Abstract
Over one-third of all proteins require metallation for function (Waldron, K. J., Rutherford, J. C., Ford, D., and Robinson, N.J. (2009) Nature 460, 823–830). As biochemical studies of most proteins depend on their isolation subsequent to recombinant expression (i.e. they are seldom purified from their host organism), there is no gold standard to assess faithful metallocofactor assembly and associated function. The biosynthetic machinery for metallocofactor formation in the recombinant expression system may be absent, inadequately expressed, or incompatible with a heterologously expressed protein. A combination of biochemical and genetic studies has led to the identification of key proteins involved in biosynthesis and likely repair of the metallocofactor of ribonucleotide reductases in both bacteria and the budding yeast. In this minireview, we will discuss the recent progress in understanding controlled delivery of metal, oxidants, and reducing equivalents for cofactor assembly in ribonucleotide reductases and highlight issues associated with controlling Fe/Mn metallation and avoidance of mismetallation.
Keywords: Bacterial Pathogenesis, Iron, Iron-Sulfur Protein, Manganese, Metal Homeostasis, Ribonucleotide Reductase, Yeast, Metallation, Mismetallation
Class I Ribonucleotide Reductases
Ribonucleotide reductases (RNRs)2 catalyze de novo biosynthesis of deoxynucleotides (Reaction 1) in almost all organisms (1). Three classes of RNRs have been identified; they all share a structurally homologous α subunit that binds the NDP(NTP) substrates and houses the two cysteines that provide the reducing equivalents for dNDP(dNTP) formation (where NDP is nucleoside diphosphate), and a third cysteine that must be oxidized transiently to a thiyl radical to initiate the reduction process (2). The class I RNRs, the focus of this review, require a second subunit β, which houses the essential metallocofactor (Fig. 1) and is required for thiyl radical formation in α in an oxidation that occurs over a 35 Å distance in an unprecedented process in biology (recently reviewed) (3). The class I RNRs have been subclassified based on their metal composition. The class Ia RNRs are found in eukaryotes (for example, humans and Saccharomyces cerevisiae) and a few prokaryotes (for example, Escherichia coli and Salmonella enterica serovar Typhimurium (S. typhimurium)). The class Ib RNRs are found in most prokaryotes (for example, E. coli, Corynebacterium ammoniagenes, Bacillus subtilis, Streptococcus sanguinis, Bacillus cereus, and Bacillus anthracis) (4). A few prokaryotes possess both Ia and Ib RNRs (5, 6). A class Ic RNR has been characterized only in vitro, and thus will not be further discussed (7). The Ia RNRs utilize a diferric-tyrosyl (FeIII2-Y•) radical cofactor, and the Ib RNRs are able to use in vitro a diferric or dimanganic-tyrosyl radical (MnIII2-Y•) cofactor.
REACTION 1.
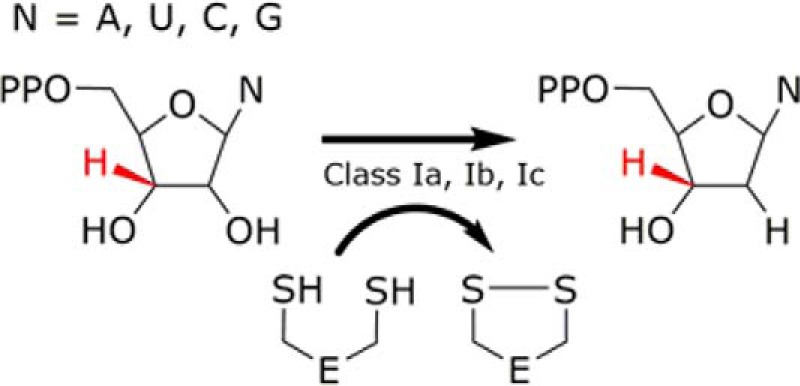
FIGURE 1.
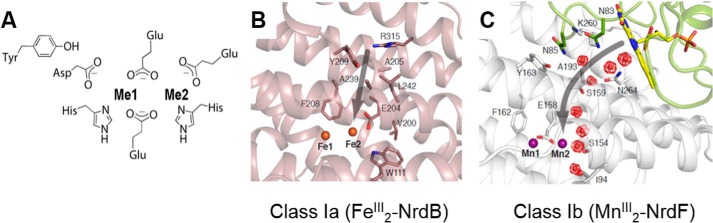
Ligands to the iron and manganese binding sites in class Ia and Ib βs and proposed pathway for oxidant access. A, the first coordination sphere ligands for iron and manganese binding are the same. The metal closest to Tyr is designated site 1 (Fe1, Mn1), and the other is site 2 (Fe2, Mn2). B, proposed route (gray arrow) for O2 (oxidant) access to the ferrous Fe2 site in the class Ia RNRs (Protein Data Bank (PDB) code 1PIY). Fe2+ ions are shown as orange spheres. C, proposed route (gray arrow) for O2⨪ (oxidant) access to the manganous Mn2 site in the class Ib RNRs (PDB 3N3A); depicted are the Mn2+ ions (purple spheres), NrdF (gray), NrdI (green), FMN (yellow), waters (red mesh spheres), and hydrophilic residues lining the oxidant channel. (Reproduced with permission from Ref. 15.).
