Abstract
The epicardium supports cardiomyocyte proliferation early in development and provides fibroblasts and vascular smooth muscle cells to the developing heart. The epicardium has been shown to play an important role during tissue remodeling after cardiac injury, making access to this cell lineage necessary for the study of regenerative medicine. Here we describe the generation of epicardial lineage cells from human pluripotent stem cells by stage-specific activation of the BMP and WNT signaling pathways. These cells display morphological characteristics and express markers of the epicardial lineage, including the transcription factors WT1 and TBX18 and the retinoic acid–producing enzyme ALDH1A2. When induced to undergo epicardial-tomesenchymal transition, the cells give rise to populations that display characteristics of the fibroblast and vascular smooth muscle lineages. These findings identify BMP and WNT as key regulators of the epicardial lineage in vitro and provide a model for investigating epicardial function in human development and disease.
The epicardium is essential for proper development of the heart and plays an important role in cardiac recovery during disease. Studies in model organisms have demonstrated that these effects are mediated either through the generation of epicardial-derived cell populations that participate in formation of the heart or through the secretion of paracrine factors by the epicardium that influence the development and proliferation of other cell types in the heart including cardiomyocytes. Given their pivotal role in normal development and disease, the epicardium and derivative cells types will have to be included as essential components of engineered heart tissue that is generated to assess drug responses and to model disease in vitro. Additionally, the ability of epicardial cells to regulate cardiomyocyte proliferation during development can be exploited to develop new strategies for replacing or regenerating functional myocardium for the treatment of cardiovascular disease. The ability to generate unlimited numbers of human pluripotent stem cell (hPSC)-derived epicardial cells through the approach described in this study provides an unprecedented opportunity to develop these applications.
The adult heart comprises three distinct cell populations—the inner endocardium, the centre myocardium and the outer epicardium—which arise during specific stages of embryonic development. The endocardial and myocardial lineages develop during the earliest stage of cardiac development in a structure known as the cardiac crescent1. The bi-lineage crescent subsequently fuses to form the heart tube, which undergoes chamber specification and looping, giving rise to the four-chambered heart. The epicardium develops during the looping stage and is derived from a distinct structure known as the proepicardial organ, which lies proximal to the heart along the septum transversum2. As the proepicardial organ buds off from the septum transversum, it migrates to and envelopes the heart to form an outer epithelial layer, the epicardium, at approximately embryonic day (E) 9.5 of mouse development3. The epicardium then undergoes an epithelial-to-mesenchymal transition (EMT) in response to various signals, including TGFβ14, 5, Wnt6, retinoic acid (RA)6, FGF7, and PDGF8, to give rise to cardiac fibroblasts and coronary vascular smooth muscle cells that invade the myocardial layer and contribute to the structural and vascular populations of the developing heart. These fibroblasts and vascular smooth muscle cells, known as epicardial-derived cells (EPDCs), constitute a substantial proportion of the non-cardiomyocyte population within the myocardial layer9. In addition to generating these cell types, the epicardium also supports the rapid proliferation of ventricular cardiomyocytes through the production of paracrine factors, including IGF and RA10. This rapid, stage-specific expansion is essential for the generation of compact ventricular myocardium.
At the molecular level, the developing epicardium can be distinguished from the myocardium and endocardium by expression of the transcription factors WT111 and TBX1812 and of the aldehyde dehydrogenase enzyme retinaldehyde dehydrogenase 2 (ALDH1A2), required for the conversion of retinol to RA13, 14. The expression of these genes defines the fetal stage of epicardial development, as their levels decrease with maturation. In the adult epicardium, myocardial infarction leads to upregulation of these genes, cell proliferation and EMT, suggesting that the epicardium is involved in the remodeling process following infarct15,16. Lineage-tracing studies have provided evidence that this activated epicardium generates new cardiomyocytes along with fibroblasts and vascular smooth muscle cells, indicating that it may contribute to the development of new myocardium17-19. However, the extent to which an epicardium-to-cardiomyocyte transition occurs is unclear.
Our understanding of epicardial lineage development and function is derived almost entirely from studies on model organisms, as access to fetal human heart tissue is limited. Studies with epicardium isolated from the adult human heart showed that the cells rapidly undergo EMT in culture, preventing detailed studies on the epithelial cell population20. The generation of epicardium in vitro from hPSCs would overcome issues of accessibility and provide an unlimited source of human cells for functional studies in vitro and in experimental models in vivo. In this study, we have addressed this issue and demonstrate that the combination of BMP and Wnt signaling promotes the generation of a WT1+TBX18+ epicardial cell-like population at the expense of cardiomyocytes from PDGFRA+ mesoderm. Following passage, these cells mature and form epithelial-like sheets with tight junctions as demonstrated by the expression of ZONA OCCLUDENS-1 (ZO1). Along with Z01, the cells upregulate expression of ALDH1A2 and exhibit aldehyde dehydrogenase activity, an indication of their ability to synthesize retinoic acid. Finally, we show that when induced with TGFB1 and BFGF, the epicardial-like cells undergo EMT and give rise to cells that display characteristics of smooth muscle cells and fibroblasts.
Results
BMP dose-dependently specifies the cardiomyocyte lineage
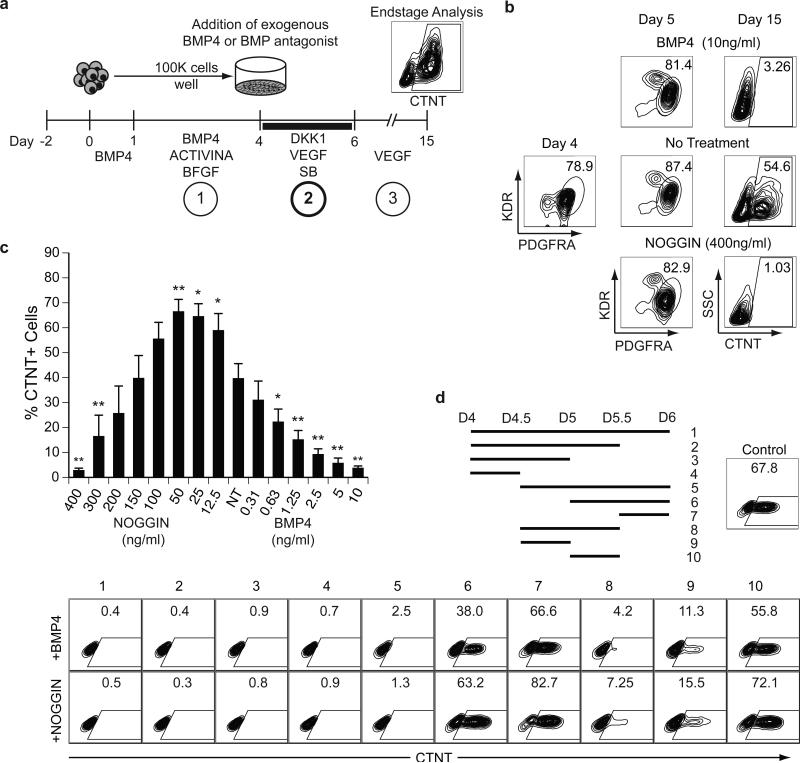
The differentiation of hPSCs towards the cardiomyocyte (CM) lineage progresses through three stages; 1) the induction of cardiovascular mesoderm defined by the expression of KDR and PDGFRA, 2) the specification of the cardiovascular mesoderm to a CM fate and 3) the maturation of the specified progenitors to functional CMs (Fig. 1a). While NODAL/ACTIVINA and BMP4 are known to be key regulators in the generation of cardiac mesoderm during the first stage of differentiation, the pathways controlling the specification towards the CM lineage during the second stage are ill-defined21. To investigate the specification step, we established a protocol that enabled us to easily manipulate signaling pathways during this stage of development in hESC differentiation cultures (Fig. 1a). With this approach, KDR+PDGFRA+ cardiovascular mesoderm is induced in embryoid bodies (EBs) with optimal concentrations of ACTIVINA (ACTA) and BMP4 as previously described21. On day (D) 4 of differentiation, the EBs are dissociated and the cells are plated as a monolayer for the specification stage and cultured in the presence of different signaling pathway agonists and antagonists for 48 hours. The cultures are then maintained in media containing VEGF and analyzed at D15 for the presence of cardiac troponin T (CTNT)+ CMs by flow cytometry.
Figure 1. BMP specifies the differentiation of cardiomyocytes from hESC-derived mesoderm.
(a) Scheme of the protocol used to differentiate hESCs towards the CM lineage highlighting the three main stages of development: 1) mesoderm induction, 2) cardiovascular specification and 3) maturation. Cells from ACTIVINA/BMP4-induced D4 EBs are plated as a monolayer on gelatin coated wells. The BMP pathway was manipulated for a 48-hour period (D4-D6) in the presence of VEGF (5 ng/ml), SB (5.4 μM) and DKK1 (150ng/ml). Following specification, the cultures were maintained in VEGF (5ng/ml) for 9 days and then analyzed for the presence of CTNT+ cells by flow cytometry.
(b) Flow cytometric analyses of the frequency of KDR+PDGFRA+ cells in D4 and D5 mesoderm populations and of CTNT+ cells in D15 populations generated from D4 mesoderm treated with: no factors (no treatment), BMP4 (10ng/ml) or NOGGIN (400ng/ml).
(c) Flow cytometric analyses of the frequency of CTNT+ cells in D15 populations generated from D4 mesoderm treated with the indicated amounts of BMP4 or NOGGIN. NT = no treatment. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 compared to NT as analyzed by Student's T-test. (d) Scheme indicating the time intervals of BMP4 or NOGGIN treatment between D4 and D6. Ten different intervals were evaluated. On D15 the resulting populations were analyzed for the presence of CTNT+ cells by flow cytometry. Control cultures were not treated with either the agonist or antagonist during the 48-hour period.
