Abstract
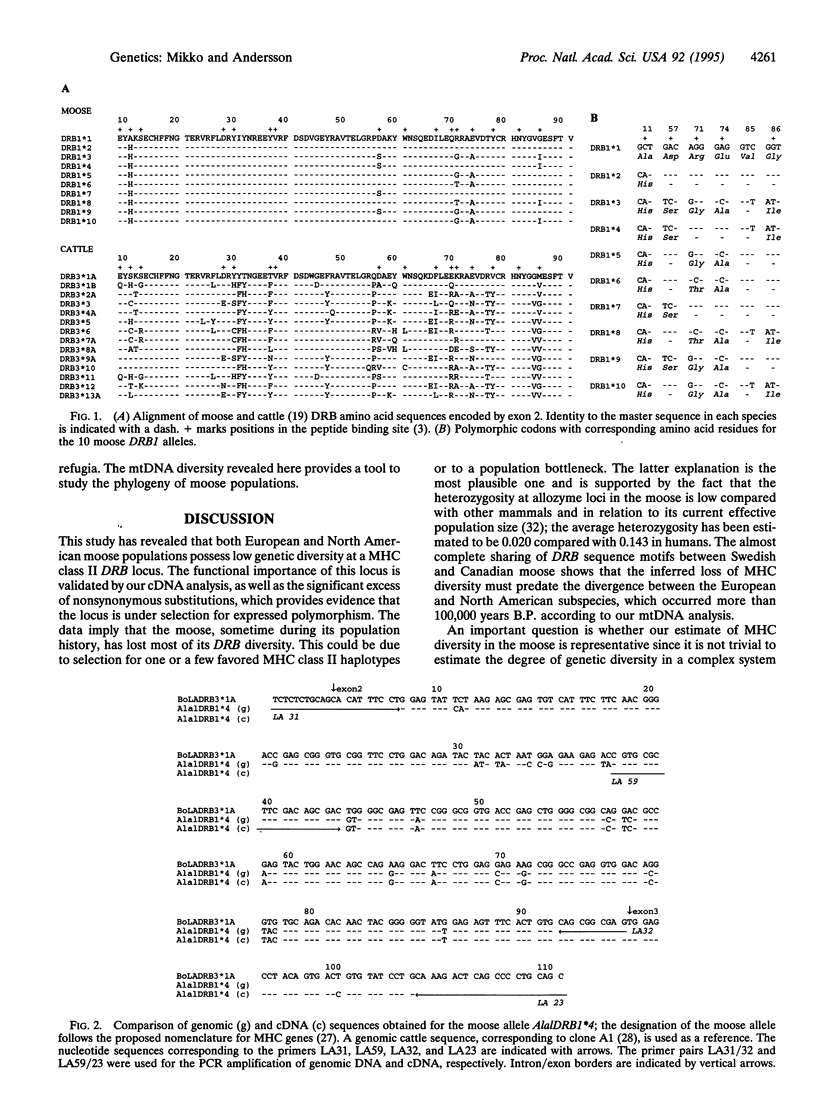
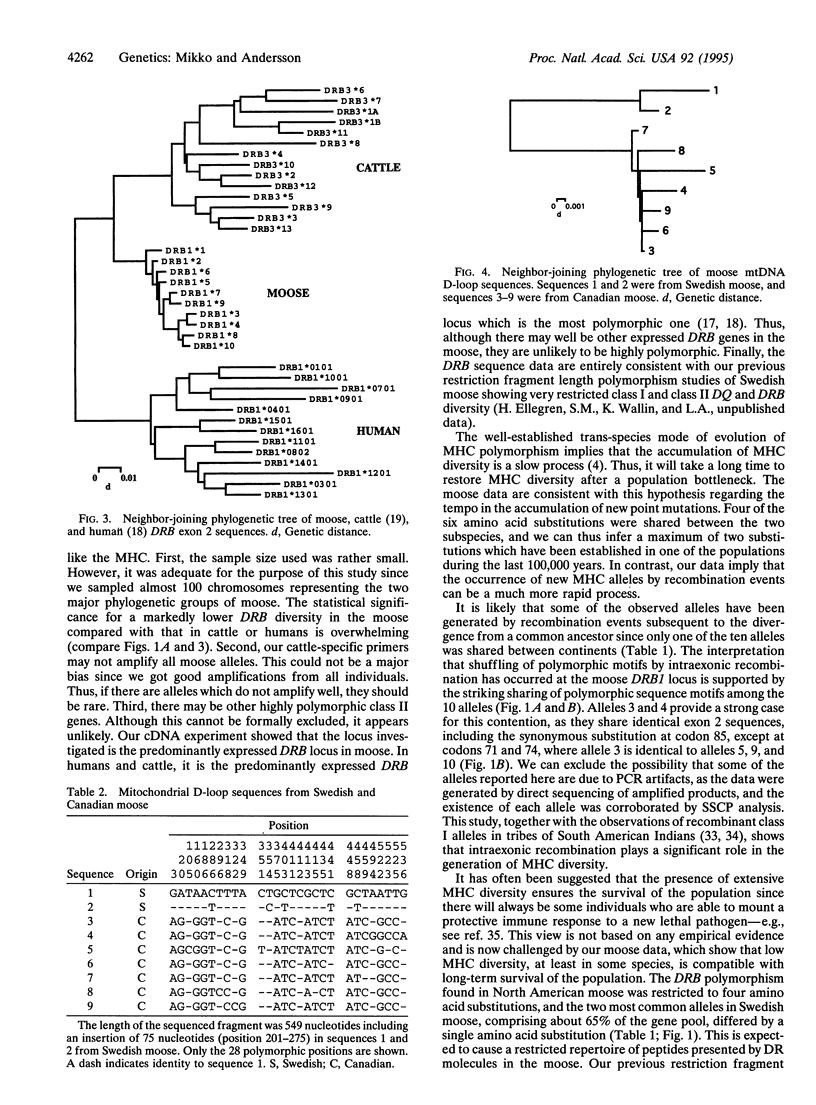
Major histocompatibility complex (MHC) genes encode cell surface proteins whose function is to bind and present intracellularly processed peptides to T lymphocytes of the immune system. Extensive MHC diversity has been documented in many species and is maintained by some form of balancing selection. We report here that both European and North American populations of moose (Alces alces) exhibit very low levels of genetic diversity at an expressed MHC class II DRB locus. The observed polymorphism was restricted to six amino acid substitutions, all in the peptide binding site, and four of these were shared between continents. The data imply that the moose have lost MHC diversity in a population bottleneck, prior to the divergence of the Old and New World subspecies. Sequence analysis of mtDNA showed that the two subspecies diverged at least 100,000 years ago. Thus, viable moose populations with very restricted MHC diversity have been maintained for a long period of time. Both positive selection for polymorphism and intraexonic recombination have contributed to the generation of MHC diversity after the putative bottleneck.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson L., Sigurdardóttir S., Borsch C., Gustafsson K. Evolution of MHC polymorphism: extensive sharing of polymorphic sequence motifs between human and bovine DRB alleles. Immunogenetics. 1991;33(3):188–193. doi: 10.1007/BF01719239. [DOI] [PubMed] [Google Scholar]
- Belich M. P., Madrigal J. A., Hildebrand W. H., Zemmour J., Williams R. C., Luz R., Petzl-Erler M. L., Parham P. Unusual HLA-B alleles in two tribes of Brazilian Indians. Nature. 1992 May 28;357(6376):326–329. doi: 10.1038/357326a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Bodmer W. F. Evolutionary significance of the HL-A system. Nature. 1972 May 19;237(5351):139–passim. doi: 10.1038/237139a0. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T., Saper M. A., Samraoui B., Bjorkman P. J., Wiley D. C. A hypothetical model of the foreign antigen binding site of class II histocompatibility molecules. Nature. 1988 Apr 28;332(6167):845–850. doi: 10.1038/332845a0. [DOI] [PubMed] [Google Scholar]
