Abstract
Self-assembly of random copolymers has attracted considerable attention recently. In this feature article, we highlight the use of random copolymers to prepare nanostructures with different morphologies and to prepare nanomaterials that are responsive to single or multiple stimuli. The synthesis of single-chain nanoparticles and their potential applications from random copolymers are also discussed in some detail. We aim to draw more attention to these easily accessible copolymers, which are likely to play an important role in translational polymer research.
Introduction
Self-assembly of polymers has been studied for many decades due to the ability of these materials to offer a rich variety of morphologies and transitions as well as their potential applications in many fields, such as biomedical, micro-electronic, photoelectric and optical materials.1–5 Significant progress has been made in the design and synthesis of a variety of polymers owing to advances in controlled polymerization techniques, such as nitroxide mediated radical polymerization (NMP), atom transfer radical polymerization (ATRP), reversible addition – fragmentation chain transfer polymerization (RAFT) and ring - opening mediated radical polymerization (ROMP).6–11 Copolymers can be broadly classified into two categories: block copolymers and random copolymers. In the case of block copolymers, the monomers are arranged systematically in the form of blocks where each block is a repetition of a certain monomer species whereas in the case of random copolymers different monomeric components of the polymer are randomly arranged where the probability of finding a given monomeric unit at any given location on the polymer is independent of the nature of the adjacent units. Until recently, much focus has been given towards understanding self-assembly of block copolymers, due to their unique and excellent assembly behaviours.12–17 However, the method involving their synthesis can be tedious and time-consuming, as it involves sequential controlled polymerization or post-polymerization treatments such as grafting, substitution, hydrolysis and “click” chemistries.18–20 Although many excellent reviews have been published on self-assembly of block copolymers,12–17 there have been no comprehensive reviews focusing on the self-assembly behaviour of random copolymers. Compared to block copolymers, preparation of random copolymers is relatively easy, as they are typically achieved in a one step co-polymerization of two (or more) different monomers. Therefore, it is intriguing to highlight the supramolecular capabilities of random copolymers in self-assembly.
In this review, we attempt to highlight the recent advances in the development of random copolymers and their applications. In the beginning, we will briefly describe some morphologies and transitions of assemblies, indicating the potential for random copolymers in this field. In this context, stimuli responsive random copolymers will also be specifically discussed. Then we will present several examples of complex aggregates, involving the use of random copolymers, to provide another aspect of application for these systems. As one specific type of assemblies from random copolymers, single-chain nanoparticles and their potential application will be discussed in some detail. Finally, we present an overview of studies on biodegradable amphiphilic copolymers. This topic is selected owing to their potential biomedical applications such as drug delivery and tissue engineering.
Self-assembly of random copolymers
Self-assembly of polymers is usually regarded as an attractive method to produce nanoscale structures with different morphologies, such as spheres, rods, vesicles and cylinders. However, most self-assembly studies have been focused on block copolymers, because their final morphologies can usually be finely controlled and even predicted by the molecular parameters, such as the molecular weight, the length of each block and the chemical nature of blocks.21–25 Compared to block copolymers that often possess excellent assembly behaviours due to their narrow dispersity in both molecular weight and block length, random copolymers are rarely employed to form assemblies because of their ill-defined properties and generally broader dispersity.
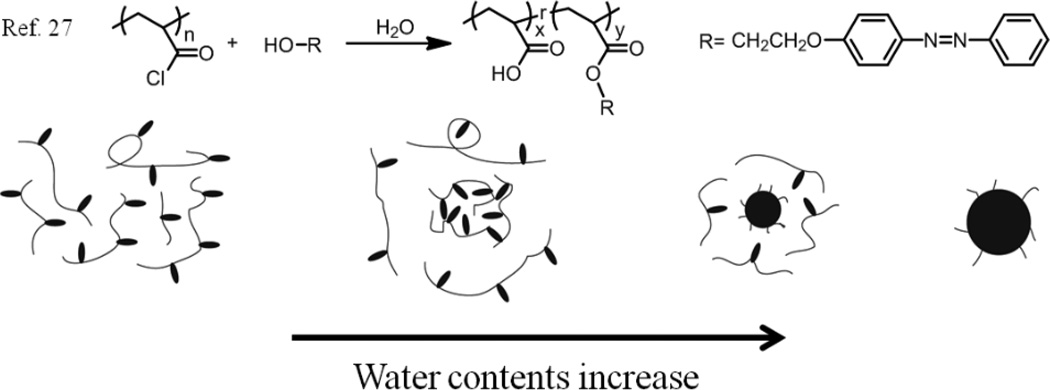
Particles, especially uniform ones, could be easily obtained from block copolymers via the water-induced micellization method.26 This process involves preparing a polymer solution initially by dissolving the block copolymer in an organic solvent. Then water is slowly added into the polymer solution, and the mixture solvent becomes progressively worse for the hydrophobic block until a certain water concentration, called as the critical water content (CWC), is reached at which point phase separation happens and the hydrophobic blocks begin to associate. The polydispersity of these particles greatly depends on the polydispersity of the polymer. Recently, colloidal spheres were prepared from an amphiphilic random copolymer poly(2-[4-(phenylazo)phenoxy]ethyl acrylate-co-acrylic acid) (PPAPE), which had a large Ð as high as 1.9 (Fig. 1).27 The random copolymer was prepared by the reaction between hydrophobic 2-[4-(phenylazo) phenoxy] ethanol and poly(acryloyl chloride), and the remaining acyl chloride groups were then converted to hydrophilic carboxyl groups via hydrolysis. According to the studies on the photoisomerization of the azobenzene units during the particle formation process, a gradual hydrophobic aggregation mechanism was determined. Similar to the water-induced micellization method, these polymeric chains are soluble in a mixture of THF and water when the water content is lower than CWC. However, because the polydispersity of the random copolymer was too high, these polymeric chains would meet their phase separation conditions at different CWC. The most hydrophobic chains aggregate first, leading to nucleation in solution at a relative low CWC. As the water content further increases, the less hydrophobic chains also starts to aggregate and gradually assemble on the surface of the nuclei. The particle sizes obtained from dynamic light scattering (DLS) experiments confirm that the colloidal size increases with increasing water content, indicating that the self-assembly process is indeed gradual. Finally, uniform colloidal spheres were prepared with cores formed from the most hydrophobic chains and coronas consisting of the most hydrophilic chains.
Fig. 1.
Schematic representation of the synthesis of the random copolymers and the sphere formation process.
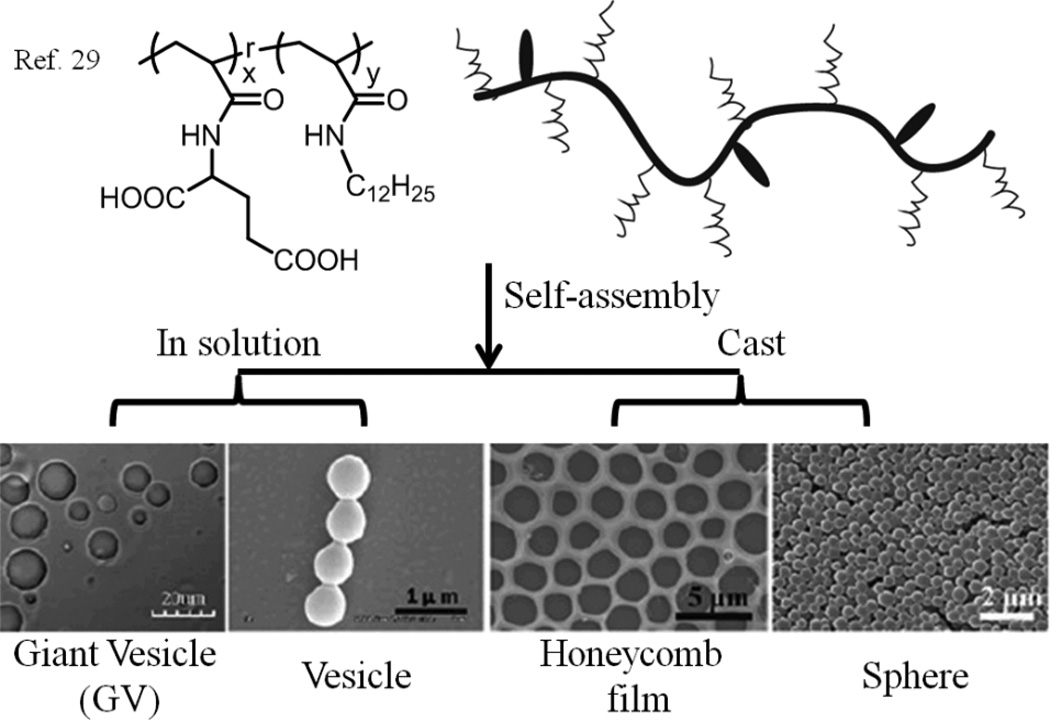
While the morphology of self-assembled block copolymer structures is often dependent on the length of the hydrophobic or hydrophilic block,28 self-assembly of random copolymers is largely dependent on their hydrophilic/hydrophobic balance (often referred as the hydrophilic-lipophilic balance (HLB)). Morphologies could greatly vary a lot with the ratio of hydrophilic side chains to hydrophobic groups. For example, amphiphilic random copolymers containing hydrophobic dodecyl (C12) chain and hydrophilic L-glutamic acid were prepared by copolymerization, in which the ratio of comonomers were easily tuned (Fig. 2).29 Vesicles could form when water was added into an ethanol solution of these polymers, because of the hydrogen bond in the side chains and the hydrophobic interaction between the long alkyl chains. The size of vesicles has been found to depend on the hydrophobic alkyl chain. While vesicles at sizes of several hundred nanometers could be achieved when the hydrophobic ratio was 76%, giant vesicles (GVs) with diameters of several micrometers were observed in a mixed solvent of ethanol and water when the content of hydrophobic alkyl chain was as high as 90%. Note that most reported polymer-based GVs are assembled from block copolymers.30–33 This size difference could be attributed to the decrease in hydrophilic L-glutamic acid groups, which leads to deficiency of hydrophilic groups on the surface to form stable vesicles with larger surface area, i.e. smaller size vesicles.
Fig. 2.
Molecular structures of copolymers and TEM images about the assemblies from these copolymers using different condition.
Another interesting point is that fusion of these GVs could be observed under certain conditions, while the smaller vesicles seem relatively stable. The authors attribute this observation to the possibility of the presence of a few hydrophobic groups along with the hydrophilic L-glutamic acid groups, since this is a random copolymer. This feature provides an opportunity for hydrophobic interactions and thus causes fusion. In the polymers that form smaller vesicles, the hydrophobic content was smaller and therefore hydrophobic interactions were unlikely to be able to effectively compete with the hydrogen bonding interactions. Furthermore, for polymers with a higher hydrophilic content, although no organized structures were formed in solution owing to the higher water solubility of these polymers, films with organized nanostructures could be obtained when the random copolymer solution was directly cast on silicon chip. The morphology was found to depend on solvent selection. Spheres were obtained with ethanol or methanol, while honeycomb-like morphologies were observed with dichloromethane as the solvent. These results show that through appropriate molecular design and preparation processes, a variety of self-assembled morphologies can be obtained with the conveniently accessible random copolymer architectures.
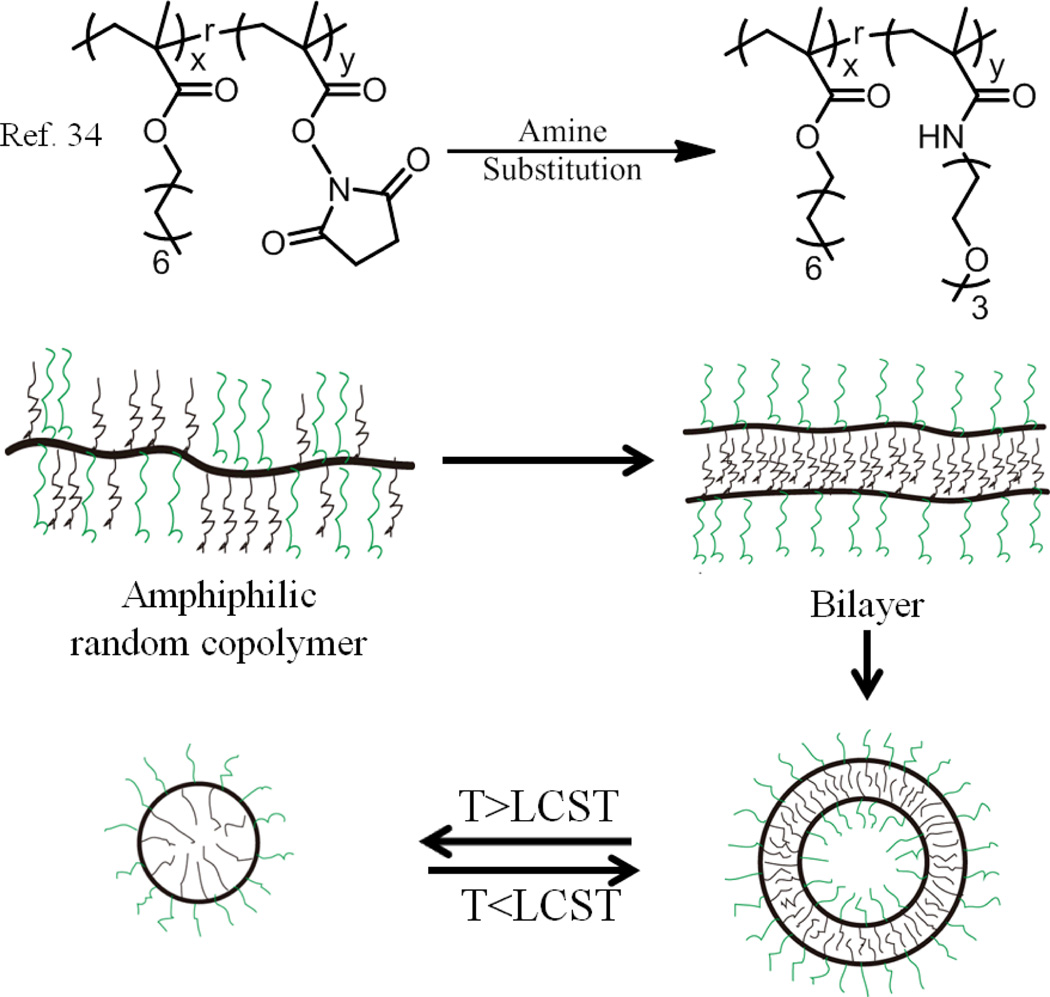
Similarly, polymersomes have been successfully achieved from self-assembly of an amphiphilic random copolymer containing methacrylate-type hydrophobic and methacrylamide-type hydrophilic repeat units.34 These polymers were synthesized through reactions between amine-containing oligooxyethylene units with random copolymers, which were prepared by a copolymerization of n-octyl methacrylate and N-hydroxysuccinimide methacrylate ester (NHSMA). As discerned by transmission electron microscopy (TEM), spherical aggregates with dark thin wall and hollow insides were observed and the aggregate size was in the range of 250–500 nm, indicating the formation of vesicular assembly in solution. Hydrophilic rhodamine 6G (R6G) could be stably encapsulated inside, further confirming that vesicles were formed. These particles undergo a thermo-responsive vesicle to micelle transition, when the temperature was higher than the lower critical solution temperature (LCST). Only dark particles of sizes around 70 nm could be observed in TEM images, suggesting a micelle type aggregation. (Fig. 3) It was suggested that the amide groups played an important role in the vesicle formation as well as in the thermo-responsive vesicle to micelle transition. This suggestion was supported by the preparation of a structurally similar control polymer, in which the amide groups were replaced by ester moieties. These polymers afford spherical micelle type aggregates with an average diameter in the range of 30–40 nm. Although they did not explicitly provide the reasons as to why the amide-based hydrogen bonding led to the specific formation of vesicles, they did demonstrate that the de-solvation of the amide groups would change the hydrophilic/liphophilic balance, resulting in the change in the aggregation morphologies. These authors have further reported that the length of oligoethylene (OE) segments could also affect the morphology of the aggregates.35 While polymers containing longer OE segments could result in the formation of vesicles, multi-micellar clusters were formed for polymers containing short OE segments. These results once again show that random copolymers could also offer a rich variety of morphologies and transitions.
Fig. 3.
Schematic representation of the synthesis of amphiphilic random copolymers and the self-assembly.