As our previous studies showed that CMs develop from KDR+PDGFRA+ mesoderm in the presence of the WNT inhibitor DKK1 and the ACTIVIN/NODAL/TGFB inhibitor SB-431542 (SB)21, these pathway antagonists were included in the culture during the specification step. Given that BMP signaling is required for cardiovascular development from mesoderm in the early embryo22, and can modulate cardiomyocyte development from hPSC-derived cardiac progenitor cells21, we focused our initial effort on further exploring the role of this pathway in the specification of the cardiovascular lineage. Manipulation of the pathway had a profound effect on cardiogenesis, as addition of BMP4 or the BMP-specific inhibitor NOGGIN during this 48 hour window completely blocked CM development as shown by the lack of CTNT+ cells at D15 (Fig. 1b). The generation of KDR+PDGFRA+ mesoderm at D5 of differentiation was not affected by these manipulations. The lack of CM development was not due to an increase in cell death as similar numbers of cells were detected in each group at D15 (Supplementary Fig. 1a). To investigate the requirement for BMP signaling during the specification of CMs, we carried out a titration of NOGGIN and BMP4 between D4 and D6 of differentiation. A clear dose dependency for BMP signaling was observed, with optimal CM specification occurring in cultures treated with levels of NOGGIN ranging from 12.5 and 50 ng/ml (Fig. 1c). Some inter-experiment variation was observed with respect to the optimal NOGGIN concentrations required for the specification of CMs and for blocking cardiogenesis. This variation typically coincided with lot changes in reagents (data not shown).
To define in more detail the temporal aspects of CM specification, we manipulated BMP signaling at different 12-hour intervals between D4 and D6 of differentiation (Fig. 1d; top panel, intervals 1-10). The untreated control cultures underwent efficient CM specification, as indicated by the high proportion of CTNT+ cells detected at D15. The inhibitory effects of BMP4 and NOGGIN were most pronounced when these factors were added between D4 and D5 of culture (groups 1-5, 8, 9). If addition of the BMP agonist/antagonist was delayed to D5 or beyond, CMs were detected in the resulting cultures (groups 6, 7 and 10). Addition of the agonist and antagonist for a 12-hour period during the D4-5 time frame was sufficient to inhibit CM development (groups 4, 8 and 9). Taken together these findings demonstrate that an optimal concentration of BMP4 is required for CM specification at D4 of differentiation and that the duration of this BMP-dependent stage is approximately 24 hours.
BMP4 treatment specifies a WT1+ epicardial population
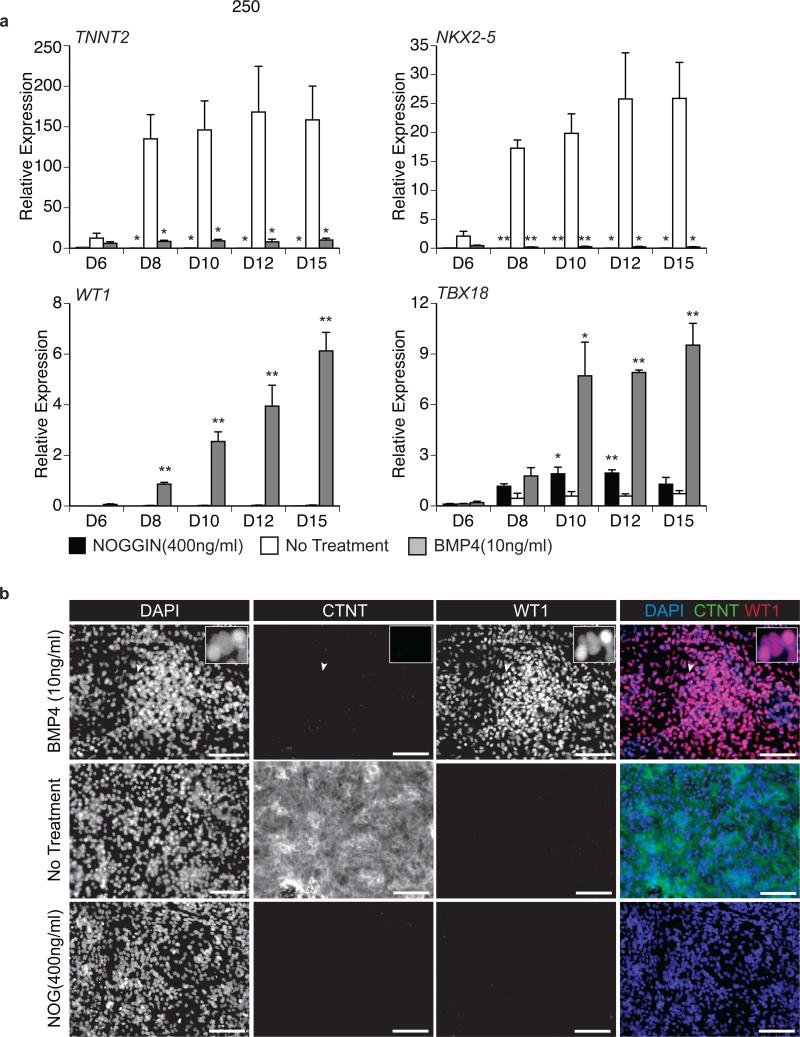
The fact that cell numbers were not significantly influenced by the manipulation of the BMP pathway indicated that other cells types, possibly of other cardiovascular lineages, were generated in the presence or absence of BMP signaling. To begin to characterize these cell populations, we assessed gene expression patterns indicative of different cardiovascular cell types, as well of other mesoderm-derived lineages, by quantitative real-time PCR (qRT-PCR) (Fig. 2a, Fig. 3b and not shown). As expected, genes associated with cardiomyocyte development, including TNNT2 and the cardiac transcription factor NKX2-5, were only expressed in the untreated control cultures. Notably, two epicardial markers, WT1 and TBX18, were expressed exclusively (WT1) or predominantly (TBX18) in the cultures treated with BMP4, indicating that these cells may represent the developing epicardial lineage (Fig. 2a). Immunostaining confirmed the qRT-PCR findings and demonstrated that the BMP4 treated cells expressed nuclear WT1 protein (Fig. 2b). WT1 was not detected in the NOGGIN treated or untreated cells, indicating that it was specifically upregulated in the BMP4-treated group. Immunostaining for CTNT was only observed in untreated cultures (Fig. 2b), confirming the qRT-PCR and FACS analyses.
Figure 2. BMP4 treatment specifies WT1+ cells.
(a) qRT-PCR-based expression analyses of the cardiac genes TNNT2 and NKX2-5 and the epicardial genes WT1 and TBX18 on the indicated days (D) of culture from cell populations generated by treatment with BMP4 (10ng/ml), NOGGIN (400ng/ml) or no additional factors. Values shown are relative to the housekeeping gene TBP. Error bars represent standard error of the mean from the values of three independent experiments (N=3); *P≤0.05, **P≤0.01 compared to non-treated cells as analyzed by Student's T-test.
(b) Fluorescent immunostaining for the presence of CTNT and WT1 protein in D15 populations treated with no additional factors, BMP4 (10ng/ml) or NOGGIN (400ng/ml). DAPI staining shows cell nuclei. Insert is high magnification showing nuclear staining of WT1 where the region of interest is indicated by the white arrowhead. Scale bars represent 100μm.
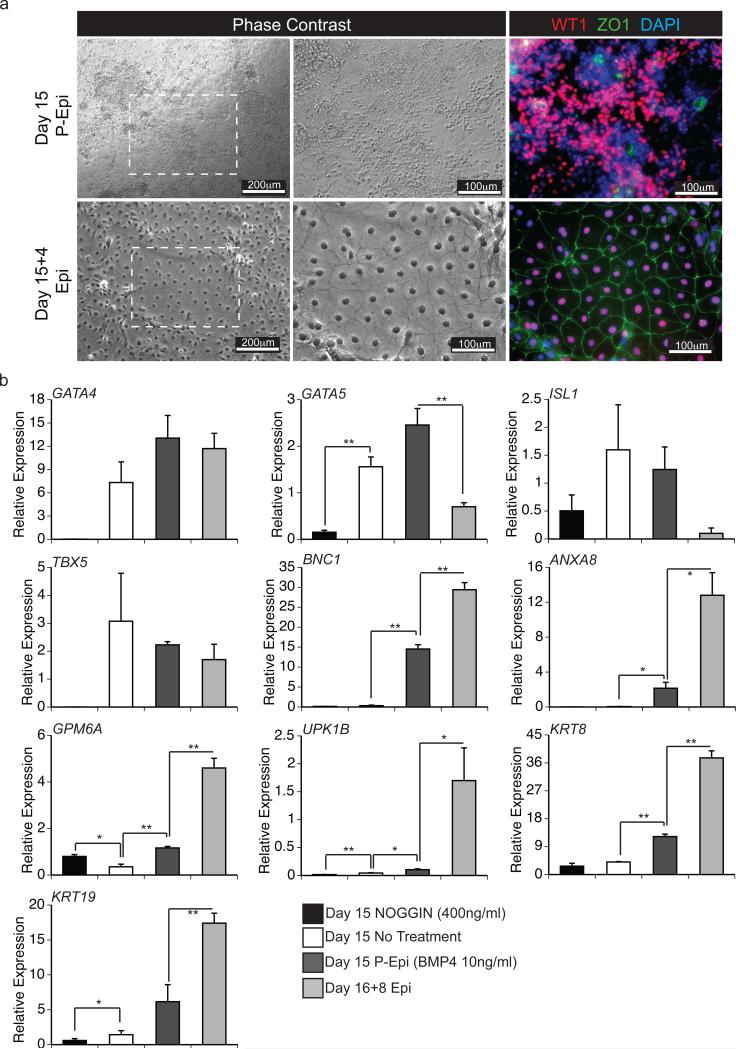
Figure 3. WT1+ cells generate epithelial sheets following passage.