E. coli Has Both Class Ia and Class Ib RNRs
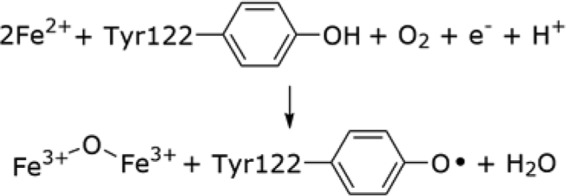
The metallocofactor in the E. coli class Ia RNR was the first one characterized. Landmark experiments identified a “stable” Y• whose formation was mediated by the adjacent non-heme di-iron cluster (8). Fortuitously, β in the apo-form can self-assemble an “active” cofactor from Fe2+, O2, and a reducing equivalent (Reaction 2) (9), providing insight for biosynthetic requirements: modulation of apo-β2 conformation and controlled metal, oxidant, and reductant deliveries. The success of the assembly process in vitro, however, is highly variable between RNRs in different organisms and when optimized gives ∼1 Y•/β2. We have recently established that the Y• is distributed equally between each β, suggesting the likelihood of 2 Y•s/β2 (5). The variability of cofactor loading and the genomic/bioinformatic discoveries that in many prokaryotes, biosynthetic pathways are organized in operons, suggested that even with a cofactor as simple as those required for class I RNR activity, dedicated machinery will be required. In this review, we briefly summarize the search for and discovery of candidate proteins involved in the assembly of the FeIII2-Y• cofactor in the class Ia RNRs from E. coli and S. cerevisiae and the discovery of the biosynthetic machinery for the long proposed (10), but elusive, MnIII2-Y• cofactor in the class Ib RNRs of E. coli, B. subtilis, and S. sanguinis. The class Ib RNRs also can self-assemble an active FeIII2-Y•, and thus this class of RNR offers a unique system to discover and understand the factors that control transition metal delivery for cofactor assembly in the cell.
REACTION 2.
The laboratory strain of E. coli is unusual in that it has both a class Ia RNR and a class Ib RNR. Its class Ia enzyme, coded for by nrdAnrdB (NrdA or α and NrdB or β), is essential for DNA replication and repair. The class Ib, on the other hand, is coded for by nrdEnrdF (NrdE or α and NrdF or β), is expressed under iron-limited and oxidative stress conditions, and cannot support growth in the absence of nrdAnrdB (6, 11). In contrast, most prokaryotic organisms only have a class Ib RNR that plays the same role as the Ia, that is, it is involved in DNA replication and repair.
A bioinformatics search for conserved genes of interest contiguous with nrdAnrdB identified yfaE, annotated as a [2Fe-2S]2+/1+-ferredoxin, within the same operon (12). Our hypothesis was that YfaE might function as the one electron donor required for cofactor assembly (Reaction 2) as well as the reductant to repair Y• reduced RNR, met-RNR, by reduction of the diferric cluster to the diferrous state, which then allows biosynthesis (Fig. 2). Biochemical studies monitoring cluster self-assembly establish that only 2 Fe2+/β are required for assembly when YfaE is present in the reaction mixture, whereas 3 Fe2+/β are required in its absence (the third Fe2+ can supply the reducing equivalent in vitro) (12). This observation supports the proposed role in cluster assembly. In a second set of experiments (13), expression of β in E. coli from 4 to 3300 μm revealed in all cases 0.1–0.3 Y•s/β2. The normal endogenous β level is 0.5 μm. Titration of [2Fe-2S]1+-YfaE into crude cell extracts followed by the addition of O2 revealed, for the first time, that 2 Y•s/β2 could be generated and as much as 250 μm β2 could be loaded. Thus, YfaE is interesting and as noted below likely plays an additional role in active cluster formation (13). Measurement of concentrations of YfaE relative to NrdB has been challenging, due to poor antibody quality, but it appears to be acting catalytically. Two unresolved issues regarding YfaE function are: 1) a number of genomes with only class Ia RNRs do not have a readily identifiable YfaE based on bioinformatic searches; and 2) the E. coli YfaE is not essential under normal growth conditions (LB). However, as noted in the next section, YfaE is essential for NrdB cluster assembly under oxidative stress (6), and a YfaE counterpart appears to play an essential role in S. cerevisiae cluster assembly (14).
FIGURE 2.
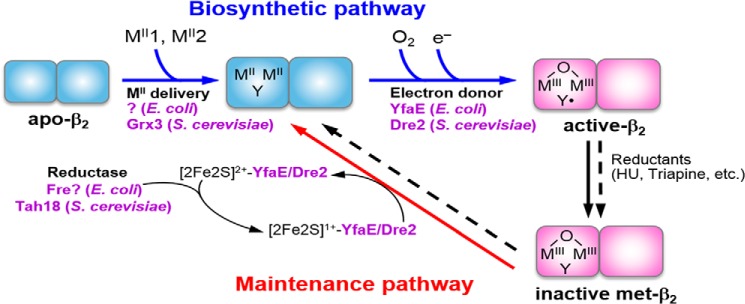
A model for the biosynthesis and maintenance pathways of the FeIII2-Y• cofactor of the class Ia RNR in E. coli and S. cerevisiae. Cofactor assembly in only one monomer of β2 is shown for simplicity. The biosynthetic pathway requires loading of two Fe2+ and one reducing equivalent per β to carry out the four-electron reduction of O2 to H2O (Reaction 2); the other three electrons come from the two Fe2+ and the Tyr residue that form the FeIII2-Y•. YfaE in E. coli and Dre2 in S. cerevisiae are proposed to supply the obligatory electron by oxidation of their [2Fe-2S]1+ to [2Fe-2S]2+, which is subsequently re-reduced to [2Fe-2S]1+ by Fre and Tah18, respectively. The maintenance pathway may use the same source of reducing equivalents to convert the inactive FeIII2-Y cluster to FeII2-Y, which subsequently reforms FeIII2-Y• in the presence of O2 via the biosynthetic pathway. HU = hydroxyurea.