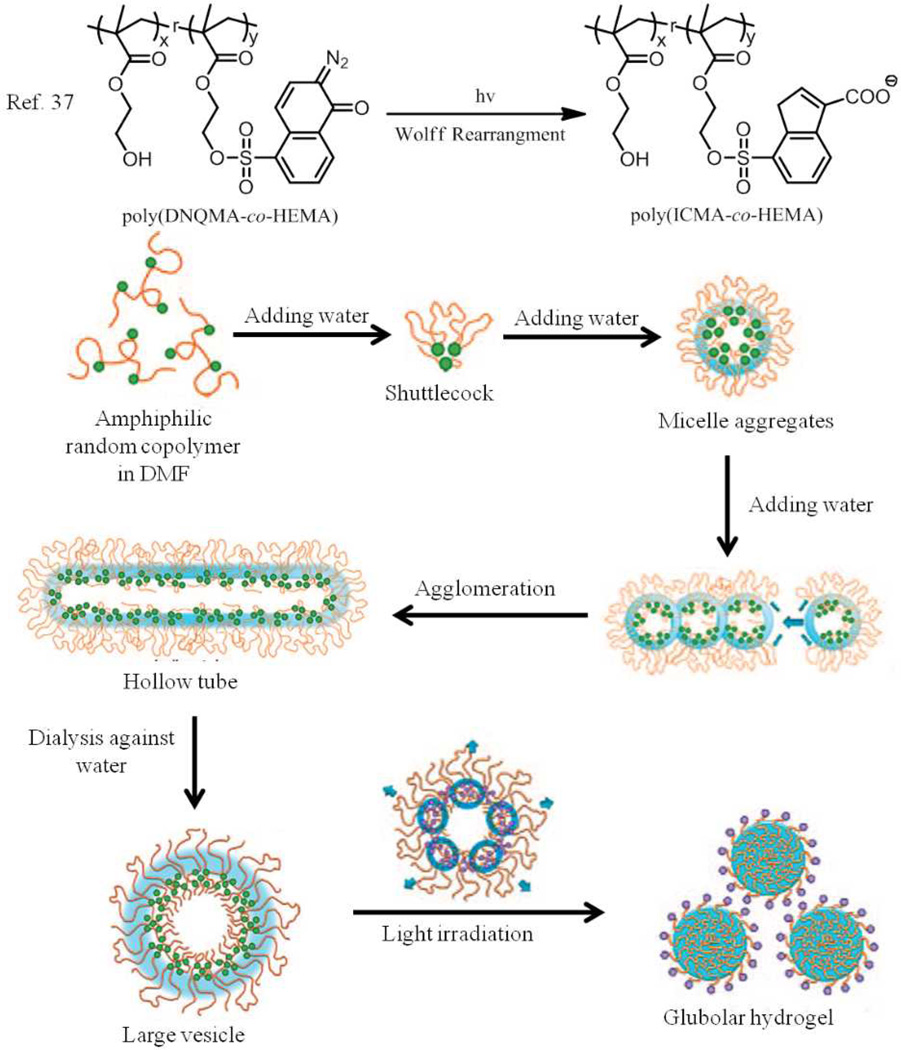
The nature of solvent used for polymeric self-assembly has a significant effect on the final morphology. The micellar morphologies of polystyrene-b-poly(ethylene oxide) have been shown to change from spheres to wormlike and finally to vesicles by adding a solvent (water or acetonitrile) that is selective to one of the components to a copolymer solution in DMF.36 Different morphologies, including spherical micelles, hollow tubes, wormlike rods, and large vesicles have been successfully achieved using amphiphilic random copolymers poly(DNQMA-co-HEMA) with a dispersity of 1.5 (Fig. 4).37 These amphiphilic random copolymers were prepared by esterification between the side chain hydroxyl groups from the hydrophilic poly(hydroxyethyl methacrylate) (PHEMA) and hydrophobic 2-diazo-1,2-naphthquinone (DNQ) molecules. First, the random copolymer poly(DNQMA-co-HEMA) was dissolved in DMF, which is a good solvent for both PHEMA and DNQ. The CWC was determined to be ∼18 wt% by turbidity measurement. When the water content was increased to 20 wt%, the “shuttlecock” morphology with a corklike hydrophobic head and a coneshaped hydrophilic tail was first formed due to the collapse of hydrophobic DNQ chains. Several shuttlecock structures assembled together to form spherical micelles with a diameter around 35 nm, presumably in an effort to reduce the interfacial energy between the polymers and water. However, these spherical assemblies were found not to be the stable state, when the water content further increased to 35 wt%. Further aggregation through the rearrangement of the original shuttlecock-like particles afforded a hollow tube, which was attributed to the lower interfacial energy between the random copolymers and water in this supramolecular arrangement. The water content was further increased up to 60 wt%, which caused the hollow tubes to change their conformation to give wormlike rods. Finally, large vesicles were observed after dialysis in water to completely remove DMF. It is believed that the relatively high molecular weight of poly(DNQMA-co-HEMA) and the suitable hydrophilicity of the polymer backbone play an important role in this consecutive morphological transition driven by simply varying the water content, since the longer chain with hydrophilic backbone provided the possibility to adjust the conformation. Furthermore, it was noticed that the morphology of the large vesicles could change due to the photosensitivity of the DNQ side chains. Upon irradiation with light at 405 nm, hydrophobic DNQ moieties underwent the Wolff rearrangement, resulting in hydrophilic 3-indenecarboxylate groups (IC). This change led to the formation of uniform globular hydrogel-like particles through a chain-rolling process in which the relatively weakly hydrophilic PHEMA backbone formed the framework of hydrogel, while the more hydrophilic IC groups would be exposed on the surface to stabilize the particles. This example indicates that random copolymers have the potential to afford assemblies with different morphologies and functions through simple solvent processing.
Fig. 4.
Schematic representation of the synthesis of the random copolymers and the consecutive morphological transitions in nanoaggregates selfassembled from photo-responsive amphiphilic random copolymer via waterdriven micellization and light-triggered dissociation.
The examples above clearly illustrate that significant progress has been made in tuning the morphologies and their transitions via self-assembling random copolymers. Among them, we also noticed that morphology of the self-assembled structures could also be changed due to the external stimuli such as temperature and light irradiation. These stimuli responsive features of self-assembled structures are of particular interest in the field of drug delivery. Polymeric drug delivery vehicles that respond to single or multiple stimuli have been extensively studied.38–42 By incorporating stimuli responsive features into these delivery vehicles, an increase in therapeutic efficacy of encapsulated drugs can be achieved by triggered release. There are several responsive nanocarrier examples based on custom-designed random copolymers.
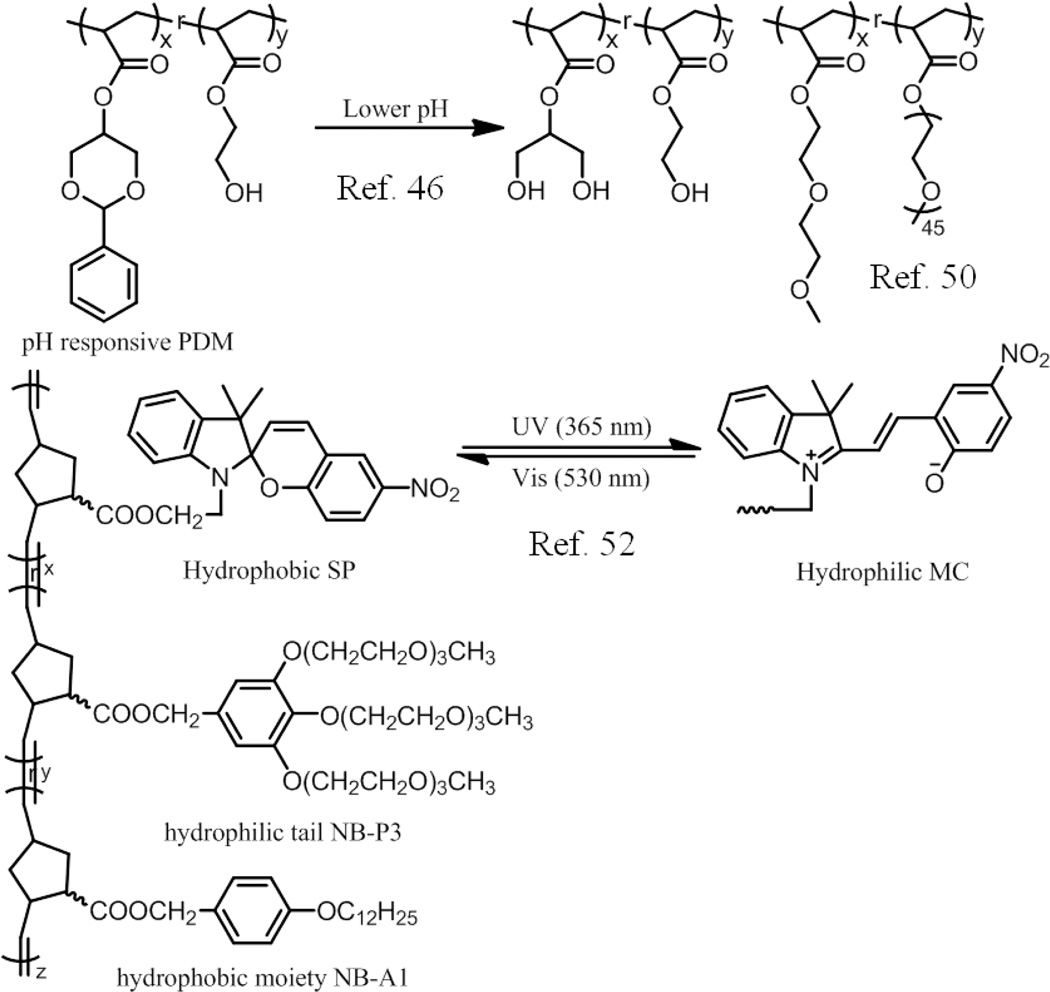
Among current responsive drug delivery system, pH is one of the most popular stimuli and has been explored extensively.43–45 For example, pH sensitive nanoparticles were prepared from amphiphilic copolymer poly(2-phenyl-1,3-dioxan-5-yl methacrylate-co-2-hydroxyethyl acrylate), poly(PDM-co-HEA), in which PDM is pH sensitive hydrophobic moiety and HEA is hydrophilic moiety (Scheme 1).46 Nanoparticles with sizes of about 167 nm (Đ =0.03) were formed in aqueous media. However, when the nanoparticles solution was adjusted to pH 5.5, rapid and remarkable swelling of nanoparticles was observed, as confirmed by DLS experiments. The reason for this phenomenon could be ascribed to the cleavage of hydrophobic, cyclic benzylidene acetal moieties of the polymer, leading to their conversion to more hydrophilic dihydroxypropyl units. Hydrophobic Nile red could be stably encapsulated inside these particles, but were released at low pH.
Scheme 1.
Schematic representation of the polymers used in ref. 46, ref. 50 and ref. 52.
Temperature is another useful stimuli, since it can be conveniently and externally controlled.47–49 A recent report illustrates an interesting example of a two-stage thermal transition of a well-defined random copolymer containing 2-(2-methoxyethxy)ethylmethacrylate (MEO2MA, Mn=188 g/mol) and poly(ethylene glycol)methylethylethermethacrylate (PEGMA, Mn=2080 g/mol) (Scheme 1).50 Taking advantage of the relationship between oligoethylene glycol chain length and temperature sensitivity, it was found that larger aggregates first formed due to the dehydration of short side chains at 27 °C, and then micelles with a compact core-shell structure could be observed upon further heating, which led to the dehydration of longer ethylene glycol segments with a higher LCST. Such multistep aggregation was previously observed with poly (methoxytri(ethylene glycol) acrylate)-b-poly(4-vinyl-benzylmethoxytri-(oxyethylene)ether poly(TEGMA-b-TEGSt) block copolymer.51
`Another interesting example shown below presents the ability to reversibly tune the morphology of the amphiphilic random copolymer in aqueous solutions with light as the stimulus.52 Here, ring-opening metathesis polymerization (ROMP) was used to prepare an amphiphilic random copolymer containing a hydrophobic moiety NB-A1, a hydrophilic tail NB-P3 and functional group NB-SP (Scheme 1), which undergoes reversible photochromic transformation between hydrophobic spiropyran (SP) and hydrophilic merocyanine (MC) moieties upon irradiation by UV and visible light. It is reported that the amphiphilic random copolymer can self-assemble to form polymeric micelles in water, which could be disrupted upon light irradiation at 365 nm, but the micellar architectures could rapidly be regenerated upon light irradiation at 530 nm.
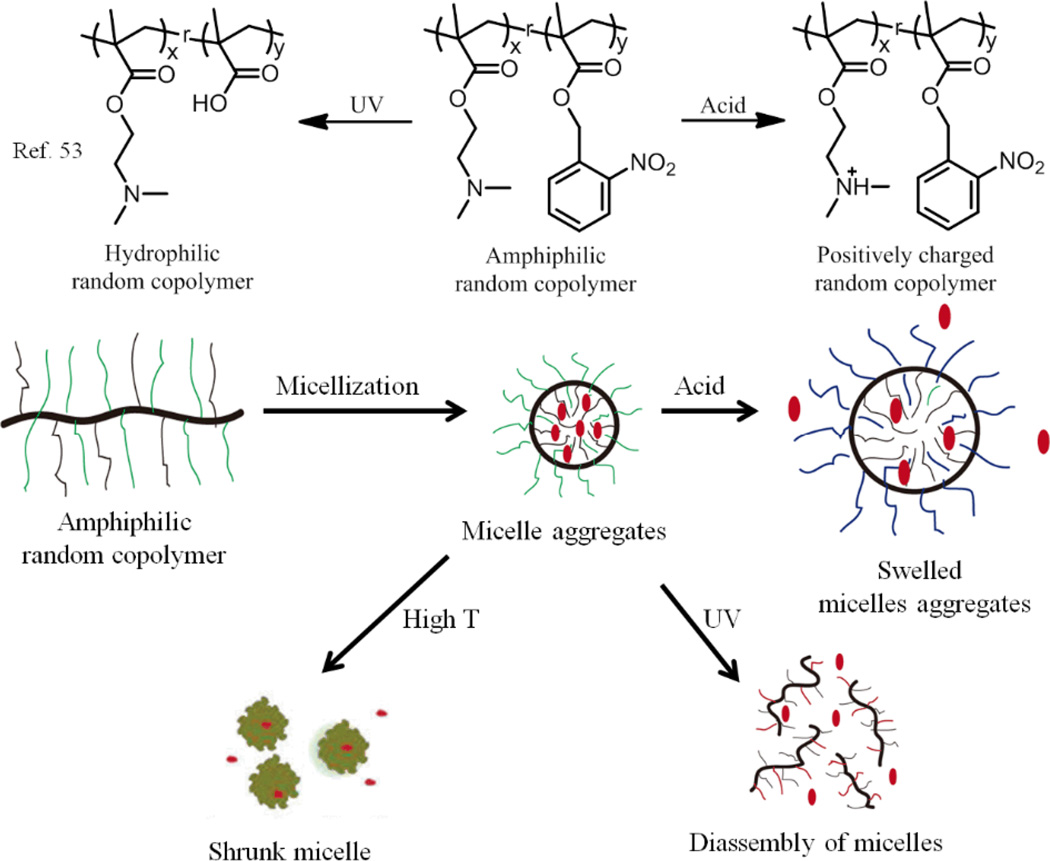
Multi stimuli-responsive assemblies based on random copolymers were also prepared by combining two or more functional groups into polymers. For example, random copolymer poly(DMAEMA-co-NBM), consisting of photo-, acid- and thermo-responsive moieties was synthesized by copolymerization of temperature/acid sensitive dimethylaminoethyl methacrylate (DMAEMA) and light-responsive 2-nitrobenzyl methacrylate (NBM) (Fig. 5).53 The random copolymers could spontaneously form polymeric micelles in water with hydrophobic NBM as cores and hydrophilic DMAEMA as shells at room temperature. The amine groups of DMAEMA could be protonated in low pH. As the pH was decreased to 3.0, the electrostatic repulsion between the protonated DMAEMA resulted in swelling of the particles to a larger size. The DMAEMA groups also change from hydrophilic to hydrophobic above the LCST of the polymer. TEM images show that the size of micelles became smaller due to the collapse of DMAEMA segments. In addition, the photocleavage of the NBM could produce 2-nitrosobenzaldehyde and make the hydrophobic NBM become hydrophilic poly(methacrylic acid) (PMA). This change breaks the hydrophilic-lipophilic balance, leading to the disassembly of the micelles. However, it is interesting to notice that there were still some smaller aggregates in solution because of the interaction between the amine groups of DMAEMA and the carboxylic acid moieties formed from the photocleavage of the NBM. Finally, the authors demonstrated that the Nile red encapsulated inside the polymer micelles could be successfully released under the influence of the triple stimuli (photo, acid and temperature).
Fig. 5.
Schematic representation of the amphiphilic random copolymer assembly which can respond to photo-, acid- and thermo-stimuli.
