Abstract
House flies (Diptera: Muscidae; Musca domestica L.) harbor and transmit a variety of human enteropathogens including E. coli O157:H7. Interactions between ingested bacteria and the fly gut directly impact bacterial persistence, survival and ultimately fly vector competence. We assessed the temporospatial fate of GFP-E. coli O157:H7 (GFP-ECO157) in house flies along with fly antimicrobial responses for 12 h post-ingestion. In flies fed GFP-ECO157, culture and microscopy revealed a steady decrease in bacterial load over 12 h, which was likely attributable to the combined effects of immobilization within the peritrophic matrix, lysis and peristaltic excretion. However, flies can putatively transmit this pathogen in excreta because intact bacteria were observed in the crop and rectum. qRT-PCR analysis of antimicrobial peptides (AMP) and lysozyme gene expression showed minimal upregulation in both the gut and carcass of house flies fed GFP-ECO157. However, these genes were upregulated in fly heads and salivary glands, and effector proteins were detected in the gut of some flies. Collectively, these data indicate that house flies can serve as reservoirs of E. coli O157:H7 for up to 12 h, and factors in addition to AMPs and lysozyme may contribute to bacteria destruction in the gut.
Keywords: Musca domestica, antimicrobial peptides, cecropin, defensin, diptericin, immunofluorescence, lysozyme, qRT-PCR, vector
Introduction
House flies (Musca domestica L.) feed and breed on septic substrates, putting them in direct contact with a multitude of pathogenic microorganisms. Since house flies are synanthropic organisms, they transport these pathogens from septic environments to domestic habitats. As a result, flies have long been implicated as agents in the spread of human disease pathogens (Hawley, 1951; West, 1951; Greenberg, 1959). Despite the recognized relevance of house flies in harboring and disseminating a wide variety of infectious agents affecting humans (Graczyk et al., 2001), only recently has focus shifted to elucidating the role house flies serve beyond their well-known capacity as a mechanical vector.
The potential of house flies to serve as competent vectors for bacteria has been investigated in studies that determined the fate of bacteria and transmission potential in experimentally-infected flies. Zurek et al. (2001) found that bacteria-fed flies harbored the zoonotic turkey pathogen Yersinia pseudotuberculosis for up to 36 h post-exposure. In 2002, Nayduch et al. demonstrated that house flies could harbor the enteropathogen Aeromonas caviae up to 8 d after feeding and that flies transmitted viable bacteria in excreta. Subsequent experiments revealed that a related species, Aeromonas hydrophila, survived in the house fly crop and was recoverable from vomit specks, which indicated that the primary mode of transmission of Aeromonas was via regurgitation (McGaughey and Nayduch, 2009). More recently, the fate of Staphylococcus aureus in house flies was investigated (Nayduch et al., 2013), and although bacteria counts declined rapidly in the fly alimentary canal, substantial numbers of this pathogen were excreted within 2 h after ingestion. Collectively, these studies demonstrate that house flies harbor a number of pathogens in the alimentary canal and can serve as significant reservoirs and transmitters of some bacterial species.
Although house flies internally harbor human and animal pathogens, they rarely become diseased themselves, showing remarkable resilience and what must be a very efficient defense response in conjunction with physical barriers against bacteria in the alimentary canal. The midgut epithelium of the house fly is protected by a type II peritrophic matrix (PM), an acellular double layer secreted by a section of the proventriculus known as the cardia (Lehane, 1997). The PM protects the epithelium from directly contacting ingested material, including microbes (Lehane, 1997). Bacteria do not traverse the PM and are sequestered therein by size exclusion, and some bacterial species appear to be immobilized within the PM by an unknown mechanism (McGaughey and Nayduch, 2009). The PM is continuous throughout the midgut and terminates at the hindgut. The hindgut and rectum epithelia are lined in cuticle, which offers protection from microbial invasion in the absence of the PM.
In addition to physical barriers, physiological defenses such as digestive processes, pH/ionic fluxes, and innate immune responses help to protect the fly gut from microbes. The dipteran innate immune system combats microbes on both a systemic (i.e. fat body) and local epithelial (e.g., the alimentary canal) level. Flies such as Drosophila melanogaster detect microbes when microbe-associated molecular patterns (MAMPs) like bacteria peptidoglycan (PGN) bind pathogen recognition receptors (PRRs) that subsequently activate signaling pathways of the humoral response (Lemaitre and Hoffmann, 2007). In the gut, small dimers of PGN are able to traverse the PM; therefore, PRRs on epithelial cells detect pathogens without directly contacting bacteria (Charroux and Royet, 2010). Activation of these pathways results in the expression of effector molecules including antimicrobial peptides (AMPs) (Lemaitre and Hoffmann, 2007). AMPs show target specificity in induction and activity, enabling an efficient innate immune response to invading microbes. In fruit flies, diptericin, attacin, drosocin, and cecropin target Gram-negative bacteria and defensin targets Gram-positive bacteria (Lemaitre and Hoffmann, 2007). However, in filth flies such as house flies and blow flies, defensin has demonstrated more broad-spectrum activity (Lambert et al. 1989; Dang et al. 2010). In addition to the antimicrobial activity of AMPs, the peptidoglycan-digesting enzyme lysozyme has shown extensive bacteriolytic activity in the house fly gut (Terra et al., 1988; Ren et al., 2009) and is constitutively expressed when flies are in contact with large amounts in bacteria (Nayduch and Joyner, 2013).