In our search for factors involved in Fe2+ delivery (iron transporter, ferritin, chaperone etc.) required for Ia cofactor assembly, isogenic strains of the WT E. coli where four Fe2+/Me2+ transporters were deleted were studied by whole cell Mössbauer spectroscopy (13, 15). Under these conditions, we found that NrdEF is induced and becomes important for E. coli viability. A search of genomes for genes contiguous to nrdEnrdF revealed nrdI, annotated as a flavodoxin. NrdI was first identified in an operon containing the Lactococcus lactis nrdEnrdF genes, suggesting its importance in RNR metabolism (16). However, until 2010 its function remained a mystery. Initially, we thought that NrdI would function as an electron donor to assemble an FeIII2-Y• cofactor in RNR Ib, that is, a YfaE equivalent under iron-limiting conditions (17). However, old biological literature suggested that there was a manganese-requiring RNR (10). Despite efforts by many investigators to load Ia or Ib RNRs with manganese followed by the addition of a wide range of oxidants (O2⨪, H2O2, HOCl), in analogy with the self-assembly of the FeIII2-Y• cofactor, no RNR activity and no Y• were detected. The missing link turned out to be NrdI. Its unusual 1e− oxidation potentials, relative to flavodoxins involved in electron transfer, suggested that it might be functioning as the missing oxidant required to assemble a MnIII2-Y• cofactor (5). In vitro, this turns out to be the case. Our recent mechanistic studies suggest that the function of the reduced form of NrdI (NrdIhq, reduced FMN) is to generate O2⨪ and to deliver it directly via channeling to manganese in the metal 2 site in NrdF in a NrdF·NrdI complex (Fig. 1C) (18). Thus, in the case of MnIII2-Y• cluster assembly, the required e− and oxidant combine, and O2⨪ is the oxidant (Reaction 3). As noted above, an FeIII2-Y• cluster can also self-assemble to give active RNRs with Mn-RNR five times more active than the Fe-RNR (19).
REACTION 3.
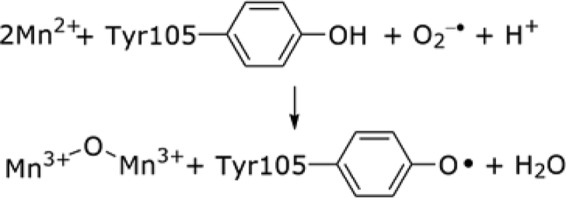
Class Ib RNRs Are Mn-RNRs In Vivo
Recent studies using different approaches have established the importance of the MnIII2-Y• NrdF in vivo in organisms with class Ia and Ib RNRs and with only class Ib RNRs. Studies by Martin and Imlay (6) in E. coli took advantage of the presence of a class III RNR (glycyl radical, anaerobic RNR) to keep ΔnrdAnrdB cells alive under anaerobic conditions, which allowed outgrowth in NrdEF+ cells under iron-limiting and iron-replete conditions in oxygenated media. In the former case, the Y• of Mn-NrdF was observed by whole cell EPR, whereas in the latter case, the Y• of Fe-NrdF was observed. The Mn-NrdF allowed cell growth, whereas the Fe-NrdF did not. These results suggest that the NrdF can self-assemble an iron cofactor in vivo. The inactivity of Fe-NrdF, however, is at odds with our experiments in vitro (20).
The Martin and Imlay studies (6) on NrdF gave insight into the function of YfaE in NrdB cluster assembly, specifically that YfaE is required to select iron over manganese loading to form active cofactor. They established that although yfaE is not essential under iron-replete growth conditions, it becomes essential in Hpx− cells. These cells lack catalase and peroxidase genes and as a result experience increase oxidative stress, including elevated levels of H2O2. Elevated peroxide up-regulates MntH, a manganese transporter. The lethality of ΔhpxΔyfaE was suppressed by deletion of MntH (6). The model is that manganese is able to compete with iron for binding to NrdB and that YfaE facilitates iron loading over manganese. Thus, their studies indicate that mismetallation occurs and that mechanisms have evolved to limit mismetallation, but the details remain to be revealed.
An alternative approach to establishing the importance of Mn-NrdF in vivo involved isolation of NrdF from its endogenous host. Studies from the Auling/Lubitz groups (21) with C. ammoniagenes and our group with E. coli and B. subtilis (20, 22, 23) all revealed Mn-NrdF and no apparent Fe-NrdF. Biochemical studies on these NrdFs have revealed that the activity of Mn-NrdF is 3.5–7 times that of the iron form and that both are active. These studies and more recent studies on NrdFs from B. anthracis, B. cereus, and S. sanguinis reveal that the issue of manganese loading remains a challenge (24–27). The reported very low specific activities are likely associated with apo-proteins in conformations that cannot be properly loaded with manganese in vitro. As in the class Ia RNRs, we are unable to deliver all of the metals (Me1 versus Me2, Fig. 1A) with the correct timing to the correct state of β. The timing of metal delivery, that is, whether to the folded or unfolded β and whether to the β (monomer) or β2 (dimer), is not known. Finally, in the case of B. subtilis, S. sanguinis, and Streptococcus pyogenes, NrdI has been shown to be essential under normal growth conditions (LB), and antibody studies reveal that NrdI acts catalytically (23, 27, 28).
Recently, a third and most comprehensive set of experiments has revealed the importance of S. sanguinis Mn-RNR to the survival of the organism in culture and in an animal model (27). This organism colonizes in biofilms in plaque deposits on teeth and during dental surgery can be dislodged into the blood, ultimately resulting in infective endocarditis due to the ability of the bacterium to colonize in the heart. As a note that reemphasizes the importance of the environmental niche of the organism being investigated, in the mouth, the concentration of manganese is 36 μm, and in the serum, it is 20 nm. S. sanguinis is also unusual in that it can tolerate 100 μm concentrations of H2O2 (29).
