Summary
The growth or virulence of Mycobacterium tuberculosis bacilli depends on homologous type VII secretion systems, ESX-1, ESX-3 and ESX-5, which export a number of protein effectors across membranes to the bacterial surface and environment. PE and PPE proteins represent two large families of highly polymorphic proteins that are secreted by these ESX systems. Recently, it was shown that these proteins require system-specific cytoplasmic chaperones for secretion. Here, we report the crystal structure of M. tuberculosis ESX-5-secreted PE25–PPE41 heterodimer in complex with the cytoplasmic chaperone EspG5. EspG5 represents a novel fold that is unrelated to previously characterized secretion chaperones. Functional analysis of the EspG5-binding region uncovered a hydrophobic patch on PPE41 that promotes dimer aggregation, and the chaperone effectively abolishes this process. We show that PPE41 contains a characteristic chaperone-binding sequence, the hh motif, which is highly conserved among ESX-1-, ESX-3- and ESX-5-specific PPE proteins. Disrupting the interaction between EspG5 and three different PPE target proteins by introducing different point mutations generally affected protein secretion. We further demonstrate that the EspG5 chaperone plays an important role in the ESX secretion mechanism by keeping aggregation-prone PE–PPE proteins in their soluble state.
Keywords: EspG, ESX, Mycobacterium tuberculosis, Type VII Secretion System
Introduction
Mycobacteria, such as the etiological agent of human tuberculosis, Mycobacterium tuberculosis, are a group of unusual Gram-positive bacteria that have remarkably complex cell envelopes. Similar to protein secretion of Gram-negative bacteria, protein transport across mycobacterial membranes requires a specialized export machinery known as the type VII secretion or ESX system (Houben et al., 2014). The genome of M. tuberculosis encodes 5 paralogous ESX systems, named ESX-1 to ESX-5. The ESX-1 secretion system is critical for virulence of M. tuberculosis (Stanley et al., 2003, Guinn et al., 2004) and Mycobacterium marinum (Gao et al., 2004, Tan et al., 2006) and is required for bacterial replication in macrophages and phagosomal escape into the cytosol (van der Wel et al., 2007, Smith et al., 2008, Simeone et al., 2012). The ESX-3 locus is involved in iron acquisition and is essential in a number of mycobacterial species (Serafini et al., 2009, Siegrist et al., 2009, Siegrist et al., 2014). Additionally, the ESX-3 system is implicated in disruption of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery and arrest of phagosome maturation (Mehra et al., 2013). In M. tuberculosis and M. marinum the ESX-5 system is associated with virulence mechanisms by modulating host immune responses to the mycobacteria (Bottai et al., 2012, Weerdenburg et al., 2012). The functions of the ESX-2 and the ESX-4 systems in mycobacteria are not yet understood. Each ESX system consists of ATPases, membrane proteins, a protease, accessory proteins, and transports secreted substrates that lack classical signal sequences (Houben et al., 2014).
The most numerous ESX substrates, common to all mycobacterial ESX systems except ancestral ESX-4, are the PE and PPE proteins. These secreted proteins are named after several highly conserved residues in their characteristic N-terminal motifs: proline-glutamic acid (PE) and proline-proline-glutamic acid (PPE) (Cole et al., 1998). The C-terminal domains of both PE and PPE proteins can be extremely long and some carry functional domains such as lipase (Deb et al., 2006, Mishra et al., 2008, Daleke et al., 2011), aspartic protease (Barathy & Suguna, 2013) and serine hydrolase (Sultana et al., 2011, Sultana et al., 2013) domains. Although the precise function of these proteins is largely unknown, it has been demonstrated that various PE/PPE proteins are associated with virulence and persistence in the host (Sampson, 2011). They are highly abundant in pathogenic mycobacteria, with 100 pe and 69 ppe genes in M. tuberculosis H37Rv (Gey van Pittius et al., 2006). Based on the phylogenetic analysis of PE and PPE families and experimental data, it has been suggested that the majority of these proteins are secreted through the ESX-5 system (Abdallah et al., 2009).
Pe/ppe genes are usually organized in bicistronic operons, although this is not universal for all PE/PPE homologs. It has been shown that M. tuberculosis PE25 and PPE41 form a heterodimer (Strong et al., 2006). In the structure these two proteins interact via a hydrophobic interface forming a four-helix bundle with two α-helices contributed by both PE25 and PPE41 (Strong et al., 2006). PE25–PPE41 heterodimer is secreted by the ESX-5 system of M. tuberculosis (Bottai et al., 2012). While M. tuberculosis PE25–PPE41 proteins do not have close homologs in M. marinum (Abdallah et al., 2006), when this protein pair has been expressed in M. marinum, it is also secreted through the ESX-5 system. Based on the fact that ESX-1 and ESX-5 systems secrete a specific subset of PE/PPE proteins, it has been suggested that PE/PPE proteins encode a secretion signal, which is recognized by their cognate ESX machinery for export (Daleke et al., 2012b, Daleke et al., 2012a). A highly conserved YxxxD/E motif has been identified in ESX-secreted proteins, including PE proteins (Daleke et al., 2012a). This motif is required for ESX-specific secretion, however it does not by itself determine through which ESX system the PE–PPE pair is transported (Daleke et al., 2012a).
Recently, the ESX-5 encoded EspG protein (EspG5) was found to form a 1:1:1 complex with PE25 and PPE41 (Daleke et al., 2012b). In contrast, EspG5 does not interact with the non-cognate PE–PPE pair secreted via the ESX-1 translocon (Daleke et al., 2012b). Reciprocally, ESX-1-encoded EspG1 recognizes only its cognate PE35–PPE68_1 heterodimer (encoded by mmar_0185-mmar_0186 locus). It has been proposed that EspG functions as a secretion chaperone by binding of the PE–PPE pair and directing it to its cognate ESX translocation machinery (Daleke et al., 2012b).
To gain insight into the functional role of EspG5 in the ESX-5 secretory mechanism, we have solved the structure of the heterotrimeric complex of EspG5–PE25–PPE41. This novel structure provides the first snapshot of interactions between one of the key components of the ESX machinery and ESX secreted proteins. Furthermore, a structure-guided mutational analysis allowed us to identify a sequence motif in the PPE protein family that is targeted by EspG5. Finally, we show that the chaperone plays an important role in preventing aggregation of PE–PPE dimers.
Results
Overall structure of PE25–PPE41–EspG5 complex
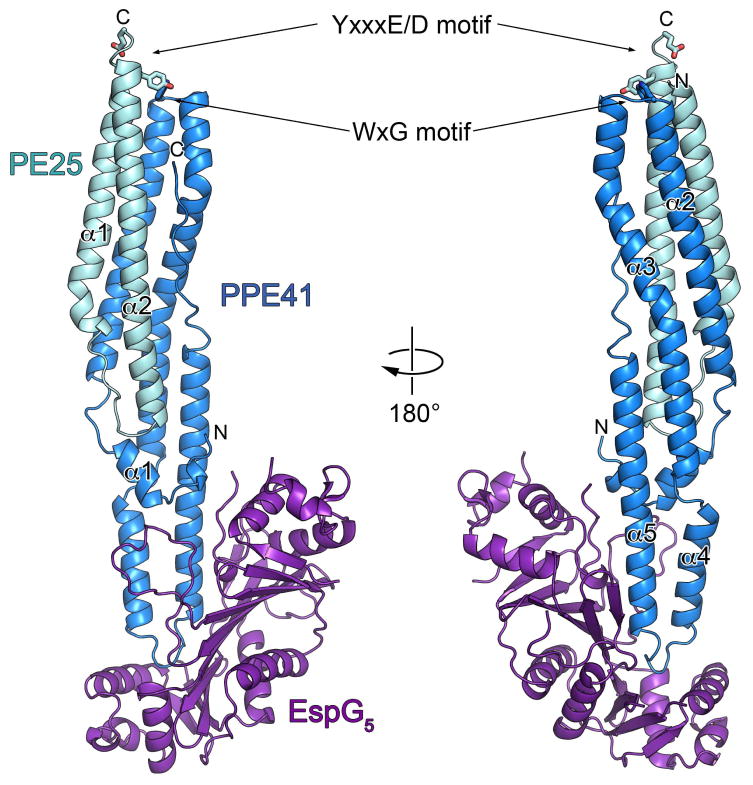
To understand how PE and PPE proteins are recognized by their cognate EspG chaperones in molecular details we determined the crystal structures of a PE25–PPE41 dimer in complex with EspG5. The constructs of full-length M. tuberculosis EspG5 and PE25–PPE41 were expressed in Escherichia coli and purified. The ternary complex yielded crystals that diffracted to 2.6 Å resolution (Table 1). The structure of the complex was solved by molecular replacement using the structure of PE25–PPE41 heterodimer (Strong et al., 2006) as a search model. We found that EspG5 interacts exclusively with the PPE41 protein at one end of the elongated PE25–PPE41 heterodimer (Fig. 1). The comparison of the PE25–PPE41 dimer bound to EspG5 with the previously reported PE25–PPE41 structure (PDB 2G38) showed that the structures are very similar, with an r.m.s.d. of 0.7 Å (PE25) and 1.2 Å (PPE41) between the dimer and the ternary complex structures. Thus, the binding of EspG5 chaperone does not cause conformational changes in the PE25–PPE41 dimer. EspG5 binds the PE25–PPE41 dimer at a location that is distal from the C-terminal regions of both PE25 and PPE41, which leaves the signature ESX secretory motif YxxxD/E of PE25 protein (Daleke et al., 2012a) completely accessible for putative interactions with the secretory machinery. This structural information is in line with the observation that these regions are not required for binding of EspG (Daleke et al., 2012b). In contrast with PE25–PPE41 heterodimer structure where the YxxxD/E motif (residues 87–91) is disordered, in our structure the YxxxD/E motif adopts a helical conformation at the C-terminal end of α2 helix of PE25. This helical conformation of YxxxD/E motif is reminiscent of recently reported crystal structures of ESX-secreted proteins: M. tuberculosis EsxO–EsxP (PDB 4GZR) and EsxA–EsxB (PDB 3FAV), and M. smegmatis EsxG–EsxH (PDB 3Q4H) where YxxxD/E motifs also form helical structures (Arbing et al., 2010, Poulsen et al., 2014). Therefore, the helical conformation of the YxxxD/E motif may be a general feature of proteins secreted by ESX systems.
