Abstract
Efforts to identify and annotate cancer driver genetic lesions have been almost exclusively focused on the analysis of protein coding genes. Here we identify a new long-range acting MYC enhancer controlled by NOTCH1, targeted by recurrent chromosomal duplications in human T-cell acute lymphoblastic leukemia (T-ALL). This highly conserved regulatory element, hereby named N-Me for NOTCH MYC enhancer, is located within a broad super-enhancer region +1.47 Mb from the MYC transcription initiating site, interacts with the MYC proximal promoter and induces orientation-independent MYC expression in reporter assays. Moreover, analysis of N-Me knockout mice demonstrates a selective and essential role of this regulatory element during thymocyte development and in NOTCH1-induced T-ALL. Altogether, these results identify N-Me as a long range oncogenic enhancer directly implicated in the pathogenesis of human leukemia and highlight the fundamental importance of the NOTCH1-MYC regulatory axis in T-cell transformation and as therapeutic target in T-ALL.
T-ALL is an aggressive hematologic malignancy resulting from the transformation of immature T-cell progenitor cells1 in which activation of transcription factor oncogenes and deregulation of transcriptional regulatory networks play particularly prominent roles1–5. In this context, activating mutations in the NOTCH1 ligand-activated transcription factor oncogene are found in over 60% of human T-ALLs6,7. Mechanistically, NOTCH1 mutations found in T-ALL result in high levels of NOTCH1 signaling, which promotes T-cell transformation by hijacking physiologic functions of the NOTCH1 receptor in the thymus, where ligand-induced NOTCH signaling drives hematopoietic progenitors into the T-cell lineage and promotes thymocyte development7–9. Both physiologic and oncogenic effects of NOTCH1 require translocation of the intracellular portion of the NOTCH1 receptor to the nucleus where it activates gene expression in association with the RBPJ DNA binding protein7,10,11. Gene expression studies have identified NOTCH1 as a key regulator of cell growth in T-ALL lymphoblasts directly controlling numerous genes involved in cell growth and metabolism12. The importance of understanding the hierarchy of the oncogenic NOTCH1 transcriptional programs is highlighted by the proposed role of small molecule γ-secretase inhibitors, which block the release of intracellular NOTCH1 from the membrane and effectively suppress NOTCH signaling, as targeted therapies for the treatment of T-ALL7.
Over the last years, numerous studies have dissected the mutational landscape of T-ALL resulting in the identification of numerous oncogenes and tumor suppressors implicated in T-cell transformation1,4,13. However, and most intriguingly, most genetic abnormalities found in human cancer are located in intergenic regions14, whose role in cancer development, if any, remains poorly understood. Here we hypothesized that recurrent cancer-associated intergenic mutations, amplifications and deletions may implicate strong transcriptional regulatory sequences responsible for the activation of oncogenic factors downstream of key T-ALL transcription factor oncogenes such as NOTCH1.
RESULTS
Amplification of a MYC NOTCH1-occupied enhancer in T-ALL
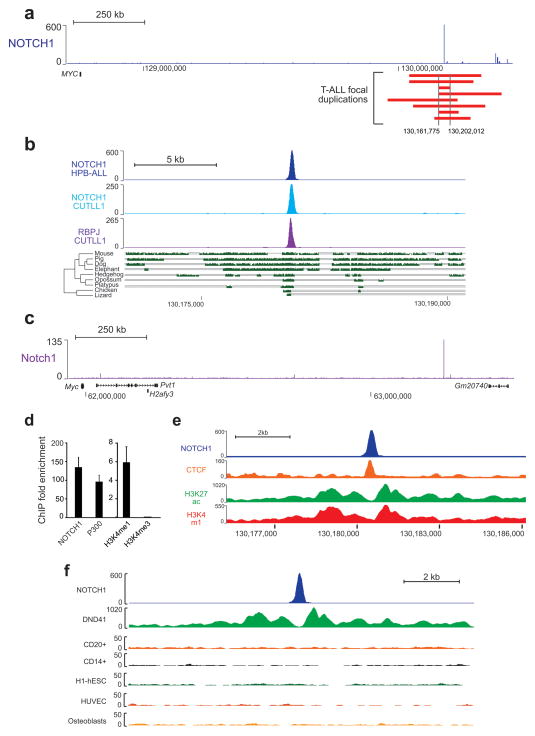
To assess the role of intergenic copy number alterations in the pathogenesis of T-ALL we analyzed array comparative genomic hybridization data from 160 T-ALL cases. This analysis identified recurrent focal duplications at chromosome 8q24 in 8/160 (5%) T-ALL cases in an area devoid of protein-coding genes located +1,427 kb downstream of MYC (Fig. 1a), a critical oncogene in the pathogenesis of NOTCH1-induced T-ALL12,15,16 (Fig. 1a, Supplementary Fig. 1 and Supplementary Tables 1–2). Notably, no duplications in this region were identified in 258 non T-ALL hematologic tumors and no germline copy number variant polymorphisms encompassing this area have been reported. Moreover, analysis of normal (remission) DNA confirmed the somatic origin of these copy number alterations in all 4 cases with available material (Supplementary Fig. 1). Interestingly, chromatin immunoprecipitation (ChIP) followed by next generation sequencing (ChIP-seq) analysis of NOTCH1 chromatin binding sites in HPB-ALL T-ALL cells revealed a prominent 1 kb NOTCH1 peak in chromosome 8q24 located within the common 40 Kb segment duplicated in all these eight leukemia cases (Fig. 1a). A survey of NOTCH1 and RBPJ ChIP-seq data generated in the CUTLL1 T-ALL cell line17 confirmed the presence of high levels of NOTCH1 and RBPJ binding in this site (Fig. 1b and Supplementary Fig. 2). Notably, multispecies DNA sequence alignment revealed remarkable conservation of this region in mammals, birds and reptiles (Fig. 1b), with 88.6% nucleotide identity between the 500 base pair human and mouse sequences centered on the NOTCH1 peak, compared with an average conservation of 44% nucleotide identity along the 1.4 Mb gene desert telomeric to MYC. Consistently, analysis of NOTCH1 ChIP-seq data in G4A2 mouse T-ALL cells18,19 revealed matching NOTCH1 and RBPJ binding peaks located +1,27 Mb from the Myc transcription initiation site (Fig. 1c and Supplementary Fig. 2).
Figure 1.
Identification of N-Me, a NOTCH-bound MYC enhancer recurrently amplified in T-ALL. (a) NOTCH1 ChIP-seq binding occupancy profile in the MYC locus in HPB-ALL T-ALL cells and schematic representation of chromosome 8q24 focal amplifications (red bars) found in human T-ALL. (b) NOTCH1 and RBPJ ChIP-seq binding in T-ALL cells and multispecies sequence conservation along a 20 kb window encompassing the HPB-ALL MYC +1.47 Mb NOTCH1 binding site. The green blocks at the bottom represent pairwise aligned regions with the height of the bars indicating the average base pair alignment score with the human sequence. (c) NOTCH1 ChIP-seq binding occupancy profile in the Myc locus in G4A2 mouse T-ALL cells. (d) ChIP analysis of NOTCH1, P300, H3K4me1 and H3K4Me3 in HPB-ALL T-ALL cells. (e) CTCF, H3K27ac and H3K4me1 ChIP-seq occupancy in DND-41 T-ALL cells along a 10 kb window containing the MYC +1.47 Mb NOTCH1 binding site. (f) ChIP-seq analysis of H3K27ac along the N-Me sequence in DND-41 T-ALL cells, B-cells (CD20+), monocytes (CD14+), embryonic stem cells (H1-hESC), endothelial cells (HUVEC) and osteoblasts. Y axis in ChIP-seq plots indicates fragment density in counts per 10 million.
