Background: Interleukin-8 (IL-8) is a potent proinflammatory cytokine.
Results: We identify many regulators of IL-8 secretion induced by the pattern recognition receptor NOD2.
Conclusion: Many diverse cellular processes, including protein ubiquitination and trafficking, specifically affect IL-8 secretion.
Significance: The identified regulators, particularly those present in inflammatory disease loci, reveal molecular targets for therapeutic modulation of IL-8.
Keywords: Cytokine, Inflammation, Inflammatory Bowel Disease (IBD), NOD-like Receptor (NLR), Secretion, IL-8, JAK2, NOD2, USP8
Abstract
NOD2 encodes an intracellular multidomain pattern recognition receptor that is the strongest known genetic risk factor in the pathogenesis of Crohn disease (CD), a chronic relapsing inflammatory disorder of the intestinal tract. NOD2 functions as a sensor for bacterial cell wall components and activates proinflammatory and antimicrobial signaling pathways. Here, using a genome-wide small interfering RNA (siRNA) screen, we identify numerous genes that regulate secretion of the proinflammatory cytokine IL-8 in response to NOD2 activation. Moreover, many of the identified IL-8 regulators are linked by protein-protein interactions, revealing subnetworks of highly connected IL-8 regulators implicated in processes such as vesicle formation, mRNA stability, and protein ubiquitination and trafficking. A TNFα counterscreen to induce IL-8 secretion in an NOD2-independent manner reveals that the majority of the identified regulators affect IL-8 secretion irrespective of the initiating stimuli. Using immortalized macrophages, we validate the ubiquitin protease, USP8, and the endosomal sorting protein, VPS28, as negative regulators of NOD2-induced cytokine secretion. Interestingly, several genes that affect NOD2-induced IL-8 secretion are present in loci associated with CD risk by genome-wide association studies, supporting a role for the NOD2/IL-8 pathway, and not just NOD2, in the pathogenesis of CD. Overall, this screen provides a valuable resource in the advancement of our understanding of the genes that regulate the secretion of IL-8.
Introduction
Cytokines are secreted immunoregulators affecting the recruitment and activation of various immune cell types. Their production is tightly controlled and deregulated and is associated with a variety of human diseases (1). The secretion of cytokines in response to microorganisms plays a critical role in host defense and is mediated by pattern recognition receptors. Indeed, polymorphisms in multiple members of the NOD-like receptor family of intracellular pattern recognition receptors are associated with various inflammatory diseases (2, 3). The NOD-like receptor family member NOD2 is one of best characterized genetic risk factors in the pathogenesis of Crohn disease (CD)3 (4, 5). NOD2, which is expressed in a variety of cell types, including intestinal epithelial cells, macrophages, and dendritic cells, functions as a sensor for bacterial peptidoglycan (6, 7). Muramyl-dipeptide (MDP), a synthetic molecule that is the minimum known component of bacterial peptidoglycan capable of activating NOD2, leads to the recruitment of the Ser/Thr/Tyr kinase RIPK2, a critical step in the activation of downstream signaling pathways (8). Subsequent ubiquitination of RIPK2 leads to the recruitment of the Ser/Thr kinase TAK1 that mediates the activation of both the MAPK and NF-κB signaling pathways (9–11) that together regulate the production of effector molecules, such as IL-8.
IL-8 (CXCL8) is a pleiotropic proinflammatory cytokine produced by many cell types in response to various stressors. IL-8 was first described as a neutrophil chemo-attractant (12) but has since been identified as a potent angiogenic factor involved in tumor growth and metastasis (13, 14) as well as being a biomarker for various chronic inflammatory conditions (15–17). Many cell types secrete IL-8 in response to NOD2 activation (18), and multiple groups have shown decreased IL-8 secretion in cells from CD patients with NOD2 polymorphisms (19, 20). Moreover, NOD2 mutations are associated with Blau syndrome and early onset sarcoidosis (21, 22), two inflammatory disorders affecting the eyes, skin, and joints. Thus, it is clear that NOD2 signaling plays an important role in maintaining inflammatory homeostasis, prompting us to systematically interrogate regulators of NOD2-induced IL-8 secretion.