E. coli O157:H7 is an important human pathogen commonly isolated from wild-caught house flies (Forster et al., 2007). Previous studies (Kobayashi et al., 1999; Sasaki et al., 2000) have demonstrated E. coli O157:H7 colonization of fly mouthparts and persistence in the alimentary canal as well as transmission, but did not observe the role of fly-microbe interactions in these phenomena. Therefore, the aims of this study were to (1) determine the temporospatial fate of E. coli O157:H7 within house flies over 12 h and to (2) assess the concurrent temporospatial expression of immune effectors (AMPs and lysozyme) mounted by the fly after ingestion of E. coli O157:H7.
Materials and Methods
Bacteria culture
Escherichia coli O157:H7 EDL 933 (ECO157) was transformed with the plasmid pGFPuv (Clontech, Mountain View, CA, USA) with an additional kanamycin resistance cassette as previously described (McGaughey and Nayduch, 2009). Stock cultures of GFP-expressing E. coli O157:H7 (GFP-ECO157) were maintained on Luria-Bertani media (Fisher Scientific, Atlanta, GA, USA) with 100 μg/ml (w/v) of ampicillin sodium and 50 μg/ml (w/v) kanamycin sulfate (LBAK agar or broth). Prior to fly feeding, bacteria were cultured in 50 ml LBAK broth for 8–9 h while shaking at 37°C, and 1 ml was sub-cultured in 25 ml LBAK broth until an OD600 of 1.00–1.20 (± 0.05) was reached.
House fly rearing and bacteria feeding
House flies were reared as described, and puparia were kept in sterile glass jars until eclosion (McGaughey and Nayduch 2009). Newly emerged (2–3 day-old), mixed-sex flies were used for all experiments. Eclosed flies were fed sterile 10% (w/v) fly food solution (40% powdered sugar, 40% powdered milk, 20% powdered egg) ad libitum for 24 h, then transferred to individual sterile glass jars where they were maintained on sterile 5% sucrose at room temperature (22–25°C) overnight. To induce feeding, flies were fasted for at least 12 h, placed in 30°C incubator for approximately 2 h, and finally placed in the 37°C incubator for 1 h before each experiment. A 2 μl droplet of GFP-ECO157 liquid culture (described above) was placed on a small (15mm × 15mm) Parafilm® M square (Fisher Scientific) in each jar, and the flies were monitored until the entire droplet was consumed. House flies that did not feed within 30 min were discarded, and the remaining flies were maintained at room temperature for the rest of the experiment.
Enumeration of GFP-ECO157 from house flies
Flies (total n = 76 in four biological replicates) were individually-fed a 2 μl droplet of GFP-ECO157 (mean CFU ± SE in droplet: 2.6 ± 0.9 × 106) and at 2, 4, 6, and 12 h post-ingestion (h PI), flies (n = 3 to 6 per replicate per time point) were immobilized by chilling at 0° C. A group of control flies fed sterile 5% sucrose (n = 5 per replicate) served as negative controls. Individual flies were homogenized in 500 μl sterile PBS (per 1 L: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, pH 7.4) and homogenate was serially diluted and plated in duplicate on LBAK agar for GFP-ECO157 recovery. Culture plates were incubated at 37°C for 24 h. The total number of CFU recovered from each fly were log10 transformed and data were analyzed using Kruskal-Wallis/Dunn’s Test (JMP ® 9; SAS Institute Inc., Cary, NC, USA). Bacterial plate counts above 300 CFU and below 30 CFU were excluded from statistical analyses, as is common convention (Madigan, 2009). Counts below 30 CFU are subject to estimation error, and above 300 CFU cannot reliably be counted on culture plates since there is an increased chance of merged colonies due to crowding.
Microscopic localization of bacteria in the house fly alimentary canal
Flies (total n = 28 in two biological replicates) were fed a 2 μl droplet of GFP-ECO157 (mean CFU ± SE in droplet: 1.8 ± 0.9 × 106) and were immobilized by chilling at 0°C at 2, 4, and 6 h PI with one replicate extending to 12 h PI (n = 4 flies per time point in each replicate). The entire alimentary canal (i.e. proventriculus, crop, midgut, hindgut and rectum) was removed by aseptic dissection and viewed with an epifluorescence microscope to visualize GFP-ECO157 (Leitz Laborlux 12 epiflourescence microscope; Wetzlar, Germany). Images of bacteria in the alimentary canal were captured using a Leica DFC 420 digital camera (Leica Microsystems Ltd., Buffalo Grove, IL, U.S.A.), while also noting bacteria location, motility and cellular integrity. A group of negative control flies (n = 5 per replicate) were individually fed 5 μl of sterile 5% sucrose, dissected, and examined via microscopy as above to confirm no contamination with the test strain.