(a) Phase contrast microscopy and fluorescent immunostaining showing morphology of D15 P-Epi and D15+4 Epi populations and the presence of ZO1 (green) and WT1 (red) protein. DAPI (blue) staining shows cell nuclei. Scale bar are as indicated. White dashed box in first panel depicts region of view at higher magnification in subsequent panels.
(b) qRT-PCR-based expression analyses of the indicated genes in D15 populations generated from untreated cells (control) or cells treated with NOGGIN (400ng/ml), or BMP4 (P-Epi, 10ng/ml) as well as in D16+8 Epi cells derived from the BMP-4 treated PEpi cells. Values shown are relative to the housekeeping gene TBP. Error bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 compared to non-treated cells or D15 P-Epi cells as analyzed by Student's T-test.
In vivo, epicardial cells form an epithelial layer that surrounds the developing heart. In addition to their distinct cobblestone morphology, epithelial cells in culture are characterized by their ability to form tight junctions that can be monitored by the presence of ZONA OCCLUDENS 1 (ZO1) along the cell membrane4. At D15 of culture the WT1-expressing cultures did not display the cobblestone morphology typical of cultured epithelial cells and ZO1 expression was not readily observed along the cell borders (Fig. 3a). However, following passage and culture in a larger format (from a 96-well to a 6-well plate) for 4 days, WT1+ cells expanded to generate a confluent monolayer with a cobblestone morphology (Fig. 3a). These cells maintained expression of WT1 and now expressed ZO1 along well-defined cell borders. Given these characteristics, the D15 pre-passaged WT1+ cells will be referred to as pro-epicardial (PEpi) cells and the post-D15 passaged WT+ cells as epicardial (Epi) cells.
Molecular analyses revealed that the P-Epi cells expressed the cardiogenic genes GATA4, GATA5, ISL1 and TBX5 at levels similar those found in the cardiomyocytes (Fig. 3b). With passage, expression of ISL1 and GATA5 was downregulated. These patterns largely reflect those in the developing epicardium in the mouse23-26. Through microarray analyses, Bochmann et al.27 identified sets of genes that were expressed at significantly higher levels in the epicardium than in the myocardium of the adult mouse. We analysed six genes from the set that showed the greatest differential in their study (BNC1, ANXA8, GPM6A, UPK1B, KRT8, and KRT19) and found that the expression of all were siginficantly upregulated in the passaged Epi cells compared to the P-Epi cells and CMs (Fig. 3b).
To further characterize the P-Epi and Epi cell populations, we analyzed them for the expression of various surface antigens by flow cytometry. The expression patterns of these populations were compared to that of D15 CMs (Supplementary Fig. 1b). The CMs and both Epi populations expressed podoplanin (PDPN), a transmembrane glycoprotein thought to be associated with cell migration and found on developing mouse cardiomyocytes and on adult mouse epicardium27, 28. Notably, both Epi populations were also positive for SIRPA, a receptor we have previously shown to be expressed on hPSC-derived and fetal CMs29. The Epi populations, but not the CMs, expressed the mesenchymal/fibroblast marker CD90, although the levels appeared to decrease with passage. PDGFRB, expressed on embryonic epicardium in the mouse, was detected at low levels in both Epi populations30. In contrast, the mesoderm progenitor marker PDGFRA was not expressed on any of the populations, indicating that it is downregulated with lineage specification. None of the populations expressed CD31, VECADHERIN or CKIT (not shown). Taken together with the above molecular analyses, these findings provide strong evidence that the hPSC-derived Epi populations represent the human epicardial lineage.
To demonstrate that the WT1-expressing cells develop from the mesodermal population present at D4, we isolated the PDGFRA+ (mesoderm) and PDGFRA- (nonmesoderm) fractions at this time point by FACS and cultured the cells either without further treatment or with BMP4 for 48 hours (Supplementary Fig. 2a). Cultures were maintained until D15 as illustrated in Figure 1a. Flow cytometry and immunostaining revealed that CTNT+ CMs (Supplementary Fig. 2b) and WT1+ cells (Supplementary Fig. 2c) were generated only from the PDGFRA+ population indicating that these cells types are of mesodermal origin. Gene expression analysis confirmed these findings and showed that that expression of WT1 and additionally TBX18 were only detected in the BMP4-treated PDGFRA+ mesoderm-derived population (Supplementary Fig. 2d).
WNT signaling co-regulates cardiac lineage specification
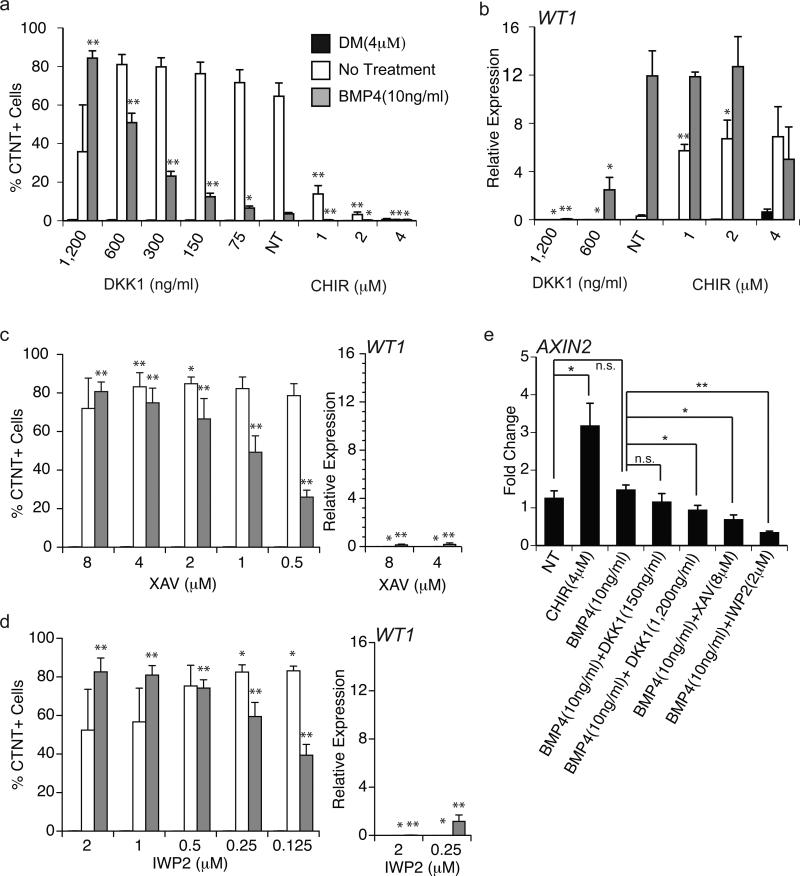
Our findings that BMP signaling can specify the CM and P-Epi lineages in the presence of WNT and ACTIVIN/NODAL/TGFB inhibitors suggest that these pathways do not play a role at this stage of cardiovascular development. This interpretation is not, however, in line with the observations that the hearts of Dkk1−/−Dkk2−/− null mice have an increased thickness of the epicardium and a thinner myocardium compared to wild type littermates. These findings suggest that Wnt signaling does, in fact, play some role in the development of this lineage31. To reconcile these differences, we further manipulated WNT signaling at this stage, specifically focusing on inhibition of the pathway by addition of increased amounts of DKK1, or the small molecule antagonists XAV939 (XAV) and IWP2. These antagonists were added to cultures containing either BMP4 (10ng/ml), the small molecule BMP inhibitor dorsomorphin32 (4μM; DM, in place of NOGGIN) or no other factors (endogenous BMP). Increasing the concentration of DKK1 in the presence of BMP4 promoted the development of CMs at the expense of P-Epi cells as indicated by the lack of WT1 expression in the population treated with 1,200 ng/ml of the antagonist (Fig. 4a and b). The addition of XAV or IWP2 had similar effects to that of high concentrations of DKK1 (Fig. 4c and d). The CM potential of the DM-treated and non-treated cultures were largely unaffected by these manipulations (Fig. 4a, c and d). As expected, activation of the canonical WNT pathway by the addition of the small molecule WNT agonist CHIR99021 (CHIR) inhibited CM development in the endogenous BMP control (Fig. 4a). Western blot analyses showed that the untreated population (endogenous BMP) contains pSMAD1/5/8 that is inhibited by the addition of DM, consistent with the interpretation that the BMP pathway is active in the cells in the absence of added ligand (Supplementary Fig. 3a). The levels of pSMAD1/5/8 were not appreciably affected by the addition of a WNT inhibitor indicating that manipulation of the WNT pathway does not impact BMP/pSMAD signaling. qRT-PCR analyses revealed that the addition of BMP4 did not significantly increase the levels of expression of the WNT/BETA-CATENIN target AXIN2 above that in the untreated population, suggesting that the BMP4 pathway does not directly regulate canonical WNT signaling (Fig. 4e). Addition of IWP2, XAV or high (1,200 ng/ml) but not low (150 ng/ml) concentrations of DKK1 led to a significant reduction in AXIN2 expression, consistent with the observation that these manipulations promoted cardiac development.
Figure 4. BMP and WNT signaling modulate cardiomyocyte and pre-epicardium specification.
(a) Flow cytometric analyses of the frequency of CTNT+ cells in D15 cultures generated from D4 mesoderm treated with BMP4 (10ng/ml), DM (4μM) or no treatment (control) in combination with the indicated amounts of DKK1 or CHIR. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 when compared to the ‘no WNT treatment’ (NT) control in the context of the indicated manipulation of the BMP pathway as analyzed by Student's T-test.