In this paper, we report the results of a genome-wide siRNA screen identifying numerous genes that regulate NOD2-induced IL-8 secretion. Many of the identified IL-8 regulators affect both NF-κB induction and IL-8 secretion, validating our previously reported regulators of NOD2-induced NF-κB activation (23). Here, however, we focus our analysis on genes that specifically affect IL-8 secretion independent of NF-κB activation. Interestingly, many of the identified IL-8 regulators are present in loci associated with CD risk. Overall, the identified IL-8 regulators provide a molecular framework for understanding how NOD2-induced IL-8 secretion is controlled and offers insight into the molecular mechanisms that may be deregulated in inflammatory diseases, such as Crohn disease.
EXPERIMENTAL PROCEDURES
Genome-wide siRNA Screen
IL-8 levels from frozen cell supernatants stored from our previously described siRNA screen (23) were measured by ELISA. Briefly, HEK293 cells stably expressing human NOD2 (HEK293-NOD2) were cultured on 384-well plates (Greiner Bio-One) in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mm glutamine, 1 mm sodium pyruvate, and 1× penicillin/streptomycin (Invitrogen). Pools of four distinct siGENOME siRNAs (Thermo Scientific) were reverse transfected at a concentration of 20 nm with Lipofectamine 2000 (Invitrogen) diluted in Opti-MEM (Invitrogen). After 48 h, to allow for gene silencing, cells were stimulated with the NOD2 agonist MDP (20 ng/ml; Bachem) using a Thermo Labsystems Multidrop plate dispenser. Cell supernatants were collected 18 h later using a Biomek FX programmable robot (Beckman), sealed, and frozen at −20 °C. Experimental conditions were chosen based on initial optimization experiments testing different siRNA transfection protocols and varying the length and dose of MDP to achieve an ∼10-fold increase in IL-8 secretion after MDP stimulation relative to unstimulated cells. The concentration of human IL-8 was measured by sandwich ELISA (R&D Systems) according to the manufacturer's recommended protocol adapted to a 384-well plate format. Cell supernatants were thawed and diluted 1:1 in assay buffer (Tris-buffered casein; Surmodics) and developed with TMB substrate (Surmodics). Concentrations of IL-8 were determined by comparison with an eight-point standard curve (in quadruplicate) prepared on each plate.
Secondary Validation
Reporter cells were reverse transfected with 40 nm ON-TARGETplus siRNA pools (Thermo Scientific) for 48 h, followed by stimulation with either MDP (20 ng/ml) or human TNFα (10 ng/ml, R&D Biosystems) for 18 h. IL-8 levels were determined as described above, and NF-κB luciferase and viability assays were carried out as described previously (23).
Data Analysis
A custom-built database (MScreen) was used for data storage, display, and analysis. In order to compare IL-8 levels across assay plates, IL-8 values were normalized relative to RIPK2-specific and non-targeting (NT) siRNA present on each assay plate. The average amount of IL-8 (in pg/ml) from multiple wells receiving RIPK2-specific siRNA was considered as 100% inhibition, whereas the average amount of IL-8 in wells that received NT siRNA was considered as 0% inhibition. Therefore, test siRNA that resulted in decreased IL-8 secretion (relative to those receiving NT siRNA) resulted in values greater than 0, whereas test siRNAs that resulted in increased IL-8 secretion (relative to that of cells treated with NT siRNA) resulted in values less than 0. For assay plates stimulated with TNFα, IL-8 levels were normalized against wells receiving RELA-specific siRNA, which was considered as 100% inhibition.
Reagents
JAK Inhibitors were purchased from EMD Millipore (pan JAK inh (catalog no. 420099) and JAK2 inh (catalog no. 420139)).
Quantitative Real-time PCR
Total RNA was isolated using an EZNA Omega Biotek kit as directed by the manufacturer. cDNA was synthesized using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer's instructions. Quantitative real-time PCR was performed using predesigned gene-specific Taqman primers and probes, PCR master mix, and the StepOne PlusTM real-time PCR system (Applied Biosystems). Expression was normalized to that of GAPDH, and expression levels were analyzed by the 2−ΔΔCt method.