Table 1.
Data collection and refinement statistics.
| PE25–PPE41–EspG5 (PDB 4KXR) | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9790 |
| Space group | P6122 |
| Cell dimensions: | |
| a, b, c (Å) | 139.10, 139.10, 171.01 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 49.24–2.60 (2.67–2.60)a |
| Rsym | 0.120 (1.535) |
| CC½b | 99.8 (66.4) |
| I / σI | 15.8 (1.8) |
| Completeness (%) | 99.9 (99.2) |
| Multiplicity | 10.9 (10.9) |
| Refinement | |
| Resolution (Å) | 49.24–2.60 |
| No. reflections (total / free) | 30602 / 1553 |
| Rwork / Rfree | 0.194 / 0.244 |
| Number of atoms: | |
| Protein | 4135 |
| Ligand/ion | 0 |
| Water | 109 |
| B-factors: | |
| PE25 | 65.6 |
| PPE41 | 50.1 |
| EspG5 | 74.9 |
| Water | 47.8 |
| All atoms | 64.6 |
| Wilson B | 60.3 |
| R.m.s. deviations: | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.266 |
| Ramachandran distributionc (%): | |
| Favored | 96.57 |
| Outliers | 0.19 |
Values in parentheses are for the highest-resolution shell.
CC1/2 correlation coefficient as defined in Karplus & Diederichs (Karplus & Diederichs, 2012) and calculated by XSCALE (Kabsch, 2010).
Calculated using the MolProbity server (http://molprobity.biochem.duke.edu) (Chen et al., 2010).
Fig. 1. Crystal structure of the M. tuberculosis PE25–PPE41–EspG5 complex.
Ribbon representation of the complex in two views related by ~180° rotation. EspG5 interacts with PPE protein on the opposite side from the conserved YxxxD/E and WxG motifs of PE25–PPE41 dimer.
Structure of EspG5 reveals a novel fold
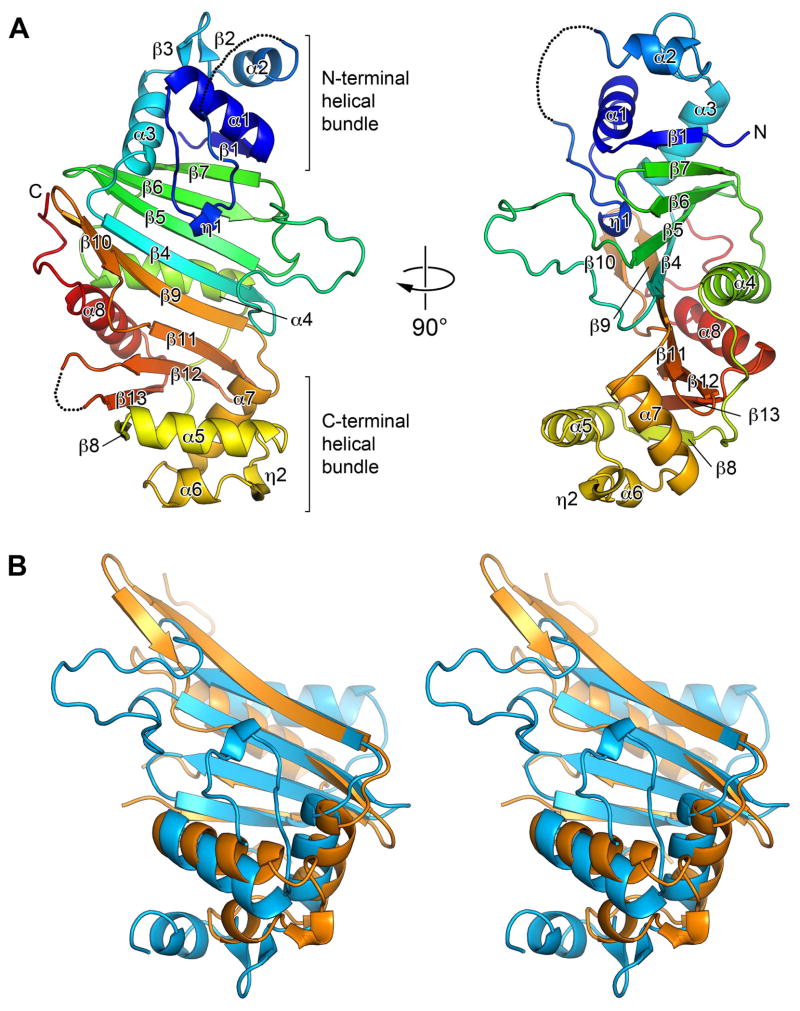
The structure of EspG5 consists of two halves, or sub-domains, that are related by a pseudo 2-fold symmetry (Fig. 2 and Fig. S1) and can be superimposed with an r.m.s.d of 3.8 Å and 13% sequence identity between 88 aligned residues (Fig. 2B), indicating that the EspG family of proteins probably evolved by intergenic duplication event. The central continuous anti-parallel β-sheet is composed of β-strands β1 and β4–β7 from the N-terminal half and β8–β13 from the C-terminal half. Two helical bundles are located on the opposite sides of the front surface of the central β-sheet. The N-terminal helical bundle contains α-helices α1–α3, a 310-helix η1 and a β-hairpin β2–β3. The C-terminal helical bundle is composed of α-helices α5–α7 and a 310-helix η2. The α-helices α4 and α8 are packed against the back surface of the central β-sheet. A search using the Dali (Holm & Rosenstrom, 2010) and PDBeFold (Krissinel & Henrick, 2004) servers did not reveal any close structural homologs of EspG5.
Fig. 2. Structure of EspG5 represents a novel fold with quasi 2-fold symmetry.
A. Ribbon representation is colored in rainbow colors from N-terminus (blue) to C-terminus (red). Secondary structure elements are labeled α1–α8 and β1–β13. The disordered loops are indicated as dashed lines. The two views are related by ~90° rotation.
B. Stereo view of the structural superposition of the N-terminal (blue) and the C-terminal (orange) sub-domains of EspG5.
Analysis of EspG5–PPE41 interface
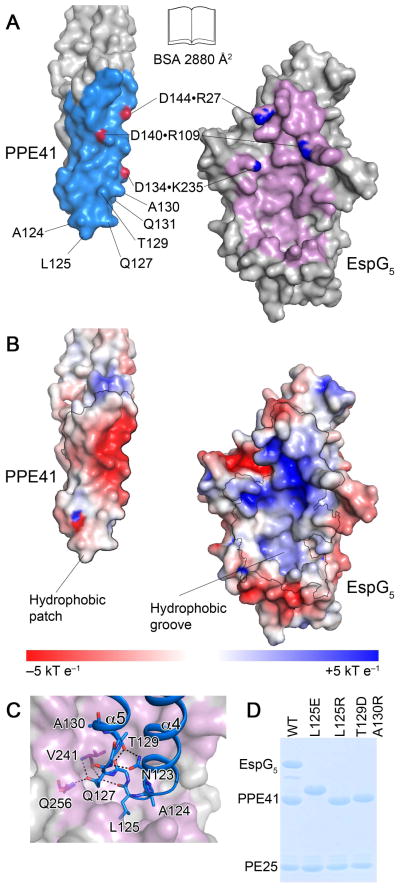
The interface between EspG5 and PPE41 buries 2,880 Å2 of solvent-accessible surface area as calculated by the PISA server (Krissinel & Henrick, 2007). Overall, 45 residues from EspG5 and 34 residues from PPE41 are engaged in multiple hydrophobic interactions, a number of hydrogen bonds and 3 salt bridges (Fig. 3). The ‘tip’ of PPE41 composed of helices α4 and α5 is inserted in a deep hydrophobic groove formed by the central β-sheet and the C-terminal α-helical bundle of EspG5. Additional interactions form between helix α5 of PPE41 and the N-terminal α-helical bundle of EspG5, and between the N-terminal ‘hook’ of PPE41 and the long β4–β5 loop strands of EspG5. Notably, the β4–β5 loop of EspG5 projects away from PPE41 forming a cleft between the two proteins. Additional interactions between the β4–β5 loop of EspG5 and PPE41 may occur in solution, because the observed ‘outward’ orientation of β4–β5 loop is stabilized by crystal contacts.
Fig. 3. The interface between EspG5 and PPE41.
A. An ‘open book’ view of the PPE41–EspG5 complex. Contact residues in the interface are colored in light blue (PPE41) and purple (EspG5). Atoms participating in intermolecular salt bridges are colored in red and blue.
B. EspG5 and PPE41 are shown in the same orientation as in panel (A). The surface is colored according to electrostatic surface potential contoured at ±5 kT e−1, with red corresponding to a negative and blue to a positive potential. The contact areas are indicated by black lines.
C. EspG5 is shown in surface representation as in panel (B), PPE41 is shown in ribbon representation (blue). Residues in α4-α5 loop are shown in stick representation. Hydrogen bonds are shown as black dashed lines.