(b) qRT-PCR-based analyses of WT1 expression in D15 cultures generated from D4 mesoderm treated BMP4 (10ng/ml), DM (4μM) or no additional factors (control) in combination with the indicated amounts of DKK1 or CHIR. Values shown are relative to the housekeeping gene TBP. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 when compared to the ‘no WNT treatment’ (NT) control in the context of the indicated manipulation of the BMP pathway as analyzed by Student's T-test.
(c) The frequency of CTNT+ cells determined by flow cytometry and the expression levels of WT1 evaluated by qRT-PCR analyses in D15 cultures generated from D4 mesoderm treated BMP4 (10ng/ml), DM (4μM) or no additional factors (control) in combination with the indicated amounts of XAV. Bars represent standard error of the mean of the values from three independent experiments (N=3) *P≤0.05, **P≤0.01 when compared to no WNT treatment (see Figure 4a and 4b) in context of specific BMP treatment as analyzed by Student's T-test.
(d) The frequency of CTNT+ cells determined by flow cytometry and the expression levels of WT1 evaluated by qRT-PCR analyses in D15 cultures generated from D4 mesoderm treated with BMP4 (10ng/ml), DM (4μM) or no additional factors (control) in combination with the indicated amounts of IWP2. Bars represent standard error of the mean of the values from three independent experiments (N=3) *P≤0.05, **P≤0.01 when compared to no WNT treatment (see Figure 4a and 4b) in context of specific BMP treatment as analyzed by Student's T-test.
(e) qRT-PCR-based expression analyses of AXIN2 in D5 populations, 24 hours following the indicated treatments. Values shown are fold change relative to D4 pre-treated cultures. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 as analyzed by Student's T-test.
The addition of high concentrations of DKK1 (600ng/ml and above) or the small molecule antagonists XAV and IWP2 decreased WT1 expression in BMP4-treated cells indicating a loss of P-Epi cells with specification of the CM lineage (Fig. 4b-d). Activation of the WNT pathway by CHIR did not impact WT1 expression in BMP4-treated cells but did result in increased expression in the endogenous BMP population. DM treated cultures showed no change in WT1 expression following these manipulations (Fig. 4b). Taken together, these observations demonstrate that BMP signaling is required for specification of both the CM and Epi lineages as neither develops in the DM treated cultures. In the context of BMP signaling, development of the Epi lineage is dependent on Wnt signaling whereas establishment of the CM lineage requires efficient inhibition of this pathway.
Studies in the chick embryo have provided evidence that bFGF signaling promotes proliferation and mesenchymal differentiation of the proepicardial organ cells in tissue explant cultures33, suggesting that this pathway may play a role in the early stages of Epi lineage development. To determine if FGF impacts P-Epi specification from hPSCs, we manipulated the pathway by the addition of BFGF or the receptor antagonist PD-173074 together with BMP4 and CHIR to D4 mesoderm. Addition of PD-173074 did not affect WT1 expression or cell number output compared to the non-treated control (Supplementary Fig. 3b). Conversely, the addition of BFGF inhibited WT1 expression indicating that signaling through this pathway is detrimental to the development of the P-Epi lineage (Supplementary Fig. 3b). Cell numbers were not affected following the addition of BFGF suggesting that signaling through this pathway promoted the development of an alternative cell lineage.
Based on the inclusion of VEGF during the protocol, we wanted to determine if this signaling pathway impacts epicardial lineage specification and maturation. Notably, the D15 P-Epi population expressed KDR (Supplementary Fig. 3c), suggesting that this population may respond to VEGF signaling. Removal of VEGF from the specification step or during the following nine days of culture had no detrimental effect on D15 KDR expression, WT1 expression or total cell output indicating that it is not required for establishment of the lineage (Supplementary Fig. 3c and 3d). The majority of passaged Epi cells maintained expression of KDR, however, addition of VEGF during the culture of the Epi population resulted in a reduction in the number cells generated eight days later (Supplementary Fig. 3e and 3f). The reduction in cell number was not associated with maturation along the endothelial lineage, however, as the cells did not upregulate CD31 (Supplementary Fig. 3e).
Given the above findings, for the remaining experiments in this study CMs were generated by the addition of BMP4 and XAV or IWP2 to D4 mesoderm whereas the combination of BMP4 and CHIR was used to induce the Epi lineage. Cells treated with the BMP inhibitor DM were used as the non-CM, non-Epi control population. With this approach it was possible to generate Epi cells from other hPSC lines including a human fibroblast-derived iPSC line (Sendai hiPSC) and the hESC line H7 (Supplementary Fig. 4a and b). The protocol used for Epi development from the H7 hESCs was identical to that for the HES2 line. For the Sendai hiPSC line we shortened the mesoderm induction stage by one day (D1-3) as we have previously shown that the kinetics of differentiation is accelerated in some iPSCs21.
hPSC-derived epicardial cells undergo EMT
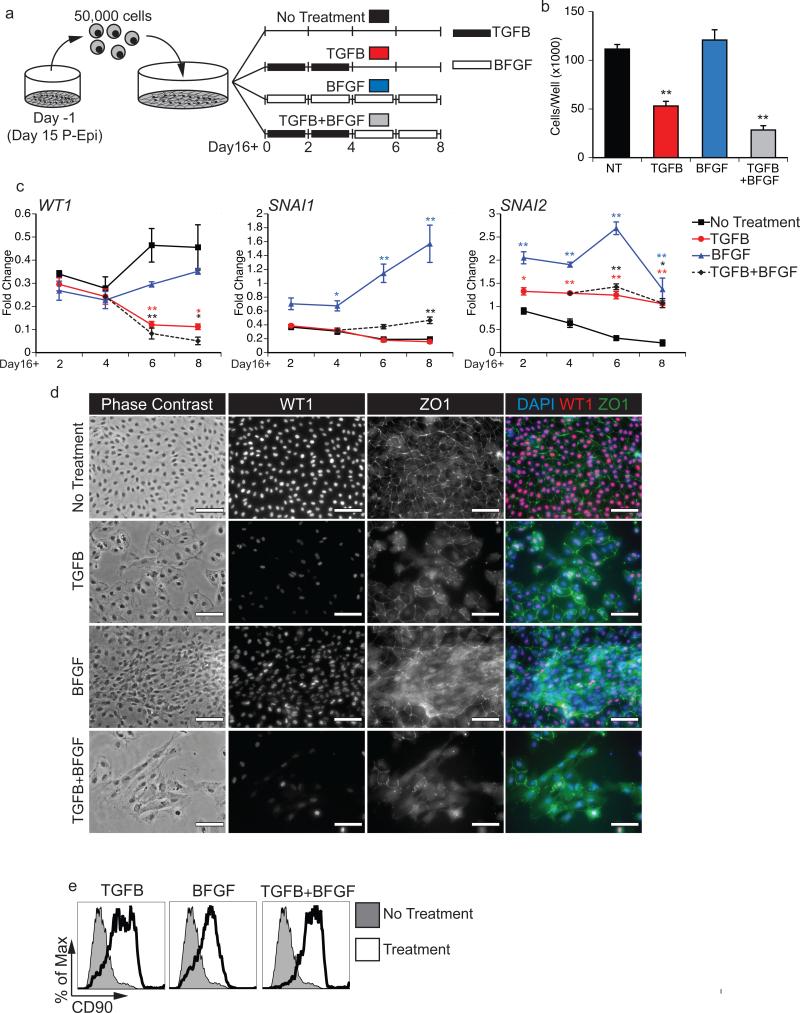
Previous studies have shown that both mouse and chick embryonic epicardial cells undergo EMT in vitro in response to factors that regulate this process in vivo including TGFβ14, 5, Wnt6, RA6, FGF7, PDGF8 and give rise to cell populations that display properties of the fibroblast and vascular smooth muscle lineages. To determine if the hESC-derived Epi cells can undergo EMT, we designed an assay in which D15 P-Epi cells are passaged, allowed one day to recover, and then treated for a total of eight days with one of four treatment regimens. The regimens consisted of: 1) TGFB1 for four days followed by four days with no additional factor (TGFB), 2) BFGF for eight days (BFGF) 3) TGFB1 for four days followed by four days with BFGF (TGFB+BFGF) or 4) no additional factors for eight days (Fig. 5a). Following culture under the different conditions, the cells were harvested and analyzed by qRT-PCR, flow cytometry and immunostaining.
Figure 5. Epicardial cells undergo EMT in response to TGFB1 and BFGF treatment.
(a) Scheme of the protocol used for the EMT analyses. D15 P-Epi cultures were passaged, allowed to settle for 1 day and then subjected to the following treatments: TGFB1 (5ng/ml) for 4 days followed by no treatment (TGFB, red), BFGF (10ng/ml) for 8 days (BFGF, blue), TGFB1 (5ng/ml) for 4 days followed by BFGF (10ng/ml) for 4 day (TGFB+BFGF, green) or no additional factors throughout the 8 day culture (control, black).
(b) Total cell numbers per well 8 days following EMT initiation with the indicated treatments. Cultures were initiated with 5x104 cells. Bars represent standard error of the mean of the values from three independent experiments (N=3); **P≤0.01 compared to the non-treated control as analyzed by Student's T-test.
(c) qRT-PCR-based expression analyses of the epicardial gene WT1 and the EMT-regulated genes SNAI1 and SNAI2 on days (D) 2, 4, 6 and 8 following EMT initiation with the indicated factors. Values are expressed as fold change relative to experiment-matched pre-passaged D15 P-Epi cultures. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 compared to non-treated control as analyzed by Student's T-test.