The S. sanguinis genome has been sequenced (30), the essential genes have been identified (31), and SsaB, a manganese transporter, has been established as a virulence factor (29, 32). Bioinformatics analysis of the S. sanguinis genome revealed that it has one aerobic RNR, a class Ib enzyme, and a class III RNR, required for anaerobic growth. In addition, three genes were annotated as NrdIs, only one of which is essential and involved in manganese cofactor assembly (27). Biochemical analysis has established that both the manganese-loaded and the iron-loaded RNRs are active with the former being 3.5 times more active than the latter (26).
Two sets of experiments have recently established that the Mn-RNR is required under normal aerobic growth conditions (6% O2/brain heart infusion medium) and in a rabbit model for infective endocarditis. To test the metallation state, several knockouts in the WT strain background were generated: ΔnrdHEKF,3 ΔnrdD, and ΔnrdI. The experimental design is based on the different O2 requirements for cluster assembly of the Fe-RNR and Mn-RNR and the class III RNR. For the Fe-RNR to be active, only NrdENrdF are required, and the organism should be viable with deletion of NrdI; the Mn-RNR requires NrdENrdF and NrdI; and the NrdD (anaerobic RNR) is inactivated in O2. The strains were prepared, and growth studies were carried out. The results indicated that under normal aerobic conditions, ΔnrdI showed a 10-fold decrease in cfu and that ΔnrdHEKF showed a similar decrease. Thus, these data reveal that a self-assembled Fe-RNR, if formed in vivo, is not sufficient to keep the organism alive and strongly support the importance of the Mn-RNR in vivo. In a rabbit model for infective endocarditis, in which a catheter was used to damage the heart valve where the bacteria colonize, the same strains, each with a distinct antibiotic resistance marker to facilitate cfu counting, were used. The WT strain and, for example, the ΔnrdI cells were mixed in a 1:1 ratio and injected into the rabbit. After an established time period, the heart was removed, and the vegetative growth of S. sanguinis was examined. The results indicated that the viability of the ΔnrdD strain was very similar to WT, and thus the anaerobic RNR is not required for colony growth in this model. The studies with ΔnrdI resulted in 104 less cfu relative to WT and indicate that NrdEF, as in the in vitro growth assays described above, is unable to support growth with Fe-NrdF. The two independent assays highlight the importance of the Mn-RNR and suggest that the virulence associated with the SsaB transporter could be related to the requirement of the organism for the Mn-RNR.
The results from the S. sanguinis studies and the E. coli studies, with Ib RNRs with very distinct functions, strongly suggest that in both cases, the Mn-RNR is the important form in vivo. The presence of a MnIII2-Y• cofactor in E. coli Ib RNR in which Ia is required for primary metabolism and in S. sanguinis where Ib is required for primary metabolism suggests that Mn-RNR can be essential for viability of organisms with different life styles and in different environments.
Class Ia RNRs of Eukaryotes: S. cerevisiae as a Model System
Eukaryotic class Ia RNRs thus far have been shown to utilize only the FeIII2-Y• cofactor. As with the bacterial systems, the cluster can self-assemble, and with the mouse RNR (33), the stoichiometry is identical to that shown in Reaction 2. Our focus has been on the model organism S. cerevisiae as it is amenable to both biochemical and genetic analyses. The RNRs from S. cerevisiae are biochemically similar to the human RNRs, but the regulation of iron homeostasis and of dNTP production is distinct. Thus, we will focus on our studies in S. cerevisiae and the similarities/differences of our results from the bacterial system.
S. cerevisiae RNR is unusual in that its β subunit is a heterodimer (ββ′) in which β′ is homologous to β, but has mutations in three of the six active site amino acids ligated to iron, preventing cluster assembly (34, 35). Thus, in vitro, only 1 Y•/ββ′ is possible. Studies in vitro have established that recombinant β and β′ form apo-homodimers, which rapidly (<1 min) interconvert to ββ′ when mixed in a 1:1 ratio (36). Structures of β2, β′2, and ββ′ have been solved, but metal loading in the oxidized and reduced states is incomplete, suggesting protein conformational heterogeneity (37, 38). As observed with the bacterial RNRs, our in vitro self-assembly studies with optimized protocols generated only ∼0.2 Y•/ββ′ (39), suggesting the importance of a biosynthetic pathway (Fig. 2). We and others have established that the active form of the S. cerevisiae β2 subunit in vivo under normal growth conditions is ββ′, and our whole cell EPR and quantitative Western analyses under normal growth conditions reveal 1 Y•/ββ′ and very high catalytic activity (36, 39, 61). In S. cerevisiae W303 strains, β′ (encoded by RNR4) is essential for dNTP production and cell growth, whereas in the S288C strains, it is not. However, the doubling time of the S288C Δrnr4 mutant is prolonged (180 min versus 90 min for the isogenic WT), and β is overexpressed 10-fold. The Δrnr4 mutant has only 0.01 Y•/β2, which surprisingly can support cell proliferation (36). High levels of RNR activity can be rapidly restored to these cells by induced expression of β′ (e.g. by using a galactose-inducible promoter-driven GalRNR4) or by supplementation of recombinant β′ to permeabilized Δrnr4 cells (14). Thus, β′ is also central to FeIII2-Y• formation and RNR activity. β′ appears to be required to maintain β in a conformation that is accessible for iron loading.