- Doherty P. C., Zinkernagel R. M. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975 Jul 3;256(5512):50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Hartman G., Johansson M., Andersson L. Major histocompatibility complex monomorphism and low levels of DNA fingerprinting variability in a reintroduced and rapidly expanding population of beavers. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8150–8153. doi: 10.1073/pnas.90.17.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., van der Poel J. J., Dijkhof R. J., Giphart M. J. The nucleotide sequence of bovine MHC class II DQB and DRB genes. Immunogenetics. 1990;31(1):37–44. doi: 10.1007/BF00702487. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., Thomson G. Evidence for balancing selection at HLA. Genetics. 1983 Jul;104(3):449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci U S A. 1989 Feb;86(3):958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Klein J., Bontrop R. E., Dawkins R. L., Erlich H. A., Gyllensten U. B., Heise E. R., Jones P. P., Parham P., Wakeland E. K., Watkins D. I. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31(4):217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Klein J., Satta Y., O'hUigin C., Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Zemmour J., Ennis P. D., Parham P. Evolution of class-I MHC genes and proteins: from natural selection to thymic selection. Annu Rev Immunol. 1990;8:23–63. doi: 10.1146/annurev.iy.08.040190.000323. [DOI] [PubMed] [Google Scholar]
- Loftus R. T., MacHugh D. E., Bradley D. G., Sharp P. M., Cunningham P. Evidence for two independent domestications of cattle. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2757–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S. G., Bodmer J. G. HLA-DRB nucleotide sequences, 1990. Immunogenetics. 1990;31(3):141–144. doi: 10.1007/BF00211548. [DOI] [PubMed] [Google Scholar]