D. SDS-PAGE analysis of co-purification of EspG5 and PE25–PPE41 mutant variant dimers. Proteins were purified by affinity chromatography from the lysate of E. coli strain expressing N-terminally His-tagged PE25, PPE41 and EspG5. Note that PPE41L125E has an altered mobility on SDS-PAGE.
In the ‘lower’ part of the interface, the loop between helices α4 and α5 of PPE41 is almost completely shielded by interactions with EspG5. Interestingly, the conformation of the α4–α5 loop of PPE41 is preserved both in the apo– and EspG5–bound structures. This conformation is stabilized by a hydrogen-bond network between the side-chains of N123 and T129, carbonyl of G126, and the main chain N of F128 of PPE41 (Fig. 3C). In addition, another hydrogen-bond network is formed in the PPE41–EspG5 complex, which involves the main chain carbonyl of L125PPE41 and the side chain of Q127PPE41, and the main chain N and carbonyl of V241EspG5 and the side chain of Q256EspG5 (Fig. 3C). While the interactions in the ‘lower’ part of PPE41–EspG5 interface are hydrophobic, the ‘upper’ part of the interface displays complementary charged areas, with 3 salt bridges formed by D134PPE41–K235EspG5, D140PPE41–R109EspG5, and D144PPE41–R27EspG5 (Fig. 3A). Therefore, hydrophobic interactions, as well as hydrogen bonds, shape, and electrostatic complementarity, all contribute to stabilization of the PPE41–EspG5 complex.
Substitutions in EspG5–PPE41 interface disrupt complex formation
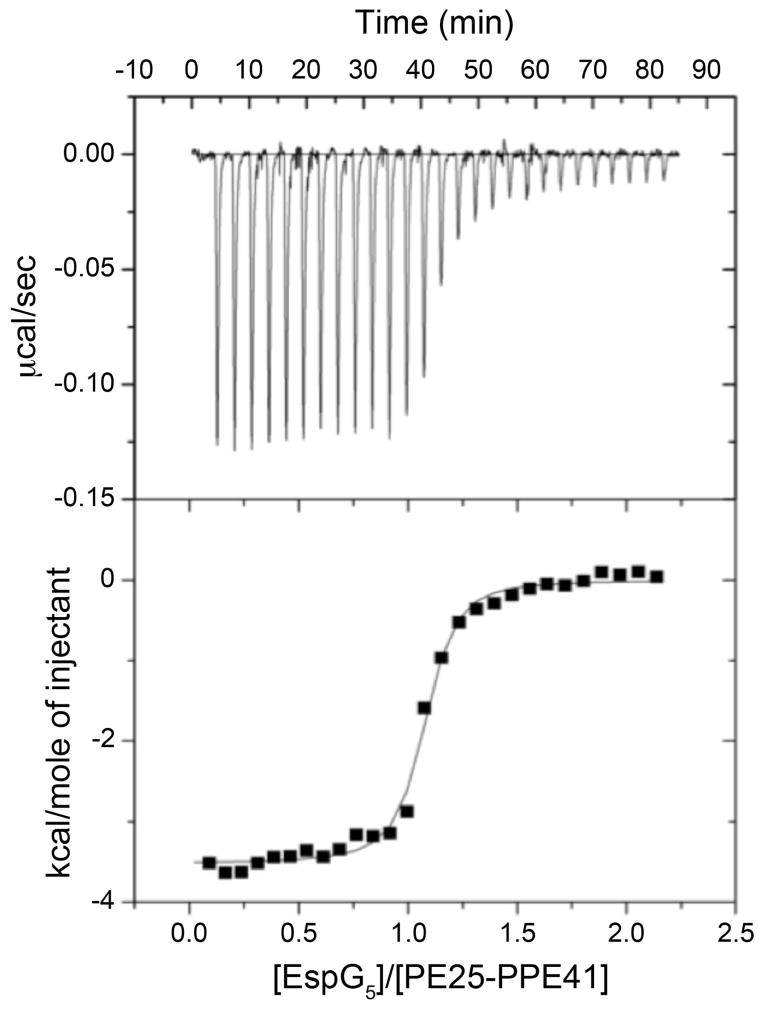
To confirm that the interface between EspG5 and PPE41 is physiologically relevant, we introduced L125E, L125R and T129D/A130R amino acid substitutions in PPE41. L125, T129 and A130 are located in the loop between helices α4 and α5 of PPE41 and are involved in interactions with EspG (Fig. 3C). The mutant variants of PE25–PPE41 and EspG5 were expressed in E. coli and employed for pull-down experiments (Fig. 3D). Using CD spectroscopy we confirmed that the introduced mutations did not alter the secondary and tertiary structural features of PE–PPE heterodimers (Fig. S3). However, our pull-down experiments confirmed that, although the structure and dimer formation was similar, these mutations abolished EspG5 binding to PE25–PPE41. In addition, we analyzed EspG5 binding to PE25–PPE41 using isothermal titration calorimetry (ITC). We found that the chaperone binds to PE25–PPE41 with high affinity with a dissociation constant of 48 nM (Fig. 4 and Table S1). However we did not detect any EspG5 binding to the PE25–PPE41 variants with the mutations in the interface. Therefore, the interface we identified in our crystal structure is relevant for complex formation in vitro.
Fig. 4. Isothermal calorimetry of PE25–PPE41 with EspG5.
EspG5 (100 μM) was injected into a solution of PE25–PPE41 (10 μM) containing the same buffer. The resulting isotherm data were fit to a single-site model, which gave an average dissociation constant of 48.1 nM. One representative data set is shown here.
Analysis of the EspG-binding region on PPE proteins
It has been shown that EspG5 does not interact with the M. marinum PE35–PPE68_1 dimer secreted via the ESX-1 system (Daleke et al., 2012b). We found that when PE35–PPE68_1 is co-expressed with EspG5, the heterodimer is insoluble and does not bind the chaperone. However when it is co-expressed with EspG1, the heterodimer is soluble and binds the chaperone (Fig. S4). We further extended this observation and analyzed co-expression of the ESX-3-specific PE–PPE pair, PE5–PPE4, either with EspG3 or with EspG5 chaperones. Similar to the ESX-1 specific heterodimer, PE5–PPE4 does not recognize EspG5, but binds the cognate chaperone (Fig. S5). These data suggest that EspG5 interacts exclusively with the ESX-5 secreted PE–PPE proteins.
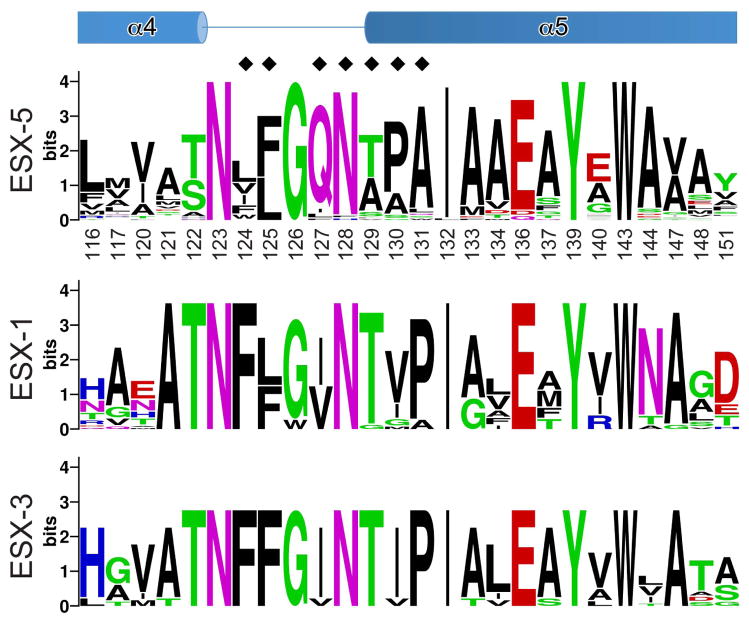
We hypothesized that each ESX-specific family of PPE proteins could contain a specific sequence, possibly the chaperone binding region, recognized by their cognate chaperone. Based on phylogenetic analysis of the PPE proteins encoded by the M. tuberculosis genome it has been predicted that ESX-3 and ESX-5 clusters may secrete 9 and more than 46 PPE proteins, respectively, whereas the ESX-1 cluster is predicted to be involved in the secretion of a single PPE68 protein in M. tuberculosis (Gey van Pittius et al., 2006). Using our EspG5–PPE41 structure as a guide and considering the sequences of the ESX-5 specific PPE proteins, we analyzed the EspG5 binding region in the PPE proteins. This region comprises 25 amino acid residues of PPE41 (Fig. 5 and Fig. S2).
Fig. 5. EspG5-binding region of PPE41.
The sequence conservation of the EspG5-binding site is displayed as a sequence logo (http://weblogo.berkeley.edu) (Crooks et al., 2004) based on the sequence alignments of ESX-5- and ESX-3-specific PPE proteins of M. tuberculosis H37Rv and ESX-1-specific PPE68 homologs from mycobacteria (Fig. S2, S6 and S7). Secondary structure elements of PPE41 are shown at the top. Black diamonds indicate residues that were subjected to mutational analysis (Table 2 and Fig. 3C, S8, S9 and S10).