To functionally characterize the potential role of this NOTCH1 binding site in gene regulation, we performed local ChIP analysis of chromatin regulatory factors and epigenetic histone marks in HPB-ALL T-ALL cells. These analyses confirmed high levels of NOTCH1 binding at this site and revealed bona fide active enhancer features associated with this region including P300 occupancy and high levels and H3K4 monomethylation (H3K4me1) with low levels of H3K4 trimethylation (H3K4me3) (Fig. 1d and Supplementary Fig. 3). Moreover, inspection of ENCODE data in this region revealed that this NOTCH1 occupied enhancer sits within a 20 kb segment containing high levels of H3K27 acetylation and H3K4 monomethylation (Fig. 1e), features characteristically associated with highly dynamic long-range superenhancer regulatory sequences20. In addition, extended examination of ENCODE data revealed that H3K27 acetylation in this area is selectively present in the DND-41 cells, a T-ALL cell line harboring a highly active mutant form of NOTCH16, but not in primary CD20+ B cells or in cell lines derived from a variety of other hematopoietic and non-hematopoietic tissues and cell lines (Fig. 1f and Supplementary Fig. 3).
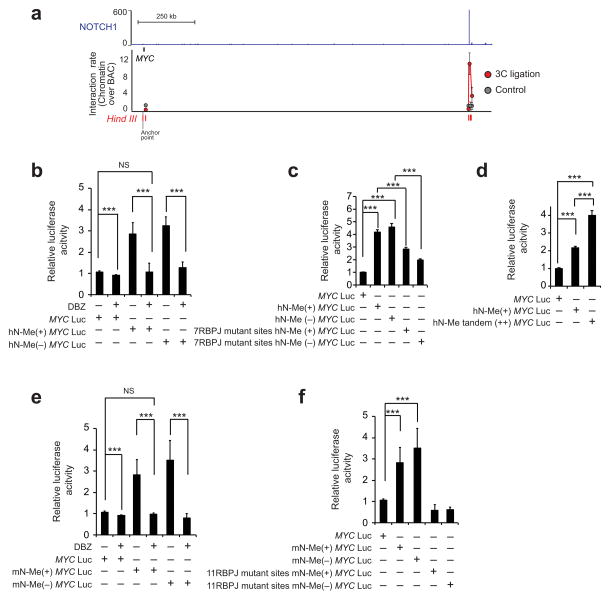
Based on these results, we proposed that this +1,4 Mb MYC NOTCH1-occupied enhancer –hereby named N-Me for NOTCH-bound MYC enhancer– could function as an important regulatory element driving the activation of MYC downstream of NOTCH1 in T-ALL. Consistent with this hypothesis, chromatin configuration 3C analysis of the MYC locus demonstrated the association of this enhancer with proximal regulatory sequences in the MYC promoter (Fig. 2a). Moreover, luciferase reporter assays showed strong, orientation-independent activation of reporter constructs containing this enhancer in association with a −2.5 kb MYC proximal promoter21 in JURKAT T-ALL cells, which express high levels of constitutively active NOTCH1 protein 22, but not in Daudi and Raji B-cell lineage cells (Fig. 2b and Supplementary Fig. 4). In addition, human N-Me driven reporter activity in JURKAT cells was effectively suppressed upon inhibition of NOTCH1 signaling with the DBZ γ-secretase inhibitor and decreased upon mutation of N-Me RBPJ binding sites (Fig 2b,c and Supplementary Fig. 5). Conversely, reporter activity in these assays was increased by the introduction of two tandem copies of the human N-Me enhancer demonstrating quantitative, dose dependent increase of MYC promoter activity by this regulatory sequence (Fig. 2d). Similar results were obtained in reporter assays testing the activity and NOTCH signaling dependency of the mouse N-Me enhancer (Fig. 2e,f and Supplementary Fig. 5). Overall, these results identify the N-Me enhancer as a recurrently duplicated long-range regulatory element involved in the control of MYC expression downstream of NOTCH1 in T-ALL.
Figure 2.
Functional characterization of the N-Me enhancer. (a) NOTCH1 ChIP-seq binding occupancy profile in the MYC locus in HPB-ALL T-ALL cells along with 3C quantitative PCR analysis of the interaction frequency between DNA sequences flanking a HindIII restriction site in the MYC promoter with indicated sites located 3′ in the MYC locus and in the vicinity of the MYC +1.47 Mb NOTCH1 binding site. PCR signal is normalized to bacterial artificial chromosome (BAC) templates. (b) Luciferase reporter activity and response to inhibition of NOTCH with the DBZ γ-secretase inhibitor in JURKAT T-ALL cells of a MYC promoter construct (MYC Luc), a MYC promoter plus human N-Me enhancer in + orientation (hN-Me(+) MYC Luc) and a MYC promoter plus human N-Me enhancer in – orientation (hN-Me(−) MYC Luc) construct. Data from five technical replicates is shown. (c) Luciferase reporter activity in JURKAT T-ALL cells of a MYC promoter construct (MYC Prom-Luc), a MYC promoter plus human N-Me enhancer in + orientation (hN-Me(+) MYC Luc), and a MYC promoter plus human N-Me enhancer in – orientation (hN-Me(−) MYC Luc), both using the wild type hN-Me sequence or a hN-Me sequence with mutations in 7 RBPJ binding sites. Data from five technical replicates is shown. (d) Luciferase reporter activity in JURKAT T-ALL cells of a MYC promoter construct (MYC Luc), a MYC promoter plus human N-Me enhancer in + orientation (hN-Me(+) MYC Luc), and a MYC promoter plus a tandem duplication of the human N-Me enhancer in + orientation (hN-Me(++) MYC Luc). Data from five technical replicates is shown. (e) Luciferase reporter activity and response to inhibition of NOTCH with the DBZ γ-secretase inhibitor in JURKAT TALL cells of a MYC promoter construct (MYC Luc), a MYC promoter plus mouse N-Me enhancer in + orientation (mN-Me(+) MYC Luc) and a MYC promoter plus mouse N-Me enhancer in – orientation (mN-Me(−) MYC Luc). Data from five technical replicates is shown. (f) Luciferase reporter activity in JURKAT T-ALL cells of a MYC promoter construct (MYC Luc), a MYC promoter plus mouse N-Me enhancer in + orientation (mN-Me(+) MYC Luc), and a MYC promoter plus mouse N-Me enhancer in – orientation (mN-Me(−) MYC Luc), both using the wild type mN-Me sequence or a mN-Me sequence with mutations in 11 RBPJ binding sites. Data from five technical replicates is shown. Bar graphs in b–f indicate mean values and error bars represent s.d. P values were calculated using the two-tailed Student’s t-test: *** P ≤ 0.005. Y axis in ChIP-seq plots indicates fragment density in counts per 10 million.