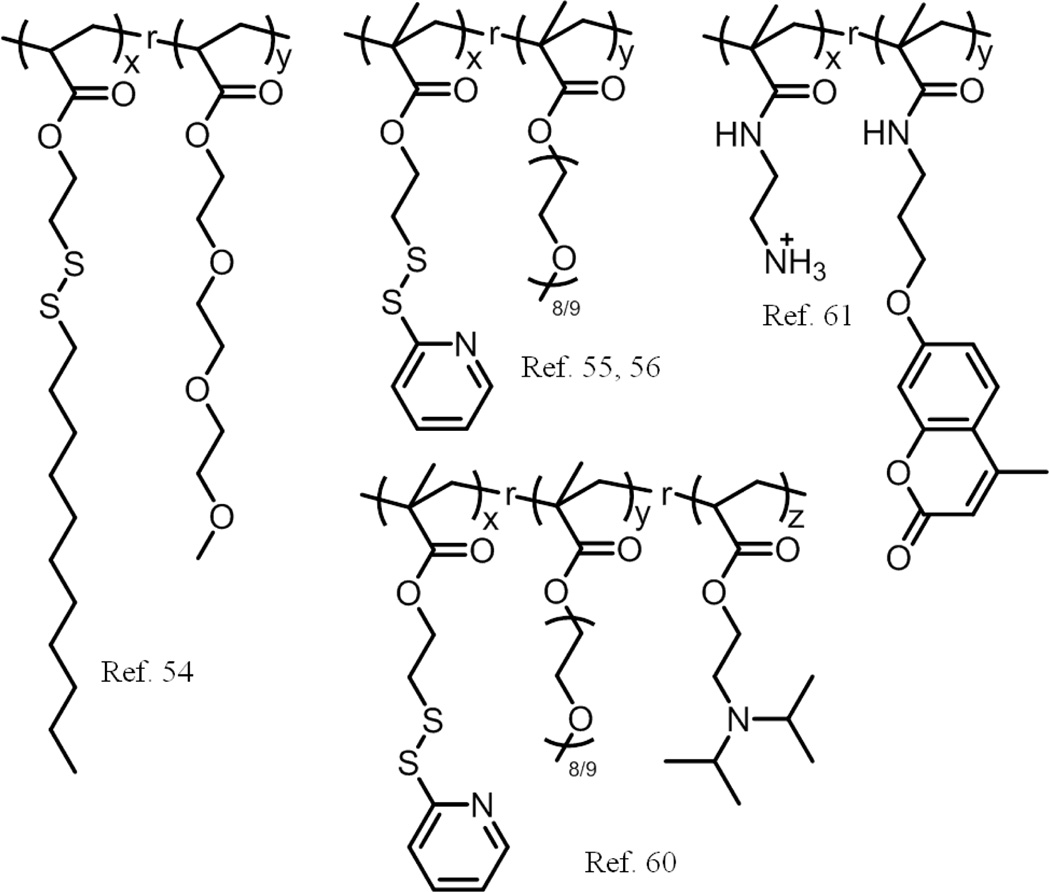
Our group has had a longstanding interest in using random copolymers for self-assembly (Scheme 2). Amphiphilic random copolymers, containing triethylene glycol as the hydrophilic part and an alkyl chain connected by disulfide bond as the hydrophobic part, were prepared by free radical polymerization.54 These polymers were able to form micelle-like nanoassemblies in water and encapsulate hydrophobic guests inside their core. These nanoassemblies disintegrate in the presence of a reducing environment, leading to a release of encapsulated guest molecules. This stimuli responsive behaviour is due to the cleavage of the disulfide bond that connects the hydrophobic moiety to the polymer backbone. Thus, under the reducing conditions, the self-assembling amphiphilic polymer is converted to a hydrophilic polymer that no longer has the ability to self-assemble leading to the release of guest molecules.
Scheme 2.
Schematic representation of the the polymers used in ref. 54, ref. 55, 56, ref. 60 and ref. 61.
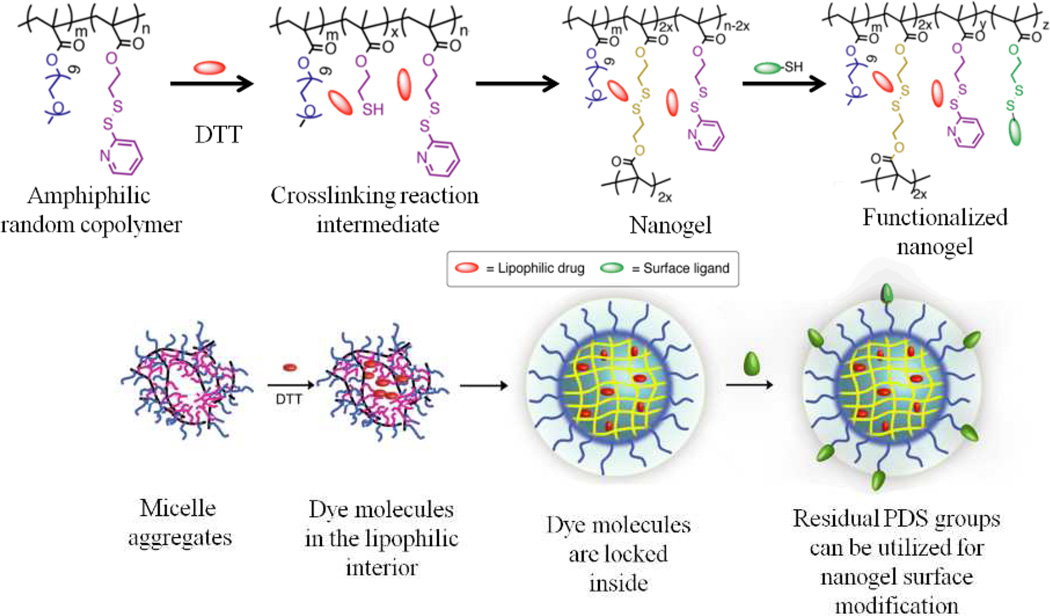
Considering that micelle-type assemblies can be destabilized upon dilution, strategies to crosslink the core of these nanoassemblies have been developed. Apart from retaining their structural integrity upon dilution, these nanogels also can stably encapsulate guest molecules and release them only in response to a redox trigger, such as glutathione (GSH).55, 56 The nanogel is based on a random copolymer that contains hydrophilic oligoethylene glycol (OEG) and hydrophobic pyridyldisulfide (PDS) units as side chain functionalities. This random copolymer forms nanoaggregates in water; addition of a deficient amount of dithiothreitol (DTT) to this solution leads to an intra/inter polymer chain disulfide exchange reaction to afford the core-crosslinked polymeric assembly (Fig. 6). The size of these polymeric aggregates could be tuned by varying the properties of the polymer, such as molecular weight of the polymer and the relative percentages of OEG units and PDS units incorporated into the polymer. In addition, external conditions such as temperature and presence of salts (Hofmeister effect) have been shown to affect the size of the nanoaggregates and thus the size of the nanogels. Using these features, the size of these nanogels have been systematically and predictably tuned from ∼10 to ∼200 nm.57 The unique advantage of these assemblies made from random copolymers is that the surface of our nanogel can be conveniently functionalized by free thiol containing molecules via thiol-disulfide exchange.58 Unlike the block copolymer assemblies where the hydrophobic units are completely buried in the interior, random copolymer aggregates have a certain portion of surface exposed functional hydrophobic PDS moieties, which have been exploited for nanogel decoration with ligands or other functional molecules. We have demonstrated that these nanogels could not only be used to deliver hydrophobic drugs, but also be used to bind proteins on their surface through electrostatic interactions, resulting in the concurrent delivery of proteins and hydrophobic small molecules.59 To achieve the ability of changing their surface properties and thus gain entry into a cell, we developed another random copolymer through a simple copolymerization of OEG, PDS and 2-diisopropyl amino (DPA) moieties.60 We showed that the pH at which the charge is generated can be adjusted by varying the percentage of PDS units in the nanogel, its preparation process and the crosslinking density. Cellular uptake of these nanogels was greatly enhanced in an acidic pH environment due to the surface charge generation. More recently, we have designed polymer nanoparticles that provide the ability to both encapsulate hydrophobic guest molecules and surface functionalization with different functional groups, based on random copolymers by copolymerization of 2-aminoethylmethacylamide and 3-(9-methylcoumarinxy) propylmethacrylamide.61 Our studies demonstrate the versatile nature of the assemblies from random copolymers to produce highly functional nanoparticles with robust tunability in their structural and the functional features.
Fig. 6.
Schematic representation of the preparation of self-crosslinking nanogels with hydrophobic guest encapsulation and surface functionalization features.
Complex polymeric aggregates
Various supramolecular assemblies have been designed to be responsive towards a variety of environmental conditions such as pH, redox, temperature, enzymes, light etc. Many of these nanostructures are designed using amphiphilic polymers that show stimuli responsive behaviour towards a specific environmental change. In order to prepare smart materials that have additional features such as stealth characteristics and stable encapsulation of guest molecules in addition to stimuli responsive behaviour towards multiple environmental triggers, multiple polymers or polymer assemblies have been integrated to form self-assembled structures.
These complex aggregates can be divided into different types depending on the interplay among the participating polymeric entities. In this section, we will discuss the design of random copolymers that lead to mixed micelle assemblies, polymer coated nanostructures and composite nanostructures with disparate morphological and self-assembly characteristics.
Mixed micelle type assemblies are typically formed from the concurrent participation of multiple polymers in a single assembly or nanostructure. These mixed micelle systems have the distinct advantage of having the potential to amalgamate the properties derived from the different types of polymers involved in the assembly. This strategy has been used to incorporate properties such as enhanced stability,62 introducing functionalities that can contribute to the stimuli responsive behaviour in response to change in environmental conditions such as temperature, pH, and redox environment.
Mixed micelles in block copolymers have been often achieved using a crosstalk between different functional groups in the polymer chain. For example, hydrogen bonding or complementary electrostatics has been utilized to obtain mixed micelles from polymers.63–66 Micelles formed from such interactions have been explored for applications in areas such as drug delivery and nucleic acid delivery.67, 68
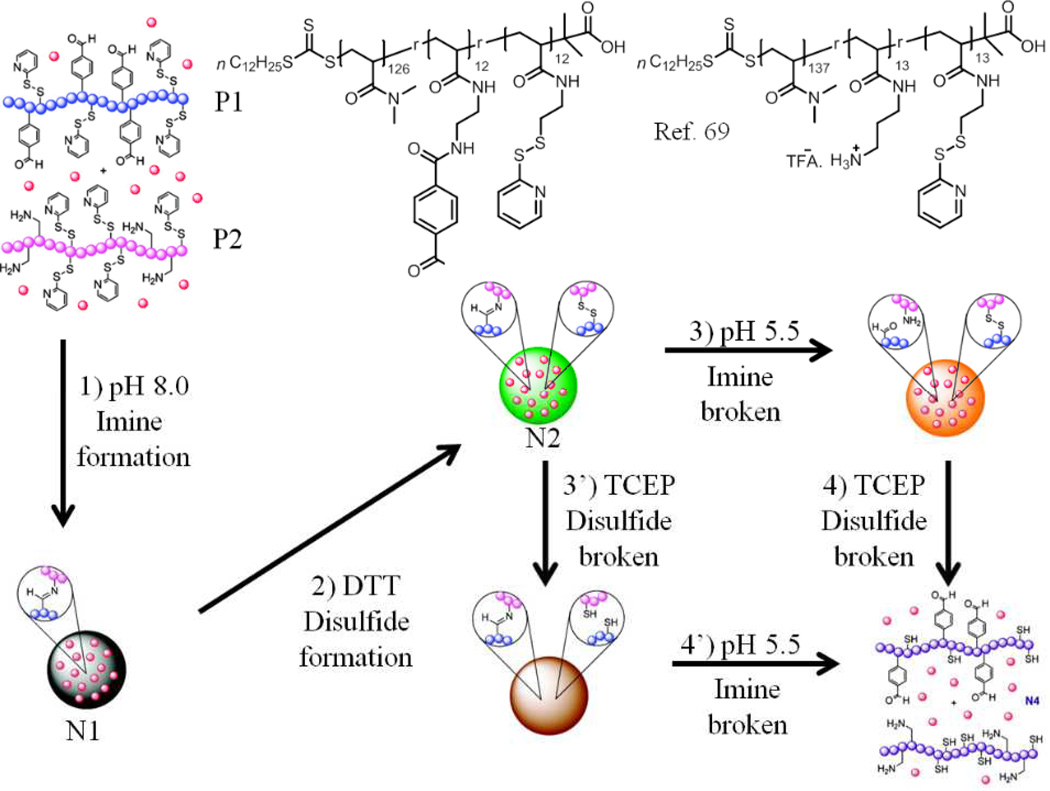
In addition to hydrogen bonding and electrostatic driven self-assemblies, multicomponent aggregates have been prepared using covalent or dynamic covalent bonds between two random copolymers. In a recent report, dynamic covalent interactions such as disulfide and imine bonds between the functional groups of different polymers have been utilized to prepare core crosslinked micelles (nanogels) as shown in Fig. 7.69 This was achieved by utilizing two different random copolymers, where aromatic carboxaldehyde and pyridyl disulfide (PDS) moieties are the key functional groups in one polymer P1, while primary amine and PDS are the key moieties in the other polymer P2. These two random copolymers form mixed micelle aggregates N1 upon mixing together at pH 8.0, which were stably crosslinked by formation of inter polymer imine bond between the aldehyde units of P1 and the primary amines of P2. In addition, a disulfide crosslinking between PDS functionalities was also achieved using a reducing agent, following an intra-aggregate crosslinking strategy that was recently reported.56 This crosslinking results in formation of nanogels N2, which have both pH-responsive imine bonds and redox-responsive disulfide bonds. Hence, in order to disassemble this nanostructure, a simultaneous application of both pH and reducing environment is required. It is shown that when one of these stimuli, either redox or acidic pH, is not present, the nanogel does not disassemble (Fig. 7). This work clearly demonstrates how multi polymeric aggregates can blend the distinct properties of the corresponding polymers involved to bring about a unique nanoassembly.
Fig. 7.
Schematic representation of the polymer structure, the assembly and disassembly behaviour.
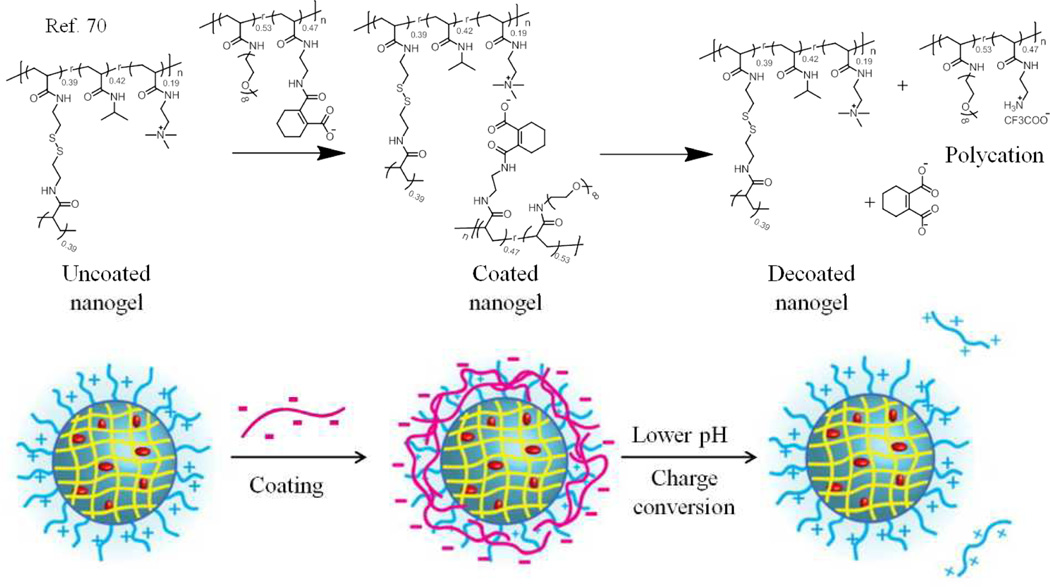
Unlike the complex aggregates discussed so far where multiple polymers contribute to form a single nanostructure, the next type of complex aggregates involves the strategy of non-covalently coating one random copolymer assembly with another random copolymer with complementary characteristics. These self-assembled structures can be either in the form of (i) polymer coating on the existing assemblies or a (ii) template driven layer by layer strategy to form assemblies. The first case of polymer-coated assemblies was mainly developed so as to mask the surface properties viz., surface charge on the nanoparticle surface and also increase the guest encapsulation stability. Our group has used this method of nanoparticle coating through electrostatic complementarity (Fig. 8).70
Fig. 8.
Schematic representation of the polymer structure , the coating and decoating process.
In this strategy, first a polycationic nanogel was achieved using our intra-aggregate self-crosslinking strategy with PDS functionalities.56 These disulfide crosslinked nanoassemblies were coated with a pH-sensitive anionic polymer through the electrostatic interactions. This resulted in masking of the positive surface charge of the nanogel and also led to an increase in the encapsulation stability of the non-covalently sequestered guest molecules inside the nanogels. The pH-sensitivity of the coating polymer is such that there is a charge conversion in the polymer in response to lower pH. The pH-induced charge conversion causes electrostatic repulsion between the coating polymer and the polymer nanogel to reveal the positive charge on the nanogel surface. Moreover, the encapsulation stability of the nanogel is also weakened due to the pH-induced decoating, which is further accentuated using a redox stimulus.
Layer by layer (LBL) techniques have been used to generate self-assembled structures, where a nanoparticle is used as a template for assembly formation. This process involves use of multiple polymers, where each constitutes a layer in the LBL assembly and the inter-polymer interactions based on hydrogen bonding or electrostatics drive the assembly formation. Recently, hydrogen bonding interactions between thiolated poly(methacrylic acid) (PMASH) and poly(vinylpyrrolidone) (PVPON) have been exploited to coat these polymers sequentially on a silica nanoparticle template.71 This LBL coating was followed by a disulfide crosslinking between different layers of PMASH through an oxidation reaction. Selective etching of the silica particle resulted in a capsule formation.