Next, the identified region was searched in the aligned sequences of the PPE proteins that belong to the ESX-3- and ESX-1-specific PPE subfamilies. Because PPE68 is the only ESX-1-specific PPE protein in M. tuberculosis, we extended our analysis by including the ESX-1-specific PPE homologues from other mycobacteria (Fig. S6). To understand the mechanism of PPE proteins recognition by EspG5, we looked for PPE residues in the identified region that distinguish ESX-5 secreted proteins from all other PPE proteins in mycobacteria. Surprisingly, we found that this region is highly conserved among PPE proteins of ESX-3 and ESX-1 families, except three residues, 127, 130 and 131 (numbering corresponds to PPE41 sequence), that are located in the α4-α5 loop of PPE41 (Fig. 5, S6 and S7). Q127 is present in 41 PPE proteins out of 46 analyzed ESX-5 specific family members. The majority of analyzed ESX-3 and ESX-1 specific PPE proteins have isoleucine in this position. The position 130 is occupied by small non-polar amino acids such as alanine or proline, and rarely by serine or glycine in the ESX-5 specific PPE proteins. In contrast, the majority of the ESX-3- and ESX-1-specific PPE proteins have isoleucine in this position. Finally, alanine most frequently occupies position 131 in the ESX-5 specific PPE proteins, whereas PPE41 has glutamine in this position. However, all ESX-3 specific PPE proteins and the majority of ESX-1 specific PPE proteins (17 out of 22 PPE proteins) have proline in this position.
To examine whether Q127I, A130I and Q131P amino acid substitutions disrupt EspG5 recognition of PPE41, we introduced single, double or triple amino acid substitutions in PPE41 (Table 2 and Fig. S8). These PE25–PPE41 mutant variants were co-expressed with EspG5 in E. coli, and analyzed by pull-down experiments. Remarkably, we found that all PPE41 mutant variants bind to EspG5. Moreover, even multiple substitutions in PPE41 that matched the ESX-1/ESX-3 PPE sequence region did not disrupt the interaction with EspG5 (Table 2 and Fig. S8).
Table 2.
Analysis of PE25–PPE41–EspG5 interactions in vitro.
| PPE41 variant | Interaction |
|---|---|
| WTa | + |
| L125E | − |
| L125R | − |
| T129D/A130R | − |
| Q127I | + |
| Q127I/F128N | + |
| A130I | + |
| Q127I/A130I | + |
| Q127I/F128N/Q131P | + |
| Q127I/F128N/A130I/Q131P | + |
| A124F/L125F/Q127I/F128N | + |
| A124F/L125F/Q127I/F128N/A130I/Q131P | + |
| A124L | + |
| A124L/L125F | + |
| A124F/L125F | + |
| A124W/L125F | + |
Interactions of PE25–PPE41 with EspG5 were analyzed using pull-down experiments presented in Fig. 3C, S8, S9 and S10.
WT — wild type.
Therefore, our data indicate that EspG5 - PPE interactions are characterized by a striking structural plasticity that allows contact points of PPE to adapt multiple amino acid substitutions. This propensity of the EspG5 – PPE interactions is likely to facilitate EspG5 specific recognition of multiple PPE members of ESX-5 family. Furthermore we found that the sequence variability in the EspG5-binding region of PPE proteins does not determine the specificity of recognition by the chaperone. This observation suggests that EspG5-binding domain is not a linear sequence, but rather a conformational motif recognized by the chaperone. The α4–α5 region of PPE proteins of ESX-1 and ESX-3 families is likely to have the distinct conformational topology that is determined by PE–PPE sequence differences located outside of this region. The differences in the 3D shape of the binding domain might preclude the accessibility of this region for non-cognate chaperone recognition.
EspG5 protects PE–PPE from self-aggregation
PE25–PPE41 heterodimer can be expressed independently of EspG5 in E. coli and purified in soluble form (Strong et al., 2006). However, we observed that other PE–PPE family proteins required co-expression of their cognate EspG chaperones for soluble expression. For example, co-expression of the ESX-1-specific M. marinum PE35–PPE68_1 heterodimer in the absence of its cognate chaperone in E. coli resulted mainly in the production of insoluble proteins. When PE35–PPE68_1 pair was co-expressed with EspG1, we obtained a highly soluble and stable heterotrimer (Fig. S4). These observations suggest that EspG promotes PE–PPE heterodimer folding/stability. Interestingly, analysis of the EspG5-binding region in 46 ESX-5-specific PPE family members revealed that PPE41 is unique among PPE proteins. All members of the PPE family, except PPE41, are characterized by large non-polar residues in positions 124 and 125 located in the loop between helices α4 and α5 of PPE41 (Fig. 5 and Fig. S2). In addition, the hydrophobic residues are also present in equivalent positions in the ESX-1- and ESX-3-specific members of PPE family (Fig. 5, S6 and S7). This observation led us to hypothesize that PPE proteins are unstable without the chaperone because of this hydrophobic motif, which we designated as the hh motif, where h is an aliphatic or aromatic amino acid. It is possible that EspG binding to PPE shields this hydrophobic patch thus preventing self-association and non-productive inter-molecular interactions in the mycobacterial cytoplasm. To test this hypothesis, we replaced PPE41 A124 and L125 with large non-polar amino acid residues that are more frequently present in other members of the ESX-5-specific PPE family. His-tagged mutant variants of PE25–PPE41 complexes with A124L, A124L/L125F, A124F/L125F or A124W/L125F substitutions were expressed in E. coli in the presence or absence of EspG5 and purified by affinity chromatography. We found that co-expression of the PE25–PPE41 mutant variants with increased hydrophobicity in the α4-α5 loop of PPE41 with EspG5 yielded soluble PE41–PPE25–EspG5 heterotrimers (Fig. S9). In contrast, expression of PE25–PPE41 mutant variants in the absence of EspG5 resulted in accumulation of significant amounts of insoluble PE25–PPE41 proteins and progressively decreasing yield of soluble heterodimer (Table 3 and Fig. S9). Moreover, examination of the soluble PE25–PPE41A124L mutant complex by size-exclusion chromatography revealed the presence of large molecular weight protein aggregates that eluted near the column’s void volume (Fig. 6). In contrast, PE25–PPE41 WT complex did not aggregate at the same protein concentration (Fig. 6). These experiments indicate that the hh motif promotes aggregation and, by increasing hydrophobicity of the hh motif, the heterodimer becomes more dependent on the EspG chaperone for folding/stability.
Table 3.
Hydrophobic substitutions in the hh motif reduce solubility of PE25–PPE41.
| PPE41 variant | Protein yield (mg×g−1) |
|---|---|
| WTa | 3.6 |
| A124L | 2.9 |
| A124L/L125F | 1.0 |
| A124F/L124F | 0.9 |
| A124W/L124F | 1.1 |
The PE25–PPE41 variants were purified under identical conditions and the total protein yield was calculated (Fig. S9).
WT — wild type.
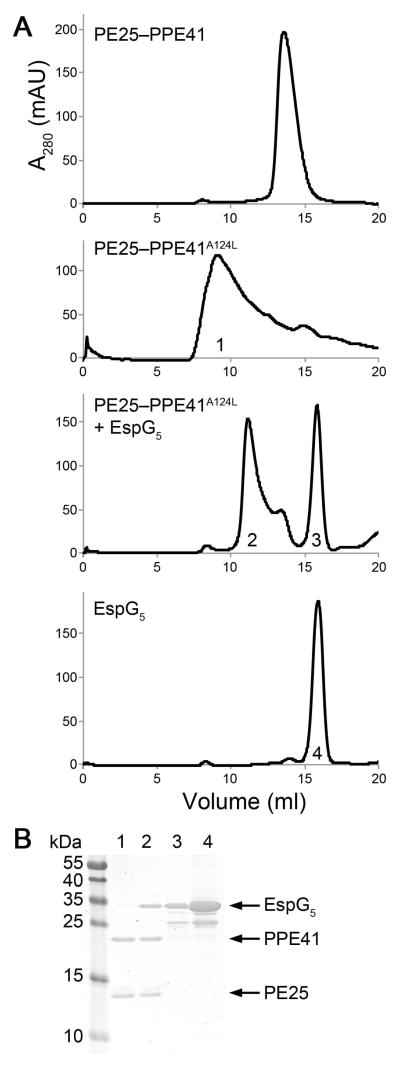
Fig. 6. EspG5 binding to PE25–PPE41A124L mutant complex induces dis-aggregation of the heterodimer.
A. Size-exclusion chromatography of PE25–PPE41, PE25–PPE41A124L mutant, EspG5 incubated with PE25–PPE41A124L mutant and EspG5.
B. SDS-PAGE analysis of the peak fractions indicated (1–4) from the size-exclusion chromatography.
As a further test, we examined whether purified EspG5 is capable of rescuing the PE25–PPE41A124L aggregates. EspG5 was incubated with PE25–PPE41A124L aggregates and the sample was analyzed by size-exclusion chromatography (Fig. 6). We found that incubation with EspG5 resulted in PE25–PPE41 disaggregation and formation of a PE25–PPE41–EspG5 complex (Fig. 6). This experiment provides further evidence that EspG5 acts as a molecular chaperone for PE–PPE dimers by binding and protecting the aggregation-prone hh motif on PPE proteins, thereby keeping the dimers in a secretion-competent state.
Effect of mutations in the PPE41 interface on PE25–PPE41 secretion
It has been reported that EspG5 is dispensable for PE25–PPE41 secretion via ESX-5 in M. tuberculosis (Bottai et al., 2012). However, we have previously demonstrated that expression of PE25 and PPE41 proteins in non-native host M. marinum resulted in their secretion via the ESX-5 system (Abdallah et al., 2006) and EspG5mm is required for this process (Abdallah et al., 2009). To determine the role of EspG5 in PE25-PPE41 secretion and the function of the hh motif we analyzed the secretion of PPE41 interface mutants in M. marinum. First, we confirmed that M. marinum EspG5 functions similarly in the in vitro binding assays with M. tuberculosis PE25–PPE41. EspG5 of M. tuberculosis is 97% identical at the amino acid level to the M. marinum EspG5 (EspG5mma). The analysis of the EspG5–PPE41 interface indicates that EspG5mma has a single amino acid substitution in the interface — M176, which is unlikely to have an effect on EspG5 binding. To confirm that L125E, L125R and T129D/A130R amino acid substitutions in PPE41 also disrupt the binding of EspG5mma, pull-down experiments were conducted with the mutant variants (Fig. S10). This analysis showed that EspG5mma very likely uses the same interface as M. tuberculosis EspG5 to bind PE25–PPE41.