Transfection of Immortalized Macrophages
Bone marrow-derived macrophages from B6 mice were immortalized as described previously (24) and were transfected using the INTERFERinTM polyplus transfection reagent (VWR) as described by the manufacturer.
Bacterial Stimulation
Staphylococcus aureus (strain 8325-4, a gift from Timothy Foster, Trinity College, Dublin, Ireland) was inoculated into brain heart infusion medium and incubated overnight at 30 °C with shaking. Overnight cultures were subcultured into fresh media and incubated at 37 °C with shaking for ∼2 h until reaching an A600 of 0.6. The bacteria were washed and diluted in sterile phosphate-buffered saline (PBS). 40,000 bacteria were added to each well, giving a bacterium/HEK293-NOD2 cell ratio of ∼1:1. After a 1-h infection, gentamicin (50 ng/μl; Invitrogen) was added to the media. Supernatants were collected after 18 h.
RESULTS
siRNA Screening Reveals Regulators of NOD2-induced IL-8 Secretion
We previously reported a genome-wide siRNA screen targeting 18,100 human genes to identify regulators of NOD2-induced NF-κB activation (23). To further examine the relationship between NOD2 signaling and IL-8 secretion downstream of NF-κB activation, we measured the levels of IL-8 present in cell supernatants by ELISA (Fig. 1A and supplemental Fig. S1). Genes whose silencing decreases IL-8 secretion were classified as positive regulators, whereas genes whose knockdown increased IL-8 secretion were categorized as negative regulators (Fig. 1B). Relative IL-8 levels were normalized against multiple positive (RIPK2) and negative siRNA controls on each assay plate (supplemental Fig. S2) and are reported in supplemental Table S1. Histogram analysis reveals a non-normal distribution skewed toward negative regulators (supplemental Fig. S3). Duplicate IL-8 measurements from independent replica plates were collected, and linear correlation analysis shows a high degree of reproducibility (supplemental Fig. S4). As previously reported, the effect of gene silencing on cell viability was quantified to control for nonspecific effects on cell number (23). As expected, genes whose knockdown significantly reduces cell viability tend to have decreased levels of IL-8 secretion, whereas gene knockdown resulting in increased viability tends to result in increased IL-8 levels (supplemental Fig. S5). Therefore, to reduce false positives related to nonspecific effects on cell number, genes whose silencing either enhances IL-8 secretion and increases viability (>120%) or decreases both IL-8 secretion and viability (<70%) were considered inconclusive with respect to their effect on NOD2-induced IL-8 secretion.
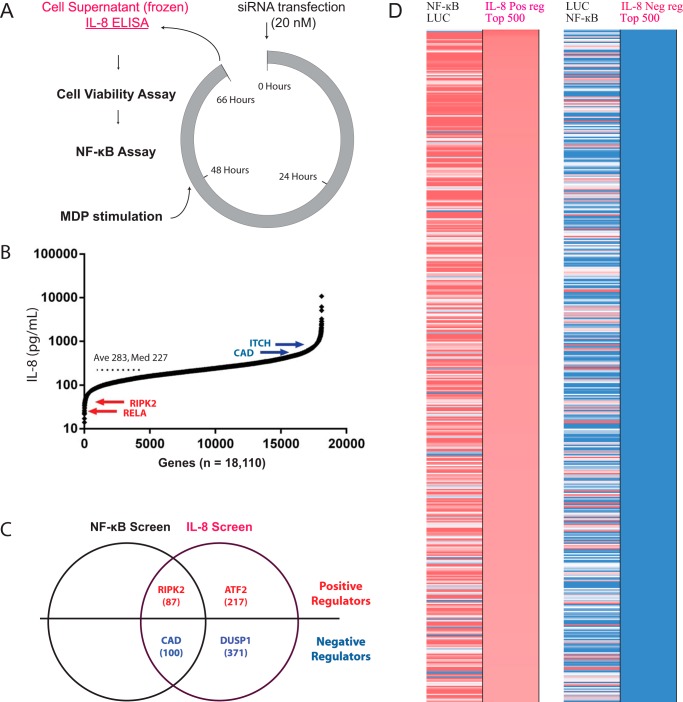
FIGURE 1.