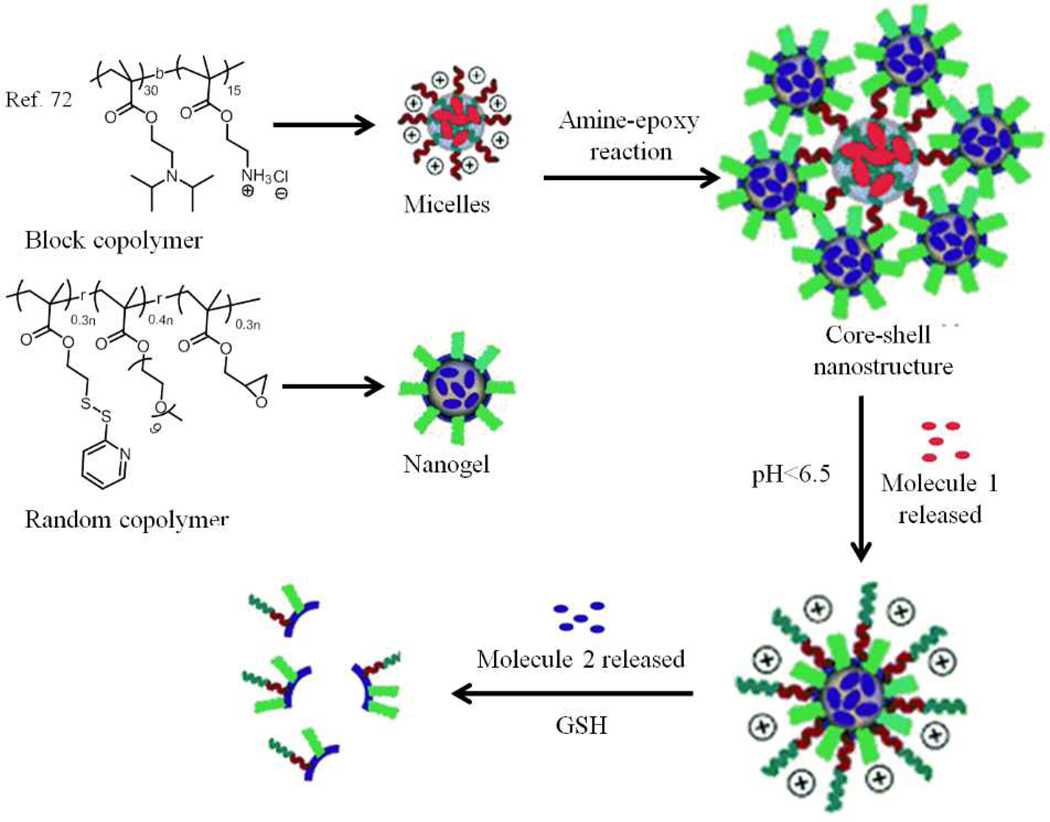
So far we have discussed about nanoassemblies that are formed by using multiple polymeric species integrated into a single structure or about a polymer coated onto a pre-formed nanoparticle. Now, we will discuss another class of nanostructures that are composite in nature in that these are formed by integration of more than one pre-formed nanostructures. Our group recently reported on such a composite nanostructures, where the combination of two independent supramolecular assemblies result in a novel, dynamic composite nanostructure.72 These composite structures are developed by using polymeric micelles from a block copolymer (poly(2-(diisopropylamino)ethylmethacrylate-b-2-aminoethyl methacrylate hydrochloride)) and nanogels formed from a random copolymer poly(oligoethyleneglycolmonomethylether methacrylate-co-glycidylmethacrylate-co-pyridyldisulfide ethyl methacrylate) (Fig. 9). We have integrated these supramolecular assemblies together by utilizing the covalent reaction between primary amines on the surface of the micellar assemblies and epoxide functionalities present on the nanogel surface to produce a composite nanostructure between the two nanoassemblies.
Fig. 9.
Schematic representation of the composite nanostructure assembly and stimuli-sensitive disassembly.
The diisopropylamine (DIPA) block, which constitutes the hydrophobic core of the polymer micelle, endows the composite assembly with pH sensitivity. The pKa of the protonated tertiary amine (DIPA) is about 6.8. Therefore, under physiologically neutral pH (around pH 7.4 or higher) the DIPA block would be mostly unprotonated and therefore would be hydrophobic. However, when the pH is lowered below 6.5, these functional groups are protonated and convert the hydrophobic core to a hydrophilic one causing a disassembly of micellar aggregates. The nanogels on the other hand are redox responsive.55 In the composite assembly, each of these assemblies retains their individual stimulus-responsive characteristics. But, there exists a synergy. Since the composite nanostructure is formed based on a reactive self-assembly event, when the micelle at the core of this composite assembly breaks, the disassembled polymer chain becomes covalently attached to the nanogel. This feature endows the nanogel with a positively charged surface that was previously unavailable on the nanogel. The pH-induced charge generation that leads to rapid cellular uptake and the possibility of encapsulating and releasing two different molecules at two different times and locations potentially lend themselves for applications in cancer therapy.73,74
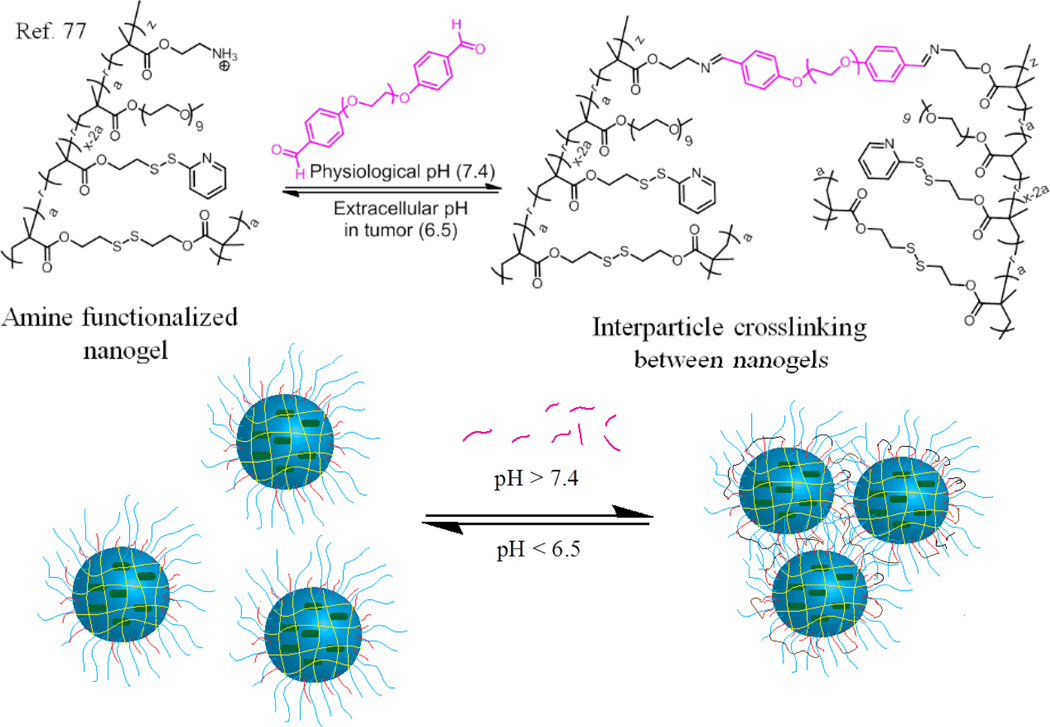
Considering the significant role that the size of drug delivery vehicles play in enhanced permeability and retention (EPR) effect based tumour targeting75, 76, it is interesting to be able to design composite nanostructures that change size in response to microenvironments that are unique to cancer tissues. For example, larger nanoparticle size is desired for tumour homing, while much smaller nanoparticles sizes are desired for tissue penetration. This combined with the fact that the tumour pH is lower; it is interesting to design a system that exhibits one size at neutral pH and reduces in size when subjected to lower pH conditions. Accordingly, we designed a system of complex aggregates that exhibit variations in size and charge in response to slight changes in pH.77
This design involves preparation of stimuli responsive nanoclusters by crosslinking multiple nanoparticles, utilizing the pH sensitive dynamic covalent imine bond between them (Fig. 10). This is achieved by reversibly crosslinking the nanoassemblies using a small molecule crosslinker. These nanogels are prepared using our nanogel system55 with a slight modification by using a random copolymer poly(oligoethyleneglycol monomethylether methacrylate-co-aminoethyl methacrylate hydrochloride-co-pyridyldisulfide ethyl methacrylate). The interparticle crosslinking between these nanogels was achieved by reacting the nanogels with a calculated amount of hexaethyleglycol dibenzaldehyde crosslinker at pH 7.4. This results in the formation of nanoclusters due to the imine formation. These nanoclusters break down into smaller nanogel particles at slightly acidic pH 6.5, presumably due to hydrolysis of imine bond. This also leads to a simultaneous exposure of free primary amines to present positive charges on the nanogel surface that accelerate cellular uptake.
Fig. 10.
Schematic representation of nanocluster formation at physiological pH and reversal at lower pH.
Single-Chain Polymeric Nanoparticles (SCNPs)
While the self-assembly strategies outlined above are based on the controlled aggregation of several polymer chains, there is also a great interest in the possibility of preparing nanoparticles through the intramolecular crosslinking, i.e. collapse of single polymer chains. This idea has biomimetic origins, because protein folding is both a classical and a sophisticated process in single molecule self-assembly in which a single-stranded polypeptide chain folds to form a well-defined three-dimensional tertiary structure. Considering the arrangement of amino acids in the sequence of polypeptide that has the ability to form a regular three-dimensional structure, random copolymers are usually used to mimic this unimolecular self-assembly process. Since a tutorial review on this topic was recently published,78 the studies discussed here will mostly include examples published after this review.
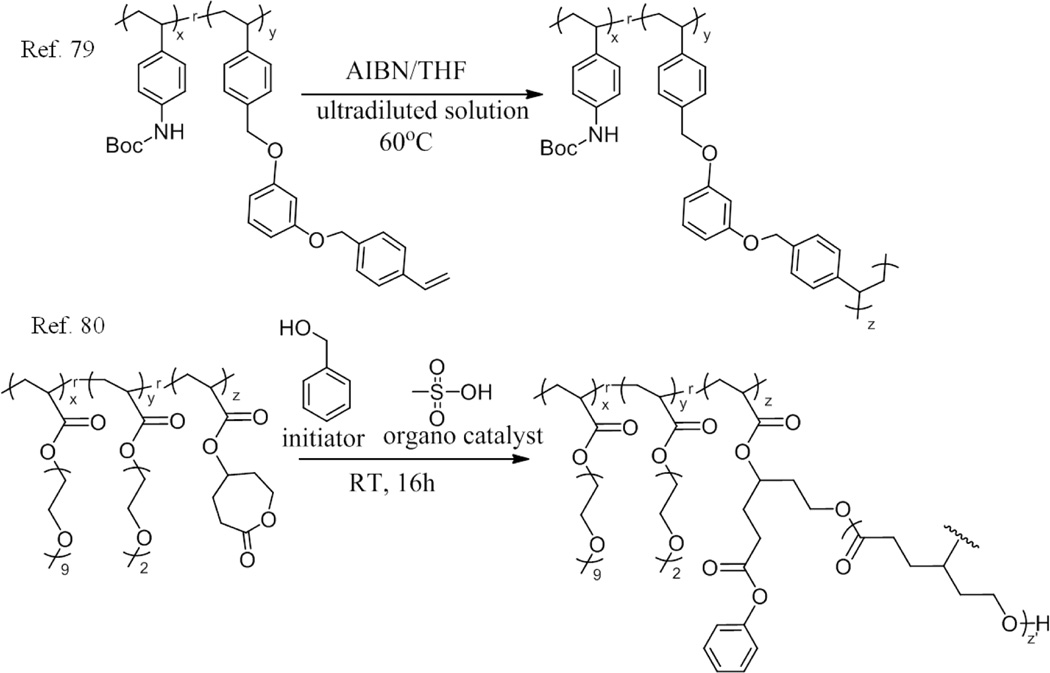
Our group reported the preparation of amine-functionalized nanoparticles via an intramolecular collapse of vinyl-functionalized random polymer following a crosslinking reaction driven by polymerization (Scheme 3).79 The vinyl-functionalized polymers was synthesized by reacting 4-[(3-hydroxyphenoxy)-methyl] styrene with random copolymers, which were achieved by RAFT polymerization of the protected amino group monomer (4-N-Boc-aminostyrene) and chloromethylstyrene using AIBN as the initiator. The “wormlike” structure was observed by atomic force microscopy (AFM) for these polymers. After refluxing the polymer THF solution of ultra low concentration in the presence of AIBN, the polymerization of the styrene moieties resulted in the crosslinking reaction, leading to the formation of the nanoparticle. From 1H NMR, it was clear that the peaks corresponding to the styrene double bond disappeared, indicating that all styrene reacted. The crosslinking reaction was determined to be intramolecular after the analysis of size-exclusion chromatography (SEC) and DLS results; both molecular weight and hydrodynamic radius of the particles decreased compared to the un-crosslinked polymers. The tert-butyl carbonate moieties could be removed from the particles by reacting with acetyl chloride in methanol, yielding the final amine-functionalized nanoparticles. We found that the size of these nanoparticles could be easily tuned by controlling the crosslinking density. The number of amino functionalities could also be tuned by adjusting the monomer ratio in the polymers.
Scheme 3.
Schematic representation of the polymer used and their crosslinking reaction in ref. 79 and ref. 80.
Similarly ring-opening polymerization (ROP) can also be used for the intramolecular crosslinking of polymers to form SCNPs. For that, random copolymers poly[(oligo-(ethylene glycol) methyl ether acrylate)-co-(di(ethylene glycol) ethyl ether acrylate)-co-(4-(acryloyloxy)-ε-caprolactone)] were synthesized by RAFT polymerization.80 Due to the polymerizable caprolactone groups in the side chains, SCNPs were easily prepared by using these polymers, which could be intramolecularly crosslinked via ROP with benzyl alcohol as the nucleophilic initiator and methanesulfonic acid as the organo catalyst (Scheme 3). The size of these SCNPs depended on the molecular weight of the polymers. Cytotoxicity studies further showed that these SCNPs were nontoxic, suggesting potential use in drug delivery.
Click chemistry has been extensively applied for SCNPs construction.81 In one example, a Diels-Alder-type cycloaddition reaction was used for SCNP formation.82 The reaction they used was a metal-free C-C click chemistry involving benzocyclobutene (BCB) functional groups, which requires activation at 250 °C. To overcome the high temperature requirement, a refined technique has been developed by introducing benzosulfone reactive groups instead of BCB moieties.83, 84 Another interesting approach in intrachain homocoupling for SCNP formation has been applied by using alkyne functional groups that were activated in a rapid and highly efficient manner at room temperature.85 Similarly, the self-assembly behaviour of linear poly(MMA-co-PgA) has been used to achieve SCNPs via metal-catalyzed C-C click covalent interactions. In addition, copper-catalyzed azide-alkyne cycloaddition, the so-called click chemistry, has also been used for the synthesis of bioconjugable poly(methyl methacrylate)-based single-chain nanoparticles.86–88
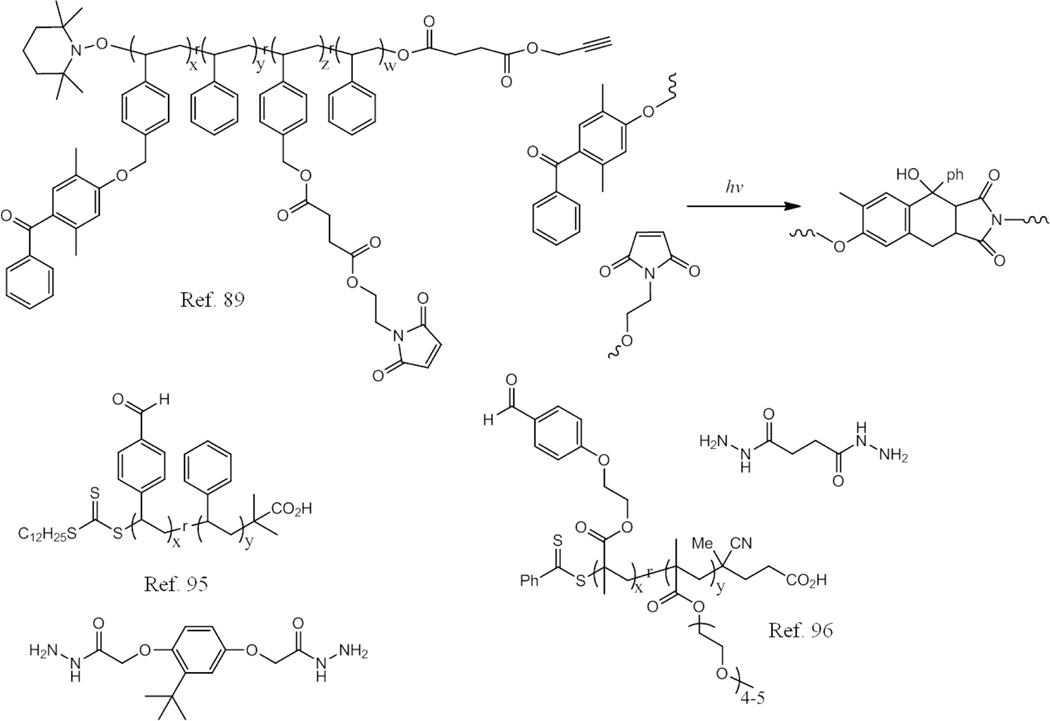
Similarly photoinduced Diels-Alder (DA) reactions have also been used for SCNP formation under ambient temperatures.89 Nitroxide-mediated radical polymerization (NMP) was used to synthesize the random copolymer containing styrene (S) and 4-chloromethylstyrene (CMS). Then, 4-hydroxy-2,5-dimethylbenzophenone (DMBP) and maleimide (Mal) functionalities were introduced into the copolymers by modification of CMS groups in a one-pot/two-step process (Scheme 4). Irradiation of this polymer with UV light afforded SCNPs due to a reaction between the maleimide groups and the o-quinidomethane type intermediate generated from the DMBP moiety.