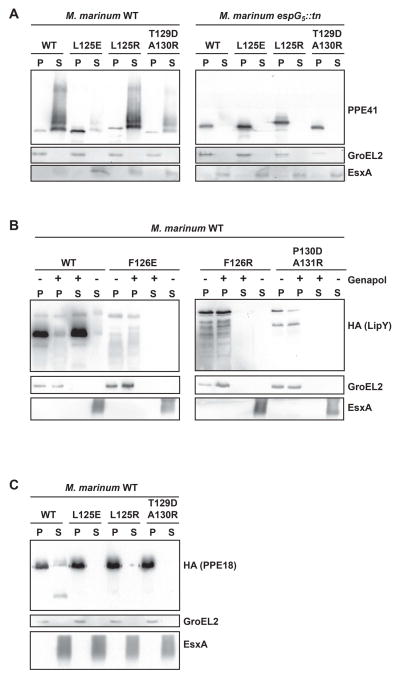
Second, we expressed PE25–PPE41 interface mutants on a plasmid in a WT strain of M. marinum. The immunoblot analysis of the culture filtrates and bacterial pellets demonstrated that PE25–PPE41 variant with L125R substitution was not secreted, but accumulated in the bacterial pellet (Fig. 7A). However, the PE25–PPE41 proteins with L125E or T129D/A130R mutations were detected in normal amounts in the supernatant, indicating that these mutations do not impair PE25–PPE41 secretion.
Fig. 7. Effect of the substitutions in the PPE41 interface on PE25–PPE41 secretion in M. marinum.
A. Immunoblot analysis of PPE41 in culture supernatant and cell pellet fractions of M. marinum WT (left panel) and its EspG5 transposon mutant (right panel). Equivalent amounts of protein were loaded. GroEL and EsxA were used as a cytoplasmic and secreted fraction controls, respectively. Analysis of LipY (panel B) and PPE18 (panel C) in culture supernatant and cell pellet fractions of M. marinum WT.
Because this finding was surprising we also examined whether the L125E and T129D/A130R PPE41 mutants still require EspG5 for export. We analyzed the secretion of these PE25–PPE41 interface mutants in the M. marinum EspG5 mutant strain (Abdallah et al., 2006). We found that none of the PE25–PPE41 mutant variants are secreted in the espG5 mutant strain (Fig. 7A), indicating that L125E and T129D/A130R mutations do not seem to uncouple the dependence of the heterodimer on EspG5 presence in mycobacteria for its export. Taking together our results show that the loss of EspG5 binding to PE25–PPE41 does not necessarily impair the heterodimer secretion, indicating that these interactions are not essential for PE25–PPE41 secretion. Nevertheless, our observation that PPE L125R mutation completely disrupts the heterodimer export indicates that the hh motif is critical for processes of secretion.
EspG5-binding domain is critical for secretion of other ESX-5 specific substrates
The observation that the interaction between EspG5 and PE25–PPE41 is not strictly required for ESX-5 dependent secretion is rather unexpected. However, PE25–PPE41 is an unusual substrate, first of all it is heterologous substrate that is not normally present in M. marinum and in addition these proteins form a soluble complex in E. coli in the absence of the EspG5 chaperone ((Strong et al., 2006); this study). We therefore examined whether EspG5-binding domain is important for secretion of other PPE proteins that are native substrates of the ESX-5 system in M. marinum. For our analysis we selected LipYmm (mmar_1547) that has an N-terminal PPE domain and PE31–PPE18 protein pair (mmar_4241-mmar_4240). LipYmm has previously been shown to be transported to the surface of M. marinum in an ESX-5 dependent manner. This protein is processed upon secretion to release the active C-terminal lipase domain (Daleke et al., 2011). PPE18 is a probable ESX-5 substrate based on proteomic analysis (Ates and Houben, submitted).
Firstly, using co-purification analysis of the wild-type PE31–PPE18 and EspG5mm we confirmed that the chaperone binds this heterodimer and is required for production of soluble complex in vitro (Fig. S11). In addition, we confirmed that the PPE18 EspG5-binding domain is essential for the interactions in vitro. We found that PPE18 variants with L125E, L125R or T129D/A130R amino acid substitutions were produced in the insoluble forms when they were co-expressed with the chaperone (Fig. S11). These data are consistent with our observation that the chaperone is required for folding/stability of PE/PPE heterodimers. We did not analyze the binding of EspG5 to LipYmm in vitro, because the putative PE partner of LipYmm is currently unknown. Then, we analyzed the effect of the substitutions in the hh motif (LipYmmF126E, LipYmmF126R, PPE18L125E, PPE18L125R) and EspG5-binding site (LipYmmP130D/A131R, PPE18T129D/A130R) on secretion of both LipYmm and PE31–PPE18 (Fig. 7B and 7C). Importantly, all substitutions in both substrates completely abolished secretion (Fig. 7B and 7C). Therefore, these data show that homologous ESX-5 substrates LipY and PE31–PPE18 are strongly dependent on EspG5 binding for secretion.
Discussion
During infection pathogenic mycobacteria depend upon several related secretion pathways, ESX-1, ESX-3, and ESX-5, that are involved in translocation of multiple protein substrates across their complex cell envelope. A majority of ESX secreted effectors identified so far are members of either the WXG100 or PE/PPE protein superfamilies that share a similar structural fold and possess a signature YxxxD/E motif required for secretion (Daleke et al., 2012a). These protein families have similar folding characteristics and both belong to the so-called EsxAB clan (http://pfam.xfam.org/clan/EsxAB). PE and PPE proteins represent two highly polymorphic protein families that are widely distributed in pathogenic mycobacteria, comprising nearly 10% of the coding capacity of M. tuberculosis and M. marinum genomes (Cole et al., 1998, Stinear et al., 2008). The ESX-5 pathway has been implicated in the export of a majority of PE and PPE proteins (Abdallah et al., 2009, Bottai et al., 2012). Furthermore, the genes encoding PE/PPE are often located next to the gene encoding the cytoplasmic protein EspG. Recent work in the field has identified EspG as a binding partner of PE–PPE dimers (Daleke et al., 2012b). In contrast, EspG does not interact with the secreted substrates that belong to WXG100 family (Daleke et al., 2012b).
In this study we report the crystal structure of the ESX-5 specific PE25–PPE41 heterodimer in complex with EspG5. A notable feature of the chaperone is a novel EspG fold with the unexpected internal quasi 2-fold symmetry. Importantly, the EspG fold is unrelated to the structures of chaperones from other bacterial secretion systems. The structure of the heterotrimer reveals that EspG5 binds PE25–PPE41 at the tip of the complex, interacting solely with PPE41. The conserved ESX secretory signal YxxxD/E is located on the opposite part of the PE25–PPE41 complex and is not obstructed by EspG5.
Structural data together with PPE41 mutant and sequence analyses of PPE proteins from different ESX clusters allowed us to identify a specific region within the ESX-5-specific PPE proteins recognized by EspG5 chaperone. Surprisingly, we found that this region is well conserved among the ESX-1, ESX-3 and ESX-5-specific PPE proteins. No amino acid residue in the region was found to define the specificity of EspG5 recognition of ESX-5-encoded PPE proteins. Nevertheless, no cross-interactions were observed between non-cognate EspG5 and PPE proteins encoded by ESX-1 and ESX-3 clusters. These data indicate that structural elements outside the binding region differentiate ESX-5-specific PPE proteins from ESX-1 and ESX-3 encoded homologs for EspG5 binding. Thus, the mechanism of substrate recognition by EspG5 may incorporate two unique features: the conservative binding region within PE–PPE dimer that is in the direct contact with the chaperone and the PE–PPE scaffold that does not contact the chaperone, but presents this region for chaperone binding and determines the specificity of recognition. The conserved nature of EspG5-binding region suggests that it may be an important determinant of ESX secretion mechanism.
What is the function of the chaperone during ESX mediated secretion? Of significance for EspG5 functioning is the fact that it binds PE25–PPE41 tightly in the nanomolar range of the affinities as determined by our ITC analysis, strongly indication that these interactions occur in vivo, an observation which is also confirmed by previous pull-down experiments (Daleke et al., 2012b). Based on the fact that EspG is strictly located in the cytosol and interacts only with cognate PE–PPE complexes, it seems likely that EspG acts as a chaperone to escort PE–PPE dimers to their cognate ESX secretion machinery where the membrane complex recognizes YxxxD/E secretory signal and releases EspG from the heterotrimer, possibly with the help of the ESX-associated ATPases. Subsequently, the PE–PPE dimer would be exported through the membrane and EspG recycled for the next round of binding. It has been suggested that EspG might provide PE–PPE with an additional secretion signal, which specifies the proper ESX pathway for export. If this hypothesis is correct, the chaperone interactions with PE–PPE should be indispensable for the dimer secretion via cognate ESX system. Our in vivo analysis indicates that EspG5 binding is required for secretion of ESX-5 specific substrates, LipY and PE31–PPE18, but is in fact not essential for PE25–PPE41 secretion in M. marinum. These data suggest that either the chaperone does not direct PE25–PPE41 dimer to the ESX-5 secretion machinery or this putative chaperone function is redundant. This finding is in line with a previous study showing that EspG5 is not essential for PE25–PPE41 secretion in M. tuberculosis (Bottai et al., 2012). Surprisingly, we found that secretion of PE25–PPE41 uncoupled from EspG5 remains dependent on EspG5 presence in M. marinum. This observation indicates that lack of PE25–PPE41 export in the EspG5 deletion mutant is likely due to a general defect in ESX-5 secretion mechanism. This notion is supported by the previous finding that not only the secretion of many PE and PPE proteins was affected in this mutant, but also the secretion of EsxN (Abdallah et al., 2009), which is not recognized by EspG5.