A genome-wide siRNA screen for regulators of NOD2-induced IL-8. A, schematic of the genome-wide siRNA screen highlighting the collection of cell supernatants for analysis of IL-8 secretion by ELISA (purple). B, a rank order plot summarizing the measured IL-8 protein levels for each of the 18,110 genes tested along with the average (Ave) and median (Med) IL-8 values across all genes. Selected genes known to affect NOD2 signaling are labeled for reference. C, a Venn diagram summarizing the overlap in positive and negative regulators revealed by the previously reported NF-κB screen and this IL-8 screen. The number of concordant (supplemental Table S2) and IL-8-specific (supplemental Table S3) regulators are shown along with examples of previously reported regulators of IL-8 secretion. D, a heat map comparing the effect of gene silencing on NF-κB induction and IL-8 secretion. Gene silencing resulted in increased (blue) or decreased (red) levels of IL-8. Genes with nonspecific effects on cell viability were removed, and only the top 500 for each of the positive and negative regulators genes were included in this analysis.
Numerous genes known to affect IL-8 production are identified as hits in the screen, thereby validating our experimental approach (Fig. 1C). For example, it is well known that IL-8 production is strongly regulated at the transcriptional level by NF-κB- and AP-1-responsive elements in the IL-8 promoter (25). Indeed, knockdown of the NF-κB transcription factor RELA or its binding partner RNF31 diminishes both NF-κB activation and IL-8 secretion, consistent with their previously reported roles in NOD2 signaling (23, 26). Furthermore, silencing the NOD2 binding protein, CAD, which has previously been implicated in the negative regulation of NOD2 signaling (27), increases both NF-κB activation and IL-8 secretion. By comparing our previously reported regulators of NOD2-induced NF-κB activation with this IL-8 data set, we were able to identify 87 positive and 100 negative regulators that concordantly increase or decrease both NF-κB activity and IL-8 secretion, respectively (Fig. 1D and supplemental Table S2). Of greater interest, we were able to identify many genes that specifically influenced IL-8 secretion without affecting NF-κB induction. These genes, which we refer to as IL-8-specific regulators, probably influence IL-8 secretion either downstream of NF-κB or via parallel pathways. For example, silencing SFPQ, an IL-8 mRNA-binding protein that negatively regulates IL-8 expression by sequestering IL-8 mRNA into nuclear paraspeckles (28), enhances IL-8 secretion without affecting NF-κB activity. Furthermore, many groups have shown a role for the MAPK pathway in the regulation of IL-8 secretion (29), and, although siRNAs targeting individual MAPKs did not significantly affect IL-8 secretion in our screen, presumably due to genetic redundancy, knockdown of the MAPK substrate, ATF-2 (activating transcription factor-2), which binds to and activates the IL-8 promoter (30, 31), was identified as an IL-8-specific positive regulator. In another case, DUSP1, a MAPK phosphatase that down-regulates MAPK activity, was identified as an IL-8-specific negative regulator, consistent with previous reports (32, 33). Furthermore, knockdown of either ITCH (34) or BSG (35), two NOD2-binding proteins known to negatively regulate NOD2 signaling, increases IL-8 secretion. In total, 218 IL-8-specific positive regulators and 391 IL-8-specific negative regulators were identified that either decrease or increase IL-8 secretion, respectively, without major effects on NF-κB activation (Fig. 1D and supplemental Table S3). These results highlight the utility of monitoring IL-8 at the protein level, thereby allowing us to interrogate genes that influence either parallel pathways (i.e. the MAPK pathway) or pathways downstream of NF-κB induction that are specific to IL-8 secretion, thereby providing additional insight into the molecular mechanisms that control NOD2 signaling.