Temporospatial qRTPCR analysis of AMP and lysozyme expression in GFP-ECO157-fed flies
Flies (total n = 95 in three biological replicates) were fed 2 μl GFP-ECO157 (mean CFU ± SE in droplet: 1.40 ± 0.30 × 106) as described above, and in each replicate 7 or 8 flies were aseptically dissected at 2, 4, 6, and 12 h PI to remove (1) the head including attached salivary glands, (2) the alimentary canal (i.e. crop, from proventriculus to rectum), and (3) the remaining carcass with fat body. LBAK broth-fed flies (n = 4) served as controls and were dissected as above. The dissected tissues were pooled for each replicate and homogenized in 500 μl TRIzol® reagent (Invitrogen, Grand Island, NY, U.S.A.). RNA was extracted using the Ambion® Ribopure™ kit (Ambion Life Technologies, Grand Island, NY, U.S.A.) following the manufacturer’s protocol. One microgram of RNA was then reverse transcribed using a Quantitect® Reverse Transcription kit (Qiagen Inc., Valencia, CA, U.S.A.) following the manufacturer’s instructions. After synthesis, cDNA was diluted 1:10 and used in qRT-PCR (quantitative reverse-transcriptase PCR) analysis. Each reaction was performed using the RealMasterMix SyBR ROX kit (5-Prime Inc., Gaithersburg, MD, USA) with 500 nM of sense and antisense primers of target (AMPs, lysozyme) and reference (rps18) genes (Table 1). The reactions were performed on an Mastercycler Realplex2 (Eppendorf, Happuage, NY, USA) with the following program: 2 min at 95°C, 32 cycles of 20 s at 61°C, 15 s at 68°C, 15 s at 95°C, and a final extension for 2 min at 68°C. Negative controls were included during each run and contained all components of the reaction except cDNA template, which was replaced with DEPC treated nuclease-free water (Ambion Life Technologies).
Table 1.
GenBank accession number, sense sequence, and antisense sequence of primers used for qRT-PCR analysis.
| Gene | Accession Number |
Sense Sequence | Antisense Sequence |
|---|---|---|---|
| rps18 | ES608249.1 | 5′-GCGTGACGATTTGGAACGCTTGAA-3′ | 5′-TTCTTGGATACACCGACAGTGCGA-3′ |
| cecropin | ES608437 | 5′-GGACAAAGTGAAGCTGGATGGTTG-3′ | 5′-GCTGGGCCACACCAATAGTTTGAA-3′ |
| defensin | ES608345 | 5′-AAATTTCGTCCATGGAGCTGACGC-3′ | 5′-ACCGCTCAACAAATCGCAAGTAGC-3′ |
| diptericin | ES608652 | 5′-AGTGCAACATTTGTGGTTGCCGAC-3′ | 5′-GCCATAACCTGCTGTGGCATCAAA-3′ |
| lysozyme | DQ384635 | 5′-TCGAATGGCTCCAACGATTACGGT-3′ | 5′-TCCAGCCTTGTTGGGACTTGATCT-3′ |
For relative expression analysis, CT values for calibrator conditions (i.e. flies fed LBAK broth) and treatment groups (i.e. flies fed bacteria) were analyzed using REST-MCS® software and Pairwise Fixed Reallocation Randomization Tests® (Pfaffl, 2002). Log2 fold-change in expression ratios of the target genes cecropin, defensin, diptericin and lysozyme (cec, def, dpt, and lyz) were compared to the calibrator condition using the reference gene rps18. In addition, the effect of time or tissue (head, gut, carcass) on effector expression was analyzed. A Shapiro-Wilk test was performed and determined that expression ratios for cec and dpt were normally distributed (P>0.05) while def and lyz were not normally distributed (P<0.05). Therefore, ANOVA with Tukey-Kramer post hoc tests were used for cec and dpt analyses, and Kruskal-Wallis with Dunn’s test was used for def and lyz analyses. All statistical tests were performed using JMP9® (SAS Institute Inc.).
Immunofluorescent detection of AMPs and lysozyme in GFP-ECO157-fed flies
Flies (total n = 40 in two biological replicates) were fed GFP-ECO157 (mean CFU ± SE: 2.36 ± 0.3 × 105) as described above. At 2, 4, 6, and 12 h PI, the alimentary canal was removed (n = 5 flies per time point) and fixed in 4% paraformaldehyde for 2 h. Tissues were dehydrated through a graded series of alcohols (50%–100%), cleared in Citrisolv™ (Fisher Scientific), pooled by time point and embedded in Paraplast® Plus (Fisher Scientific). Serial sections (5 μm) were affixed to slides (Superfrost®, Fisher Scientific) and rehydrated and blocked with StartingBlock™ T20 Blocking Buffer (Thermo Scientific) for one hour. Slides were incubated individually in one of four polyclonal antibodies overnight at room temperature (RT) (Genscript, Piscataway, NJ, U.S.A.) at the following concentrations in 0.1% bovine serum albumin (BSA): rabbit anti-cecropin (20 μg/ml), rat anti-lysozyme (43.2 μg/ml), chicken anti-defensin (5 μg/ml), mouse anti-diptericin (6.69 μg/ml). Slides were washed in PBS (pH 7.4) for 5 min at RT and incubated with appropriate Alexa Fluor® (Invitrogen) fluorescent secondary antibodies (2 μg/ml) overnight at RT: Alexa Fluor® 568 goat anti-rabbit, Alexa Fluor® 568 goat anti-rat, Alexa Fluor® 488 goat anti-chicken, or Alexa Fluor® 488 goat anti-mouse. Technical controls consisted of at least two slides per time point that were incubated with secondary antibodies only. Biological controls were adult flies (n=10) that were individually-fed 2 μl of sterile LBAK broth and dissected at 5 h PI and processed as above. Images were captured using a Leica DFC420 microscope camera (Leica Microsystems Ltd.).