(d) Phase contrast and fluorescent immunostaining showing cell morphology and the presence of ZO1 and WT1 proteins in Epi derived cultures 8 days following EMT initiation with the indicated factors. DAPI staining shows cell nuclei. Scale bar represents 100μm. (e) Flow cytometric analyses for the expression of CD90 in the cultures 8 days following EMT initiation with the indicated factors.
Cultures maintained in the absence of factors or in the presence of BFGF led to a doubling of cell numbers over the eight-day period (Fig. 5b). In contrast, cell numbers in cultures treated with TGFB alone or TGFB+BFGF remained at input or lower than input levels. Expression levels of WT1 in the control cells were downregulated immediately following passage and then gradually upregulated over the eight-day culture period (Fig. 5c). Cells treated with either TGFB or TGFB+BFGF showed steady decreases in WT1 expression over time, indicating the transition towards an EPDC phenotype (Fig. 5bc. In contrast, the levels of WT1 expression did not decline below those of the control in the BFGF treated cells. Expression of the EMT markers SNAI1 and SNAI2 were also increased in the treated populations, although the levels varied depending on the cytokine combination. TGFB, TGFB+BFGF and BFGF all led to increases in SNAI2 expression while only BFGF induced the expression of SNAI1 (Fig. 5c). The recent findings by Casanova et al.34 demonstrating that SNAI1 is dispensable for epicardial EMT may explain why its expression levels did not change in the TGFB treated cultures. Immunostaining illustrated that ZO1 was lost from cell borders and possibly internalized following TGFB or BFGF treatment (Fig. 5d). Expression of WT1 by immunostaining was consistent with transcript expression determined by qRT-PCR. The most significant change in WT1 and ZO1 staining was observed in TGFB+BFGB treated cells, although they were largely morphologically indistinguishable from those treated with only TGFB. Flow cytometry showed that cells in all groups had upregulated the mesenchymal marker CD90 compared to the untreated control (Fig. 5e), supporting the interpretation that TGFB and BFGF had initiated EMT. Taken together, these findings indicate that the Epi cells are capable of undergoing EMT following activation of the TGFB and BFGF pathways and as such provide further evidence that they represent the in vitro equivalent of the developing epicardium.
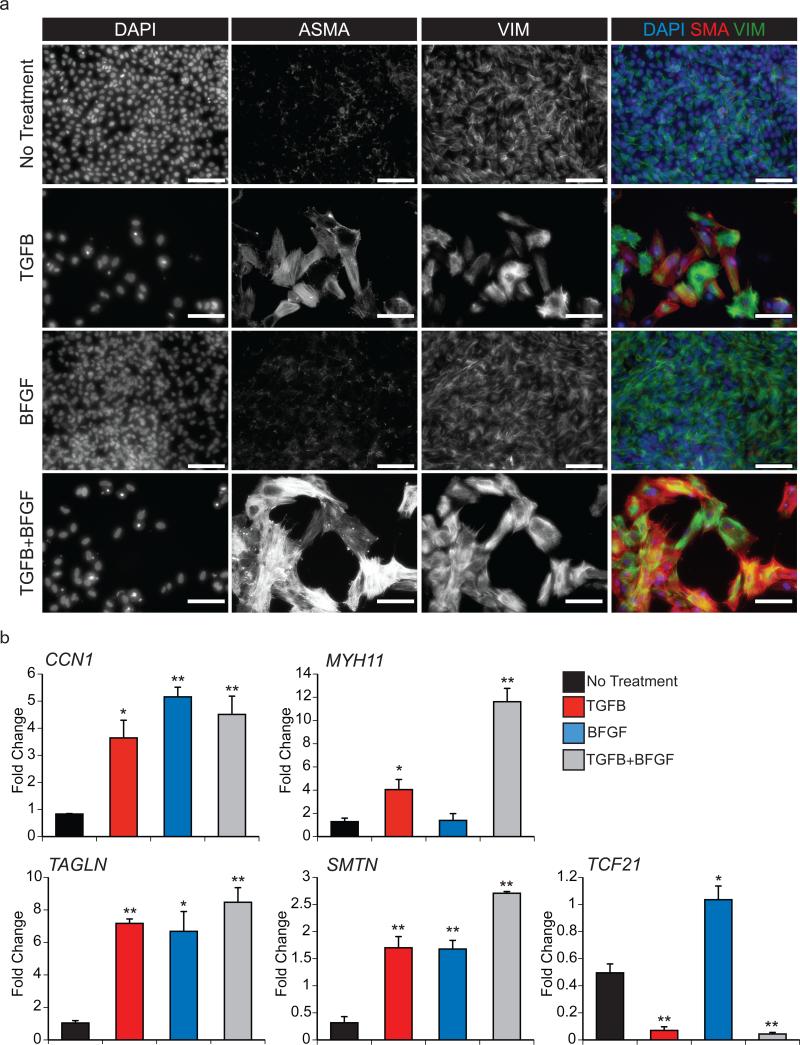
To identify the cell types being induced by EMT, the derivative populations were analyzed by immunostaining for the presence of the mesenchymal marker VIMENTIN (VIM) and the smooth muscle marker ALPHA-SMOOTH MUSCLE ACTIN (ASMA) and by qRT-PCR for expression of the smooth muscle genes CNN1, MYH11, TAGLN and SMTN35 and the epicardium-derived fibroblast marker TCF2136. While VIM was expressed to some degree in all populations, substantially brighter staining was observed in TGFB and TGFB+BFGF induced cells than in those cultured with BFGF alone (Fig. 6a). ASMA was also detected at higher levels in the TGFB and TGFB+BFGF induced cells compared to those induced with BFGF or those cultured in the absence of factors. These patterns suggest that cells induced with either TGFB or TGFB+BFGF are progressing along the vascular smooth muscle lineage (Fig 6a). In support of this is the observation that these cells upregulated expression of CNN1, MYH11, TAGLN and SMTN but not TCF21 (Fig. 6b). In contrast, the BFGF induced population expressed TCF21 in addition to CNN1, TAGLN and SMTN. These cells did not express substantial levels of MYH11 (Fig. 6b). Collectively, these findings indicate that TGFB specifies the Epi cells towards a smooth muscle-like fate whereas BFGF promotes the development of fibroblast-like cells. BFGF treatment following TGFB appeared to enhance the smooth muscle-like fate as observed by the increased expression of CNN1, MYH11, TAGLN and SMTN in these cells.
Figure 6. Epicardium-derived cells display characteristics of fibroblasts and vascular smooth muscle cells.
(a) Fluorescent immunostaining for ASMA and VIM proteins in the cultures 8 days following EMT initiation with the indicated treatments. DAPI staining shows cell nuclei. Scale bar represents 100μm.
(b) qRT-PCR-based expression analyses of the smooth muscle genes CCN1, MYH11, TAGLN and SMTN and the epicardial/cardiac fibroblast gene TCF21 in cultures 8 days following EMT initiation with the indicated factors. Values are expressed as fold change relative to experiment-matched pre-passaged D15 P-Epi cells. Bars represent standard error of the mean of the values from three independent experiments (N=3); *P≤0.05, **P≤0.01 compared to the non-treated cells as analyzed by Student's T-test.
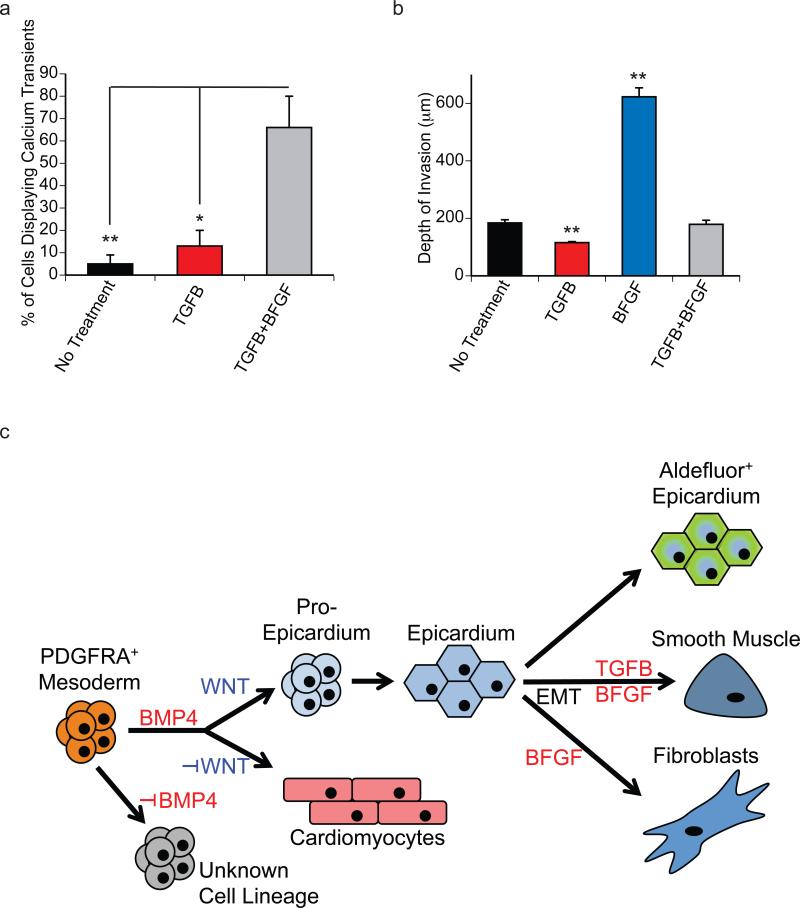
To test the contractile function of the smooth muscle-like cells generated following the EMT induced with TGFB and TGFB+BFGF, we measured calcium transients following stimulation with norepinephrine (NE) and phenylephrine (PE) using previously described methods37 (Supplementary Fig. 5a, 5b and Supplementary Video 1). The proportion of cells displaying calcium transients was highest (70%) in the population induced by TGFB+BFGF indicating that this combination of signaling pathways efficiently promoted the development of smooth muscle cells capable of contraction (Fig. 7a). Of the cells that exhibited calcium transients, similar rates of cycling were observed in the TGFB and TGFB+BFGF induced cells following PE stimulation. Cells from both populations exhibited faster calcium cycling rates than those in the non-induced control cultures (Fig. Supplementary Fig. 5c). The amplitude of calcium response was greatest in PE-treated TGFB cells compared to control cultures (Supplementary Fig. 5d). Finally, the duration of calcium transients following PE-treatment were significantly longer in the TGFB+BFGF induced cells than in those treated with TGFB or those in the control population (Supplementary Fig. 5e). Taken together these findings demonstrate that induction of the Epi cells with the combination of TGFB+BFGF promotes the development of smooth muscle cells capable of responding to agonists that result in increased calcium handling that may facilitate smooth muscle action potentials and contractility.