In M. tuberculosis deletion of the ESX-1-specific chaperone EspG1 resulted in substantially lower amounts of the ESX-1 encoded PPE, PPE68, in their cell lysates (Bottai et al., 2011), indicating that EspG1 is necessary for PPE68 folding or stability. Consistent with this notion, our in vitro data show that EspG1 is essential for folding/stability of the ESX-1-specific PE–PPE dimer. Moreover, we observed that EspG5 is necessary for production of soluble ESX-5 specific dimer, PE31–PPE18, in vitro. By contrast, the complex of PE25–PPE41 is highly soluble and does not require EspG5 for stability. Mapping of PE25–PPE41 surface hydrophobicity revealed a hydrophobic patch, the hh motif, representing a part of the EspG5 binding region. Intriguingly, sequence analysis of ESX-1-, ESX-3- and ESX-5-specific PPE proteins indicated that the majority of PPE proteins contain more hydrophobic residues in the hh motif than PPE41 does. Substitution of PPE41 hh motif residues to these more hydrophobic amino acids resulted in PE25–PPE41 self-association which could be rescued by EspG5. Apparently, the primary function of EspG is to shield this aggregation-prone hh motif on PPE proteins. Thus, PPE41 is probably an exception within the PPE family that has lost dependence on EspG5 binding for folding/stability and secretion. This observation is in agreement with a previous study showing that most PE/PPE proteins cannot be expressed as soluble proteins in E. coli (Strong et al., 2006).
Based on phylogenetic analysis it is predicted that pe/ppe genes encoded within ESX-1 and ESX-3 clusters are the most ancestral representatives of the families (Gey van Pittius et al., 2006). The ESX-5 pathway denotes the most recently evolved ESX system from which a large number of pe/ppe genes have expanded to the rest of the mycobacterial genome (Gey van Pittius et al., 2006). It is plausible that during duplication/recombination events some PPE proteins lost a requirement for the chaperone binding. Given the high abundance of ESX-5-dependent PPE proteins in pathogenic mycobacteria, inadequate concentrations of EspG5 chaperone might result in the build-up of large quantities of protein aggregates in the mycobacterial cytoplasm. Perhaps, uncoupling EspG5 from this essential function drove the extensive expansion of pe/ppe genes from the ESX-5 cluster to the rest of the genome.
In other bacterial protein secretion systems, different classes of energy-independent chaperones participate in secretory processes including Sec (Castanie-Cornet et al., 2014) and chaperone/usher (Geibel & Waksman, 2014) pathways, type III secretion system (Izore et al., 2011) and related flagellar export system (Minamino, 2014), and type IV secretion system (Christie et al., 2014). These chaperones play multiple functions in secretion processes such as stabilizing the hydrophobic translocon-forming proteins, preventing premature self-polymerization of flagellar and fimbrial proteins, directing substrates through the correct secretion system, and regulating a hierarchy in secretion. All of these functions ultimately serve to keep secreted substrates in a secretion-competent state before their export. Intriguingly, although structurally different, EspG chaperones have a function in the secretory mechanism that is similar to SycE and SycO, the chaperones of type III secretion system in Yersinia (Birtalan et al., 2002, Letzelter et al., 2006). SycE and SycO chaperones mask the aggregation-prone regions of secreted effectors, YopE and YopO, respectively. The aggregation-prone regions of YopE and YopO represent the hydrophobic membrane domains that are essential for proper localization of the effectors in the host cell (Birtalan et al., 2002, Letzelter et al., 2006). It is possible that the hydrophobic hh motif in PPE proteins also has a functional role in the ESX secretory mechanism. In line with this hypothesis we identified one mutation in the hh motif of PPE41 that completely blocks the heterodimer secretion. Further experiments are required to decipher conclusively the function of the motif.
In conclusion, the current study presents the first picture of the ESX secretion system’s chaperone in complex with secreted substrates. This structure reveals the molecular determinants of chaperone binding specificity and provides evidence for the importance of chaperone function in the ESX secretory mechanism. The structure of the PE25–PPE41–EspG5 complex provides an important tool for future investigations into the architecture, assembly, and secretory mechanism of this sophisticated mycobacterial secretion system. It could also serve as a basis for the development of therapeutic compounds designed to disrupt the chaperone binding to PE–PPE, which in turn could prevent PE–PPE secretion and diminish mycobacterial pathogenicity.
Experimental Procedures
Bacterial strains and growth conditions
M. marinum strain E11 (Puttinaowarat et al., 2000) and the EspG5 deletion mutant (Abdallah et al., 2009) were grown at 30 °C with shaking at 90 rpm, in Middlebrook 7H9 medium (Difco-BD Biosciences), supplemented with Middlebrook ADC and 0.05% Tween-80 (Sigma-Aldrich). Electroporation of M. marinum was performed as described (Daleke et al., 2011) and transformants were selected on Middlebrook 7H10 agar supplemented with Middlebrook OADC and appropriate antibiotics.
E. coli strains including TOP10, DH5α, BL21(DE3) and Rosetta (DE3) were grown in Luria-Bertani (LB) medium or on LB agar at 37 °C. When required, antibiotics were included at the following concentrations: ampicillin at 100 μg ml−1 for E. coli; chloramphenicol at 10 μg ml−1 for E. coli; streptomycin at 50 μg ml−1 for E. coli; kanamycin at 50 μg ml−1 for E. coli; hygromycin at 100 μg ml−1 for E. coli and 50 μg ml−1 for M. marinum.
Expression of EspG5, EspG1 and EspG3
The EspG5 expression plasmid for crystallization experiments was constructed as follows. The espG5 gene was PCR-amplified from M. tuberculosis H37Rv genomic DNA and cloned into a modified pRSF-Duet1 vector (EMD Millipore). EspG5, EspG1 and EspG3 expression plasmids for pull-down experiments were constructed based on a modified pCDF-Duet1 vector (EMD Millipore), creating pCDF-Duet1-EspG5, pCDF-Duet1-EspG1 and pCDF-Duet1-EspG3, respectively.
EspG5 was expressed with N-terminal His6 and SUMO3 tags for ITC experiments. EspG5 gene was amplified from M. tuberculosis genomic DNA using primers AP-324 (5′-ATATATACCGGTGGAATGGATCAACAGAGTACCCG-3′) and AP-325 (5′ ATATATATGCGGCCGCTCATACTCTGCTGTGTGTTTTCC-3′). The PCR product was digested and ligated in pETM11-SUMO3GFP (obtained from Dr. Hüseyin Besir). The resultant pETHIS6SUMO3-EspG5 plasmid was transferred into competent E. coli BL21(DE3) cells (Novagen).
Expression of PE25–PPE41, PE35–PPE68_1 and PE5–PPE4 heterodimers
To construct a plasmid for co-expression of PE25 and PPE41 proteins in E. coli, the DNA fragments corresponding to pe25 and ppe41 were PCR-amplified from M. tuberculosis genomic DNA and cloned separately into a modified pET-28b vector (EMD Millipore) creating a bi-cistronic operon under control of the T7 promoter with an N-terminal His6-tag followed by a tobacco etch virus (TEV) protease cleavage site. The resulting pET-PE25-PPE41 plasmid was used as a template for generating mutated variants of PPE41.
The PE35 and PPE68_1 proteins were expressed using a previously described pET29b(+)::PE35-PPE68_1-His vector (Daleke et al., 2012b), which contains the M. marinum PE35/PPE68 homologs, mmar_0185 and mmar_0186. The construct was modified to remove the C-terminal HA-tag from PE35.
In order to co-express M. smegmatis PE5 and PPE41–180, an optimized DNA sequence corresponding to the protein sequences of PE5 and PPE4 were obtained from Invitrogen. The resulting plasmid was designated pET-PE5-PPE4.
For co-expression of EspG5, PE25, and PPE41 proteins, E.coli Rosetta(DE3) cells were co-transformed with pCDF-Duet1-EspG5 and pET-PE25-PPE41 plasmids. Similarly, for co-expression of EspG1, PE35, and PPE68_1 proteins, E.coli Rosetta(DE3) was co-transformed with pCDF-Duet1-EspG1 and pET-PE35-PPE68_1 plasmids. For co-expression of EspG3, PE5 and PPE4 proteins, E.coli Rosetta (DE3) was co-transformed with pCDF-Duet1-EspG3 and pET-PE5-PPE4 plasmids.
Plasmids for M. marinum expression of PE25-PPE41were derived from the pSMT3::PE25-HA-PPE41 vector (Daleke et al., 2012a) which encodes PE25 modified with a C-terminal HA epitope as well as PPE41. This vector was used as a template for generating mutated versions of PE25–PPE41.