Orthologonal Data Sets Support Numerous IL-8 Regulators
Given that protein-protein interactions (PPIs) mediate the assembly and regulation of signal transduction pathways, we examined proteins that interact with known core components of the NOD2/IL-8 signaling pathway using the STRING (36) and BIOGRID (37) PPI databases. Many NOD2-, RIPK2-, RELA-, and MAPK-associated proteins affect IL-8 secretion (Fig. 2 and supplemental Fig. S6). In addition, we asked whether gene knockdown of proteins reported to interact with NOD2 in the literature (supplemental Table S4) influenced IL-8 secretion. Under the conditions used in the screen, 8 of 44 reported NOD2-interacting proteins affect IL-8 secretion, and numerous other proteins that interact with these known NOD2-associated proteins affect IL-8 secretion (supplemental Fig. S7). Finally, using various sources, including recently reported NOD1 (39), NOD2 (40), and NF-κB (41) siRNA screens (supplemental Table S5), we were able to identify supporting evidence in the literature for many of the identified IL-8 regulators. Overall, the supporting evidence provided by overlaying information from PPI networks and other relevant “omics” data sets increases the confidence in the validity of the identified IL-8 regulators.
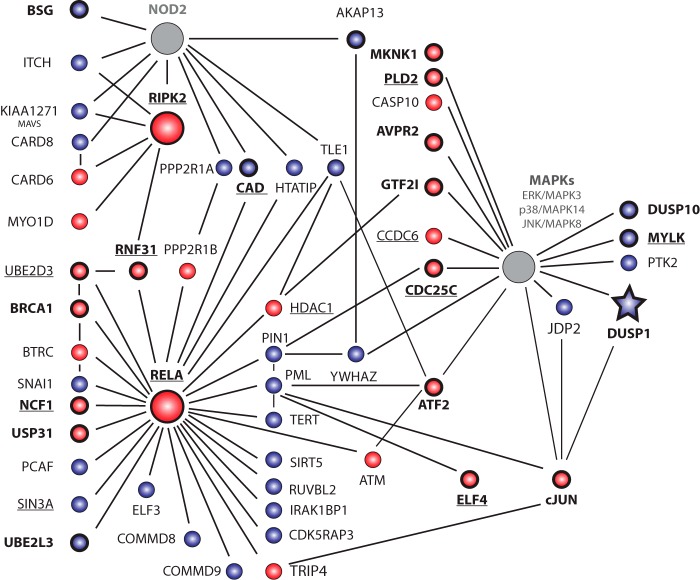
FIGURE 2.
Numerous proteins that interact with core components of the NOD2 and MAPK signaling pathways were identified as regulators of IL-8 secretion. Positive (red) or negative (blue) regulators of IL-8 secretion known to interact with core components of the NOD2 signaling pathway, as determined using STRING and/or BIOGRID PPI databases, are shown. NOD2, RIPK2, RELA, and the indicated MAPKs were selected as core components and are highlighted by large circles. Gray shading indicates genes whose knockdown did not significantly affect IL-8 protein levels. The knockdown of underlined genes affected both IL-8 secretion and NF-κB activation. Genes in boldface type were validated using independent siRNA pools. Normalized IL-8 data is included in supplemental Fig. S6.
Secondary Validation and TNFα Counterscreening
To minimize false positives due to off-target effects, a secondary validation screen was performed using alternative siRNA pools. In total, 554 genes whose silencing affected MDP-induced IL-8 secretion in the primary screen were assessed in triplicate, validating 53 positive and 83 negative regulators (supplemental Table S6). For example, RIPK2, RELA, and ELF4, which encodes a transcription factor that binds to the IL-8 promoter (42), were each confirmed as positive regulators of IL-8 secretion, whereas silencing TNFAIP3, a known negative regulator of NOD2 signaling (43), enhanced IL-8 secretion. In addition, knockdown of MKNK1 (MAPK-interacting Ser/Thr kinase 1), which acts downstream of p38 to increase protein translation (44, 45), decreased IL-8 secretion, whereas ARRB2 silencing increased IL-8 secretion, consistent with previous reports implicating it as a negative regulator of other proinflammatory cytokines (46, 47). In summary, the use of independent siRNA reagents confirmed several known regulators, highlighted above, and provides a list of dozens of high confidence IL-8 regulators for future in vivo characterization.