Figure 7. Functional assessment of EPDCs.
(a) The total proportion of actively cycling cells in EMT-induced cultures was measured for the indicated treatments. Treatment with TGFB+BFGF generated populations with the largest proportion of actively cycling cells in response to agonists. Bars represent standard error of the mean; N=3/group; *P<0.05, **P<0.01 compared by one-way ANOVA with Tukey post hoc test.
(e) Maximum matrigel invasion depth on D8 following EMT initiation. Bars represent standard error of the mean of the values from three independent experiments (N=3); **P≤0.01 compared to non-treated controls as analyzed by Student's T-test.
(e) Model highlighting the specification of PDGFRA+ mesoderm to the cardiomyocyte and pro-epicardial lineages by BMP4 and canonical WNT signaling. When passaged in the absence of factors, the pro-epicardial cells form epithelial sheets resembling the epicardium that can be identified through the Aldefluor assay. In the presence of TGFB+BFGF or BFGF, the passaged cells undergo EMT and give rise to smooth muscle-like and fibroblast-like cells.
It is well established that during heart development EPDCs, and in particular cardiac fibroblasts, invade the myocardial layer9. To assess this potential of the hPSC- derived Epi cells, we measured their ability to invade a 3D layer of Matrigel following induction of EMT with the different factors. To enable us to easily track the migration of the cells, we generated the Epi population from GFP expressing hESCs38. Matrigel invasion was monitored eight days following the induction of EMT by confocal microscopy and evaluated using 3D image reconstruction (Supplementary Fig. 6a and 6a’). The cells induced with BFGF alone were the most migratory and invaded the matrigel to the greatest depth (Fig. 7b), supporting the interpretation that they are fibroblastic in nature. Along with invasion, BFGF treatment also led to an increase in total cell number within the regions of interest (ROI). None of the other groups showed this expansion (no treatment, 73.8±6.1 cells per ROI; TGFB, 56.8±9.5 cells per ROI; BFGF, 379.7±40.5 cells per ROI, p=0.0017; TGFB+BFGF, 80.3±17.1 cells per ROI). Notably, the population induced with TGFB alone showed little capacity to invade the Matrigel, even less than the non-treated control that may contain some cells that have undergone spontaneous EMT to the fibroblast lineage (white arrow heads). Cells induced with the combination of TGFB+BFGF behaved similarly to the control population and were considerably less invasive than those induced with BFGF alone. To further quantify the degree of invasion, we calculated the proportion of cells in each population that migrated to different depths. As shown in Supplementary Figure 6b, virtually all of the cells in the non-induced, the TGFB-induced and TGFB+BFGF-induced populations were detected within the first 200 μm of the gel. In contrast, approximately half of the cells in the BFGF-induced population migrated beyond this depth, some as far as 600μm. Collectively, these findings demonstrate that the BFGF-induced population displays migratory behavior consistent with that predicted for EPDC in vivo. The observation that the cells with smooth muscle characteristics do not show this potential suggests that the maturation of this lineage likely occurs following migration into the tissue.
hESC-derived epicardium has aldehyde dehydrogenase activity
It is well accepted that the epicardium produces retinoic acid during development and following cardiac injury through the upregulation of the retinaldehyde dehydrogenase 2 (ALDH1A2). At D15 of differentiation P-Epi cells did not express ALDH1A2 (Supplementary Fig. 7a), nor did they stain positive for Aldefluor by flow cytometry, a marker of aldehyde dehydrogenase activity (Supplementary Fig. 7b). Following passage, however, this population showed steady increases in ALDH1A2 expression (Supplementary Fig. 7c). The retinaldehyde dehydrogenases 1 and 3 (ALDH1A1 and ALDH1A3), also involved in the synthesis of retinoic acid but not associated with the epicardium, were expressed at significantly lower levels than ALDH1A2 (Supplementary Fig. 7a). Eight days following passage, the Epi cell population had substantially upregulated aldehyde dehydrogenase activity as measured by Aldefluor staining to the stage at which greater than 75% were positive (Supplementary Fig. 7d). Cultures in which EMT had been induced with TGFB, BFGF or TGFB+BFGF showed dramatically lower levels of ALDH1A2 expression and Aldefluor staining, consistent with the interpretation that they have lost their Epi identity.
Taken together, our results show that the stage-specific activation of BMP4 and WNT signaling regulates the specification of P-Epi cells from hPSC-derived mesoderm. Once passaged, Epi cells have the ability to undergo EMT towards smooth muscle–like and fibroblast-like cells in response to TGFB and BFGF. In the absence of an EMT-inducing signal, Epi cells acquire a cobblestone morphology and aldehyde dehydrogenase activity through the upregulation of ALDH1A2, indicating their ability to synthesize RA (Fig. 7c). Based on immunostaining, we estimate that more than 85% (87.3%±7.1% N=4, standard deviation of the mean) of the P-Epi population is WT1+on D15. The frequency of WT1+ cells increased to > 90% (94.3±5.3%; N=3, standard deviation of the mean) 8 days following passage (D16+8). Given that we observe a 5 fold increase in cell numbers between D4 and D16+8, we estimate an output of 4-5 Epi cells per D4 mesoderm input cell.
Discussion
This study demonstrates the efficient and reproducible differentiation of hPSCs to epicardial-like cells that display many properties of the epicardium in other species, including the ability to undergo EMT to give rise to fibroblastic and vascular smooth muscle cell types. Our approach will enable future studies aimed at establishing the role of epicardial cells in cardiomyocyte growth and differentiation in vitro and in repair after cardiac injury in vivo. Access to stage-specific progenitors has enabled us to identify the BMP and canonical WNT pathways as key regulators of cardiomyocyte and epicardial specification from cardiogenic mesoderm (Fig. 8e). Our findings show an absolute requirement for BMP signaling for the generation of both lineages as neither develops when the pathway is inhibited. In the presence of BMP signaling, the decision to differentiate towards the cardiomyocyte or epicardial lineages is mediated by the canonical WNT pathway. The cardiomyocyte fate is specified in the absence of WNT signaling, whereas the epicardial lineage is generated by activation of the WNT pathway. Our data also suggest that these pathways act independently, as the levels of pSMAD1/5/8 were not affected by inhibition of WNT, and AXIN2 expression did not increase following the addition of BMP4. One interpretation of these findings is that these pathways act sequentially, with BMP inducing a Wnt responsive population that displays both epicardial and cardiomyocyte potential. Whether or not these pathways do indeed act sequentially remains to be determined.
In contrast to the BMP and WNT pathways, BFGF and NODAL/ACTIVIN signaling do not play a role in epicardial specification as the lineage can be initiated in the presence of antagonists of both pathways. In fact, FGF signaling between D4 and D6 of differentiation does not appear to be compatible with epicardial specification as addition of BFGF at this stage blocked development of the lineage. Kruithof et al.33 have provided evidence that bFGF signaling promotes proliferation and mesenchymal differentiation of proepicardial organ cells in chick tissue explant cultures. These findings are not necessarily contradictory to ours, as the stage of development assayed in their study is likely most similar to the day 15 P-Epi cells that we identified, a stage considerably later than the D4 mesoderm. In this regard, their observation that bFGF promotes mesenchymal differentiation is consistent with our findings that signaling through this pathway induces EMT in the hESC-derived Epi cells.
A pivotal role for BMP signaling in cardiac specification shown here is consistent with findings from studies in the mouse that demonstrate a block in heart development following the conditional knockout of Bmpr1a in cells expressing Mesp1-cre22. Additionally, the observation that inhibition of WNT signaling is required for specification of the cardiomyocyte lineage is in line with a large number of studies showing that stage-specific inhibition of the pathway is required for cardiogenesis in vivo and in hPSC differentiation cultures39-41. A specific requirement for this pathway in the specification of the epicardial fate is supported by in vivo studies showing that the hearts of mice lacking the Wnt inhibitors Dkk1 and Dkk2 have a thicker epicardium and thinner myocardium compared to wild type littermates. The efficient generation of epicardial lineage cells, even in the presence of endogenous levels of WNT signaling or suboptimal concentrations of DKK1 shown here, provides an explanation for the strict stage specific requirement for inhibiting this pathway for generating cardiomyocytes and suggests that under suboptimal conditions cultures may be contaminated with epicardial cells or EPDCs.