The N-Me enhancer is required for thymocyte development
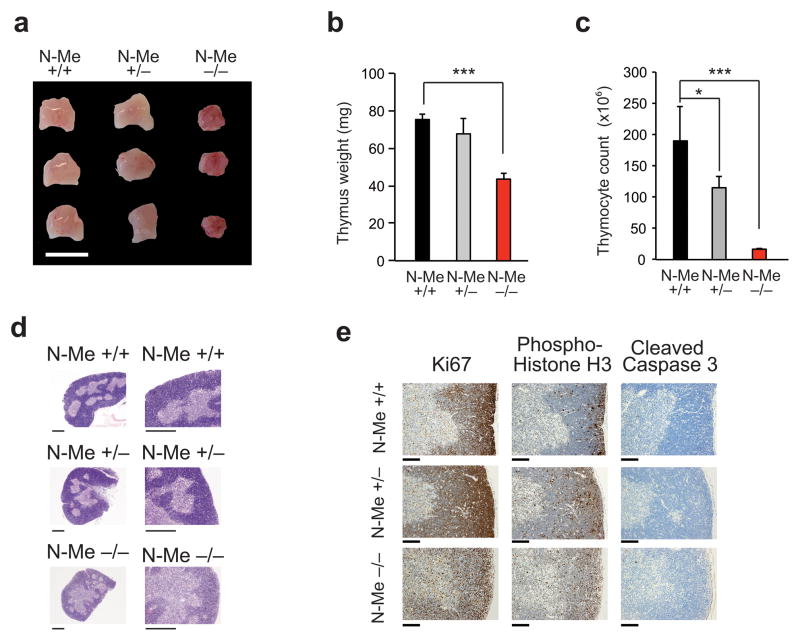
Next, and to test the specificity and functional relevance of the N-Me enhancer in T-cell development and transformation, we used homologous recombination in mouse embryonic stem cells to generate N-Me knockout and conditional knockout mice (Supplementary Fig. 6). N-Me knockout (N-Me−/−) mice were born with mendelian frequencies (Supplementary Table 3), survived perinatally and showed normal postnatal development without apparent behavioral abnormalities or defects in body size (Supplementary Fig. 7). Notably, anatomical and histological analysis of mice at 6 weeks of age revealed marked selective reduction in thymus size and cellularity in N-Me knockout animals compared to littermate controls (Fig. 3a–d). Interestingly, and despite preservation of thymus size and architecture, thymocyte counts were partially reduced in N-Me heterozygous knockout mice (Fig. 3a–d). Mechanistically, immunohistochemical analysis of N-Me knockout thymi showed marked reductions in proliferation markers (Ki67 and phospho-histone H3) with no apparent increase in apoptosis (cleaved caspase 3) (Fig. 3e). In addition, analysis of T cell populations in spleen and lymph nodes of N-Me knockout animals showed a marked reduction in the number mature CD4 and CD8 T cells (Supplementary Fig. 7). Analysis of bone marrow hematopoietic populations revealed normal numbers of B-cell precursors, myeloid progenitors, megakaryocytes and hematopoietic stem cells (Supplementary Fig. 7). Moreover, histological analysis of multiple tissues including those in which NOTCH signaling plays important developmental roles such as breast epithelium, skin, and intestine showed no alterations (Supplementary Fig. 7) and RT-PCR analysis showed no effects of N-Me deletion in Myc expression (Supplementary Fig. 7).
Figure 3.
Histological analysis of the thymus in N-Me knockout mice. (a–d) Thymus morphology (a), weight (b), cellularity (c) and histology (d) in 6-week old N-me wild type (N-Me+/+) heterozygous (N-Me+/−) and knockout (N-Me−/−) mice (n = 3 per genotype). (e) Immunohistochemical analysis of Ki67, phopho-histone H3 and cleaved caspase 3 expression in thymi from 6-week old N-me wild type (N-Me+/+), heterozygous (N-Me+/−) and knockout (N-Me−/−) mice. Scale bar in (a) indicates 1 cm. Scale bar in (d) indicates 100 μm. Scale bar in (e) indicates 200 μm. Representative images of three independent samples are shown in d,e. P values were calculated using the one-tailed Student’s t-test:: * P ≤ 0.05; *** P ≤ 0.005.
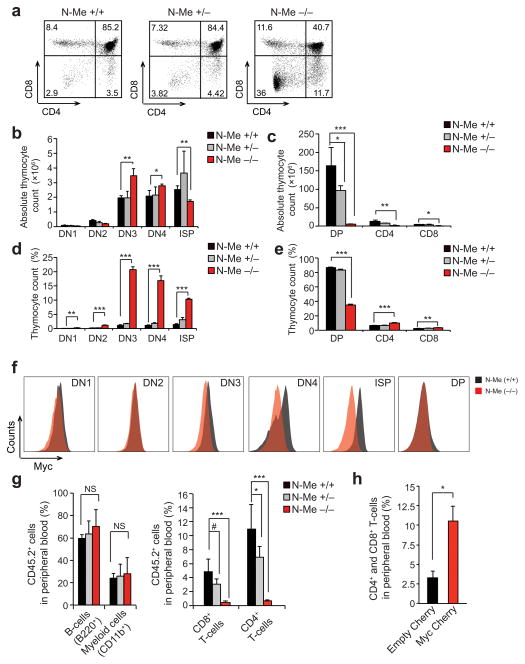
Detailed immunophenotypic analysis of thymic populations in N-Me knockout mice demonstrated accumulation of double negative 3 (DN3) T cells and marked reductions in double positive and CD4+ and CD8+ single positive cells (Fig. 4a–e). Mechanistically, this phenotype was associated with a marked reduction in Myc expression in DN3, DN4 and intermediate single positive cells (Fig. 4f). Consistently, transplantation of CD45.2 N-Me knockout bone marrow hematopoietic progenitors into lethally irradiated CD45.1 mice effectively reconstituted B-cell and myeloid lineages, but not T-cell development (Fig. 4g). Moreover, retroviral expression of MYC in N-Me knockout hematopoietic progenitors restored their capacity to restore T-cell development when transplanted into immunodeficient NRG mice (Fig. 4h).
Figure 4.
Characterization of T-cell development in N-Me knockout mice. (a) Representative flow cytometry plots of thymocyte populations stained with anti-CD4 and anti-CD8 in N-me wild type, heterozygous and knockout mice. Percentage populations are indicated in each quadrant. (b,c) Quantification of intrathymic T-cell populations in N-Me wild type, heterozygous and null mice in absolute numbers. (d,e) Quantification of intrathymic T-cell populations in N-Me wild type, heterozygous and null mice in relative numbers. (f) Quantification of intracellular Myc levels in immature T-cell populations from N-Me wild type and knockout mice as measured by flow cytometry. (g) Myeloid, B-cell and T-cell levels in peripheral blood of CD45.1 lethally irradiated mice 6 weeks after transplantation with CD45.2 bone marrow progenitors from N-Me knockout mice. (h) T-cell levels in peripheral blood of NRG mice 3 weeks after transplantation with bone marrow progenitors from N-Me knockout mice infected with retroviruses expressing mCherry (Empty Cherry) or Myc plus mCherry (Myc Cherry). Bar graphs in (b–d and g–h) indicate mean values across triplicate samples and error bars represent s.d. P values in were calculated using the two-tailed Student’s t-test: # P = 0.07; * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.005.