Results
Enumeration of GFP-ECO157 from house flies
Culture recovery of GFP-ECO157 from whole house flies revealed a 96.3% reduction in fly bacterial load from 0 to 12 h PI, decreasing from mean CFU ± SE fed: 2.65 ± 0.9 × 106 to 9.83 ± 3.1 × 104 (n = 12), respectively (Fig. 1). The percentage of the fed CFU remaining in flies steadily declined over this time interval: at 2 h PI, 56% of the initial dose was recovered (mean CFU ± SE: 1.38 ± 0.3 × 106, n = 16); at 4 h PI, 35% of initial dose (mean CFU ± SE: 9.53 ± 2.5 × 105, n = 15); at 6 h PI, 18% of initial dose (mean CFU ± SE: 4.9 ± 1.1 × 105, n = 14); at 12 h PI, 3.7% of the initial dose (9.83 ± 3.1 × 104, n = 12). The mean CFUs recovered from flies at 12 h PI differed from the amount fed as well as the mean CFUs recovered at 2 and 4 h PI (P ≤ 0.0097), but other time points did not differ significantly from one another or the amount fed in the droplet (P > 0.05). GFP-ECO157 was not recovered from control flies fed sterile 5% sucrose. A total of 17 flies across all time points and replicates had colony counts outside the countable range (i.e., 30–300 CFU). Of those, 3 were above the countable range and 14 below the countable range. These flies were not included in the enumerations above. Additionally, across all replicates 2 flies died during the course of the experiment, and were not included in the analyses.
Figure 1. Recovery of GFP-Escherichia coli O157:H7 from the house fly.
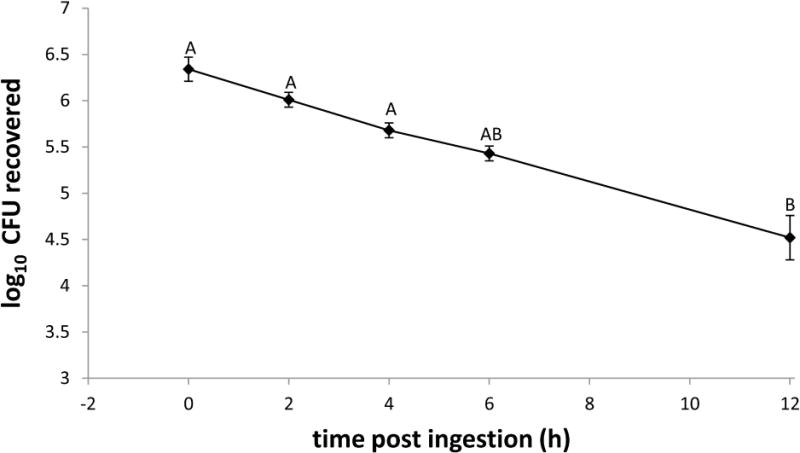
Flies (total n = 76 in four biological replicates) were fed an average of 2.65 ± 0.9 × 106 CFU (mean ± SE) GFP-expressing E. coli O157:H7. Mean recoveries from whole house flies are shown. Different letters denote statistical significance difference (P < 0.05). Error bars are standard error.
GFP-ECO157 visualization in the house fly gut
Although intact GFP-ECO157 persisted in the alimentary canal up to 12 h PI, as soon as 2 h PI, aggregates of immobilized, yet intact, bacteria were visible in the crop and midgut (Fig. 2A; Table 2). In the midgut, bacteria were confined within the PM (Fig. 2). Immotile, intact cells also were observed in the rectum as soon as 2 h PI (Table 2). At 4 h PI, both free GFP (likely due to lysis of GFP-ECO157) and intact, immotile bacteria were observed in food boluses in the midgut of all flies (Fig. 2B; Table 2). Large numbers of intact cells were observed in the crop in over 50% of flies, but all bacteria were immotile.
Figure 2. GFP-Escherichia coli O157:H7 in the alimentary canal of the house fly.
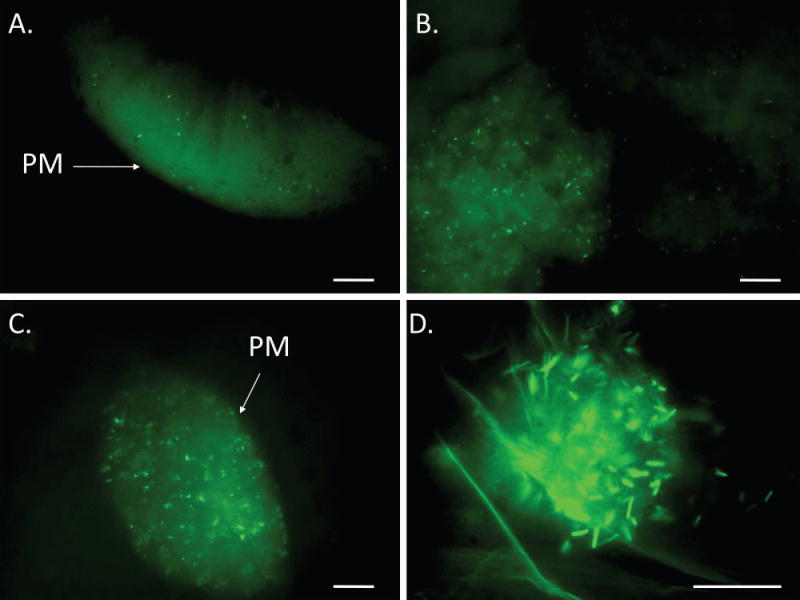
House flies (total n = 28 in three biological replicates) were fed 1.8 ± 0.9 × 106 CFU (mean ± SE) GFP-expressing E. coli O157:H7. Bacterial cells (green rods) were visible in the midgut at all time points including 2 h PI (A), 4 h PI (B), and 6 h PI (C). Clumps of intact cells were visible in the crop at all time points including 12 h PI (D) in one replicate. PM, peritrophic matrix. Scale bar is 10 μm.