Source of Iron
As noted with bacterial systems, the source of iron (iron importers, storage proteins, labile iron pool, chaperone proteins) (40) has not been established in any case. To address this issue in eukaryotes, experiments in humans and S. cerevisiae have been carried out by several groups (41–45). Recent studies by the Philpott group in human cells suggest that poly(rC)-binding proteins (PCBPs) function as iron chaperones that are required for iron delivery to some iron-requiring proteins including ferritin (41), hypoxia-inducible factor (HIF) prolyl hydroxylases (42), and deoxyhypusine hydroxylase (43). However, because S. cerevisiae does not have a poly(rC)-binding protein counterpart, this protein will not be further discussed. Recent studies (44) have shown that the cytosolic monothiol glutaredoxins, Grx3 and Grx4, function in intracellular iron sensing and trafficking. Grx3 and Grx4 form homo- or heterodimers with a labile, glutathione-ligated [2Fe-2S] cluster at the dimer interface (45). Cells depleted of Grx3/Grx4 are impaired in iron-requiring reactions in the mitochondria, cytoplasm, and nucleus including proteins with Fe-S clusters, hemes, and di-iron clusters such as RNR. It is unclear whether and how Grx3/4 directly or indirectly participates in iron delivery into these diverse iron-requiring proteins. However, simultaneous deletion of both GRX3 and GRX4 is viable in the S288C strain background, whereas it is lethal in the W303 strain background. Thus, additional protein(s) or small peptides must be able to take the place of Grx3/4 as the iron source in the viable Δgrx3Δgrx4 mutant cells.
Whether a protein with an Fe-S cluster can deliver only iron and function as a chaperone protein remains to be established. Nevertheless, from studies on Fe-S-containing bacterial transcription factors, it is clear that oxidation of an Fe-S cluster can result in labilization of iron (46), and studies on Fe-S-requiring biotin synthase (47) and lipoate synthase (48) have demonstrated that an Fe-S cluster can deliver sulfur with loss of iron during this process. It remains to be determined whether the [2Fe-2S] cluster in Grx3/4 or a less stable version of this cluster can perform the function of iron delivery for RNR.
Source of Electrons
Our search for eukaryotic electron donors in RNR cofactor assembly was inspired by studies of the E. coli YfaE. The only yeast ferredoxin ortholog identified to date (Yah1) and its reductase (Arh1) are localized predominantly in the mitochondria. Studies by Netz et al. (49) and us (14, 50) have suggested that a protein complex formed between Dre2, containing a [2Fe-2S]2+/1+ and a [4Fe-4S] cluster (the redox state of the [4Fe-4S] is unclear), and Tah18, an NADPH-FMN/FADH2 oxidoreductase, functions as the cytosolic counterpart of Yah1-Arh1. Dre2-Tah18 constitutes an electron transfer chain in which electrons from NADPH are transferred via the flavins to the [2Fe-2S]2+ cluster of Dre2. Both DRE2 and TAH18 are essential genes. By using a conditional promoter-driven GalDRE2, we found that depletion of Dre2 greatly diminishes cellular RNR activity (50). In a separate assay, we used a GalRNR4Δcrt1 system in which β is constitutively overexpressed (due to removal of CRT1-mediated transcription repression), and thus formation of ββ′ and assembly of FeIII2-Y• are solely dependent on induction of β′ by turning on the galactose-inducible promoter. Under these conditions, we monitored reconstitution of Y• and ββ′ activity upon β′ induction in the WT and two tah18 temperature-sensitive mutant strains under the non-permissive temperature. We found that the tah18 mutants exhibited significant defects in both the kinetics and the maximum levels of Y• and ββ′ activity relative to the WT. These results support a model in which Dre2-Tah18 serves as the electron donor to RNR cofactor assembly. Importantly, Dre2 interacts physically with β, consistent with the notion of Dre2 directly delivering an electron to β.
In addition to RNR cofactor assembly, Dre2-Tah18 is also required at an early step(s) in the cytosolic Fe-S cluster assembly (CIA) pathway (49). Target proteins of the CIA machinery include the three major replicative DNA polymerases (α, ϵ, and δ) and several DNA helicases/nucleases (51, 52). As such, deficiency in Dre2-Tah18 leads to pleiotropic defects in genome stability due to their roles in cluster assembly both in the DNA replication and repair enzymes and in the RNR supplying the dNTP pools. Thus, genetic and biochemical analyses need to be combined to dissect the complex contributions of Dre2-Tah18 in these processes.
Link of RNR to Iron Metabolism
Physical interactions have been reported between Dre2 and Grx3 (53, 54), suggesting that electron transfer or perhaps Fe-S transfer can occur between these two proteins (Fig. 3). The findings that Grx3/4 and Dre2-Tah18 are required for both RNR cofactor formation and the CIA pathway have raised the interesting perspective that two distinct types of non-heme iron cofactors share the same sources of iron as well as reducing equivalents. A mechanism for how iron could be delivered to apo-protein from Grx3 (if in fact this occurs directly) remains unknown.
FIGURE 3.