Similar to the epicardium in vivo, the hPSC-derived epicardial cells can undergo EMT in vitro and give rise to cells that display characteristics of fibroblasts and vascular smooth muscle cells. Cardiac fibroblasts are necessary for proper cardiomyocyte alignment42, electrical propagation43 and extracellular matrix deposition42. They are also one of the primary cell types involved during cardiac remodeling following injury16. Given the function of these cells in vivo, it is likely that they will play an important role in the development of engineered heart tissue constructs designed to create functional myocardium in vitro. In addition, as the epicardium is the primary source of coronary vascular smooth muscle cells, it is possible that the smooth muscle-like cells generated from the hESC-derived epicardial cells do represent this population. Access to these cells from epicardial progenitors could have clinical implications as alterations in the function of the vascular smooth muscle population is thought to be important in the development of atherosclerotic plaques44, 45. The generation of patient-specific smooth muscle cells through this approach will provide a model system to study the early changes associated with atherosclerosis and cardiovascular disease.
In summary, the findings presented here identify critical signaling pathways that regulate the specification of the cardiomyocyte and epicardial lineages from hPSC-derived mesoderm. With these insights, it is now possible to efficiently generate epicardial lineage cells from hPSCs, enabling us to define, in detail, their role in cardiac development, proliferation and maturation in vitro. Additionally, it will now be possible to investigate their function following transplantation into experimental animals in vivo, similar to the strategy used to test the effect of adult heart derived EPDCs46, 47.
Methods
hPSC culture maintenance and differentiation
hESCs and hiPSCs were maintained as previously described21, 38, 48. The hESC lines HES2 and H7 were obtained from WiCell and were determined to be mycoplasma negative by The Hospital for Sick Children Department of Paediatric Laboratory Medicine (Toronto, Canada). Sendai hiPSCs were kindly provided by Dr. Toshi Araki and Dr. Ben Neel (Ontario Cancer Institute, Toronto, Canada) and determined to be mycoplasma negative by qRT-PCR (Agilent Technologies MycoSensor qPCR Assay Kit, cat.no. 302109). Briefly, hESCs were maintained on irradiated mouse embryonic fibroblast feeders (MEFs) and feeder-depleted by passage onto plates coated with Matrigel (VWR, cat.no. 354230) for 2 days. On day 0, hESCs were dissociated and EBs were generated in StemPro-34 media (Life Technologies. cat.no 10640019) in the presence of recombinant human (rh)BMP4 (0.5ng/ml, R&D, cat.no. 314-BP). After 24 hours, EBs were cultured in StemPro-34 supplemented with rhBMP4 (3ng/ml), rhACTIVINA (2ng/ml, R&D, cat.no. 338-AC), and rhBFGF (5ng/ml, R&D, cat.no. 233-FB) to induce mesoderm formation. On day 4 of differentiation (day 3 for hiPSCs as indicated in Supplementary Fig. S3b), EBs were dissociated using TrypLE (Invitrogen) and plated in monolayer on gelatin-coated plates at 1x105 cells per well in StemPro-34 supplemented with rhDKK1 (150ng/ml, R&D. cat.no. 5439-DK/CF), the ACTIVIN/NODAL inhibitor SB-431542 (5.4μM, Sigma, cat.no. S-4317) and rhVEGF (5ng/ml) unless otherwise indicated. On day 6, media was changed to StemPro-34 supplemented with rhVEGF (5ng/ml) and cultures were maintained in this media until day 15 of differentiation with media changes every 2 days. For experiments involving WNT signaling, CHIR-99021 (Stemgent, cat.no. 04-004), XAV-939 (R&D, cat.no. 3748, or IWP2(R&D, cat.no. 3533) were used at the indicated concentrations. For experiments involving the inhibition of FGF signaling, PD-173074 (Tocris, cat.no 3044) was used at 100μM.
Monolayer passage and EMT induction
At day 15, WT1+ monolayers were lifted off of the plate by treatment with collagenase B (Roche, cat.no. 11088831001) for 1 hour. Monolayers were then harvested and dissociated with TrypLE and washed with IMDM. Dissociated cells were plated on matrigel coated plates at a density of 5x104 cells per well in StemPro-34 supplemented with the ROCK inhibitor Y-27632 Dihydrochloride Hydrate (Toronto Research Chemicals, cat.no Y100500). After 24 hours, media was changed to StemPro-34 and treated with rhTGFB1 (R&D, cat.no. 240-B) and rhBFGF as indicated. Media was changed every 2 days until analysis.
Flow cytometry and cell sorting
For cell-surface antigens, staining was carried out in PBS with 3% FCS. For intracellular antigens, staining was carried out on cells fixed with 4% paraformaldehyde in PBS. Staining was done in PBS with 3% FCS and 0.5% saponin (Sigma). Cells were stained at a concentration of 2.5 × 106 cells/ml with anti-KDR- APC (R&D Systems; 1:10) and anti-PDGFRA– PE (R&D Systems; 1:20), anti-SIRPA–PE-Cy7 (clone SE5A5; BioLegend; 1:500), anti-Podoplanin-PE (BioLegend, 1:200), anti-CD90-APC (BD Pharmingen, 1:2000), anti-PDGFRβ-PE (BD Pharmingen, 1:200), anti-EPCAM-PE (BD Pharmingen, 1:200), anti-CD31-PE (BD Pharmingen, 1:10), anti-CTNT (clone 13-11; Thermo NeoMarkers; 1:400), goat anti-mouse IgG–APC(BD; 1:200). Cells assayed for Aldefluor (STEMCELL Technologies) were prepared based on manufacturers instructions. Incubation with the Aldefluor reagent was 45 minutes. Stained cells were analyzed on an LSRII flow cytometer (BD Biosciences). For FACS, the cells were sorted at a concentration of 106 cells/ml in IMDM/5% FCS using a FACSAriaTMII (BD Biosciences) cell sorter (SickKids-UHN Flow Cytometry Facility). Data were analyzed using FlowJo software (Treestar).
Immunostaining and microscopy
Immunostaining was performed as previously described29 using the following primary antibodies: mouse anti-CTNT (Thermo NeoMarkers; 1:100), rabbit anti-WT1 (Abcam Epitomics, 1:100), mouse anti-ZO1 (Invitrogen, 1:100), rabbit anti-α-Smooth Muscle Actin (Abcam, 1:100) and mouse anti-Vimentin (Sigma-Aldrich, 1:100). Secondary antibodies used were: donkey anti-rabbit IgGCy3 (Jackson ImmunoResearch; 1:300), donkey anti-mouse IgG-Alexa 488 (Invitrogen; 1:300). DAPI (Life Technologies, SlowFade Gold) was used to counterstain nuclei. The stained cells were visualized using a fluorescence microscope (Leica CTR6000) and images captured using the Leica Application Suite software.
Quantitative real-time PCR
qRT-PCR was performed as previously reported29, 49. Total RNA was prepared with the RNAqueous-Micro Kit (Ambion) and treated with RNase-free DNase (Ambion). RNA (500 ng to 1 μg) was reverse transcribed into cDNA using random hexamers and Oligo(dT) with Superscript III Reverse Transcriptase (Invitrogen). qPCR was performed on a MasterCycler EP RealPlex (Eppendorf) using QuantiFast SYBR Green PCR Kit (Qiagen). Expression levels were normalized to the housekeeping gene TATA binding protein (TBP). Primer sequences are listed in Supplementary Table 1.
Western Blotting
D4 cells were harvested and treated with the indicated reagents for 30 minutes. Nuclear protein extracts were harvested using the NE-Per Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific, cat.no. 78835) according to manufacturers instructions. Lysates were separated by SDS-PAGE gel and transferred to PVDF membranes for immunoblotting. Immunoblots were incubated with anti-pSMAD1/5/8 (1:1000, Cell Signaling, cat.no. 9516) and anti-βactin (1:1000, Cell Signaling, cat.no. 3700) overnight at 4°C. Immunoblots were performed using infrared fluorescence-conjugated secondary antibodies and analyzed using the Odyssey infrared imaging system (LiCor Biotechnology).
Calcium Imaging
Cells were grown to near confluence on 8-well glass chamber slides coated with Matrigel and treated according to the EMT assay. Calcium imagining was performed as previously described37. Cells were incubated with 5μM of the membrane permeable calcium dye Fluo 4-AM (Invitrogen) and 0.1% (w/v) Plurionic F-127 (Invitrogen) for 35min at 37°C. Cells were washed with Tyrode's solution consisting of 140mM NaCl, 4mM KCl, 2mM CaCl2, 1mM MgCl2, 10mM HEPES, 10mM glucose pH 7.4. Calcium imaging was performed using an Olympus FV1000 confocal microscope with an excitation at 488nm at 0.5% power to reduce photo-bleaching. Images were continuously acquired at a resolution of 512x512 pixels using a 40X objective and were stitched together into a continuous sequence over a 12min period. All recordings were performed using the same excitation and acquisition settings. Each recording consisted of 4min baseline and by equivalent 4min recording periods following the addition of 20μM of norepinephrine (NE, Sigma) and 15μM of phenylephrine (PE, Sigma). These agonists were prepared from stock concentrations and diluted in Tyrode's buffer. Three independent experiments were performed for each condition, and within each field of view, regions of interest (ROIs) corresponding to the cytoplasmic area of independent cells were analyzed. Representative ROIs demonstrate the arbitrary fluorescence signal (F) generated as an average pixel intensity for the ROI over the course of the 12min recording, normalized by dividing the average ROI intensity during the first minute of non-calcium cycling baseline (F/F0). The proportion of cycling cells was determined after counting cells that exhibited at least one calcium transient and dividing by the total number of cells in the field of view. The frequency response was calculated by counting the number of cycles during each 4min stage of the 12min recording with a normalized fluorescence intensity (F/F0) greater than 1.5. The summed total of the frequency response for each group represents the average number of cycles for all actively cycling cells analyzed for each group and is expressed in number of cycles per minute. The duration of calcium cycling for all active ROIs was manually calculated by determining the number of frames during which a calcium cycle spiked above baseline. The result was multiplied by the frame duration of 1.109s. Calcium cycle duration was only measured for cycles that occurred at the start of the 4-min period corresponding to the addition of NE or PE. All data represent 3 independent experiments, with n=6-10 cells/field, and data expressed as mean +/− SEM. *P≤0.05, **P≤0.01 compared by one-way ANOVA with Tukey post hoc test.