Table 2.
Temporal location of GFP-E. coli O157:H7 in the house fly alimentary canal. Time point is hours after ingesting bacteria (fed amount was 1.8 ± 0.9 × 106 CFU (mean ± SE)), n is the sample size at that time point, location of bacteria indicates the number of flies with bacteria in the described region at that time point as observed using epifluorescence microscopy (Fig. 2).
| Location of Bacteria | |||||
|---|---|---|---|---|---|
|
| |||||
| Time point (h PI) |
n | Crop | Midgut | Hindgut | Rectum |
| 2 | 8 | 5 | 7 | 0 | 1 |
| 4 | 8 | 3 | 8 | 0 | 0 |
| 6 | 8 | 4 | 8 | 4 | 4 |
| 12 | 4 | 2 | 0 | 2 | 2 |
At 6 h PI, GFP-ECO157 were visible throughout the midgut of all flies and in the crop, hindgut and/or rectum of approximately 50% of flies (Fig. 2C; Table 2). When compared to earlier observations, fewer bacteria were visible in the gut at 6 h PI, and all bacteria were nonmotile, irrespective of location (Table 2). To determine the fate of GFP-ECO157 in house flies past 6 h PI, one biological replicate was extended to 12 h. At this time point, no GFP-ECO157 were observed in the midgut of any flies (n=4), and only small numbers of immotile bacteria were visible in the hindgut and rectum of two flies, as well as the crop (Fig. 2D; Table 2).
Analysis of temporospatial expression of AMPs and lysozyme by qRT-PCR
qRTPCR analysis of AMP and lysozyme expression revealed that the most immune-responsive tissues in flies fed GFP-ECO157 were the head and salivary glands. In these tissues, cec and lyz were significantly upregulated from baseline expression levels at all time points (P ≤ 0.05; Fig. 3), and likewise def and dpt were significantly upregulated at 2, 6 and 12 h PI (P < 0.05) but not at 4 h PI (P > 0.05). Interestingly, AMP and lyz expression in the gut and carcass was not significantly different from baseline levels except for dpt in the gut at 6 h PI (Fig. 3, Panel C; P < 0.05).
Figure 3. Expression of antimicrobial peptides and lysozyme in house flies after ingestion of GFP-Escherichia coli O157:H7.
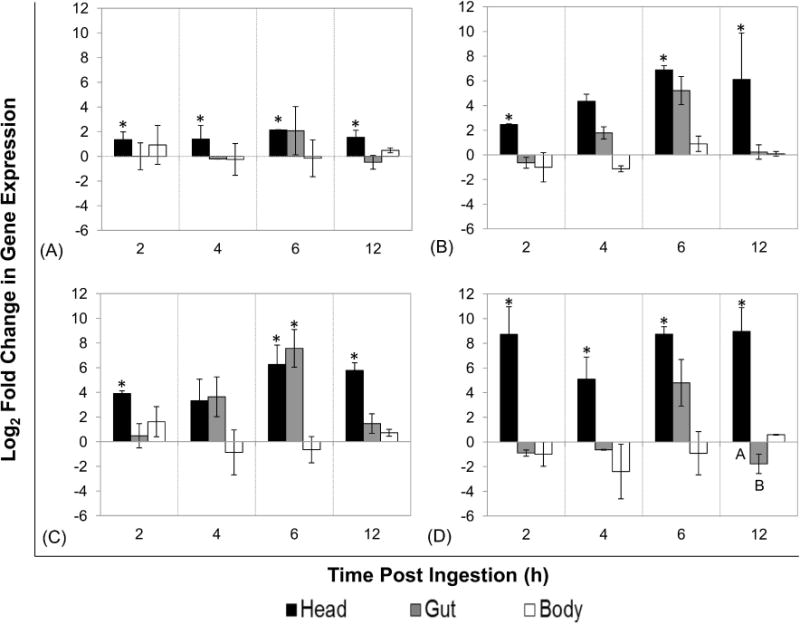
Flies (total n = 95 in three biological replicates) were fed GFP-expressing E. coli O157:H7 (mean CFU ± SE: 1.40 ± 0.30 × 106) and aseptically dissected at 2, 4, 6, and 12 h post-ingestion (n=7–8 per time point) to separate heads (with salivary glands), guts (i.e. proventriculus to rectum), and carcasses. qRT-PCR analysis was used to determine relative tissue-specific expression of cecropin (A), defensin (B), diptericin (C), and lysozyme (D) in house fly tissues (pooled per tissue within replicate). Log2 fold change in expression was determined using the REST-MCS® software, with calibration to matched tissues from broth-fed control flies and the rps 18 reference gene. The mean expression of three biological replicates along with standard error is shown. Different letters denote statistical significance (P < 0.04) between tissues within time point. Asterisks denote statistical significance from broth-fed control flies (P < 0.05)
The change in effector gene expression over time was also evaluated. Unexpectedly, there was no significant change in the temporal expression of cec, dpt, def, or lyz after ingestion of GFP-ECO157 within any of the tissues examined (P > 0.05) despite the observed increases above baseline expression. Further, comparative analyses of gene expression across tissues, but within time points, only showed a significant difference in lyz expression between head/salivary glands and gut tissues at 12 h PI (P < 0.04; Fig. 3, Panel D).