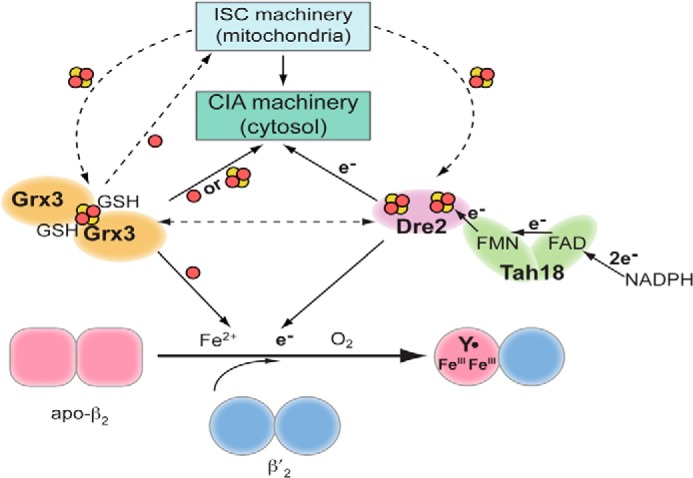
Connection between cofactor assembly pathways of the RNR and the mitochondrial (ISC) and cytosolic (CIA) iron-sulfur protein biosynthetic pathways in S. cerevisiae. Apo-β2 and -β′2 rapidly form apo-ββ′. Assembly of FeIII2-Y• in ββ′ is facilitated by β′, which stabilizes β in a conformation that allows iron binding. The [2Fe-2S]-bridged homodimer of Grx3 (Grx4) or heterodimer of Grx3/Grx4 is proposed to be required directly or indirectly for iron delivery to proteins involved in Fe-S cluster biosynthesis. The [2Fe-2S]2+ cluster of Dre2 receives an electron from NADPH via the FAD/FMN in Tah18 and is proposed to deliver it to β in RNR and to early step(s) in CIA (between Cfd1 and Nbp35). Assembly of the [2Fe-2S] clusters in Grx3/4 and Dre2 requires the mitochondria ISC, but not the cytosolic ISC machinery. Genetic and physical interactions have been observed between Dre2 and Grx3, although a mechanistic understanding of these interactions is lacking.
Finally, both Grx3 and Dre2-Tah18 have counterparts in humans (PICOT and CIAPIN1-NDOR1). CIAPIN1-NDOR1 and CIAPIN1-PICOT form complexes as well, and the former can substitute for the essential function of their yeast counterparts in yeast cells (49, 55). Their potential roles in Fe-S protein biogenesis and RNR cofactor assembly in human cells remain to be addressed. Given the surprisingly short half-life of the human FeIII2-Y• (30 min at 37 °C) relative to the length of the S phase (5–6 h) of the cell cycle, it is likely that FeIII2-Y• biosynthesis and repair play an important role in regulating human RNR activity. Further study of these proteins is thus warranted.
Issue of Mismetallation
Previous studies with prokaryotic and eukaryotic RNRs in vitro have established that Mn2+ can compete with Fe2+ for binding and prevent FeIII2-Y• cluster formation (56, 57). Given that mismetallation has been demonstrated for yeast mitochondrial manganese-superoxide dismutase when Fe2+ levels are elevated inside the mitochondrion (58, 59), it is likely that when the Mn2+/Fe2+ ratios are altered, similar mismetallation could occur in RNR, leading to reduced ability to generate dNTPs. The Culotta and Hoffman groups have recently shown that cells lacking the Golgi Mn2+/Ca2+ transporter Pmr1 have up to a 10-fold increase of intracellular manganese levels, which can be increased further by Mn2+ supplementation in the medium (60). Using this information, the Δpmr1GalDRE2 strain was created to investigate RNR. We found that the levels of Fe/β drop from 2 to 0.7 in the Δpmr1GalDRE2 mutant grown under Gal-off and Mn2+-supplemented conditions.4 Thus, in cells where Dre2 is depleted and Mn2+ levels are elevated, iron loading into β is compromised, possibly due to mismetallation by Mn2+. It remains to be determined whether Dre2, like the E. coli YfaE, can facilitate the choice of iron over manganese in RNR cofactor assembly in yeast cells. Consistent with a role in metal selection, YfaE is always found in bacterial genomes that contain both an Fe-RNR (Ia) and a Mn-RNR (Ib). Although eukaryotes only have Ia RNRs, it is conceivable that a YfaE-like specificity factor is needed to deal with complex changes in cellular iron and manganese levels in different cell types under various physiological conditions.
Summary and Future Prospects
Studies in the past few years have shown that under perturbed environmental conditions (often genetically), mismetallation in metalloenzymes can occur, raising the possibility that it may occur to some extent under normal growth conditions as well. Using RNR as the example for consideration of other metalloproteins, the extent of mismetallation will be governed by the binding constants for Fe2+ versus Mn2+ loading, the ratios of Fe2+/Mn2+ in the cytosol where cofactor assembly occurs, the speciation of both metals, the concentration of β, and the rate constants for Fe2+ and Mn2+ binding, dissociation, and cofactor assembly. Biochemical and biophysical determinations of these parameters present many technical challenges. It is remarkable how little quantitative information is available for any metalloprotein about these fundamental steps in cluster biosynthesis. In eukaryotic systems, the issues are even more complex as metal compartmentalization also comes into play. Each organism has distinct regulatory mechanisms for metal sensing, uptake, efflux, storage, etc. Finally, with RNR, the studies described herein on cluster assembly highlight another regulatory mechanism for controlling dNTP pools: cluster assembly and repair. The recent discovery (51, 52) of Fe-S clusters in polymerases, helicases, DNA repair enzymes, and the links between the CIA pathway and di-iron-cluster in RNR in eukaryotes makes iron homeostasis and nucleic acid biosynthesis intricately intertwined and deconvolution especially challenging. Because many of the proteins involved in cluster assembly are essential for cell survival, investigations of their in vivo functions have to utilize a protein depletion experiment using a conditional promoter shut-off method. The extent of the depletion depends on the half-life of the protein and whether it acts catalytically. Prolonged depletion may also trigger cellular stress responses that further complicate interpretation of the downstream events. New and more sensitive methods will be essential for future progress in both in vitro and in vivo studies addressing these critical problems.
This work was supported, in whole or in part, by National Institutes of Health Grants GM81393 (to J. S.) and CA125574 (to M. H.). This is the second article in the Thematic Minireview series “Metals in Biology 2014.”
nrdK is a non-essential gene of unknown function in the S. sanguinis class Ib RNR operon (31). Our biochemical studies suggest that it is not a manganese chaperone protein (unpublished data (O. Makhlynets and J. Stubbe)).