N-Me is required for NOTCH1-induced T-cell leukemogenesis
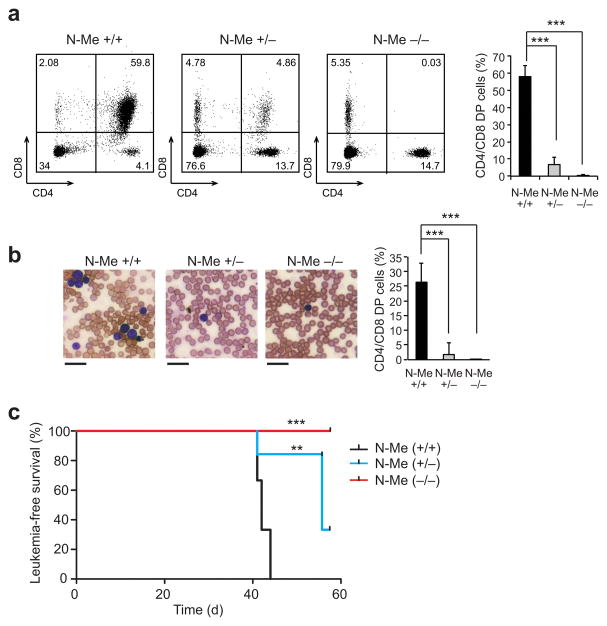
Given the important role of NOTCH1 induced MYC upregulation in the pathogenesis of T-ALL, we hypothesized that deletion of the N-Me enhancer could disrupt NOTCH1 induced leukemogenesis. To test this possibility we transplanted isogenic C57Bl6 mice with N-Me wild type, heterozygous and homozygous knockout hematopoietic progenitors infected with retroviruses driving the expression of an oncogenic constitutively active form of NOTCH1 (ΔE-NOTCH1)10. Consistent with previous reports23, mice transplanted with ΔE-NOTCH1 infected N-Me wild type cells showed a transient wave of CD4 CD8 double positive cells in peripheral blood three weeks post-transplant (Fig. 5a), which anticipated the development of overt T-ALL 3 weeks later (Fig. 5b,c). Mice transplanted with ΔE-NOTCH1-expressing N-Me heterozygous progenitors showed a blunted CD4 CD8 double positive cell wave and delayed tumor development (P <0.01) (Fig. 5a–c), while animals transplanted with ΔE-NOTCH1 N-Me knockout cells failed to develop a CD4 CD8 double positive wave and remained leukemia free 9 weeks post-transplant (P <0.001)(Fig. 5a–c).
Figure 5.
Analysis of T-ALL tumor initiation in N-Me knockout mice. (a) Representative flow cytometry plots of peripheral blood stained with anti-CD4 and anti-CD8 from mice transplantated with ΔE-NOTCH expressing N-Me wild type, heterozygous or knockout progenitors 3 weeks post-transplant. Percentage populations are indicated in each quadrant. Quantification of CD4 CD8 double positive population is shown on the right (n = 6 per genotype). (b) Representative blood smear preparations from mice transplanted with ΔE-NOTCH expressing N-Me wild type, heterozygous or knockout progenitors at 6 weeks post-transplant. Quantification of CD4 CD8 double positive population is shown on the right (n = 6 per genotype). (c) Kaplan-Meier survival curves of mice transplanted with ΔE-NOTCH-infected N-Me wild type, heterozygous or knockout hematopoietic progenitors (n = 6 per genotype). Bar graphs in (a) indicate mean values and error bars represent s.d. Scale bar in (b) indicates 100μm. P values in (a) were calculated using the two-tailed Student’s t-test: *** P ≤ 0.005. P values in (c) were calculated using the log-rank test: ** P ≤ 0.01; *** P ≤ 0.005.
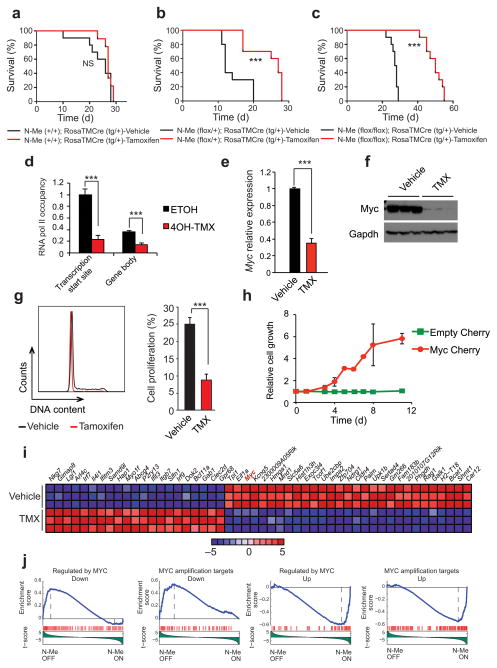
Next, and to explore the pathogenic role of N-Me-mediated Myc expression in NOTCH induced leukemia tumor maintenance, we generated ΔE-NOTCH1-induced T-ALL tumors from N-Me wild type (Rosa26TMCre N-Me+/+) and tamoxifen-inducible conditional heterozygous (Rosa26TMCre N-Meflox/+) and homozygous (Rosa26TMCre N-Meflox/flox) N-Me knockout hematopoietic progenitors as before. In these experiments, mice transplanted with ΔE-NOTCH1-expressing N-Me wild type and tamoxifen-inducible heterozygous and homozygous conditional N-Me knockout cells developed NOTCH1-induced T-ALLs with identical kinetics and immunophenotype (Supplementary Fig. 8). To test the effects of N-Me deletion in these established leukemias we then injected N-Me wild type and tamoxifen-inducible heterozygous and homozygous conditional N-Me knockout T-ALL lymphoblasts into secondary recipients and treated them with vehicle only or tamoxifen three days after transplant. In these experiments tamoxifen-induced activation of Cre recombinase had no effect in leukemia progression in mice harboring N-Me wild type tumor cells compared with controls (Fig. 6a). In contrast, tamoxifen-induced deletion of one copy of the N-Me enhancer in Rosa26TMCre N-Meflox/+ tumors or two copies in Rosa26TMCre N-Meflox/flox leukemias resulted in overt antileukemic effects and improved survival (Fig. 6b,c). Interestingly, analysis of Rosa26TMCre N-Meflox/+ tumor cells recovered upon disease progression after tamoxifen treatment showed heterozygous deletion of the N-Me enhancer (Supplementary Fig. 9), while analysis of tamoxifen-treated progressing Rosa26TMCre N-Meflox/flox tumors demonstrated retention of one (2/9) or both (5/9) copies of the N-Meflox allele (Supplementary Fig. 9). Finally, we performed limited dilution transplantations of conditional NOTCH1-induced N-Me knockout tumor cells and analyzed the effects N-Me inactivation via tamoxifen treatment in leukemia initiating cell activity. In this experiment, genetic inactivation of the N-Me enhancer resulted in a ~170 fold reduction in leukemia initiating cell frequency (Supplementary Fig. 10). Overall these results show a strong dose dependent effect of the N-Me enhancer in NOTCH1-induced tumor initiation and maintenance and in leukemia initiating cell activity.
Figure 6.