Expression and purification of EspG, PE–PPE and EspG–PE–PPE
The proteins were expressed in E.coli Rosetta(DE3) or (EspG5, PE25–PPE41, PE35–PPE68_1, PE5–PPE4, EspG1–PE35–PPE68_1 and EspG3–PE5–PPE41–180) or in E. coli BL21(DE3) (His6-SUMO3-EspG5) cells by induction with 0.5 mM IPTG for 4 h at 18 °C (EspG5, PE25–PPE41, PE35–PPE68_1, PE5–PPE41–180, EspG1–PE35–PPE68_1 and EspG3–PE5–PPE41–180) or for 4 h at 24 °C (PE25–PPE41 and EspG5–PPE25–PPE41) or 20 h at 28 °C (His6-SUMO3-EspG5). Cells were harvested by centrifugation.
His6-SUMO3-EspG5 protein for ITC experiments was purified as follows: Cells were resuspended in lysis buffer A [20 mM HEPES, 300 mM NaCl, 10mM imidazole, 0.25mM tris(2-carboxyethyl)phosphine (TCEP), 10% (w/v) glycerol (pH 7.5)] containing 1/100 protease inihibitor mix HP (Serva) and disrupted by lysozyme treatment followed by sonication. The protein was purified via Ni-NTA (Qiagen) affinity chromotography. Following the cleavage of His6-SUMO3 tag by SenP2 protease, the protein was further purified on a Phenyl Sepharose HP column (GE Biosciences), then concentrated and injected into a Superdex200 16/60 size-exclusion chromatography column (GE Biosciences) pre-equilibrated 20 mM HEPES, 300 mM NaCl, 0.25mM TCEP, 10% (w/v) glycerol (pH 7.5) for removal of aggregated protein.
The complex of EspG5, PE25 and PPE41 for crystallization was purified as follows: Cells expressing EspG5 were mixed with cells expressing PE25–PPE41 and resuspended in lysis buffer B (20 mM Tris-HCl pH 8.4, 300 mM NaCl, and 20 mM imidazole). The resuspended cells were lysed using an EmulsiFlex-C5 homogenizer (Avestin) and proteins were purified via Ni-NTA. Following the cleavage of His6-tag by TEV protease, proteins were purified via size-exclusion on a Superdex200 column (GE Biosciences) in buffer containing 20 mM Hepes pH 7.5 and 100 mM NaCl.
To isolate EspG–PE–PPE complexes for pull-down experiments, cells that have co-expressed EspG, PE, PPE proteins were lysed with lysozyme (0.25 mg/ml) in BugBuster buffer (EMD Millipore) supplemented with Benzonase Nuclease (EMD Millipore). The proteins were purified via a Ni-NTA column (Qiagen).
Crystallization, data collection and structure solution
The initial crystallization conditions were identified using JCSG Suites I–IV screens (Qiagen). The optimized hexagonal rod-shaped crystals were obtained by the vapor diffusion method using 0.1 M Tris-HCl pH 8.6, 0.2 M NaCl, 8 % PEG8000 as precipitant. Crystals were transferred to crystallization solution supplemented with 20 % ethylene glycol and flash-cooled in liquid N2 prior to data collection. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Data were processed and scaled using XDS and XSCALE (Kabsch, 2010). The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) and the structure of PE25–PPE41 (PDB 2G38 (Strong et al., 2006)) as a search model. The electron density modification was performed using Parrot (Cowtan, 2010) and the initial model was built using Buccaneer (Cowtan, 2006). The initial model was improved by iterative cycles of refinement and rebuilding using REFMAC5 and Coot (Murshudov et al., 2011, Emsley et al., 2010). The final structure was refined with REFMAC5 using translation, libration and screw-rotation (TLS) groups identified by the TLSMD server (Painter & Merritt, 2006). Data collection and refinement statistics are listed in Table 1. The quality of the final model was assessed using Coot and the MolProbity server (Chen et al., 2010). The structural figures were generated using PyMol (www.pymol.org).
Expression and secretion of PE25–PPE41 proteins in M. marinum
Secretion of PPE41 proteins was carried out as previously described (Daleke et al., 2012a). Briefly, M. marinum strains were grown to mid-logarithmic phase in Middlebrook 7H9 supplemented with 0.2% glycerol, 0.2% dextrose and 0.05% Tween, at which bacteria were pelleted by centrifugation. Secreted proteins were precipitated with 10% trichloroacetic acid (wt/vol) added to the supernatant. Cells were lysed by sonication. Proteins were separated on 10–16% SDS-PAGE gel, transferred to nitrocellulose membrane, and immunostained with rabbit polyclonal sera recognizing PPE41.
Sequence analysis of PPE proteins
The sequence alignments were done using ClustalW (Larkin et al., 2007) and rendered using the ESPript server (Gouet et al., 2003).
Isothermal titration calorimetry (ITC) analysis
All measurements were conducted at 30 °C on a MicroCal VP-ITC calorimeter with a reference cell filled with MilliQ water. 100 μM EspG5 was injected into the sample cell containing 10 μM PE25–PPE41. Experiments were performed in triplicate and corrected for dilution enthalpy. Raw data were analyzed with MicroCal Origin 7.0. Data were fit with a single-site binding model. The experiments were performed in triplicate.
CD spectroscopy
Circular dichroism spectra were recorded using 7–9.5 μM protein in ITC buffer on a Chirascan spectrometer (Applied Photophysics) with a 0.5 mm cuvette. The CD signal was converted to the mean residue ellipticity using the protein concentration determined by absorbance measurements and the calculated molar extinction coefficient at 280 nm. All CD data presented are the averages of at least three measurements.
Supplementary Material
Fig. S1. Sequence alignment of the EspG5 family members.
Sequences with >98 % identity were excluded from the alignment. Identical residues are highlighted in red. Blue triangles at the top of the alignments indicate residues that are in contact with PPE41 in the M. tuberculosis PE25–PPE41–EspG5 complex. The secondary structure elements of M. tuberculosis EspG5 are shown at the top of the alignment. Black dashed lines indicate disordered residues.
Fig. S2. Sequence alignment of the ESX-5-specific PPE proteins from M. tuberculosis H37Rv. Only the core PPE domain sequences (~180 residues) are aligned. Identical residues are highlighted in red. The PPE and WxG motifs are highlighted in blue. Residues that are in contact with EspG5 and PE25 in the M. tuberculosis PE25–PPE41–EspG5 complex are labeled by purple and cyan triangles, respectively. Purple stars indicate two hydrophobic residues in the α4–α5 loop of PPE proteins that comprise the hh motif. The secondary structure elements of PPE41 are shown at the top of the alignment.
Fig. S3. Circular dichroism analysis of the wild-type and mutant PE25–PPE41 complexes. Spectra between wavelengths 200 nm and 260 nm are plotted against mean residue ellipticity. The mutant complexes exhibit a similar secondary structure content as the wild-type PE25–PPE41.
Table S1. Summary of isothermal calorimetry data on interaction of EspG5 with PE25–PPE41.
Fig. S4. The ESX-1-specific PE35–PPE68_1 dimer interacts with EspG1 but not with EspG5. A. M. marinum PE35, PPE_68-His and EspG1 were co-expressed in E. coli and purified using immobilized Ni2+ affinity and size-exclusion chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W), eluted proteins (E) and peak fraction from size-exclusion chromatography (SEC) were analyzed by SDS-PAGE. B. M. marinum PE35, PPE_68-His and EspG5 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. Fractions were analyzed by SDS-PAGE and immunoblotting with anti-His tag antibodies. PPE_68-His was found in the insoluble fraction.
Fig. S5. The ESX-3-specific PE5–PPE4 dimer interacts with EspG3 but not with EspG5. A. M. smegmatis PE5-His, the core domain of PPE4 (residues 1–180) and EspG3 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W) and eluted proteins (E) were analyzed by SDS-PAGE.
B. M. smegmatis PE5-His, PPE41–180 and M. marinum EspG5 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. While PE5–PPE41–180 dimer could be purified, it is unstable and precipitates in the absence of cognate EspG3 chaperone.
Fig. S6. Sequence alignment of PPE68 homologs from Mycobacteria.
Only the core PPE domain sequences (~180 residues) are aligned. The PPE and WxG motifs are highlighted in blue. The predicted secondary structure elements are shown at the top of the alignments. Purple stars indicate the hh motif. The majority of mycobacterial genomes contain a single PPE68 homolog, whereas M. marinum and M. kansasii have several PPE68 homologs.
Fig. S7. Sequence alignment of the ESX-3-specific PPE proteins from M. tuberculosis H37Rv. Only the core PPE domain sequences (~180 residues) are aligned. The PPE and WxG motifs are highlighted in blue. The predicted secondary structure elements are shown at the top. The hh motif is indicated by purple stars.
Fig. S8. Co-purification of EspG5 and PE25–PPE41 mutant variant dimers.
PE25–PPE41 dimers were co-expressed in E. coli with either M. tuberculosis EspG5 (EspG5mtu) or M. marinum EspG5 (EspG5mma). Proteins were purified using immobilized Ni2+ affinity chromatography. Notably, the excess of EspG5mma chaperone leads to complete solubility of PE25–PPE41 with no dimer present in the insoluble fraction.
Fig. S9. Effect of increased hydrophobicity in the hh motif of PPE41 on protein solubility. PE25–PPE41 dimers were expressed in E. coli in the absence of chaperone or co-expressed with M. marinum EspG5 (EspG5mma). Proteins were purified using immobilized Ni2+ affinity chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W) and eluted proteins (E) were analyzed by SDS-PAGE. Notably, the excess of EspG5mma chaperone leads to complete solubility of PE25–PPE41 with no dimer present in the insoluble fraction.
Fig. S10. Effect of disruptive mutations in the PPE41 interface on the binding of PE25–PPE41 heterodimer to M. marinum EspG5.
PE25–PPE41 variants were co-expressed with EspG5mma and purified via immobilized Ni2+ affinity chromatography. The results are comparable with M. tuberculosis EspG5 binding (Fig. 3D).