H. Li, M. Huang, and J. Stubbe, unpublished data.
- RNR
- ribonucleotide reductase
- CIA
- cytosolic iron-sulfur cluster assembly
- ISC
- iron-sulfur cluster
- FeIII2-Y•
- diferric-tyrosyl radical
- MnIII2-Y•
- dimanganic-tyrosyl radical.
REFERENCES
- 1. Hofer A., Crona M., Logan D. T., Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stubbe J., van Der Donk W. A. (1998) Protein radicals in enzyme catalysis. Chem. Rev. 98, 2661–2662 [DOI] [PubMed] [Google Scholar]
- 3. Minnihan E. C., Nocera D. G., Stubbe J. (2013) Reversible, long-range radical transfer in E. coli class la ribonucleotide reductase. Acc. Chem. Res. 46, 2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lundin D., Torrents E., Poole A. M., Sjöberg B.-M. (2009) RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cotruvo J. A., Stubbe J. (2011) Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin J. E., Imlay J. A. (2011) The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80, 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang W., Yun D., Saleh L., Bollinger J. M., Jr., Krebs C. (2008) Formation and function of the manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Biochemistry 47, 13736–13744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sjöberg B. M., Reichard P., Gräslund A., Ehrenberg A. (1977) Nature of the free-radical in ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 252, 536–541 [PubMed] [Google Scholar]
- 9. Atkin C. L., Thelander L., Reichard P. (1973) Iron and free radical in ribonucleotide reductase. J. Biol. Chem. 248, 7464–7472 [PubMed] [Google Scholar]
- 10. Willing A., Follmann H., Auling G. (1988) Ribonucleotide reductase of Brevibacterium ammoniagenes is a manganese enzyme. Eur. J. Biochem. 170, 603–611 [DOI] [PubMed] [Google Scholar]
- 11. Gon S., Faulkner M. J., Beckwith J. (2006) In vivo requirement for glutaredoxins and thioredoxins in the reduction of the ribonucleotide reductases of Escherichia coli. Antioxid. Redox Signal. 8, 735–742 [DOI] [PubMed] [Google Scholar]
- 12. Wu C. H., Jiang W., Krebs C., Stubbe J. (2007) YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry 46, 11577–11588 [DOI] [PubMed] [Google Scholar]
- 13. Hristova D., Wu C. H., Jiang W., Krebs C., Stubbe J. (2008) Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry 47, 3989–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., Liu L., Wu X., An X., Stubbe J., Huang M. (2011) Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 and Dre2 proteins. J. Biol. Chem. 286, 41499–41509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu C. H. (2007) In Vivo Cofactor Biosynthesis and Maintenance in the Class Ia Ribonucleotide Reductase Small Subunit of Escherichia coli. Ph.D. thesis, Massachusetts Institute of Technology [Google Scholar]
- 16. Jordan A., Pontis E., Aslund F., Hellman U., Gibert I., Reichard P. (1996) The ribonucleotide reductase system of Lactococcus lactis: characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271, 8779–8785 [DOI] [PubMed] [Google Scholar]
- 17. Cotruvo J. A., Jr., Stubbe J. (2008) NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib RNR. Proc. Natl. Acad. Sci. U.S.A. 105, 14383–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boal A. K., Cotruvo J. A., Jr., Stubbe J., Rosenzweig A. C. (2010) Structural basis for activation of class Ib ribonucleotide reductase. Science 329, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cotruvo J. A., Jr., Stubbe J. (2012) Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4, 1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cotruvo J. A., Jr., Stubbe J. (2010) An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib RNR. Biochemistry 49, 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox N., Ogata H., Stolle P., Reijerse E., Auling G., Lubitz W. (2010) A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 132, 11197–11213 [DOI] [PubMed] [Google Scholar]
- 22. Cotruvo J. A., Jr., Stubbe J. (2011) Escherichia coli class Ib RNR contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 50, 1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Stubbe J. (2011) Bacillus subtilis class Ib RNR is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry 50, 5615–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crona M., Torrents E., Røhr A. K., Hofer A., Furrer E., Tomter A. B., Andersson K. K., Sahlin M., Sjöberg B.-M. (2011) NrdH-redoxin protein mediates high enzyme activity in manganese-reconstituted ribonucleotide reductase from Bacillus anthracis. J. Biol. Chem. 286, 33053–33060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammerstad M., Hersleth H. P., Tomter A. B., Røhr A. K., Andersson K. K. (2014) Crystal structure of Bacillus cereus class Ib ribonucleotide reductase di-iron NrdF in complex with NrdI. ACS Chem. Biol. 9, 526–537 [DOI] [PubMed] [Google Scholar]
- 26. Makhlynets O., Boal A. K., Rhodes D. V., Kitten T., Rosenzweig A. C., Stubbe J. (2014) Streptococcus sanguinis class Ib ribonucleotide reductase: high activity with both iron and manganese cofactors and structural insights. J. Biol. Chem. 289, 6259–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rhodes D. V., Crump K. E., Makhlynets O., Snyder M., Ge X., Xu P., Stubbe J., Kitten T. (2014) Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis. J. Biol. Chem. 289, 6273–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roca I., Torrents E., Sahlin M., Gibert I., Sjöberg B. M. (2008) NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J. Bacteriol. 190, 4849–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crump K. E., Bainbridge B., Brusko S., Turner L. S., Ge X., Stone V., Xu P., Kitten T. (2014) The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol. Microbiol. 92, 1243–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu P., Alves J. M., Kitten T., Brown A., Chen Z., Ozaki L. S., Manque P., Ge X., Serrano M. G., Puiu D., Hendricks S., Wang Y., Chaplin M. D., Akan D., Paik S., Peterson D. L., Macrina F. L., Buck G. A. (2007) Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189, 3166–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu P., Ge X., Chen L., Wang X., Dou Y., Xu J. Z., Patel J. R., Stone V., Trinh M., Evans K., Kitten T., Bonchev D., Buck G. A. (2011) Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 1, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das S., Kanamoto T., Ge X., Xu P., Unoki T., Munro C. L., Kitten T. (2009) Contribution of lipoproteins and lipoprotein processing to endocarditis virulence in Streptococcus sanguinis. J. Bacteriol. 191, 4166–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ochiai E., Mann G. J., Gräslund A., Thelander L. (1990) Tyrosyl free-radical formation in the small subunit of mouse ribonucleotide reductase. J. Biol. Chem. 265, 15758–15761 [PubMed] [Google Scholar]