Analysis of leukemia maintenance in N-Me knockout cells. (a) Kaplan-Meier survival curves of mice harboring N-Me wild type Rosa26TMCre NOTCH1 induced leukemias treated with vehicle only (n = 10) or tamoxifen (n = 9). (b) Kaplan-Meier survival curves of mice harboring N-Me conditional heterozygous Rosa26TMCre NOTCH1 induced leukemias treated with vehicle only (n = 10) or tamoxifen (n = 10). (c) Kaplan-Meier survival curves of mice harboring N-Me conditional homozygous knockout Rosa26TMCre NOTCH1 induced leukemias treated with vehicle only (n = 10) or tamoxifen (n = 10). (d) ChIP analysis of RNApol II binding at the Myc locus 48 hours after treating N-Meflox/flox; RosaTMCretg/+ tumor cells in vitro with ethanol or 4-OH-tamoxifen, to induce N-Me deletion. Data from three technical replicates is shown. (e) Quantitative RT-PCR analysis of Myc expression in tumor cells isolated from N-Me conditional knockout-leukemia bearing mice treated with vehicle only (n = 4) or tamoxifen (n = 5) in vivo. (f) Western-blot analysis of Myc expression in tumor cells isolated from N-Me conditional knockout-leukemia bearing mice treated with vehicle only (n = 3) or tamoxifen (n = 3) in vivo. (g) Representative DNA content analysis and quantification of cell proliferation (S+G2/M fraction) of N-Me conditional knockout Rosa26TMCre NOTCH1 induced leukemias treated with vehicle only (n = 4) or tamoxifen (n = 5) in vivo. (h) Proliferation curve of 4-OH-tamoxifen treated N-Meflox/flox; RosaTMCretg/+ tumor cells infected with an MSCV-Empty cherry virus or an MSCV-Myc cherry virus, relative to ethanol treated controls. (i) Heat map representation of the top 50 differentially expressed Entrez Genes genes (P <0.001) between control and tamoxifen treated N-Me conditional knockout Rosa26TMCre NOTCH1 induced leukemias. The scale bar shows color coded differential expression with red indicating higher levels and blue lower levels of expression. (j) GSEA analysis of genes regulated by MYC in vehicle only versus tamoxifen treated N-Me conditional knockout Rosa26TMCre NOTCH1 induced leukemia cells in vivo. Bar graphs in (d,e,g) indicate mean values and error bars represent s.d. P values in (a–c) were calculated using the log-rank test: *** P ≤ 0.005. P values in (d, e, g) were calculated using the two-tailed Student’s t-test: *** P ≤ 0.005.
To better assess the mechanisms mediating the antileukemic effects of N-Me inactivation we then analyzed the cellular and transcriptional phenotypes of N-Me conditional inducible knockout T-ALL cells after tamoxifen treatment. In this setting, N-Me deletion in T-ALL cells (Supplementary Fig. 11) resulted in unloading of RNA polymerase II from the Myc transcription start site (Fig. 6d) and markedly reduced Myc RNA and protein expression levels (Fig. 6e,f). At the cellular level N-Me inactivation caused decreased cell proliferation with G1 cell cycle arrest (Fig. 6g). Notably, retroviral expression of MYC rescued the growth defect associated with genetic deletion of N-Me in NOTCH1-driven N-Me conditional knockout tumor cells (Fig 6h). Finally, gene expression profiling analysis showed that N-Me inactivation results in effective downregulation of Myc induced gene expression programs (Fig. 6i,j) and of gene expression signatures associated with cell growth and proliferation (Supplementary Table 4).
DISCUSSION
NOTCH1 plays a central role in the pathogenesis of T-ALL24 and drives an oncogenic transcriptional program that promotes cell growth proliferation and survival in T-ALL lymphoblasts. Importantly, the oncogenic effects of NOTCH1 are closely linked to activation of the MYC oncogene12,15,16. Thus, NOTCH1 activation upregulates MYC expression in T-ALL and NOTCH1 and MYC have been shown to regulate overlapping transcriptional programs in a feed-forward loop transcriptional circuitry that amplifies the oncogenic effects of NOTCH112. However, the specific mechanisms that drive MYC expression downstream of NOTCH1 in TALL remain incompletely understood. Here we report the identification and functional characterization of N-Me, a T-cell specific long-range regulatory sequence driving MYC expression downstream of NOTCH1 in T-ALL. This regulatory element, located +1,427 kb downstream of MYC in humans and +1.27 Kb downstream of the Myc locus in mice, has histone marks associated with active enhancers, interacts with MYC proximal regulatory sequences and sits within a broad area of H3K27 acetylation recently characterized as a T-cell specific super-enhancer25.
Transcriptional deregulation of MYC expression plays an important role in the pathogenesis of multiple cancers and the chromosome 8q24 region proximal to the MYC gene is a well established risk locus for epithelial tumors26–30. Most of the genetic variants associated with cancer risk at 8q24 are located in a 500 kb gene desert region located approximately 200 kb centromeric of the MYC gene. This region contains multiple enhancer regulatory sequences31, which physically associate the MYC promoter via long range chromatin loops32. Most notably, rs6983267, an enhancer-associated colon cancer risk allele variant in this region, has been shown to influence MYC expression downstream of beta-catenin-TCF433. Moreover, genetic deletion of the enhancer containing this regulatory element in the gut results in partial reduction of Myc expression in intestinal crypts and confers protection from intestinal tumorigenesis in APCmin mutant mice34. In contrast, no cancer-associated single nucleotide variants have been mapped to the larger gene-free region telomeric to the MYC gene where N-Me resides. However, a recent report has identified a broad regulatory area of about 100 kb located 1.7 Mb telomeric to the Myc gene, 400 kb downstream of N-Me35. This Myc regulatory region contains multiple enhancers active in myeloid cells, but not thymus, and is duplicated in about 3% of acute myeloid leukemias35.
Genetic deletion of N-Me in mice resulted in marked reduced thymic cellularity and cell proliferation demonstrating a critical role of this regulatory element in the homeostasis of immature T-cells. N-Me null mice showed severe reduction in the number of CD4 CD8 double positive cells and subsequent single positive CD4 and CD8 populations. These phenotypes perfectly recapitulate those reported in T-cell specific Myc conditional knockout mice (Lck-Cre Mycf/f)36. Given the proposed role of NOTCH1 signaling in hematopoietic stem cells and in non lymphoid tissues such as breast epithelium we thoroughly examined a potential role for N-Me in the control of Myc expression in these tissues. However, and consistent with a T-cell specific role of N-Me in the control of Myc expression, we detected no alterations in any cellular compartment other than T-cells. These results support that as yet to be identified NOTCH1 controlled enhancers may regulate Myc expression in breast epithelium and hematopoietic stem cells.
Strikingly, the requirement for N-Me mediated upregulation of Myc expression downstream of Notch1 was even more patent in the context of leukemia initiation, where loss of one and two copies of N-Me delayed and completely abrogated tumor development by oncogenic NOTCH1, respectively. In addition, N-Me was also required for maintenance of NOTCH1 induced leukemias as secondary deletion of one copy of N-Me in established tumors resulted in a marked delay in tumor progression and loss of two copies effectively abrogated leukemia propagation and leukemia initiating cell activity. These results are consistent with the well established quantitative effects of MYC expression in other tumor settings. Thus, loss of one copy of Myc has been shown to slow down intestinal tumorigenesis37 and homozygous deletion of Myc completely abrogates tumor development induced by loss of Apc in the gut38.
Several lines of evidence support a critical role for loss of Myc expression as the primary driver of the developmental and tumor phenotypes associated with N-me loss. In this regard, we observed marked reductions in Myc expression in developing T-cells from N-Me knockout mice and in T-ALL lymphoblast upon N-Me inactivation. Moreover, retroviral expression of Myc restored T-cell lymphopoiesis from N-Me deficient hematopoietic progenitors and rescued the defects in leukemia cell growth induced by secondary deletion on N-Me in NOTCH1-induced TALL cells. Finally, MYC inactivation has been associated with a global decrease in transcriptional activity with a particularly pronounced downregulation of genes involved in growth, proliferation and metabolism39–41, which were highly enriched in the gene expression signatures associated with N-Me loss in T-ALL.
The generation of the N-Me conditional knockout model presented here also served to analyze the specific role of this enhancer in transcriptional control. Thus, even though enhancer-promoter interactions have been implicated in the regulation of transcription by promoting the release of RNA polymerase II pausing in some cases42, deletion of N-Me in T-ALL lymphoblasts resulted in unloading of RNA polymerase II at the Myc transcription initiation site without any apparent increase in RNA polymerase II pausing.
Overall, the results shown here identify the N-Me regulatory sequence as a critical mediator of NOTCH1 induced MYC expression required for T-cell development and T-cell transformation and substantiates a pathogenic role for chromosomal duplications targeting this enhancer in the pathogenesis of T-ALL.