Scheme 4.
Schematic representation of the polymers used in ref. 89, ref. 95, and ref. 96.
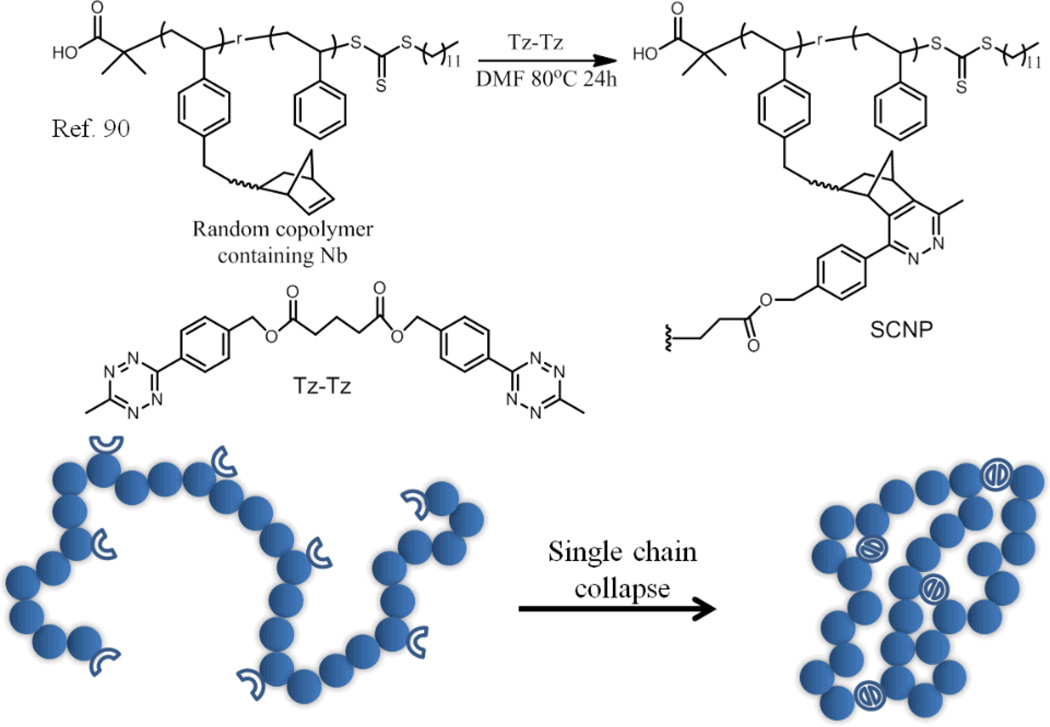
More recently, Diels-Alder type reaction between tetrazine and norbornene moieties has been used for single polymer chain collapse (Fig. 11).90 This reaction is fast, high-yielding and easily carried out. No special experimental conditions, additional catalyst or stimulus are needed to achieve near-quantitative conversions at room temperature.91–93 Random copolymers PS(Nb) were prepared by RAFT copolymerization of styrene and monomer containing norbornene functionality. These copolymer chains could be collapsed in DMF due to the low solubility of the polymer in DMF. When comparing the molecular weights of linear polymers and the corresponding SCNPs by SEC, the extent of the apparent molecular weight decrease was higher with increasing the Nb content. In order to rule out the possibility that the observed changes in apparent molecular weight are in fact due to the Tz-Nb reaction based collapse and not due to changes in hydrophobicity, they prepared a model polymer by the reaction of linear polymer with a monofuctional tetrazine (Tz-COOEt), analogous to half of the Tz-Tz crosslinker. There is no significant difference between the SEC of the model polymer and the linear polymer, which further confirmed that the reaction of Tz-Nb induced the self-assembly of the polymers. Beside these efficient click reactions, other controllable reactions can also be used. For example, a ring closing metathesis (RCM) reaction was used as the driving force for the collapse of single polymer chain.94
Fig. 11.
Formation of polystyrene SCNPs using Tz?–Nb crosslinking. The adduct formed is likely to be a mixture of dihydropyradizine isomers and pyradizines, for clarity only the pyradizine adduct is shown.
The previously discussed dynamic covalent imine bonds have also been used recently for SCNP synthesis.95 Random copolymers poly(VB-co-St) were prepared via copolymerization of vinylbenzaldehyde (VB) and styrene. Linear chains of these copolymers could be intramolecularly crosslinked through the formation of dynamic covalent imine bond, resulting in the formation of SCNPs. Later, these authors prepared another copolymer by copolymerization of the monomers OEGMA300 and poly(2-methacry-loxyethoxy)benzaldehyde (MAEBA).96 The addition of dihydrazide to the polymer solution led to an intramolecular crosslinking through the formation of dynamic covalent acrylhydrazone bonds. The formation of the SCNPs was also confirmed by the increasing retention time in gel permeation chromatography. One interesting point in this work is that these SCNPs were able to become chemically crosslinked hydrogels after increasing the temperature of the solution above their LCST because of the capacity of dynamic covalent acylhydrazone bonds to undergo component exchange processes. This transition was determined to be reversible, since the hydrogel would disassemble and turn into SCNPs again, if the temperature was lowered to room temperature.
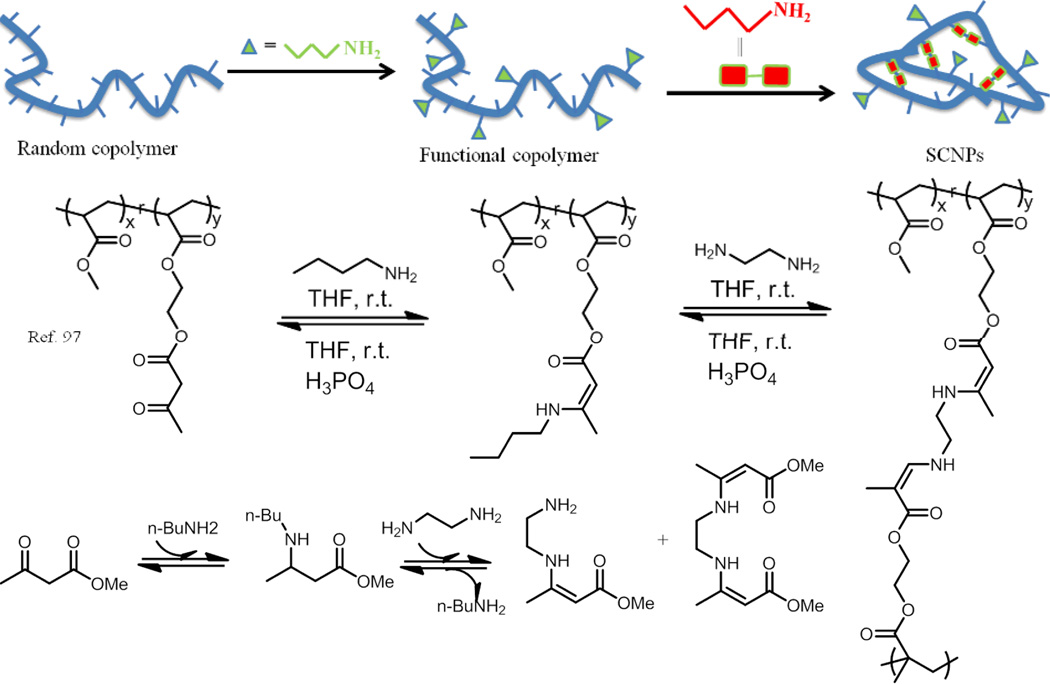
In their further study, the authors reported a method to prepare pH responsive single chain polymer nanoparticles using dynamic covalent enamine bonds (Fig. 12).97 Random copolymers poly(MMA-co-AEMA) were synthesized via copolymerization of methyl methacrylate (MMA) and (2-acetoacetoxy)ethyl methacrylate (AEMA). They first tested the enamine bond formation between this polymer containing reactive carbonyl groups and monofunctional amine butylamine in THF solution. Ethylene diamine was then used as a crosslinker for the collapse of the linear polymers. SEC results indicated that only intrachain crosslinking reaction took place, as a significant increase in retention times and reduction in molecular weight were observed. The hydrodynamic size also decreased from 7.8 nm to 5.3 nm after the SCNP formation. The pH responsive behaviour of the nanoparticles was demonstrated using phosphoric acid to trigger the disassembly of SCNP to presumably form the linear polymer chain.
Fig. 12.
Schematic representation of assembly–disassembly of single-chain polymer nanoparticles (SCNPs) by means of dynamic covalent enamine bonds. The formation of an enamine from the condensation of methyl acetoacetate and n-butylamine, and its component exchange reaction with ethylenediamine to form a bis-enamine were also shown.
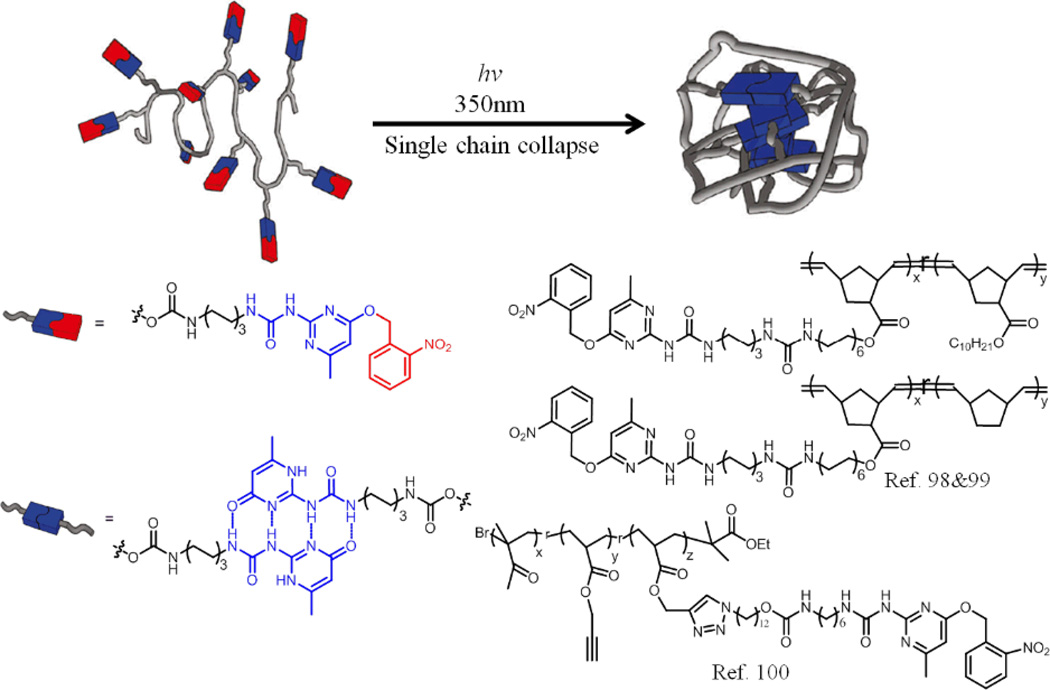
The SCNP examples above were achieved either under ultrahigh dilution conditions or through chain collapse driven by solvophobic interactions. An interesting supramolecular approach to SCNP formation involves intramolecular collapse of single polymer chains induced by hydrogen bonding interactions (Fig. 13).98–100 Ureido-pyrimidinone (UPy) groups, which are well-known for their ability to form strong and reversible hydrogen bonds, have been incorporated onto poly(norbornenes).98 Here, the terminal carbonyl moiety of the UPy groups were protected with o-nitrobenzyl moieties to avoid premature UPy dimerization. GPC results showed that the retention time of the polymers increase a bit after irradiation with 350 nm light, indicating a decrease in the molecular weight, which is taken to be an indicator of SCNP formation. These results were further supported by AFM and DLS results. Moreover, the authors have shown that the size of these supramolecular single chain polymer nanoparticles can be tuned by varying the molecular weight of the polymers. However, it is not clear whether the rather large nanoparticles obtained through tuned molecular weight are truly single-chain based nanoparticles.99
Fig. 13.
UV irradiation induced collapse of a single polymer chain into a nanoparticle via the supramolecular crosslinking of the UPy.
Since the hydrogen-bonding between the UPy groups could be disrupted by acidification, the reversibility of these particles has also been demonstrated, where the polymers expanded from particles into a coiled state in response to pH change. To further extend the scope and utility of this approach, these authors have designed another polymer based on poly(methyl methacrylate) (PMMA) using a combination of living radical polymerization and click chemistry.100 Formation of the SCNP has been confirmed by both GPC and AFM.
More recently, intramolecular hydrophobic interactions have been demonstrated to be useful in the preparation of SCNPs.101 Amphiphilic random methacrylate copolymers containing poly(ethylene glycol) (PEG) and alkyl pendent groups were synthesized by the ruthenium-catalyzed living radical copolymerization of a PEG methacrylate (PEGMA) and an alkyl methacrylate (RMA; R, −CnH2n+1, n = 1−18), where copolymer composition, degree of polymerization, and hydrophobic R- group size could be easily varied. They found that copolymers with 20–40 mol% hydrophobic units were able to form unimeric micelles, which were confirmed to be SCNPs via SEC and DLS measurement. The retention time in SEC increased, while both molecular weight and hydrodynamic size of these assemblies decreased compared to the single polymer chains. These SCNPs were dynamic andreversible in water. For example, addition of methanol can lead to disassembly.
Apart from spherical SCNPs, non-spherical SCNPs have also been reported. The random copolymers were firstly synthesized by Ru-catalyzed living radical polymerization (LPR) of benzene-1,3,5-tricarboxamidefunctionalized methacrylate (BTAMA) and oligo(ethylene glycol) methyl ether methacrylate (OEGMA, 8−9 oxyethylene units). SCNPs could be formed in water due to the self-assembly of BTA groups in water.102 Through small-angle neutron scattering experiments, it was found that polymer chain had an asymmetric ellipsoidal shape.103 Further it was also found that polymer length could affect the shape of these particles. The cross-section R of the SCNP would be constant while the major radius increased linearly with the increasing length.104 The authors also claim an interesting observation where even at high concentration (up to 100 mg/mL), most particles in solution were still SCNPs. This single chain character at such high concentrations could be very useful to improve the scalability of these nanoparticles in future application.