- 34. Huang M., Elledge S. J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P. J., Chabes A., Casagrande R., Tian X. C., Thelander L., Huffaker T. C. (1997) Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 17, 6114–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perlstein D. L., Ge J., Ortigosa A. D., Robblee J. H., Zhang Z., Huang M., Stubbe J. (2005) The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44, 15366–15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sommerhalter M., Voegtli W. C., Perlstein D. L., Ge J., Stubbe J., Rosenzweig A. C. (2004) Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers. Biochemistry 43, 7736–7742 [DOI] [PubMed] [Google Scholar]
- 38. Voegtli W. C., Ge J., Perlstein D. L., Stubbe J., Rosenzweig A. C. (2001) Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer. Proc. Natl. Acad. Sci. U.S.A. 98, 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ortigosa A. D., Hristova D., Perlstein D. L., Zhang Z., Huang M., Stubbe J. (2006) Determination of the in vivo stoichiometry of tyrosyl radical per ββ′ in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry 45, 12282–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lisher J. P., Giedroc D. P. (2013) Manganese acquisition and homeostasis at the host-pathogen interface. Front. Cell. Infect. Microbiol. 3, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nandal A., Ruiz J. C., Subramanian P., Ghimire-Rijal S., Sinnamon R. A., Stemmler T. L., Bruick R. K., Philpott C. C. (2011) Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 14, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frey A. G., Nandal A., Park J. H., Smith P. M., Yabe T., Ryu M. S., Ghosh M. C., Lee J., Rouault T. A., Park M. H., Philpott C. C. (2014) Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 111, 8031–8036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E., Lillig C. H., Lill R. (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H., Mapolelo D. T., Randeniya S., Johnson M. K., Outten C. E. (2012) Human glutaredoxin 3 forms 2Fe-2S-bridged complexes with human BolA2. Biochemistry 51, 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crack J. C., Green J., Cheesman M. R., Le Brun N. E., Thomson A. J. (2007) Superoxide-mediated amplification of the oxygen-induced switch from 4Fe-4S to 2Fe-2S clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. U.S.A. 104, 2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fugate C. J., Stich T. A., Kim E. G., Myers W. K., Britt R. D., Jarrett J. T. (2012) 9-Mercaptodethiobiotin is generated as a ligand to the [2Fe-2S]+ cluster during the reaction catalyzed by biotin synthase from Escherichia coli. J. Am. Chem. Soc. 134, 9042–9045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lanz N. D., Pandelia M. E., Kakar E. S., Lee K. H., Krebs C., Booker S. J. (2014) Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase. Biochemistry 53, 4557–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Netz D. J. A., Stümpfig M., Doré C., Mühlenhoff U., Pierik A. J., Lill R. (2010) Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y., Li H., Zhang C., An X., Liu L., Stubbe J., Huang M. (2014) Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Netz D. J. A., Stith C. M., Stümpfig M., Köpf G., Vogel D., Genau H. M., Stodola J. L., Lill R., Burgers P. M. J., Pierik A. J. (2012) Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White M. F., Dillingham M. S. (2012) Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 22, 94–100 [DOI] [PubMed] [Google Scholar]
- 53. Saito Y., Shibayama H., Tanaka H., Tanimura A., Matsumura I., Kanakura Y. (2011) PICOT is a molecule which binds to anamorsin. Biochem. Biophys. Res. Commun. 408, 329–333 [DOI] [PubMed] [Google Scholar]
- 54. Tarassov K., Messier V., Landry C. R., Radinovic S., Serna Molina M. M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. (2008) Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28, 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Atta M., Nordlund P., Åberg A., Eklund H., Fontecave M. (1992) Substitution of manganese for iron in ribonucleotide reductase from Escherichia coli: spectroscopic and crystallographic characterization. J. Biol. Chem. 267, 20682–20688 [PubMed] [Google Scholar]
- 57. Rova U., Goodtzova K., Ingemarson R., Behravan G., Gräslund A., Thelander L. (1995) Evidence by site-directed mutagenesis supports long-range electron-transfer in mouse ribonucleotide reductase. Biochemistry 34, 4267–4275 [DOI] [PubMed] [Google Scholar]
- 58. Aguirre J. D., Culotta V. C. (2012) Battles with iron: manganese in oxidative stress protection. J. Biol. Chem. 287, 13541–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C. (2009) The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284, 22633–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McNaughton R. L., Reddi A. R., Clement M. H. S., Sharma A., Barnese K., Rosenfeld L., Gralla E. B., Valentine J. S., Culotta V. C., Hoffman B. M. (2010) Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 15335–15339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chabes A., Domkin V., Larsson G., Liu A., Gräslund A., Wijmenga S., Thelander L. (2000) Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. U.S.A. 97, 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]