ONLINE METHODS
Cell lines
HPB-ALL and JURKAT were obtained from DSMZ. DAUDI and RAJI were obtained from American Type Culture Collection. Cells were cultured in standard conditions in RPMI media supplemented with 10% FBS and 1% Penicillin/Streptomycin.
NOTCH1 inhibition
We inhibited NOTCH1 in JURKAT cells with 250 nM DBZ ((S)-2-(2-(3,5-Difluorophenyl)acetamido)-N-((S)-5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl) propanamide) (Syncom) for 48h as previously described43.
Genomic analysis of primary T-ALL samples
A total of 160 T-ALL cases from adult and pediatric patients referred to St-Louis hospital, Paris, France were analyzed for copy-number abnormalities using array-comparative genomic hybridization with informed consent under the supervision of the Institutional Review Board of the Institut Universitaire d’Hématologie, Université Paris-Diderot. Sureprint G3 human CGH 180K, 244K, 400K or 1M arrays (Agilent technologies) were used and copy number alterations were identified using the Genomic Workbench software and the ADM-2 algorithm (Agilent Technologies) as previously described44.
T-ALL Oncogenic subtype was determined based on gene expression profiling as previously reported45, expression of T-ALL oncogene transcripts and presence of specific chromosomal translocations. NOTCH1 and FBXW7 mutations were analyzed as previously described46.
Chromatin immunoprecipitation (ChIP)
We performed chromatin immunoprecipitations using the Agilent Mammalian ChIP-on-chip chromatin immunoprecipitation protocol as described before12,47. Briefly, 108 HPB-ALL cells were fixed with formaldehyde in a final concentration of 1% for 10 minutes at room temperature. We quenched the crosslinking reaction with a 2.5 M glycine solution and the cells were then centrifuged, washed once with ice cold phosphate saline buffer and lysed. Cell nuclei were resuspended in lysis-buffer (10 mM Tris-HCl, pH 8.0; 100 mM NaCl; 1 mM EDTA; 0.5 mM EGTA; 0.1% sodium deoxycholate; 0.5% N-lauroylsarcosine) and fragmented to a 150–300 bp size using a Bioruptor sonicator (Diagenode). We incubated fragmented chromatin)] overnight at 4°C with magnetic beads (Dynal) loaded with 1 mg of the following specific antibodies: anti-NOTCH C-20 (sc-6014; Santa Cruz Biotechnology); anti NOTCH1 Val1744 (#2421; Cell Signaling); anti p300 (sc-585; Santa Cruz Biotechnology); anti H3K4me1 (ab8895; Abcam) and anti H3K4me3 (ab1012; Abcam).
For RNApol II ChIP, N-Me (flox/flox); RosaTMCre (tg/+) tumor cells were treated in vitro with ethanol or tamoxifen, to induce N-Me deletion. 48 hours later, 107 cells from both conditions were processed the same way as described before, and the fragmented chromatin was incubated overnight at 4°C with magnetic beads loaded with 5mg of the RNApol II antibody (sc-899, Santa Cruz Biotechnology).
We washed chromatin-antibody-bead complexes seven times with RIPA buffer (50 mM Hepes-KOH, pH 7.6; 0.5 M LiCl; 1 mM EDTA; 1% Igepal and 0.7% sodium deoxycholate), and once with TE-buffer containing 50 mM NaCl. Then we eluted the chromatin from the beads with elution buffer (50 mM Tris-HCl, pH 8.0 10 mM EDTAand 1%SDS) and reversed the cross-link by incubation at 65°C overnight. We purified DNA fragments with phenol-chloroform and ethanol precipitation. We tested enrichment of specific immunoprecipitated DNA fragments by quantitative PCR as previously described12,47.
ChIP-seq
Chip-seq for NOTCH1 in HPB-ALL cells was performed using chromatin immunoprecipitates generated as described above and further fragmented to 100 bp average size using a Covaris sonicator. All preparations of libraries for sequencing and the sequencing itself were performed according to the SOLiD3™ System protocols. For library preparation we performed adaptor ligation and purification, and the resulting DNA was amplified for 15 cycles. The libraries were then sequenced using 50 bp read lengths on a SOLiD3™ instrument (Life Technologies).
We recovered 17,530,449 reads and 9,591,524 unique reads (defined by a unique sequencing read start site) that mapped exactly once to the hg19 human genome from the NOTCH1 ChIP-seq library as well as 4,954,605 reads and 4,894,225 unique reads from the input control DNA library.
ChIP-seq and conservation analysis
We used HOMER (Hypergeometric Optimization of Motif EnRichment) software suite48 to estimate the ChIP-fragment density at each position in the human genome for HBPALL NOTCH1 ChIP-Seq data. We also analyzed ENCODE generated ChIP- seq data49 and the following publicly available ChIP-seq results: CUTLL1 NOTCH1 ChIP-seq (Gene Expression Omnibus GSM732903)18; CUTLL1 RBPJ ChIP-seq (Gene Expression Omnibus GSM732905)18; G4A2 NOTCH1 ChIP-seq (Gene Expression Omnibus GSM732916)18; G4A2 RBPJ ChIP-seq (Gene Expression Omnibus GSM732917)18.
We used MACS algorithm50 to call ChIP-Seq peaks and the top scoring intergenic peak was detected at chr8:130,180,065–130,180,972 (hg19), which aligns to the mouse genome (mm10) at chr15:63,255,515–63,256,347 (80.2% of bases, 100.0% of span).
The conservation track in Figure 1b shows a subset of the 100 vertebrates genomic sequence alignment as produced by the UCSC Genome Browser based on the MULTIZ algorithm 51.
Quantitative 3C analysis
We performed 3C analysis of chromatin interactions as previously described52 with some modifications. Briefly, HPB-ALL cells were crosslinked with formaldehyde in a final concentration of 1% for 10 minutes at room temperature. We quenched the crosslinking reaction with a 2.5 M glycine solution and the cells were then centrifuged, washed once with ice cold phosphate saline buffer and lysed. We digested cell nuclei overnight at 37°C with the HindIII restriction enzyme. After digestion the resulting fragments were highly diluted and ligated first 4 h at 16°C and then for 30 min at 37°C. After ligation the DNA was treated with proteinase K and de-crosslinked overnight at 65°C. We cleaned and purified the DNA fragments by phenol chloroform extraction and ethanol precipitation and resuspended them in 10 mM Tris pH 7.5. The resulting 3C template was used to perform quantitative PCR using primers flanking the HindIII restriction sites located in the regions of interest within the MYC locus and in the vicinity of the N-Me enhancer in 7300 quantitative PCR instrument (Applied Biosystems).
Luciferase reporter assays
We performed reporter assays using a pBV-Luc −2.5 kb MYC promoter luciferase construct21 alone and coupled with upstream human and mouse N-Me enhancer sequences cloned in the forward and reverse orientation. In these assays, we electroporated in JURKAT cells with a Gene Pulser MXcell electroporator (Bio-Rad) using MYC luciferase reporter constructs together with a plasmid driving the expression of the Renilla luciferase gene (pCMV-Renilla) used as internal control and treated them with DMSO or DBZ (250 nM) to inhibit NOTCH signaling. We measured luciferase activity 48 h after electroporation with the Dual-Luciferase Reporter Assay kit (Promega).
Mice and animal procedures
All animals were maintained in specific pathogen-free facilities at the Irving Cancer Research Center at Columbia University Medical Campus. Animal procedures were approved by the Columbia University Institutional Animal Care and Use Committee.