Immunofluorescent detection of effector molecules in the house fly gut
Immunofluorescence microscopy of alimentary tissues showed the presence of effector molecules at 2 and 6 h PI in only one of the two replicates examined. Diptericin and lysozyme were both expressed on the apical portion of midgut cells in flies from one replicate at 2 h PI (Fig. S1A, S1C). Lysozyme was detected in the apical and basal portion of the midgut and in the caudal (midgut proximal) region of the proventriculus of flies in one replicate at 6 h PI. Control house flies did not show AMP expression (Fig. S1B), however lysozyme expression was observed in the midgut (Fig. S1D), but not proventriculus of LBAK broth-fed flies.
Discussion
The current study determined that house flies can serve as short-term reservoirs for E. coli O157:H7 by simultaneously monitoring the location and number of GFP-ECO157 within the fly after ingestion. It was found that E. coli O157:H7 formed clumps, adhered to the inner PM, and that many bacteria were lysed in the gut of flies that were fed bacteria. Similar observations of immobilization, clumping and apparent adherence to the PM have been previously observed in flies fed A. hydrophila (McGaughey and Nayduch, 2009). However despite these effects, the current study detected intact GFP-ECO157 persisting in the crop, hindgut, and rectum of some flies up to 12 h post-ingestion, which implies that house flies could feasibly excrete by vomit and defecation.
Consistent with microscopy observations, GFP-ECO157 abundance steadily decreased within flies over the same time interval. These findings are similar to those of Kobayashi et al. (1999) that showed a gradual decrease of bacterial load over 4 days in flies that had been fed a comparable dose E. coli O157:H7. However, in Kobayashi et al. (1999), approximately 104 cells/fly were recovered at 24 h PI, and in our study, a similar amount of bacteria was recovered at 12 h PI. The more rapid clearing of bacteria in the current study may be attributable to the use of teneral flies in our study as opposed to fully mature adult flies in Kobayashi et al. (1999), as well the method of feeding a single droplet as opposed to allowing the flies to feed ad libitum. Since teneral flies tend to have low bacteria counts in the gut, their gut defense, digestive and immune status can likely differ from fully-developed adults. Further, feeding bacteria ad libitum increases the possibility of multiple small ingestion events as opposed to one large ingestion event of a single bolus, and this difference can potentially affect the temporospatial location of bacteria. It is important to point out that mean recoveries in the current study are likely an overestimate, since culture plates that were outside of the countable range (30–300 CFU) were excluded. Also, two flies died during the course of the experiment (probably due to fasting), and this resulted in 19/76 flies being excluded from the data set. Of these 19 flies, 73% had too few colonies to count (<30 CFU).
For the duration of the current study, the mean bacterial load within flies remained well above the potential infectious dose of 10–100 cells (Todar, 2008). Taken in conjunction with previous data (Kobayashi et al., 1999; Sasaki et al., 2000), this indicates that house flies potentially serve as significant reservoirs for this pathogen for at least 12 h. Interestingly, recent work by Schuster et al. (2013) has shown that house fly larvae that fed on E. coli-contaminated substrate carried bacteria trans-stadially, and newly-eclosed flies harbored as many as 50 CFU. The possibility of trans-stadial survival of E. coli O157:H7 deserves further study, as this would complicate the epidemiological picture. A more thorough examination of how larval exposure to bacteria impacts bacterial load, persistence and dissemination in adult house flies is a focus for future studies.
Despite the steady decrease of GFP-ECO157 within the fly after exposure, the current study did not detect a robust effector response against the ingested bacteria. Although effectors were upregulated in some fly tissues, there were no significant changes of cec, dpt, def, or lyz gene expression over time. Similarly there was little observable local immune expression and the gut only showed significant upregulation of dpt at 6 h PI. This was reflected at the protein level with the exception of diptericin being observed at 2 h PI instead of 6 h PI, and the presence of lysozyme at levels equal to that of broth fed controls. This low level of lysozyme expression in adult flies exposed to ubiquitous bacteria has been reported previously (Nayduch and Joyner, 2013). qRT-PCR and immunofluorescence analyses indicate a lack of utilization of the effectors observed here against E. coli O157:H7. This is in contrast to previous experiments with S. aureus-fed flies (Nayduch et al., 2013), which showed significant upregulation of def in response to bacterial ingestion. Similarly, flies fed P. aeruginosa significantly upregulated AMPs on both the mRNA and protein level in the gut (Joyner et al., in press). In this study, the steady decrease of ingested GFP-ECO157 can be a result of numerous other house fly gut defenses, including: lectin-mediated immobilization and/or adherence to the PM, peristalsis and excretion, shifting pH and osmolarity, production of reactive oxygen species (ROS), and/or lysis by other AMPs or digestive enzymes (Peters et al., 1983; Terra et al., 1988; Ryu et al., 2006; Lemaitre and Hoffmann, 2007).