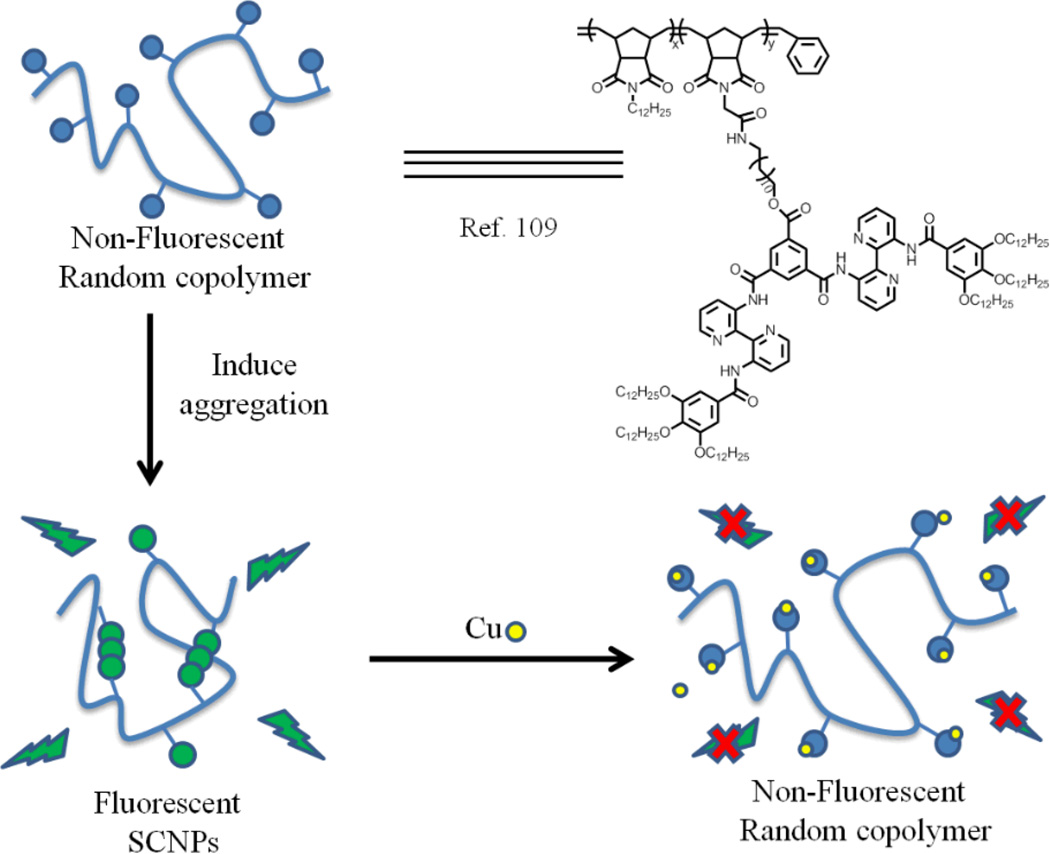
Additionally, as more and more methods have been developed, the application of these SCNPs is also reported. Recent progress has been achieved for the use of well-defined single-chain nanoparticles in some promising fields, such as nanomedicine, sensing and catalysis.102, 105–108 For example, SCNPs have been explored as compartmentalised sensors for metal ions (Fig. 14).109 Random copolymers containing 3, 3’-bis(acylamino)-2, 2’-bipyridine substituted benzene-1,3,5-tricarboxamides (BiPy-BTAs) were prepared by ROMP. This copolymer self-assembles into SCNPs via an intramolecular crosslinking reaction, which was confirmed by static and dynamic light scattering experiments. They used absorption and fluorescence spectroscopy to determine that π-π interactions between BiPy-BTAs played an important role in the aggregation. However, the interaction between BiPy-BTAs were presumably disrupted by the addition of metal ions, such as Fe(III), Cr(III), V(III), Mn(III), Zr(III) and Cu(II), since these ions bind to the bipyridine units. This disassembly leads to a decrease in the green fluorescent emission at 520 nm. Specifically, the authors noticed that their polymers show a preference for Cu(II) as a 3 fold increase in the fluorescence quenching was observed with this ion.
Fig. 14.
Schematic representation of the sensing function of the BiPy-BTA functional polymers.
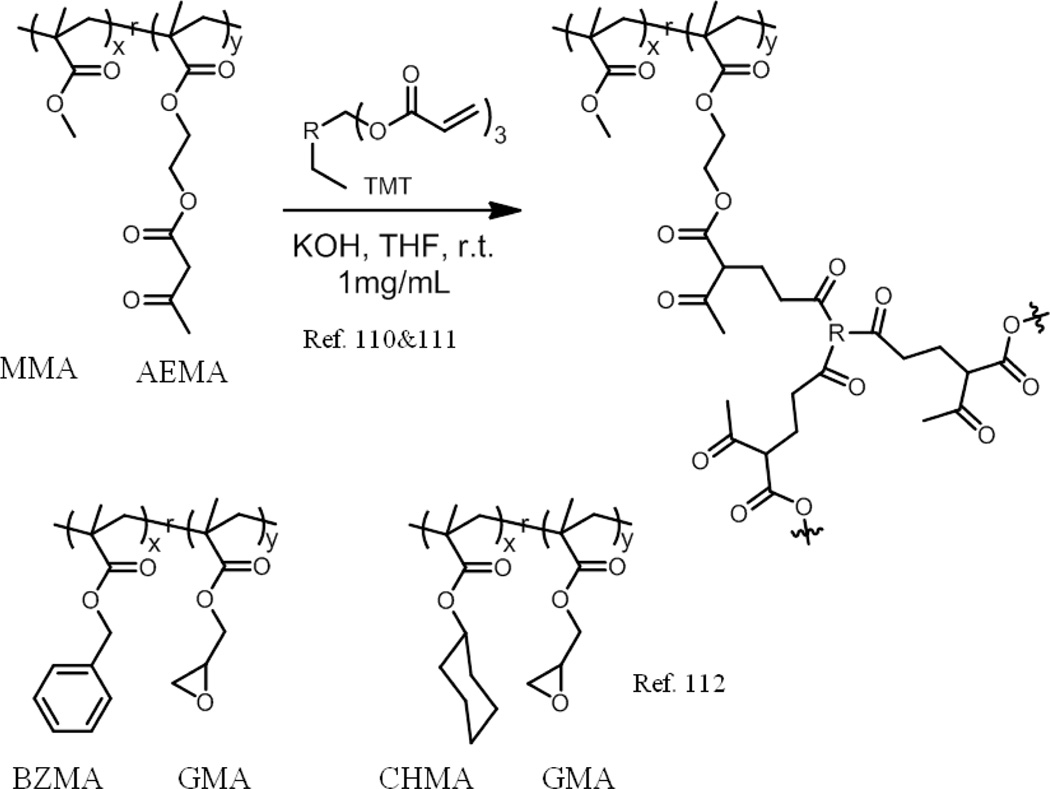
Inspired by the behaviour of natural transient-binding disordered proteins, SCNPs that could act as “Michael” nanocarriers for the delivery of vitamin B9 have been developed.110 Random copolymers poly(MMA-co-AEMA) of high molecular weight and relatively narrow size dispersity were firstly synthesized by RAFT copolymerization of methyl methacrylate and (2-acetoacetoxy)ethyl methacrylate), which are of similar reactivity ratios. Aggregates were then formed via multidirectional self-assembly of polymeric chains driven by multiple intrachain Michael addition reactions. The formation of SCNPs was supported by SEC, TEM and small-angle neutron scattering (SANS) measurements, combined with MD simulations. Vitamin B9 could be slowly released at neutral pH due to the morphology change from the dry state to solution state. Later, simultaneous delivery of folic acid or vitamin B and hinokitiol was also achieved by using the same carriers.111
In another example, SCNPs with enzyme-mimetic activity have been attempted.112 Two different types of random copolymers poly(BZMA-co-GMA) and poly(CHMA-co-GMA) were synthesized by copolymerization of glycidyl methacrylate (GMA) with benzyl methacrylate (BZMA) and cyclohexyl methacrylate (CHMA), respectively. The self-assembly of these copolymers was driven by B(C6F5)3-catalyzed ring-opening polymerization (ROP) of glycidylic groups, which was confirmed by the complete disappearance of bands corresponding to glycidylic protons in the 1H NMR. The formation of SCNPs was ascertained using SEC, SLS, and SANS experiments. Small angle neutron scattering (SANS) experiments also indicated that no multichain aggregates were formed during the crosslinking reaction. NMR studies suggested that the catalyst B(C6F5)3 still remains inside the particles. Thermal gravimetric analysis (TGA) data was used to estimate that each particle contained around 165 borane units. The reason for the retention of B(C6F5)3 is likely due to the binding interaction between Boron and oxygen-containing functional groups (ether and carbonyl). Finally, these SCNPs have been used as the catalyst in the reduction of diketones to silyl-protected 1, 2-diols, which the authors suggest as having an enzyme-like activity.
Amphiphilic random copolymers based on bio-derived polymers
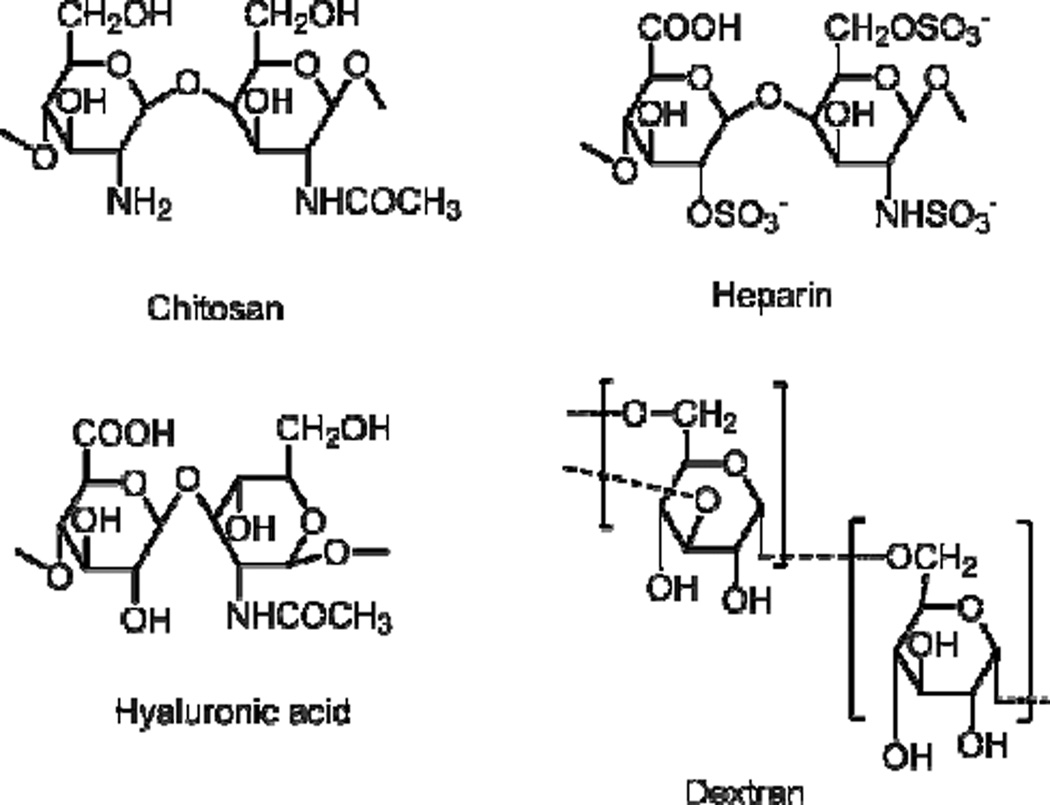
Biopolymers such as polysaccharides have been extensively studied for drug delivery applications over past few decades due to potential advantages such as non-toxicity, biocompatibility and biodegradability. Polysaccharide based materials are abundant in nature, renewable, and have size tenability.113 Some commonly studied polysaccharides are chitosan, hyaluronan, dextran and heparin (Scheme 6). These biopolymers are generally water-soluble and do not show any self-assembling properties. However, these hydrophilic polymers can be grafted onto with hydrophobic segments, resulting in amphiphilic polymers. We regard them as random copolymers since the chemical modification is likely to be random in the polymer backbone. These amphiphilic copolymers would be able to self assemble in an aqueous media, which can trap hydrophobic drug inside the hydrophobic core of the polymer.
Scheme 6.
Structures of common polysaccharides
One of the earlier works of hydrophobically modifying polysaccharides involved hydrophilic pullulan that were modified using hydrophobic cholesterol.114 Similarly, supramolecular assemblies have been achieved from chitosan polymers, where chitosan has been modified with hydrophilic PEG and hydrophobic pthalymido groups to impart amphiphilic character into the system.115 Using SEM, it was demonstrated that chitosan without modification showed irregular flakes, while pthalylated chitosan showed a spherical shape and this spherical morphology is even more pronounced when it is further modified with m-PEG. The sizes of the particles ranged from 80–400 nm depending on the molecular weight of m-PEG grafted to chitosan. Using a similar strategy, chitosan has been modified with linoleic acid through an EDC coupling between the carboxylic acid of the fatty acid and the amine of chitosan to form nanostructures of 200–600 nm.116
Aliphatic alcohols such as octanol, dodecanol and hexadecanol have been grafted onto sodium alginates to introduce hydrophobicity to the hydrophilic alginates thus forming amphiphilic sodium alginate (SA) aggregates in water.117 Pyrene is used as a fluorescent probe to analyze the self-aggregation behaviour of SA-Cn. The authors were able to demonstrate that the CMC value of the aggregates decreased with increasing in chain length. They also showed that as the hydrophobic chain length increases, the micelle size decreases owing to increased hydrophobic interaction that associate closely (from about 600 to 200 nm). The spherical morphology of the aggregates was ascertained by TEM.
Stimuli responsive drug delivery carriers are of great interest, because they can release the drug in response to a biologically relevant stimulus.118 Polysaccharides have been designed to form such assemblies by introducing functional side chains in their backbone. For example, click chemistry has been used as a tool to synthesize amphiphilic chitosan-graft-poly(2-(2-ethoxy)ethylmethacrylate-co-oligo(ethylene glycol) methacrylate) (CS-g-Poly(MEO2MA-co-OEGMA)) copolymers.119 The self-assembly behaviour of the amphiphilic random copolymer was characterized by DLS and SLS. Using transmittance measurements, the authors show that the ratio of MEO2MA and OEGMA can be used to tune the LSCT behaviour of CS-g-Poly (MEO2MA-co-OEGMA) copolymer.
Besides polysaccharides, biodegradable polyesters such as poly-lactide (PLA), poly(ε-caprolactone) (PCL), poly (lactide-co-glycolide) (PLGA) and poly (γ-valerolactone) (PVL) have also been widely used in controlled drug delivery.120, 121 They can be degraded by hydrolytic or enzymatic degradation under physiological conditions.
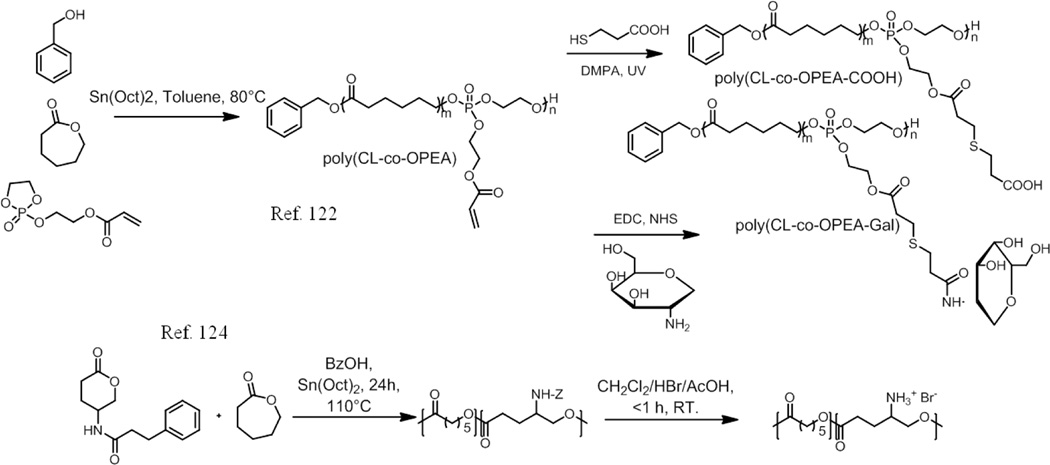
Random copolymers were easily prepared via ring-opening polymerization (ROP) of monomers in a one-pot process. For example, a galactosamine-conjugated biodegradable poly-(3-caprolactone-co-phosphoester) random copolymer, [poly(CL-co-OPEA)], was synthesized via ROP reaction ofε-CL and 2-(2-oxo-1,3,2-dioxaphospholoyloxy)ethyl acrylate (OPEA) using benzyl alcohol (BnOH) as the initiator and Sn(Oct)2 as the catalyst.122 Then liver-targeting galactosamine (Gal) was conjugated to the hydrophilic polyphophoester segments to prepare the amphiphilic copolyesters poly(CL-co-OPEA-Gal). The critical aggregation concentration (CAC) values of the copolymers were first measured using pyrene as the fluorescence probe. In water, this random copolymer was able to self assemble into micelles with hydrophobic PCL segments as the core and hydrophilic polyphosphoester parts as the shell, which was further confirmed by TEM and DLS measurements. Through the MTT assays using HEpG2 cells and HeLA cells, the copolymer has been shown to exhibit low toxicity, where the cell viabilities are still higher than 80% at concentrations up to 200 mg/mL, indicating that these random copolymers had excellent biocompatibility. The hydrophobic core was able to encapsulate the anticancer drug DOX, which could be released in an acidic environment (pH 5.0) due to the acid accelerated hydrolytic degradation of polyphosphoester. Cell uptake experiments show that Gal greatly improved the specific cell binding and cellular uptake.