Matrigel Invasion Assay
Growth factor reduced BD Matrigel (BD Sciences) was thawed overnight at 4 °C and 40 μL/well was added to a 96 well plate. The plates were then centrifuged at 600 RPM for 5 min at room temperature, followed by a 30 min incubation at 37 °C. Epicardial cells were seeded on the gels at a density of 15,000 cells/well with 150 μL/well of StemPro-34 media, and maintained at 37 °C / 5 % CO2. After 24 hours, media was changed to StemPro-34 and treated with rhTGFB1 and rhBFGF The media was changed every 2 days thereafter. After 8 days, cells were imaged using confocal microscopy (Olympus). High resolution Z-stack images were taken with a 1 μm step size. For quantification of cell number and migration, Z-stacks of each gel were imaged with a 10 μm step size, and analyzed using Imaris BitPlane software. Four regions of interest (ROI, 317.5 μm x 317.5 μm each) within each gel were used for quantification of migration, with the distance between the highest and lowest cell taken as the migration distance per ROI. The average of the four ROI’s was taken as the migration distance per gel, with a total of four gels per treatment group. The experiment was repeated with three independent cell batches for accuracy.
Statistical Analysis
All data represents 3 biological replicates or more unless otherwise indicated in the figure legend. Error bars represent standard error of the mean and Two-tailed Student's t-tests were performed for all analyses unless otherwise indicated in the figure legend.
Supplementary Material
Acknowledgments
We thank T. Araki and B. Neel for providing the Sendai hiPSC line, O. El-Mounayri and M. Husain for their advice on epithelial-to-mesenchymal transition, Mark Gagliardi for his assistance in the culture of hESC-derived epicardial cells and the Sick Kids/UHN Flow Cytometry Facility for their assistance with cell sorting. We thank members of the Keller lab for their advice on the studies and comments on the manuscript. This work was supported by the Canadian Institute of Health Research (MOP-84524; MOP-119507; CPG-127793), the Natural Sciences and Engineering Research Council of Canada (CHRPJ 446379-13) and the National Institute of Health (5U01 HL100405). This work was funded in part by VistaGen Therapeutics, Inc.
Footnotes
Author Contributions
A.D.W contributed to designing the study, designing and performing experiments, analyzing the data and writing and editing the manuscript. A.M. contributed to designing, performing and analyzing experiments concerning calcium imaging and editing the manuscript. R.Y.T. contributed to designing, performing and analyzing experiments concerning matrigel invasion. S.A.F. contributed to designing, performing and analyzing experiments concerning matrigel invasion. A.M. contributed data acquisition concerning cell quantification and editing the manuscript. M.S.S. contributed to designing experiments concerning matrigel invasion. R.K.L. contributed to designing experiments concerning calcium imaging. S.J.K. contributed to designing the study and editing the manuscript. G.K. contributed to designing the study, writing and editing the manuscript.
Competing Financial Interests
G.K. is on the scientific advisory board and a shareholder of VistaGen Therapeutics, which partially funded this work. A.D.W., S.J.K. and G.K. are co-inventors on a patent application covering the generation of human pluripotent stem cell-derived epicardial cells described here.
References
- 1.DeRuiter MC, Poelmann RE, VanderPlas-de Vries I, Mentink MM, Gittenberger-de Groot AC. The development of the myocardium and endocardium in mouse embryos. Fusion of two heart tubes? Anatomy and embryology. 1992;185:461–473. doi: 10.1007/BF00174084. [DOI] [PubMed] [Google Scholar]
- 2.Limana F, Capogrossi MC, Germani A. The epicardium in cardiac repair: from the stem cell view. Pharmacology & therapeutics. 2011;129:82–96. doi: 10.1016/j.pharmthera.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anatomy and embryology. 1987;176:183–189. doi: 10.1007/BF00310051. [DOI] [PubMed] [Google Scholar]
- 4.Austin AF, Compton LA, Love JD, Brown CB, Barnett JV. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev Dyn. 2008;237:366–376. doi: 10.1002/dvdy.21421. [DOI] [PubMed] [Google Scholar]
- 5.Bax NA, et al. In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFbeta-signaling and WT1. Basic research in cardiology. 2011;106:829–847. doi: 10.1007/s00395-011-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Gise A, et al. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Developmental biology. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Developmental biology. 2001;234:204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- 8.Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circulation research. 2011;108:e15–26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lie-Venema H, et al. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. ScientificWorldJournal. 2007;7:1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christoffels V. Regenerative medicine: Muscle for a damaged heart. Nature. 474:585–586. doi: 10.1038/474585a. [DOI] [PubMed] [Google Scholar]
- 11.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development (Cambridge, England) 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 12.Haenig B, Kispert A. Analysis of TBX18 expression in chick embryos. Development genes and evolution. 2004;214:407–411. doi: 10.1007/s00427-004-0415-3. [DOI] [PubMed] [Google Scholar]
- 13.Moss JB, et al. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Developmental biology. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 14.Xavier-Neto J, Shapiro MD, Houghton L, Rosenthal N. Sequential programs of retinoic acid synthesis in the myocardial and epicardial layers of the developing avian heart. Developmental biology. 2000;219:129–141. doi: 10.1006/dbio.1999.9588. [DOI] [PubMed] [Google Scholar]
- 15.Huang GN, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science (New York, N.Y. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 17.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Tuyn J, et al. Epicardial cells of human adults can undergo an epithelial-to mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem cells. 2007;25:271–278. doi: 10.1634/stemcells.2006-0366. [DOI] [PubMed] [Google Scholar]
- 21.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell stem cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacNeill C, French R, Evans T, Wessels A, Burch JB. Modular regulation of cGATA-5 gene expression in the developing heart and gut. Developmental biology. 2000;217:62–76. doi: 10.1006/dbio.1999.9539. [DOI] [PubMed] [Google Scholar]
- 25.Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Developmental biology. 2008;323:98–104. doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Stainier DY. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circulation research. 2010;106:1818–1828. doi: 10.1161/CIRCRESAHA.110.217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochmann L, et al. Revealing new mouse epicardial cell markers through transcriptomics. PloS one. 2010;5:e11429. doi: 10.1371/journal.pone.0011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahtab EA, et al. Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: Correlation with abnormal epicardial development. Dev Dyn. 2008;237:847–857. doi: 10.1002/dvdy.21463. [DOI] [PubMed] [Google Scholar]
- 29.Dubois NC, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature biotechnology. 2012;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellgren AM, et al. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circulation research. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips MD, Mukhopadhyay M, Poscablo C, Westphal H. Dkk1 and Dkk2 regulate epicardial specification during mouse heart development. International journal of cardiology. 2011;150:186–192. doi: 10.1016/j.ijcard.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu PB, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nature chemical biology. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruithof BP, et al. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Developmental biology. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 34.Casanova JC, Travisano S, de la Pompa JL. Epithelial-to-mesenchymal transition in epicardium is independent of Snail1. Genesis. 2012;51:32–40. doi: 10.1002/dvg.22353. [DOI] [PubMed] [Google Scholar]
- 35.Cheung C, Bernardo AS, Trotter MW, Pedersen RA, Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nature biotechnology. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development (Cambridge, England) 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Mounayri O, et al. Serum-free differentiation of functional human coronary-like vascular smooth muscle cells from embryonic stem cells. Cardiovascular research. 2013;98:125–135. doi: 10.1093/cvr/cvs357. [DOI] [PubMed] [Google Scholar]
- 38.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 39.Lian X, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nature cell biology. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 42.Weeke-Klimp A, et al. Epicardium-derived cells enhance proliferation, cellular maturation and alignment of cardiomyocytes. Journal of molecular and cellular cardiology. 2010;49:606–616. doi: 10.1016/j.yjmcc.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Gaudesius G, Miragoli M, Thomas SP, Rohr S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circulation research. 2003;93:421–428. doi: 10.1161/01.RES.0000089258.40661.0C. [DOI] [PubMed] [Google Scholar]
- 44.de la Cuesta F, et al. Deregulation of smooth muscle cell cytoskeleton within the human atherosclerotic coronary media layer. Journal of proteomics. 2013;82:155–165. doi: 10.1016/j.jprot.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 45.Jonasson L, Holm J, Skalli O, Bondjers G, Hansson GK. Regional accumulations of T cells, macrophages, and smooth muscle cells in the human atherosclerotic plaque. Arteriosclerosis (Dallas, Tex. 1986;6:131–138. doi: 10.1161/01.atv.6.2.131. [DOI] [PubMed] [Google Scholar]
- 46.Winter EM, et al. Preservation of left ventricular function and attenuation of remodeling after transplantation of human epicardium-derived cells into the infarcted mouse heart. Circulation. 2007;116:917–927. doi: 10.1161/CIRCULATIONAHA.106.668178. [DOI] [PubMed] [Google Scholar]
- 47.Winter EM, et al. A new direction for cardiac regeneration therapy: application of synergistically acting epicardium-derived cells and cardiomyocyte progenitor cells. Circulation. Heart failure. 2009;2:643–653. doi: 10.1161/CIRCHEARTFAILURE.108.843722. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke RL, et al. The expression of Sox17 identifies and regulates haemogenic endothelium. Nature cell biology. 2013;15:502–510. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.