- Merza M., Larsson E., Steen M., Morein B. Association of a retrovirus with a wasting condition in the Swedish moose. Virology. 1994 Aug 1;202(2):956–961. doi: 10.1006/viro.1994.1418. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Roelke M. E., Marker L., Newman A., Winkler C. A., Meltzer D., Colly L., Evermann J. F., Bush M., Wildt D. E. Genetic basis for species vulnerability in the cheetah. Science. 1985 Mar 22;227(4693):1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sigurdardóttir S., Borsch C., Gustafsson K., Andersson L. Cloning and sequence analysis of 14 DRB alleles of the bovine major histocompatibility complex by using the polymerase chain reaction. Anim Genet. 1991;22(3):199–209. doi: 10.1111/j.1365-2052.1991.tb00670.x. [DOI] [PubMed] [Google Scholar]
- Slade R. W. Limited MHC polymorphism in the southern elephant seal: implications for MHC evolution and marine mammal population biology. Proc Biol Sci. 1992 Aug 22;249(1325):163–171. doi: 10.1098/rspb.1992.0099. [DOI] [PubMed] [Google Scholar]
- Steen M., Rehbinder C. Nervous tissue lesions caused by elaphostrongylosis in wild Swedish moose. Acta Vet Scand. 1986;27(3):326–342. doi: 10.1186/BF03548147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M., Sherry S. T., Redd A. J., Vigilant L. New approaches to dating suggest a recent age for the human mtDNA ancestor. Philos Trans R Soc Lond B Biol Sci. 1992 Aug 29;337(1280):167–175. doi: 10.1098/rstb.1992.0094. [DOI] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990 Apr;124(4):967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R. H., Frazier B. L., Dew-Jager K., Päbo S. Extensive mitochondrial diversity within a single Amerindian tribe. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins D. I., Garber T. L., Chen Z. W., Toukatly G., Hughes A. L., Letvin N. L. Unusually limited nucleotide sequence variation of the expressed major histocompatibility complex class I genes of a New World primate species (Saguinus oedipus). Immunogenetics. 1991;33(2):79–89. doi: 10.1007/BF00210819. [DOI] [PubMed] [Google Scholar]
- Watkins D. I., McAdam S. N., Liu X., Strang C. R., Milford E. L., Levine C. G., Garber T. L., Dogon A. L., Lord C. I., Ghim S. H. New recombinant HLA-B alleles in a tribe of South American Amerindians indicate rapid evolution of MHC class I loci. Nature. 1992 May 28;357(6376):329–333. doi: 10.1038/357329a0. [DOI] [PubMed] [Google Scholar]