Amphiphilic co-polyelectrolytes are of particular interest, because both hydrophobicity and electrostatic interaction can be combined to give an enhanced solubility of the hydrophobic drug and thus greater loading capacity. This possibility has been demonstrated by encapsulating clofazimine in a hydrophobized poly(methylvinylether-alt-maleic acid) (PMVEMAc).123 Later, these authors extended this concept to biodegradable random amphiphilic polycations.124 They prepared copolymers of of 5-Z-amino-δ-valerolactone (5-NHZVL) and ε- caprolactone (ε-CL). Poly(ε-CL-co-NH3+-VL) and Poly(NH3+-VL) with different compositions. The copolymers with more than 12% ammonium groups were soluble in water. Only the ones with 100% ammonium groups were soluble in buffer of pH 7.4. The explanation for this was that the partial screening of the protonated amines by the salts in the buffer renders a decrease in hydrophilicity. The CAC’s of the copolymers were measured by conductance and was found to be 0.05 % (w/v) in water. This was further confirmed by Zeta potential measurements. Interpolymer aggregation occurs when the charges are located near the backbone (d ≈ 1.5 to 2 Å). On the other hand intrapolymer aggregation takes place when the charges are located further from the backbone (d ≈ 8 to 9 Å). In this case there is only one covalent bond between the ammonium group and the polymer backbone, thus it forms interpolymer aggregates. Aggregate size was measured by DLS, which was found to have two sizes of 30–50 and 100–250 nm. Hydrophobic compounds like pyrene, 17α-ethinylestradiol (EE) and flufenamic acid (FA) were encapsulated. They showed that the solubility of hydrophobic guests in every case increased with the hydrophobic composition of the polymer. They tested the effect of electrostatic interactions on solubility by encapsulating 1-pyrenecarboxylic acid. However, the results indicate that electrostatic interactions played only a minor role in solubilization compared to hydrophobic effect. Poly(NH3+-VL) showed very low level of hemotoxicity in buffer and in plasma. They also demonstrated that Poly(NH3+-VL) showed about 60% biocompatibility below 1 mg/ml. However the compatibility of poly(NH3+-VL), and poly(ε-CL-co-NH3+-VL) reduced with increase in the ammonium group composition. Nevertheless, the biocompatibility was significantly better than poly-L-lysine hydrobromide, which was used as a positive control.
Polypeptides are another class of biodegradable polymer, attracting increasing attention in controlled delivery. Peptide based polymers have many advantages over conventional synthetic polymers since they are able to hierarchically assemble into stable, ordered conformations. Depending on the substituents of the amino acid side chain, polypeptides are able to adopt a multitude of conformationally stable, regular secondary structures. The development of polymerizations of amino acid-N-carboxyanhydrides (NCAs) provides the possibility for large scale preparation of high molecular weight polypeptides.125–128
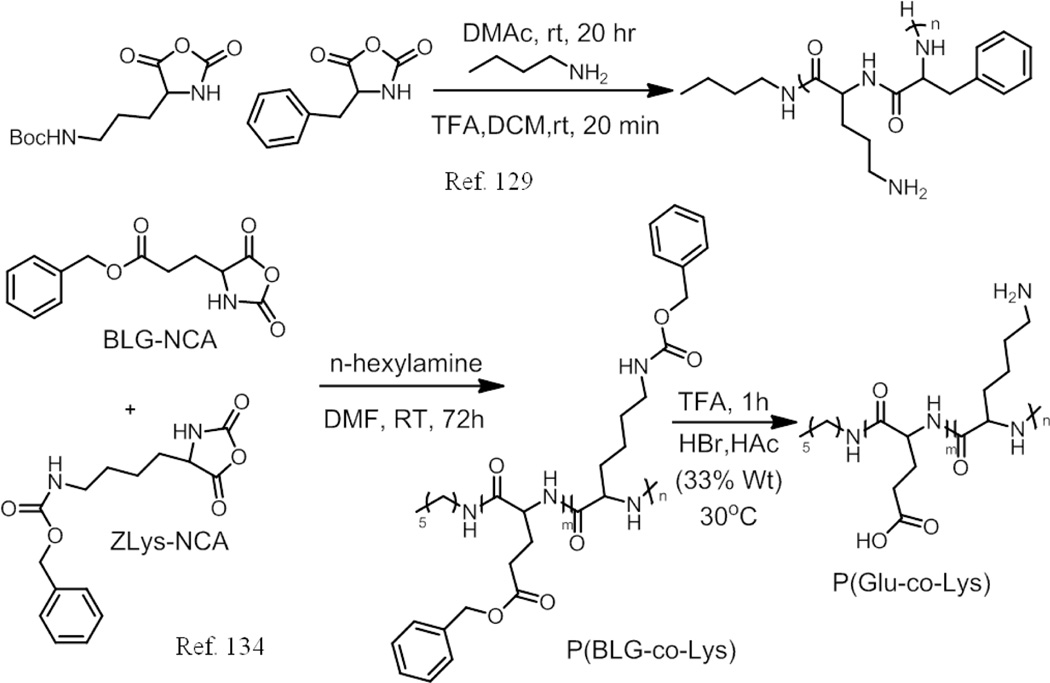
Recently, a series of amphiphilic, biodegradable polypeptide copolymers consisting of L-ornithine and L-phenylalanine were prepared for the delivery of siRNA (short interfering ribonucleic acid).129 The molecular weight could be tuned from 11 kDa to 40 kDa. The ratio of L-ornithine and L-phenylalanine was set at 4:1 because of their similar hydrophilic to hydrophobic ratio with the poly(vinylether) polymers, which were reported to have endosomal escape capabilities. These cationic polymers could form conjugates with negatively changed siRNA via electrostatic interactions. When the ratio of polymer to siRNA was the same, polymer with a higher molecular weight would have a better efficacy in vivo. This polymer also showed a stronger ability to deliver siRNA in the animal studies. Although the toxicity of these polymer conjugates increased with the molecular weight, there was no toxicity when the concentration was up to 3 mg/kg, which could lead to more than 90% mRNA knockdown. The authors claimed that this type of polymer might be broadly applied for siRNA delivery.
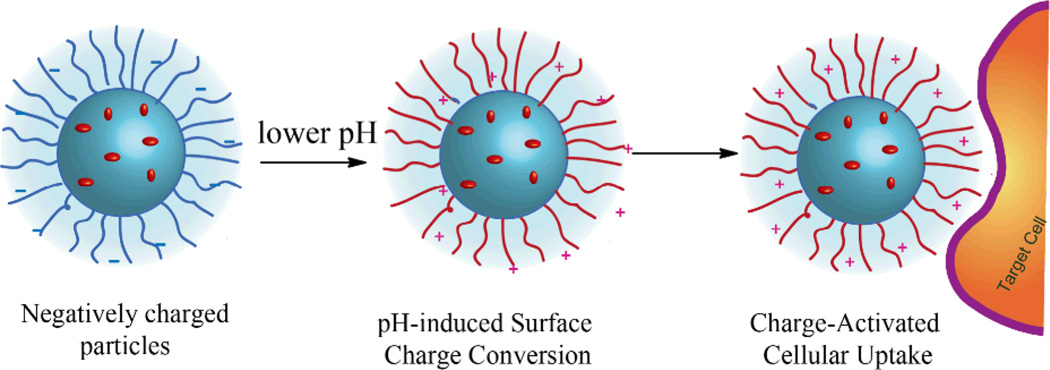
pH as a stimulus for responsive drug delivery has been explored extensively.130, 131 It is interesting to note the pH difference in blood (pH 7.4) and extracellular tumour environment (pH 6.5–7.2).132, 133. This difference can be exploited to use for systemic drug release from the carriers. Nanocarriers with negative surface charge are known to have longer circulation times. However, they suffer from poor cellular internalization presumably due to the negatively charged cellular membrane. On the other hand, positively charged nanocarriers are rapidly internalized by cells, but are known to be rapidly cleared from the body due to non-specific interactions with serum proteins during circulation and often suffer from high toxicity. Thus, in order to have better in vivo applicability, it is desirable that the carriers have a negative or a neutral surface charge during circulation which can increase the circulation time. However, once it reaches the target site (e.g. extracellular fluid of tumour), it should obtain a positive surface charge which can facilitate a faster uptake by the tumour cells (Fig. 15). Non-biodegradable polymers have been studied to incorporate the charge conversional feature for efficient cellular uptake.60 However, this feature has not been extensively studied for biodegradable polymers.
Fig. 15.
Schematic representation of the pH-induced surface charge conversion for charge-activated cellular uptake.
Poly(Glutamic acid-co-lysine) was synthesized through the random copolymerization of BLG (γ-benzyl-L-glutamate)-NCA and ZLys-NCA, followed by loading cisdiaminodichloroplatinum(II) (CDDP).134 The self-assembly in pH 7.4 is attributed to the electrostatic interaction between negatively charged Glu units and positively charged Lys units owing to their pKa’s of 4.05 and 10.54, respectively. Because of the presence of carboxyl group and amino group on the poly(Glu-co-Lys) copolymers, the aggregates exhibit pH-responsive charge conversional features. Using zeta potential measurements, the authors showed that the pH at which the charge transition occurs can be controlled by the feed ratio of BLG-NCA and ZLys in the NCA polymerization. With increase in the lysine content, the surface charge of the aggregates reversed at a higher pH. They also demonstrated that CDDP affects the pH dependent charge reversal as the amine groups of Lys can compete with platinum ions to complex carboxyl groups of poly(Glu-co-Lys). They optimized the ratio of BLG-NCA and ZLys NCA with CDDP to get charge conversion from −4.9 mV at pH 7.4 to 4.2 mV at pH 6.8. The in vitro inhibition of the proliferation of HeLa cells by CDDP/poly(Glu-co-Lys) nanoparticles were greater at pH 6.8 than pH 7.4. This was further confirmed by cellular uptake experiments.
Conclusions and outlook
In this feature article, we have attempted to draw attention to the application of random copolymers to prepare nanostructures with different morphologies and nanomaterials with single- or multi- stimuli responsive behaviours. A key advantage of using random copolymers is that the synthesis of these copolymers is simple, as they are usually prepared by a one-step copolymerization or a one-pot post-polymerization treatment. Considering this feature, these random copolymer based materials have excellent prospects in terms of end applications, as these can be conveniently scaled up. The multiple examples cited in this review unambiguously highlight the versatility of random copolymer based self-assembly in general.
For many years, the field of copolymer self-assembly has been dominated by block copolymers. A distinct advantage of block copolymers is that there exists a structure-property correlation that provides the guidelines for the type of assembly that one would anticipate from the type of amphiphilic blocks used and the molecular weights of the blocks. This type of an understanding is certainly lacking in random copolymer based self-assembly. The simplicity in synthesis and versatility in function highlighted in this review will bring both experimentalists and theoretical modelers to develop a similar structure-morphology correlation model. In addition, the facile access to random copolymers and the resulting nanostructures also provide translational opportunities in a variety of research areas, especially biomedical applications such as drug delivery, diagnostics, and sensing.
Scheme 5.
Chemical structure of random copolymer used and synthesis of the SCNPs in ref. 110, 111 and 112.
Scheme 7.
Scheme 8.
Acknowledgements
Partial supports through funding from the National Institutes of Health (GM-065255), Army Research Office (63889-CH), and the National Science Foundation (CHE-1307118) are acknowledged.
Notes and references
- 1.Ikkala O, ten Brinke G. Science. 2002;295:2407–2409. doi: 10.1126/science.1067794. [DOI] [PubMed] [Google Scholar]
- 2.Janata J, Josowicz M. Nat. Mater. 2003;2:19–24. doi: 10.1038/nmat768. [DOI] [PubMed] [Google Scholar]
- 3.Zotti G, Vercelli B, Berlin A. Acc. Chem. Res. 2008;41:1098–1109. doi: 10.1021/ar8000102. [DOI] [PubMed] [Google Scholar]
- 4.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rösler A, Vandermeulen GWM, Klok H-A. Adv. Drug Delivery Rev. 2012;64:270–279. [Google Scholar]
- 6.Nakatani K, Ogura Y, Koda Y, Terashima T, Sawamoto M. J. Am. Chem. Soc. 2012;134:4373–4383. doi: 10.1021/ja211436n. [DOI] [PubMed] [Google Scholar]
- 7.Hawker CJ, Bosman AW, Harth E. Chem. Rev. 2001;101:3661–3688. doi: 10.1021/cr990119u. [DOI] [PubMed] [Google Scholar]
- 8.Moad G, Rizzardo E, Thang SH. Aust. J. Chem. 2012;65:985–1076. [Google Scholar]
- 9.Hilf S, Kilbinger AFM. Nat. Chem. 2009;1:537–546. doi: 10.1038/nchem.347. [DOI] [PubMed] [Google Scholar]
- 10.Matyjaszewski K, Tsarevsky NV. J. Am. Chem. Soc. 2014;136:6513–6533. doi: 10.1021/ja408069v. [DOI] [PubMed] [Google Scholar]
- 11.Rosebrugh LE, Marx VM, Keitz BK, Grubbs RH. J. Am. Chem. Soc. 2013;135:10032–10035. doi: 10.1021/ja405559y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JK, Yang SY, Lee Y, Kim Y. Prog. Polym. Sci. 2010;35:1325–1349. [Google Scholar]
- 13.Zhang Q, Ko NR, Oh JK. Chem. Commun. 2012;48:7542–7552. doi: 10.1039/c2cc32408c. [DOI] [PubMed] [Google Scholar]
- 14.Ge Z, Liu S. Chem. Soc. Rev. 2013;42:7289–7325. doi: 10.1039/c3cs60048c. [DOI] [PubMed] [Google Scholar]
- 15.Ruzette A-V, Leibler L. Nat. Mater. 2005;4:19–31. doi: 10.1038/nmat1295. [DOI] [PubMed] [Google Scholar]
- 16.Mai Y, Eisenberg A. Chem. Soc. Rev. 2012;41:5969–5985. doi: 10.1039/c2cs35115c. [DOI] [PubMed] [Google Scholar]
- 17.Kim H-C, Park S-M, Hinsberg WD. Chem. Rev. 2009;110:146–177. doi: 10.1021/cr900159v. [DOI] [PubMed] [Google Scholar]
- 18.Quémener D, Davis TP, Barner-Kowollik C, Stenzel MH. Chem. Commun. 2006;48:5051–5053. doi: 10.1039/b611224b. [DOI] [PubMed] [Google Scholar]
- 19.Wong C-H, Zimmerman SC. Chem. Commun. 2013;49:1679–1695. doi: 10.1039/c2cc37316e. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JA, Lu YY, Burts AO, Lim Y-H, Finn MG, Koberstein JT, Turro NJ, Tirrell DA, Grubbs RH. J. Am. Chem. Soc. 2010;133:559–566. doi: 10.1021/ja108441d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moughton AO, O’Reilly RK. Chem. Commun. 2010;46:1091–1093. doi: 10.1039/b922289h. [DOI] [PubMed] [Google Scholar]
- 22.Blanazs A, Madsen J, Battaglia G, Ryan AJ, Armes SP. J. Am. Chem. Soc. 2011;133:16581–16587. doi: 10.1021/ja206301a. [DOI] [PubMed] [Google Scholar]
- 23.Kamps AC, Cativo MHM, Fryd M, Park S-J. Macromolecules. 2014;47:161–164. [Google Scholar]
- 24.Bang J, Jain S, Li Z, Lodge TP, Pedersen JS, Kesselman E, Talmon Y. Macromolecules. 2006;39:1199–1208. [Google Scholar]
- 25.Bhargava P, Tu Y, Zheng JX, Xiong H, Quirk RP, Cheng SZD. J. Am. Chem. Soc. 2007;129:1113–1121. doi: 10.1021/ja0653019. [DOI] [PubMed] [Google Scholar]
- 26.Shen H, Zhang L, Eisenberg A. J. Am. Chem. Soc. 1999;121:2728–2740. [Google Scholar]
- 27.Li Y, Deng Y, Tong X, Wang X. Macromolecules. 2006;39:1108–1115. [Google Scholar]
- 28.Yu K, Eisenberg A. Macromolecules. 1996;29:6359–6361. [Google Scholar]
- 29.Zhu X, Liu M. Langmuir. 2011;27:12844–12850. doi: 10.1021/la202680j. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Yan D. Angew. Chem. Int. Ed. 2004;43:4896–4899. doi: 10.1002/anie.200460325. [DOI] [PubMed] [Google Scholar]
- 31.Howse JR, Jones RAL, Battaglia G, Ducker RE, Leggett GJ, Ryan AJ. Nat. Mater. 2009;8:507–511. doi: 10.1038/nmat2446. [DOI] [PubMed] [Google Scholar]
- 32.Marguet M, Edembe L, Lecommandoux S. Angew. Chem. Int. Ed. 2012;51:1173–1176. doi: 10.1002/anie.201106410. [DOI] [PubMed] [Google Scholar]
- 33.Battaglia G, Ryan AJ. J. Am. Chem. Soc. 2005;127:8757–8764. doi: 10.1021/ja050742y. [DOI] [PubMed] [Google Scholar]
- 34.Dan K, Bose N, Ghosh S. Chem. Commun. 2011;47:12491–12493. doi: 10.1039/c1cc15663b. [DOI] [PubMed] [Google Scholar]