Figure S11. Effect of disruptive mutations in the PPE18 on the binding of PE31–PPE18 heterodimer to M. marinum EspG5. PE31–PPE18 variants were co-expressed with EspG5mma and purified via immobilized Ni2+ affinity chromatography.
Acknowledgments
Authors thank Vivian Pogenberg and the SPC Facility at EMBL Hamburg for technical assistance; Lalita Ramakrishnan (University of Washington) for providing M. marinum M genomic DNA; Carol Beach (University of Kentucky) for mass-spectrometry analysis; Charles J. Waechter and Jonathan M. Wagner (University of Kentucky) for critical comments on the manuscript; the Protein Expression and Purification Core Facility at EMBL Heidelberg for providing pETM11-SUMO3GFP; staff members of Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, for assistance during data collection. This study was supported in part by the European Commission grant PITN-GA2009-238423 to DF, a VIDI grant from the Netherlands Organization of Scientific Research to ENGH, and NIH/NIGMS grant P20GM103486 to KVK. We acknowledge the University of Kentucky Proteomics Core and Protein Analytical Core that are supported by NIH/NCRR grant P20RR020171 and by NIH/NIGMS grant P20GM103486. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Accession numbers
The coordinates and structure factors were deposited to the Protein Data Bank with accession code 4KXR.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- Arbing MA, Kaufmann M, Phan T, Chan S, Cascio D, Eisenberg D. The crystal structure of the Mycobacterium tuberculosis Rv3019c–Rv3020c ESX complex reveals a domain-swapped heterotetramer. Protein Sci. 2010;19:1692–1703. doi: 10.1002/pro.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barathy DV, Suguna K. Crystal structure of a putative aspartic proteinase domain of the Mycobacterium tuberculosis cell surface antigen PE_PGRS16. FEBS Open Bio. 2013;3:256–262. doi: 10.1016/j.fob.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–980. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, et al. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol. 2012;83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- Bottai D, Majlessi L, Simeone R, Frigui W, Laurent C, Lenormand P, et al. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J Infect Dis. 2011;203:1155–1164. doi: 10.1093/infdis/jiq089. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Bruel N, Genevaux P. Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim Biophys Acta. 2014;1843:1442–1456. doi: 10.1016/j.bbamcr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, et al. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J Biol Chem. 2011;286:19024–19034. doi: 10.1074/jbc.M110.204966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, et al. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A. 2012a;109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, et al. Specific chaperones for the type VII protein secretion pathway. J Biol Chem. 2012b;287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Geibel S, Waksman G. The molecular dissection of the chaperone-usher pathway. Biochim Biophys Acta. 2014;1843:1559–1567. doi: 10.1016/j.bbamcr.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, et al. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben EN, Korotkov KV, Bitter W. Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta. 2014;1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Izore T, Job V, Dessen A. Biogenesis, regulation, and targeting of the type III secretion system. Structure. 2011;19:603–612. doi: 10.1016/j.str.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, et al. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, et al. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog. 2013;9:e1003734. doi: 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. Protein export through the bacterial flagellar type III export pathway. Biochim Biophys Acta. 2014;1843:1642–1648. doi: 10.1016/j.bbamcr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Mishra KC, de Chastellier C, Narayana Y, Bifani P, Brown AK, Besra GS, et al. Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. Infect Immun. 2008;76:127–140. doi: 10.1128/IAI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Poulsen C, Panjikar S, Holton SJ, Wilmanns M, Song YH. WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern. PLoS One. 2014;9:e89313. doi: 10.1371/journal.pone.0089313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttinaowarat S, Thompson KD, Adams A. Mycobacteriosis: detection and identification of aquatic Mycobacterium species. Fish Vet J. 2000;5:6–21. [Google Scholar]
- Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Steigedal M, Ahmad R, Mehra A, Dragset MS, Schuster BM, et al. Mycobacterial Esx-3 requires multiple components for iron acquisition. mBio. 2014;5:e01073–01014. doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, et al. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Tanneeru K, Guruprasad L. The PE-PPE domain in mycobacterium reveals a serine alpha/beta hydrolase fold and function: an in-silico analysis. PLoS One. 2011;6:e16745. doi: 10.1371/journal.pone.0016745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Vemula MH, Banerjee S, Guruprasad L. The PE16 (Rv1430) of Mycobacterium tuberculosis is an esterase belonging to serine hydrolase superfamily of proteins. PLoS One. 2013;8:e55320. doi: 10.1371/journal.pone.0055320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 2006;8:1417–1429. doi: 10.1111/j.1462-5822.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Weerdenburg EM, Abdallah AM, Mitra S, de Punder K, van der Wel NN, Bird S, et al. ESX-5-deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cell Microbiol. 2012;14:728–739. doi: 10.1111/j.1462-5822.2012.01755.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence alignment of the EspG5 family members.
Sequences with >98 % identity were excluded from the alignment. Identical residues are highlighted in red. Blue triangles at the top of the alignments indicate residues that are in contact with PPE41 in the M. tuberculosis PE25–PPE41–EspG5 complex. The secondary structure elements of M. tuberculosis EspG5 are shown at the top of the alignment. Black dashed lines indicate disordered residues.
Fig. S2. Sequence alignment of the ESX-5-specific PPE proteins from M. tuberculosis H37Rv. Only the core PPE domain sequences (~180 residues) are aligned. Identical residues are highlighted in red. The PPE and WxG motifs are highlighted in blue. Residues that are in contact with EspG5 and PE25 in the M. tuberculosis PE25–PPE41–EspG5 complex are labeled by purple and cyan triangles, respectively. Purple stars indicate two hydrophobic residues in the α4–α5 loop of PPE proteins that comprise the hh motif. The secondary structure elements of PPE41 are shown at the top of the alignment.
Fig. S3. Circular dichroism analysis of the wild-type and mutant PE25–PPE41 complexes. Spectra between wavelengths 200 nm and 260 nm are plotted against mean residue ellipticity. The mutant complexes exhibit a similar secondary structure content as the wild-type PE25–PPE41.
Table S1. Summary of isothermal calorimetry data on interaction of EspG5 with PE25–PPE41.
Fig. S4. The ESX-1-specific PE35–PPE68_1 dimer interacts with EspG1 but not with EspG5. A. M. marinum PE35, PPE_68-His and EspG1 were co-expressed in E. coli and purified using immobilized Ni2+ affinity and size-exclusion chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W), eluted proteins (E) and peak fraction from size-exclusion chromatography (SEC) were analyzed by SDS-PAGE. B. M. marinum PE35, PPE_68-His and EspG5 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. Fractions were analyzed by SDS-PAGE and immunoblotting with anti-His tag antibodies. PPE_68-His was found in the insoluble fraction.
Fig. S5. The ESX-3-specific PE5–PPE4 dimer interacts with EspG3 but not with EspG5. A. M. smegmatis PE5-His, the core domain of PPE4 (residues 1–180) and EspG3 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W) and eluted proteins (E) were analyzed by SDS-PAGE.
B. M. smegmatis PE5-His, PPE41–180 and M. marinum EspG5 were co-expressed in E. coli and purified using immobilized Ni2+ affinity chromatography. While PE5–PPE41–180 dimer could be purified, it is unstable and precipitates in the absence of cognate EspG3 chaperone.
Fig. S6. Sequence alignment of PPE68 homologs from Mycobacteria.
Only the core PPE domain sequences (~180 residues) are aligned. The PPE and WxG motifs are highlighted in blue. The predicted secondary structure elements are shown at the top of the alignments. Purple stars indicate the hh motif. The majority of mycobacterial genomes contain a single PPE68 homolog, whereas M. marinum and M. kansasii have several PPE68 homologs.
Fig. S7. Sequence alignment of the ESX-3-specific PPE proteins from M. tuberculosis H37Rv. Only the core PPE domain sequences (~180 residues) are aligned. The PPE and WxG motifs are highlighted in blue. The predicted secondary structure elements are shown at the top. The hh motif is indicated by purple stars.
Fig. S8. Co-purification of EspG5 and PE25–PPE41 mutant variant dimers.
PE25–PPE41 dimers were co-expressed in E. coli with either M. tuberculosis EspG5 (EspG5mtu) or M. marinum EspG5 (EspG5mma). Proteins were purified using immobilized Ni2+ affinity chromatography. Notably, the excess of EspG5mma chaperone leads to complete solubility of PE25–PPE41 with no dimer present in the insoluble fraction.
Fig. S9. Effect of increased hydrophobicity in the hh motif of PPE41 on protein solubility. PE25–PPE41 dimers were expressed in E. coli in the absence of chaperone or co-expressed with M. marinum EspG5 (EspG5mma). Proteins were purified using immobilized Ni2+ affinity chromatography. Total cell lysate (T), soluble fraction (S), insoluble fraction (I), flow-through fraction (FT), wash fraction (W) and eluted proteins (E) were analyzed by SDS-PAGE. Notably, the excess of EspG5mma chaperone leads to complete solubility of PE25–PPE41 with no dimer present in the insoluble fraction.
Fig. S10. Effect of disruptive mutations in the PPE41 interface on the binding of PE25–PPE41 heterodimer to M. marinum EspG5.
PE25–PPE41 variants were co-expressed with EspG5mma and purified via immobilized Ni2+ affinity chromatography. The results are comparable with M. tuberculosis EspG5 binding (Fig. 3D).
Figure S11. Effect of disruptive mutations in the PPE18 on the binding of PE31–PPE18 heterodimer to M. marinum EspG5. PE31–PPE18 variants were co-expressed with EspG5mma and purified via immobilized Ni2+ affinity chromatography.