevention and Research Institute of Texas (CPRIT) for sharing preliminary data, reagents and discussion of this manuscript. We are grateful to John Shelton, Claire Klimko, the Simmons Comprehensive Cancer Center Microarray Core and the McDermott Sequencing Core at UT Southwestern for technical assistance. We thank Takashi Tsukamoto for sharing BPTES. This study was supported by grants from CPRIT (RP101251-P06 and RP120732-P3 to R.K.; RP130272-02 to R.J.D.); the National Institutes of Health (R01 CA157996 to R.J.D.). Metabolomics was supported in part by a gift from Jerry and Emy Lou Baldridge. R.K. is a John L. Roach Scholar in Biomedical Research and a CPRIT Scholar in Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes five supplemental figures, four supplemental tables, supplemental experimental procedures and supplemental references.
CONTRIBUTIONS
N.S. performed and analyzed most of the experiments, R.K.K. performed all computational analyses related to ChIP-seq and expression profiling, D.K.S., J.S., Z.H., H.N. performed and analyzed experiments, S.W. and C.G.H. performed experiments, R.C. and K.E.H. analyzed data, R.J.D. designed and analyzed the metabolomic experiments, and wrote the manuscript. R.K. conceived, designed and directed this study, analyzed data, and wrote the manuscript.
REFERENCES
- Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AS, Thorburn AM, Shenolikar S, Mumby MC, Feramisco JR. Regulation of cell cycle progression and nuclear affinity of the retinoblastoma protein by protein phosphatases. Proc Natl Acad Sci U S A. 1993;90:388–392. doi: 10.1073/pnas.90.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews MT, Squire TL, Bowen CM, Rollins MB. Low-temperature carbon utilization is regulated by novel gene activity in the heart of a hibernating mammal. Proc Natl Acad Sci U S A. 1998;95:8392–8397. doi: 10.1073/pnas.95.14.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni D, Yang H, Martelli F, Hofmann F, ElShamy WM, Ganesan S, Scully R, Livingston DM. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol Cell. 2003;12:735–746. doi: 10.1016/s1097-2765(03)00355-1. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–w373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnun GD, Chen L, Silvaggi J, Drapkin R, Chirieac LR, Padera RF, Upadhyay R, Vafai SB, Weissleder R, Mahmood U, et al. Regression of drug-resistant lung cancer by the combination of rosiglitazone and carboplatin. Clin Cancer Res. 2008;14:6478–6486. doi: 10.1158/1078-0432.CCR-08-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011;25:1716–1733. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness MJ, Kraus A, Harris RA, Sugden MC. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes. 2000;49:775–781. doi: 10.2337/diabetes.49.5.775. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297:E578–E591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Xie Y, Lee W, Bookout AL, Girard L, Raso G, Behrens C, Wistuba II, Gadzar AF, Minna JD, et al. Research resource: Diagnostic and therapeutic potential of nuclear receptor expression in lung cancer. Mol Endocrinol. 2012;26:1443–1454. doi: 10.1210/me.2011-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Zhou J, Hua S, Ma L, Liu Y, Pendleton E, Cheng C, Gerstein M, White KP. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. The nuclear receptor PPARgamma - bigger than fat. Curr Opin Genet Dev. 1998;8:576–581. doi: 10.1016/s0959-437x(98)80014-2. [DOI] [PubMed] [Google Scholar]
- Koeffler HP. Peroxisome proliferator-activated receptor gamma and cancers. Clin Cancer Res. 2003;9:1–9. [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr., Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–195. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, Hansen JC, Curthoys NP. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand DW, Jiang M, Murphy TA, Yi Y, Konvinse KC, Franco OE, Wang Y, Young JD, Hayward SW. PPARgamma isoforms differentially regulate metabolic networks to mediate mouse prostatic epithelial differentiation. Cell Death Dis. 2012;3:e361. doi: 10.1038/cddis.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavelu K, Pan CQ, Karlberg T, Balaji G, Uttamchandani M, Suresh V, Schuler H, Low BC, Sivaraman J. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc Natl Acad Sci U S A. 2012;109:7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2010;79:1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Willson TM, Lambert MH, Kliewer SA. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu Rev Biochem. 2001;70:341–367. doi: 10.1146/annurev.biochem.70.1.341. [DOI] [PubMed] [Google Scholar]
- Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer research. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.