To generate N-Me conditional knockout mice, we introduced LoxP sites at mouse genomic (GRCm38/mm10) positions 63.255.283 and 63.256.404 flanking the mouse N-Me enhancer together with a neomycin selection cassette flanked by Frt sites by homologous recombination in C57BL/6 embryonic stem cells. We generated chimeras in C57BL/6 albino blastocysts using three independent targeted embryonic stem cell clones identified by PCR analysis and verified by Southern blot. We verified germline transmission in the offspring of highly chimeric male mice crossed with C57BL/6 albino females. To remove the neomycin selection cassette we crossed mice harboring the targeting construct with a Flp germline deleter line (B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ, the Jackson Laboratory). To generate mice carrying a N-Me deletion we first crossed mice harboring this resulting conditional N-Me targeted allele with a Cre germline deleter line (B6.C-Tg(CMV-cre)1Cgn/J, the Jackson Laboratory) and then crossed the resulting mice with wild type C57BL/6 animals to breed out the CMV-cre allele. To generate conditional inducible N-Me knockout mice we bred animals harboring the conditional N-Me targeted allele with ROSA26TMCretg/+ mice, which express a tamoxifen-inducible form of the Cre recombinase from the ubiquitous Rosa26 locus53. Phenotypic characterization of N-Me knockout mice included male and female mice and isogenic age and gender matched controls.
Mammary epithelial cell isolation
To determine the specific estrus stage and thus the concomitant mammary gland developmental phase, we analyzed female mice for the presence of leukocytes, cornified epithelial cells, and nucleated epithelial cells in vaginal smears as previously described54. To purify mammary epithelial cells (MECs), mammary glands were dissected, mechanically disrupted and digested using a digestion solution composed by DMEM-F12, 100 units/mL Penicillin-Streptomycin, 2mg/mL collagenase-A and 100 units/mL hyaluronidase, for four hours at 37°C. After digestion we washed mammary epithelial cells in Phosphate buffered saline solution and recovered them by centrifugation.
Mammary gland whole mount preparations and carmin red staining
For whole mount preparations we dissected mouse mammary glands, fixed them in 4% paraformaldehyde for 1 hour and stained them with carmine red (C1022, Sigma-Aldrich) overnight at room temperature. Following carmin red staining we dehydrated the stained tissues in increasing concentrations of ethanol (70, 95 and 100%) and ultimately fixed them in xylene.
Immunohistochemistry
We performed immunohistochemical stainings following standard protocols using antibodies against Ki67 (15580, Abcam), phosphorylated-histone H3 (9701, Cell Signaling Technology) and Cleaved Caspase 3 (9664, Cell Signaling Technology) were used.
Generation of NOTCH1 induced leukemias
To generate NOTCH1-induced T-ALL tumors in mice, we performed retroviral transduction of bone marrow cells enriched in Lineage negative cells isolated from female mice using magnetic beads (Lineage Cell Depletion Kit, Miltenyi Biotec) with an activated form of the NOTCH1 oncogene (ΔE-NOTCH1)10 and transplanted them via intravenous injection into lethally irradiated female isogenic recipients as previously described55.
Flow cytometry analysis of hematopoietic populations
Single-cell suspensions of total bone marrow cells from 6-week old wild-type and N-Me heterozygous and homozygous knockout littermates were prepared by crushing the leg bones and passing cells through a 40 μm filter. Single-cell suspensions of thymocytes were obtained by pressing one thymic lobe through a 40 μm filter. For flow cytometry based analysis of bone marrow stem and progenitor populations, and discrete stages of T cell development, cells were stained with anti-mouse fluorochrome-conjugated antibodies as follows (clone names are provided in brackets). CD8α PE-Cy7 (53-6.7), CD16/CD32 Alexa Fluor 700 (93), CD41 PE-Cy7 (eBioMWReg30), CD44 PerCP-Cy5.5 (IM7), CD48 eFluor 450 (HM48-1), CD105 eFluor 450 (MJ7/18), CD117 PerCP-eFluor 710 (2B8), Ly6A/E (Sca-1) PE-Cy7 (D7), biotin-CD11b (M1/70), -Ly-6G (Gr1; RB6-8C5), -NK1.1 (PK136), -Ter119 (TER119), -CD8α (53-6.7), -CD4 (GK1.5), -CD19 (eBio1D3), -CD45R (B220; RA3-6B2) and Streptavidin PE were purchased from eBioscience. CD150 APC, CD150 PE (both TC15-12F12.2) and Ly-6A/E (Sca-1) PE (D7) were purchased from BioLegend. CD4 APC-Cy7 (GK1.5), CD25 APC-Cy7 (PC61) and Streptavidin APC-Cy7 were purchased from BD Biosciences. Flow cytometry analysis of the different populations [HSCs and bone marrow progenitor subsets: KLS total HSCs (Lin− CD117+ Sca-1+), LT-HSC (Lin− CD117+ Sca-1+ CD150+ CD40−), ST-HSC (Lin− CD117+ Sca-1+ CD150+ CD40+), MPP (Lin− CD117+ Sca-1+ CD150− CD40+/−), CMP total (Lin− CD117+ Sca-1−), CLP (Lin− CD117+ Sca-1int CD135+), MkP (Lin− CD117+ Sca-1− CD150+ CD41+) and GMP (Lin− CD117+ Sca-1− CD150− CD41− CD16/32+); T cell immature subsets in the thymus: ETP (Lin− CD117hi CD44+), DN1 (Lin− CD44+ CD25−), DN2 (Lin− CD44+ CD25+), DN3 (Lin− CD44− CD25+) and DN4 (Lin− CD44− CD25−)] was performed using a BD LSRII and a BD LSRFortessa cell analyzer (BD Bioscience).
For the analysis of B cell differentiation in bone marrow, we performed surface staining with the following anti-mouse fluorochrome and biotin-conjugated antibodies: B220-PE-Cy5 (BD Pharmingen-553091), CD19-biotin (eBioscience-13-0193), CD25-APC-Cy7 (BD Pharmingen-557658), IgM-APC (eBioscience-17-5790), and IgD-PE (eBioscience-12-5993). CD19-biotin antibody was detected using PE-Cy7-conjugated streptavidin (eBiosciences-25-4317). Flow cytometry analysis of the different populations [preproB (B220+ CD19−), proB (B220+ CD19+ CD25− IgM− IgD−), preB (B220+ CD19+ CD25+ IgM− IgD−) and immature B cells (B220+ CD19+ CD25− IgM+ IgD−)] was performed using a FACSCanto Flow Cytometer (BD Bioscience).
For the analysis of T cell populations in spleen and lymph nodes we collected tissues from 6-week old wild-type and N-Me knockout littermates and processed through a 70 μm mesh to obtain single cell suspensions. We removed red cells in spleen samples by incubation with red blood cell lysis buffer (155 mM NH4Cl; 12 mM KHCO3; 0.1 mM EDTA) 5 minutes at room temperature. We stained cells with APC-conjugated antibodies against mouse CD4 (BD Pharmingen-553051) and PE-conjugated antibodies against mouse CD8a (BD Pharmingen-553032).
For the analysis of megakaryocytes in bone marrow and spleen and platelets in peripheral blood, we collected tissues from 6-week old wild-type and N-Me heterozygous and homozygous knockout littermates and processed through a 70 μm mesh to obtain single cell suspensions. We removed red cells samples by incubation with red blood cell lysis buffer (155 mM NH4Cl; 12 mM KHCO3; 0.1 mM EDTA) 5 minutes at room temperature. We stained cells with a FITC-conjugated antibody against mouse CD41 (eBioscience-11-0411-81). Flow cytometry analyses were performed in a FACSCanto flow cytometer (BD Biosciences).