In contrast to the gut, the house fly head and salivary gland tissues showed significant upregulation of AMPs and lyz at almost all time points post-ingestion. Production of AMPs in the salivary glands has previously been observed in other species such as D. melanogaster (Tzou et al., 2000), Aedes aegypti (Valenzuela et al., 2002), Anopheles gambiae (Dimopoulos et al., 1998) and Stomoxys calcitrans (Wang et al., 2009). Though the current study could not distinguish between expression in the salivary glands and expression in the head (including mouthparts) in this study, it is possible that AMP production from these locations can contribute to the lysis of EC-O157 by being swallowed and passing into the gut and crop. The continued upregulation of AMPs in the salivary glands and head may indirectly indicate persistence of bacteria in the mouthparts of the fly after feeding, as has been previously observed up to at least 24 h post-ingestion (Kobayashi et al. 1999). Further protein-level analyses are needed to detect AMP production from the salivary glands, and to distinguish expression in the salivary glands from expression in the rest of the head or mouth parts.
Supplementary Material
House flies (total n = 40 in two biological replicates) were fed 2.36 ± 0.3 × 105 CFU (mean ± SE) GFP-expressing E. coli O157:H7. Immune effectors (AMPs and lysozyme) were detected in the alimentary canal using custom polyclonal primary antibodies and fluorescently labeled secondary antibodies. Diptericin (Alexa Fluor® 488 goat anti-chicken; green) in the midgut at 2 h PI in a bacteria-fed fly (A) and a control fly (B). Lysozyme (Alexa Fluor® 568 goat anti-Rat; red) in the midgut at 2 h PI in a bacteria-fed fly (C) and control fly (D). Cell nuclei stained with DAPI (blue). L, gut lumen. Scale bar is 10 μm.
Acknowledgments
We thank C. Evett for assistance with qRTPCR optimization. Special thanks to Dr. Brian Weiss at Yale University for the pGFPuv-kanamycin plasmid construct. This work was supported by R15 Academic Research Enhancement Award (AREA) 1R15AI084029-01 from the National Institutes of Health awarded to D.N. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- Callaway TR, Carr MA, Edrington TS, Anderson RC, Nisbet DJ. Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Current Issues in Molecular Biology. 2009;11:67–79. [PubMed] [Google Scholar]
- Charroux B, Royet J. Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly. 2010;4:40–47. doi: 10.4161/fly.4.1.10810. [DOI] [PubMed] [Google Scholar]
- Dang XL, Wang YS, Huang YD, Yu XQ, Zhang WQ. Purification and characterization of an antimicrobial peptide, insect defensin, from immunized house fly (Diptera: Muscidae) Journal of Medical Entomology. 2010;47:1141–5. doi: 10.1603/ME10016. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. The EMBO Journal. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk TK, Knight R, Gilman RH, Cranfield MR. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes and Infection / Institut Pasteur. 2001;3:231–235. doi: 10.1016/s1286-4579(01)01371-5. [DOI] [PubMed] [Google Scholar]
- Greenberg B. Persistence of bacteria in the developmental stages of the housefly. IV. Infectivity of the newly emerged adult. The American Journal of Tropical Medicine and Hygiene. 1959;8:618–622. doi: 10.4269/ajtmh.1959.8.618. [DOI] [PubMed] [Google Scholar]
- Greenberg B, Kowalski JA, Klowden MJ. Factors affecting the transmission of salmonella by flies: natural resistance to colonization and bacterial interference. Infection and Immunity. 1970;2:800–809. doi: 10.1128/iai.2.6.800-809.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner C, Mills MK, Nayduch D. Pseudomonas aeruginosa in Musca domestica L.: temporospatial examination of bacteria population dynamics and house fly antimicrobial responses. PLOS ONE. 2013 doi: 10.1371/journal.pone.0079224. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Sasaki T, Saito N, Tamura K, Suzuki K, Watanabe H, Agui N. Houseflies: not simple mechanical vectors of enterohemorrhagic Escherichia coli O157:H7. The American Journal of Tropical Medicine and Hygiene. 1999;61:625–629. doi: 10.4269/ajtmh.1999.61.625. [DOI] [PubMed] [Google Scholar]
- Lambert J, Keppi E, Dimarcq JL, Wicker C, Reichhart JM, Dunbar B, Lepage P, Van Dorsselaer A, Hoffmann J, Fothergill J, et al. Insect immunity: isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:262–266. doi: 10.1073/pnas.86.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ. Peritrophic matrix structure and function. Annual Review of Entomology. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review of Immunology. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Madigan M. Microbial Growth. In: Madigan M, Martinko J, Dunlap P, Clark D, editors. Brock Biology of Microorganisms. Pearson Education, Inc; San Francisco, CA: 2009. pp. 154–155. [Google Scholar]