- 35.Dan K, Rajdev P, Deb J, Jana SS, Ghosh S. J. Polym. Sci., Part A: Polym. Chem. 2013;51:4932–4943. [Google Scholar]
- 36.Bhargava P, Zheng JX, Li P, Quirk RP, Harris FW, Cheng SZD. Macromolecules. 2006;39:4880–4888. [Google Scholar]
- 37.Tian F, Yu Y, Wang C, Yang S. Macromolecules. 2008;41:3385–3388. [Google Scholar]
- 38.Bae Y, Fukushima S, Harada A, Kataoka K. Angew. Chem. Int. Ed. 2003;42:4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 39.Ma N, Li Y, Xu H, Wang Z, Zhang X. J. Am. Chem. Soc. 2009;132:442–443. doi: 10.1021/ja908124g. [DOI] [PubMed] [Google Scholar]
- 40.Zhuang J, Gordon MR, Ventura J, Li L, Thayumanavan S. Chem. Soc. Rev. 2013;42:7421–7435. doi: 10.1039/c3cs60094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gohy J-F, Zhao Y. Chem. Soc. Rev. 2013;42:7117–7129. doi: 10.1039/c3cs35469e. [DOI] [PubMed] [Google Scholar]
- 42.Doncom KEB, Hansell CF, Theato P, O’Reilly RK. Polym. Chem. 2012;3:3007–3015. [Google Scholar]
- 43.Nowag S, Haag R. Angew. Chem. Int. Ed. 2014;53:49–51. doi: 10.1002/anie.201308619. [DOI] [PubMed] [Google Scholar]
- 44.Du J, Armes SP. J. Am. Chem. Soc. 2005;127:12800–12801. doi: 10.1021/ja054755n. [DOI] [PubMed] [Google Scholar]
- 45.Du J, Fan L, Liu Q. Macromolecules. 2012;45:8275–8283. [Google Scholar]
- 46.Lu J, Li N, Xu Q, Ge J, Lu J, Xia X. Polymer. 2010;51:1709–1715. [Google Scholar]
- 47.Kim SH, Tan JPK, Fukushima K, Nederberg F, Yang YY, Waymouth RM, Hedrick JL. Biomaterials. 2011;32:5505–5514. doi: 10.1016/j.biomaterials.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 48.Lutz J-F, Akdemir Ö, Hoth A. J. Am. Chem. Soc. 2006;128:13046–13047. doi: 10.1021/ja065324n. [DOI] [PubMed] [Google Scholar]
- 49.Ward MA, Georgiou TK. Polymers. 2011;3:1215–1242. [Google Scholar]
- 50.Peng B, Grishkewich N, Yao Z, Han X, Liu H, Tam KC. ACS Macro Lett. 2012;1:632–635. doi: 10.1021/mz300135x. [DOI] [PubMed] [Google Scholar]
- 51.Hua F, Jiang X, Zhao B. Macromolecules. 2006;39:3476–3479. [Google Scholar]
- 52.Feng K, Xie N, Chen B, Zhang L-P, Tung C-H, Wu L-Z. Macromolecules. 2012;45:5596–5603. [Google Scholar]
- 53.Wu H, Dong J, Li C, Liu Y, Feng N, Xu L, Zhan X, Yang H, Wang G. Chem. Commun. 2013;49:3516–3518. doi: 10.1039/c3cc39043h. [DOI] [PubMed] [Google Scholar]
- 54.Ryu J-H, Roy R, Ventura J, Thayumanavan S. Langmuir. 2010;26:7086–7092. doi: 10.1021/la904437u. [DOI] [PubMed] [Google Scholar]
- 55.Ryu J-H, Chacko RT, Jiwpanich S, Bickerton S, Babu RP, Thayumanavan S. J. Am. Chem. Soc. 2010;132:17227–17235. doi: 10.1021/ja1069932. [DOI] [PubMed] [Google Scholar]
- 56.Ryu J-H, Jiwpanich S, Chacko R, Bickerton S, Thayumanavan S. J. Am. Chem. Soc. 2010;132:8246–8247. doi: 10.1021/ja102316a. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Ryu J-H, Thayumanavan S. Langmuir. 2012;29:50–55. doi: 10.1021/la3033463. [DOI] [PubMed] [Google Scholar]
- 58.Ryu J-H, Bickerton S, Zhuang J, Thayumanavan S. Biomacromolecules. 2012;13:1515–1522. doi: 10.1021/bm300201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.C. D, Gonzaález-Toro, Ryu J-H, Chacko RT, Zhuang J, Thayumanavan S. J. Am. Chem. Soc. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Raghupathi K, Yuan C, Thayumanavan S. Chem. Sci. 2013;4:3654–3660. [Google Scholar]
- 61.Wang H, Zhuang J, Thayumanavan S. ACS Macro Lett. 2013;2:948–951. doi: 10.1021/mz4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo C-L, Lin S-J, Tsai H-C, Chan W-H, Tsai C-H, Cheng C-HD, Hsiue G-H. Biomaterials. 2009;30:3961–3970. doi: 10.1016/j.biomaterials.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Gebhart CL, Kabanov AV. J. Controlled Release. 2001;73:401–416. doi: 10.1016/s0168-3659(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 64.Nisha CK, Manorama SV, Ganguli M, Maiti S, Kizhakkedathu JN. Langmuir. 2004;20:2386–2396. doi: 10.1021/la035737r. [DOI] [PubMed] [Google Scholar]
- 65.De Smedt SC, Demeester J, Hennink WE. Pharm. Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 66.Cheng S, Zhang M, Dixit N, Moore RB, Long TE. Macromolecules. 2012;45:805–812. [Google Scholar]
- 67.Lee Y, Kataoka K. Soft Matter. 2009;5:3810–3817. [Google Scholar]
- 68.Naito M, Ishii T, Matsumoto A, Miyata K, Miyahara Y, Kataoka K. Angew. Chem. Int. Ed. 2012;51:10751–10755. doi: 10.1002/anie.201203360. [DOI] [PubMed] [Google Scholar]
- 69.Jackson AW, Fulton DA. Macromolecules. 2012;45:2699–2708. [Google Scholar]
- 70.Zhuang J, Chacko R, Amado Torres DF, Wang H, Thayumanavan S. ACS Macro Lett. 2013;3:1–5. doi: 10.1021/mz400515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zelikin AN, Li Q, Caruso F. Chem. Mater. 2008;20:2655–2661. [Google Scholar]
- 72.Yuan C, Raghupathi K, Popere BC, Ventura J, Dai L, Thayumanavan S. Chem. Sci. 2014;5:229–234. [Google Scholar]
- 73.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popović Z, Jain RK, Bawendi MG, Fukumura D. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2426–2431. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, Sasisekharan R. Nature. 2005;436:568–572. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 75.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J. Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 76.Maeda H, Fang J, Inutsuka T, Kitamoto Y. Int. Immunopharmacol. 2003;3:319–328. doi: 10.1016/S1567-5769(02)00271-0. [DOI] [PubMed] [Google Scholar]
- 77.Raghupathi K, Li L, Ventura J, Jennings M, Thayumanavan S. Polym. Chem. 2014;5:1737–1742. [Google Scholar]
- 78.Altintas O, Barner-Kowollik C. Macromol. Rapid Commun. 2012;33:958–971. doi: 10.1002/marc.201200049. [DOI] [PubMed] [Google Scholar]
- 79.Jiang J, Thayumanavan S. Macromolecules. 2005;38:5886–5891. [Google Scholar]
- 80.Wong EHH, Lam SJ, Nam E, Qiao GG. ACS Macro Lett. 2014;3:524–528. doi: 10.1021/mz500225p. [DOI] [PubMed] [Google Scholar]
- 81.Sanchez-Sanchez A, Pérez-Baena I, Pomposo JA. Molecules. 2013;18:3339–3355. doi: 10.3390/molecules18033339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harth E, Horn BV, Lee VY, Germack DS, Gonzales CP, Miller RD, Hawker CJ. J. Am. Chem. Soc. 2002;124:8653–8660. doi: 10.1021/ja026208x. [DOI] [PubMed] [Google Scholar]
- 83.Croce TA, Hamilton SK, Chen ML, Muchalski H, Harth E. Macromolecules. 2007;40:6028–6031. [Google Scholar]
- 84.Dobish JN, Hamilton SK, Harth E. Polym. Chem. 2012;3:857–860. [Google Scholar]
- 85.Sanchez-Sanchez A, Asenjo-Sanz I, Buruaga L, Pomposo JA. Macromol. Rapid Commun. 2012;33:1262–1267. doi: 10.1002/marc.201200180. [DOI] [PubMed] [Google Scholar]
- 86.de Luzuriaga AR, Ormategui N, Grande HJ, Odriozola I, Pomposo JA, Loinaz I. Macromol. Rapid Commun. 2008;29:1156–1160. [Google Scholar]
- 87.Oria L, Aguado R, Pomposo JA, Colmenero J. Adv. Mater. 2010;22:3038–3041. doi: 10.1002/adma.201000243. [DOI] [PubMed] [Google Scholar]
- 88.Beck JB, Killops KL, Kang T, Sivanandan K, Bayles A, Mackay ME, Wooley KL, Hawker CJ. Macromolecules. 2009;42:5629–5635. doi: 10.1021/ma900899v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Altintas O, Willenbacher J, Wuest KNR, Oehlenschlaeger KK, Krolla-Sidenstein P, Gliemann H, Barner-Kowollik C. Macromolecules. 2013;46:8092–8101. [Google Scholar]
- 90.Hansell CF, Lu A, Patterson JP, O’Reilly RK. Nanoscale. 2014;6:4102–4107. doi: 10.1039/c3nr06706h. [DOI] [PubMed] [Google Scholar]
- 91.Carboni RA, Lindsey Rv., Jr J. Am. Chem. Soc. 1959;81:4342–4346. [Google Scholar]
- 92.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J. Am. Chem. Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bai Y, Xing H, Vincil GA, Lee J, Henderson EJ, Lu Y, Lemcoff NG, Zimmerman SC. Chem. Sci. 2014;5:2862–2868. [Google Scholar]
- 95.Murray BS, Fulton DA. Macromolecules. 2011;44:7242–7252. [Google Scholar]
- 96.Whitaker DE, Mahon CS, Fulton DA. Angew. Chem. Int. Ed. 2013;52:956–959. doi: 10.1002/anie.201207953. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez-Sanchez A, Fulton DA, Pomposo JA. Chem. Commun. 2014;50:1871–1874. doi: 10.1039/c3cc48733d. [DOI] [PubMed] [Google Scholar]
- 98.Foster EJ, Berda EB, Meijer EW. J. Am. Chem. Soc. 2009;131:6964–6966. doi: 10.1021/ja901687d. [DOI] [PubMed] [Google Scholar]
- 99.Foster EJ, Berda EB, Meijer EW. J. Polym. Sci., Part A: Polym. Chem. 2011;49:118–126. [Google Scholar]
- 100.Berda EB, Foster EJ, Meijer EW. Macromolecules. 2010;43:1430–1437. [Google Scholar]
- 101.Terashima T, Sugita T, Fukae K, Sawamoto M. Macromolecules. 2014;47:589–600. [Google Scholar]
- 102.Terashima T, Mes T, De Greef TFA, Gillissen MAJ, Besenius P, Palmans ARA, Meijer EW. J. Am. Chem. Soc. 2011;133:4742–4745. doi: 10.1021/ja2004494. [DOI] [PubMed] [Google Scholar]
- 103.Gillissen MAJ, Terashima T, Meijer EW, Palmans ARA, Voets IK. Macromolecules. 2013;46:4120–4125. [Google Scholar]
- 104.Stals PJM, Gillissen MAJ, Paffen TFE, de Greef TFA, Lindner P, Meijer EW, Palmans ARA, Voets IK. Macromolecules. 2014;47:2947–2954. [Google Scholar]
- 105.Njikang G, Liu G, Hong L. Langmuir. 2011;27:7176–7184. doi: 10.1021/la2006887. [DOI] [PubMed] [Google Scholar]
- 106.Perez-Baena I, Loinaz I, Padro D, García I, Grande HJ, Odriozola I. J. Mater. Chem. 2010;20:6916–6922. [Google Scholar]
- 107.Wulff G, Chong B-O, Kolb U. Angew. Chem. Int. Ed. 2006;45:2955–2958. doi: 10.1002/anie.200503926. [DOI] [PubMed] [Google Scholar]
- 108.Artar M, Terashima T, Sawamoto M, Meijer EW, Palmans ARA. J. Polym. Sci., Part A: Polym. Chem. 2014;52:12–20. [Google Scholar]
- 109.Gillissen MAJ, Voets IK, Meijer EW, Palmans ARA. Polym. Chem. 2012;3:3166–3174. [Google Scholar]
- 110.Sanchez-Sanchez A, Akbari S, Etxeberria A, Arbe A, Gasser U, Moreno AJ, Colmenero J, Pomposo JA. ACS Macro Lett. 2013;2:491–495. doi: 10.1021/mz400173c. [DOI] [PubMed] [Google Scholar]
- 111.Sanchez-Sanchez A, Akbari S, Moreno AJ, Verso FL, Arbe A, Colmenero J, Pomposo JA. Macromol. Rapid Commun. 2013;34:1681–1686. doi: 10.1002/marc.201300562. [DOI] [PubMed] [Google Scholar]
- 112.Perez-Baena I, Barroso-Bujans F, Gasser U, Arbe A, Moreno AJ, Colmenero J, Pomposo JA. ACS Macro Lett. 2013;2:775–779. doi: 10.1021/mz4003744. [DOI] [PubMed] [Google Scholar]
- 113.Oh JK, Lee DI, Park JM. Prog. Polym. Sci. 2009;34:1261–1282. [Google Scholar]
- 114.Akiyoshi K, Deguchi S, Moriguchi N, Yamaguchi S, Sunamoto J. Macromolecules. 1993;26:3062–3068. [Google Scholar]
- 115.Yoksan R, Matsusaki M, Akashi M, Chirachanchai S. Colloid Polym. Sci. 2004;282:337–342. [Google Scholar]
- 116.Chen X-G, Lee CM, Park H-J. J. Agric. Food Chem. 2003;51:3135–3139. doi: 10.1021/jf0208482. [DOI] [PubMed] [Google Scholar]
- 117.Yang JS, Zhou QQ, He W. Carbohydr. Polym. 2013;92:223–227. doi: 10.1016/j.carbpol.2012.08.100. [DOI] [PubMed] [Google Scholar]
- 118.Chen S, Li Y, Guo C, Wang J, Ma J, Liang X, Yang L-R, Liu H-Z. Langmuir. 2007;23:12669–12676. doi: 10.1021/la702049d. [DOI] [PubMed] [Google Scholar]
- 119.Li X, Yuan W, Gu S, Ren J. Mater. Lett. 2010;64:2663–2666. [Google Scholar]
- 120.Dash TK, Konkimalla VB. J. Controlled Release. 2012;158:15–33. doi: 10.1016/j.jconrel.2011.09.064. [DOI] [PubMed] [Google Scholar]
- 121.Tian H, Tang Z, Zhuang X, Chen X, Jing X. Prog. Polym. Sci., 2010. 2012;37:237–280. [Google Scholar]
- 122.Tao Y, He J, Zhang M, Hao Y, Liu J, Ni P. Polym. Chem. 2014;5:3443–3452. [Google Scholar]
- 123.Hernandez-Valdepeña I, Domurado M, Coudane J, Braud C, Baussard J-F, Vert M, Domurado D. Eur. J. Pharm. Sci. 2009;36:345–351. doi: 10.1016/j.ejps.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 124.Nottelet B, Patterer M, Francçois B, Schott M-A, Domurado M, Garric X, Domurado D, Coudane J. Biomacromolecules. 2012;13:1544–1553. doi: 10.1021/bm300251j. [DOI] [PubMed] [Google Scholar]
- 125.Deming TJ. Adv Drug Deliver Rev. 2002;54:1145–1155. doi: 10.1016/s0169-409x(02)00062-5. [DOI] [PubMed] [Google Scholar]
- 126.Rhodes AJ, Deming TJ. J Am Chem Soc. 2012;134:19463–19467. doi: 10.1021/ja308620h. [DOI] [PubMed] [Google Scholar]
- 127.Huang J, Heise A. Chem. Soc. Rev. 2013;42:7373–7390. doi: 10.1039/c3cs60063g. [DOI] [PubMed] [Google Scholar]
- 128.Lu H, Wang J, Song Z, Yin L, Zhang Y, Tang H, Tu C, Lin Y, Cheng J. Chem. Commun. 2014;50:139–155. doi: 10.1039/c3cc46317f. [DOI] [PubMed] [Google Scholar]
- 129.Barrett SE, Abrams MT, Burke R, Carr BA, Crocker LS, Garbaccio RM, Howell BJ, Kemp EA, Kowtoniuk RA, Latham AH, Leander KR, Leone AM, Patel M, Pechenov S, Pudvah NT, Riley S, Sepp-Lorenzino L, Walsh ES, Williams JM, Colletti SL. Int. J. Pharm. 2014;466:58–67. doi: 10.1016/j.ijpharm.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 130.Lee ES, Gao Z, Bae YH. J. Controlled Release. 2008;132:164–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schmaljohann D. Adv. Drug Delivery Rev. 2006;58:1655–1670. doi: 10.1016/j.addr.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 132.Du JZ, Sun TM, Song WJ, Wu J, Wang J. Angew. Chem. Int. Ed. 2010;49:3621–3626. doi: 10.1002/anie.200907210. [DOI] [PubMed] [Google Scholar]
- 133.Sethuraman VA, Bae YH. J. Controlled Release. 2007;118:216–224. doi: 10.1016/j.jconrel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang Y, Tang Z, Zhang X, Yu H, Sun H, Pang X, Chen X. Biomacromolecules. 2013;14:2023–2032. doi: 10.1021/bm400358z. [DOI] [PubMed] [Google Scholar]