Bone marrow reconstitution experiments
We isolated bone marrow lineage negative cells from N-Me+/+, N-Me+/− and N-Me−/− CD45.2 expressing mice using magnetic beads (Lineage Cell Depletion Kit, Miltenyi Biotec) following the manufacturer’s guidelines, and then transplanted into lethally irradiated CD45.1 C57BL6 female mice (Taconic). 6 weeks post-transplant, hematologic reconstitution of myeloid lineage, B-cell lineage and T-cell lineage was assessed by peripheral blood staining with the specific antibodies mentioned above, and analyzed in a FACSCanto flow cytometer using FACSDiva (BD Biosciences).
For the Myc rescue experiment in vivo we isolated bone marrow lineage negative cells from N-Me−/− mice as before, infected them with either MSCV-IRES-mCherry or MSCV-Myc-IRES-mCherry retroviruses and then transplanted into male and female NRG (NOD.Cg-Rag1 tm1Mom Il2rg tm1Wjl/SzJ) mice (The Jackson Laboratory). We assessed T-cell lineage reconstitution 3 weeks post-transplant in peripheral blood by flow cytometry quantitation of cells stained with specific antibodies against CD4-APC (BD Pharmingen-553051) and CD8-FITC (BD Pharmingen-553030) in a BD LSRFortessa flow cytometer (BD Biosciences) using FACSDiva software (BD Biosciences).
Flow cytometry analysis of Myc expression
We collected cells from the thymi of 6-week old male and female wild type and N-Me knockout mice and performed surface membrane stainings using the following antibodies: CD3-Alexa Fluor 488 (BD Pharmingen-557666), CD8a-PE-Cy7 (eBioscience-53-6.7), CD44-PerCP-Cy5.5 (eBioscience-IM7), CD25-APC-Cy7 (BD Pharmingen-557658) and CD4-APC (BD Pharmingen-553051). After surface staining we fixed the cells and permeabilized them with BD Cytofix/Cytoperm solution (BD Biosciences), washed them with BD Perm/Wash buffer (BD Biosciences), and stained them with a mouse anti-Myc antibody (sc-40, Santa Cruz) and an Alexa Fluor 546-conjugated donkey anti-mouse IgG antibody (A11036, Invitrogen). We analyzed the intracellular levels of Myc in a FACSCanto flow cytometer (BD Biosciences) using FACSDiva software (BD Biosciences).
Microarray gene expression profiling
We isolated and snap froze spleen samples from of N-Me conditional knockout NOTCH-induced T-ALL bearing mice 40 hours after treatment with vehicle only (corn oil, SIGMA) or tamoxifen (3 mg/Kg in corn oil, SIGMA) administered by intraperitoneal injection. RNA was isolated, labeled and hybridized to the MouseRef-8 v2.0 Expression BeadChip (Illumina) using standard procedures. Raw gene expression data were log2 transformed and quantile-normalized using MATLAB. Differentially expressed transcripts were identified by t-test. Enrichment of MYC signature genes56,57 were analyzed by GSEA using the t-test metric and 10,000 permutations of the gene list58. Differentially expressed genes induced by tamoxifen deletion of the N-Me enhancer (P < 0.001, fold change >1.5) were analyzed for functional annotation enrichment using DAVID Bioinformatics web tools59.
Quantitative real time PCR
We performed reverse transcription reactions with the ThermoScript RT-PCR system (Invitrogen) and analyzed resulting complementary DNA products by quantitative real-time PCR (FastStart Universal SYBR Green Master Mix (Roche) using a 7300 Real-Time PCR System (Applied Biosystems). Relative expression levels were normalized using Gapdh as a reference control.
Western Blotting
Western blot was performed using standard procedures. Antibodies against Myc (sc-764, Santa Cruz), and Gapdh (5174, Cell Signaling) were used.
Statistical analyses
We performed statistical analysis by Student’s t-test. We assumed normality and equal distribution of variance between different groups analyzed. Survival in mouse experiments was represented with Kaplan-Meier curves and significance was estimated with the log-rank test (Prism GraphPad). We analyzed serial limited dilution leukemia-initiating cell data using the ELDA software60.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grant CA120196 to A.A.F.), the Stand Up To Cancer Innovative Research Award (A.A.F.), The Pershing Square Sohn Foundation Award (A.A.F), the Swim Across America Foundation (A.A.F), the Canceropole IDF (J.S.), the program CIT from the Ligue Contre le Cancer (J.S.), the ERC St Grant Consolidator 311660 (J.S.), and the Saint-Louis Institute program ANR-10-IBHU-0002 (J.S.). D.H. is a Leukemia and Lymphoma Society postdoctoral fellow. L.B. is a postdoctoral researcher supported by the Lymphoma Research Foundation. We are grateful to P. Sumazin for assistance on the analysis of ChIP-seq data, T. Ludwig for the ROSA26Cre-ERT2/+ mouse, R. Kopan for the Δ-E NOTCH1 construct and R. Baer for helpful discussions and revision of the manuscript.
Footnotes
Author Contributions
D.H. performed most of the experiments and wrote the manuscript. T.P. performed the ChIP-seq and 3C studies. A.A.I. performed bioinfomatic analyses. S.A.S. performed in vitro studies. A.A.W. and L.B. analyzed hematopoietic phenotypes, L.X. performed some animal studies with D.H., M.C. and D.L.N. performed histological analyses, C.C.C. supervised histological analyses, J.C and E.C. performed genomic analyses of human leukemias, A.A.F designed the study, supervised research and wrote the manuscript.
References
- 1.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. The Journal of clinical investigation. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 3.De Keersmaecker K, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nature medicine. 2010;16:1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Della Gatta G, et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nature medicine. 2012;18:436–440. doi: 10.1038/nm.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanda T, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 7.Tzoneva G, Ferrando AA. Recent advances on NOTCH signaling in T-ALL. Current topics in microbiology and immunology. 2012;360:163–182. doi: 10.1007/82_2012_232. [DOI] [PubMed] [Google Scholar]
- 8.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annual review of cell and developmental biology. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 9.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 10.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 12.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nature reviews Genetics. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes development. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Molecular and cellular biology. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palomero T, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashworth TD, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Sulis ML, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. The Journal of experimental medicine. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology/the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2009:353–361. doi: 10.1182/asheducation-2009.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nature genetics. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 27.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nature genetics. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nature genetics. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia L, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS genetics. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nature genetics. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Molecular and cellular biology. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sur IK, et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 35.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dose M, et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108:2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Athineos D, Sansom OJ. Myc heterozygosity attenuates the phenotypes of APC deficiency in the small intestine. Oncogene. 2010;29:2585–2590. doi: 10.1038/onc.2010.5. [DOI] [PubMed] [Google Scholar]
- 38.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 39.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 43.Real PJ, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nature medicine. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clappier E, et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. The Journal of experimental medicine. 2011;208:653–661. doi: 10.1084/jem.20110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soulier J, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 46.Clappier E, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24:2023–2031. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- 47.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature medicine. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome research. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonis M, et al. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nature methods. 2009;6:837–842. doi: 10.1038/nmeth.1391. [DOI] [PubMed] [Google Scholar]
- 53.Guo K, et al. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology. 2007;148:3987–3997. doi: 10.1210/en.2007-0261. [DOI] [PubMed] [Google Scholar]
- 54.Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PloS one. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. The Journal of clinical investigation. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim YH, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–138. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 57.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome biology. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.