- McGaughey J, Nayduch D. Temporal and spatial fate of GFP-expressing motile and nonmotile Aeromonas hydrophila in the house fly digestive tract. Journal of Medical Entomology. 2009;46:123–130. doi: 10.1603/033.046.0116. [DOI] [PubMed] [Google Scholar]
- Moriya K, Fujibayashi T, Yoshihara T, Matsuda A, Sumi N, Umezaki N, Kurahashi H, Agui N, Wada A, Wantanabe H. Verotoxin-producing Escherichia coli O157:H7 carried by the housefly in Japan. Medical and Veterinary Entomology. 1999;13:214–216. doi: 10.1046/j.1365-2915.1999.00161.x. [DOI] [PubMed] [Google Scholar]
- Nayduch D, Cho H, Joyner C. Staphylococcus aureus in the house fly: temporospatial fate of bacteria and expression of the antimicrobial peptide defensin. Journal of Medical Entomology. 2013;50:171–178. doi: 10.1603/me12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayduch D, Joyner C. Expression of lysozyme in the life history of the house fly (Musca domestica L.) Journal of Medical Entomology. 2013;50:847–852. doi: 10.1603/me12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayduch D, Noblet GP, Stutzenberger FJ. Vector potential of houseflies for the bacterium Aeromonas caviae. Medical and Veterinary Entomology. 2002;16:193–198. doi: 10.1046/j.1365-2915.2002.00363.x. [DOI] [PubMed] [Google Scholar]
- Peters W, Kolb H, Kolb-Bachofen V. Evidence for a sugar receptor (lectin) in the peritrophic membrane of the blowfly larva, Calliphora erythrocephala Mg. (Diptera) Journal of Insect Physiology. 1983;29:275–280. [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Zhao X, Wang J. Molecular characterization and expression analysis of a chicken-type lysozyme gene from housefly (Musca domestica) Journal of Genetics and Genomics. 2009;36:7–16. doi: 10.1016/S1673-8527(09)60002-3. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, Lee DG, Kim J, Lee D, Lee WJ. An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. The EMBO Journal. 2006;25:3693–3701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Kobayashi M, Agui N. Epidemiological potential of excretion and regurgitation by Musca domestica (Diptera: Muscidae) in the dissemination of Escherichia coli O157: H7 to food. Journal of Medical Entomology. 2000;37:945–949. doi: 10.1603/0022-2585-37.6.945. [DOI] [PubMed] [Google Scholar]
- Schuster GL, Donaldson JR, Buntyn JO, Duoss HA, Callaway TR, Carroll JA, Falkenberg SM, Schmidt TB. Use of bioluminescent Escherichia coli to determine retention during the life cycle of the housefly, Musca domestica (Diptera: Muscidae, L) Foodborne Pathogens and Disease. 2013;10:442–447. doi: 10.1089/fpd.2012.1326. [DOI] [PubMed] [Google Scholar]
- Terra WR. Physiology and biochemistry of insect digestion: an evolutionary perspective. Brazilian Journal of Medical and Biological Research. 1988;21:675–734. [PubMed] [Google Scholar]
- Terra WR, Ferreira C. Insect digestive enzymes: properties, compartmentalization and function. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1994;109:1–62. [Google Scholar]
- Todar K. Pathogenic E. coli. Todar’s Online Textbook of Bacteriology. 2008 Retrieved 8/30/2013 from http://www.textbookofbacteriology.net/e.coli_4.html.
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffman JA, Imler JL. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- Hawley JE, Penner LR, Wedberg SE, Kulp W. The role of the house fly, Musca domestica, in the multiplication of certain enteric bacteria. The American Journal of Tropical Medicine and Hygiene. 1951;31:572–582. doi: 10.4269/ajtmh.1951.s1-31.572. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IMB, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochemistry and Molecular Biology. 2002;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Wang X, Ribeiro JMC, Broce AB, Wilkerson MJ, Kanost MR. An insight into the transcriptome and proteome of the salivary gland of the stable fly, Stomoxys calcitrans. Insect Biochemistry and Molecular Biology. 2009;39:607–614. doi: 10.1016/j.ibmb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LS. The Housefly, its Natural History, Medical Importance, and Control. Comstock Publishing Co; Ithaca, N.Y.: 1951. [Google Scholar]
- Zurek L, Denning SS, Schal C, Watson DW. Vector competence of Musca domestica (Diptera: Muscidae) for Yersinia pseudotuberculosis. Journal of Medical Entomology. 2001;38:333–335. doi: 10.1603/0022-2585-38.2.333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
House flies (total n = 40 in two biological replicates) were fed 2.36 ± 0.3 × 105 CFU (mean ± SE) GFP-expressing E. coli O157:H7. Immune effectors (AMPs and lysozyme) were detected in the alimentary canal using custom polyclonal primary antibodies and fluorescently labeled secondary antibodies. Diptericin (Alexa Fluor® 488 goat anti-chicken; green) in the midgut at 2 h PI in a bacteria-fed fly (A) and a control fly (B). Lysozyme (Alexa Fluor® 568 goat anti-Rat; red) in the midgut at 2 h PI in a bacteria-fed fly (C) and control fly (D). Cell nuclei stained with DAPI (blue). L, gut lumen. Scale bar is 10 μm.