Although many different stimuli are capable of inducing IL-8 secretion, we were interested in distinguishing IL-8 regulators that specifically affect NOD2-induced IL-8 secretion from those that affect IL-8 secretion in response to other stimuli. Therefore, IL-8 production in response to TNFα stimulation was measured using the same set of 554 genes described above. As expected, knockdown of RELA inhibited IL-8 production in response to stimulation with either MDP or TNFα, whereas RIPK2 silencing specifically inhibited MDP-induced IL-8, and RIPK1 knockdown specifically inhibited TNFα-induced IL-8 (supplemental Fig. S8). Overall, the vast majority of the genes tested affected both MDP- and TNFα-induced IL-8 secretion (supplemental Table S6), suggesting that most of the identified regulators function downstream of RIPK2 and act as general regulators of IL-8 secretion independent of the activating signal.
Subnetworks of Interacting Proteins Regulate IL-8 Secretion
PPIs taken from publically available databases reveal highly connected complexes of the IL-8 regulators, many of which could be linked to NOD2 (Fig. 3). These interactions provide biochemical support for and mechanistic insight into how these gene products might affect IL-8 secretion. The COP1 complex represents one notable example because multiple members of this complex were identified and validated as negative regulators of IL-8 secretion. The COP1 complex plays a role in coating intracellular vesicles that function in retrograde transport from the Golgi to the endoplasmic reticulum (48). Interestingly, the COP1 complex can be linked to NOD2 via PPIs with other identified IL-8 regulators also involved in vesicle transport (STX8, VTI1A, and the CD risk factor VAMP3). A second PPI subnetwork implicated in vesicle fusion and protein trafficking contained two 14-3-3 proteins, YWHAZ and YWHAG. Additional subnetworks of interacting IL-8 regulators were also revealed using PPI data (supplemental Figs. S9 and S10). For instance, six components of the integrator complex, a group of proteins implicated in the production of small nuclear RNAs and splicing (49), were identified as IL-8-specific negative regulators, whereas multiple components of the Mediator complex, a multiprotein complex that functions as a transcriptional co-activator (50), affected IL-8 secretion. Also of note, knockdown of multiple genes associated with Hermansky-Pudlak syndrome, a rare disorder associated with defects in protein trafficking (51), affected IL-8 secretion. Interestingly, a subset of Hermansky-Pudlak syndrome patients exhibit intestinal granulomas and inflammation similar to CD, supporting an overlap in the pathways underlying these conditions (52). Together, these data indicate that NOD2-induced IL-8 secretion is regulated by many distinct complexes of genes involved in a variety of cellular processes, such as RNA processing and protein trafficking.
FIGURE 3.
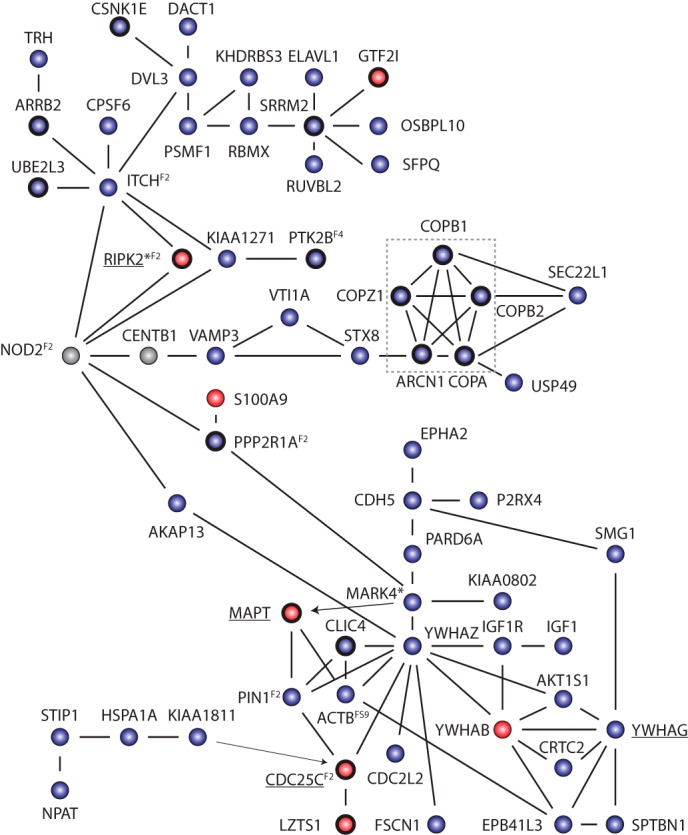
Genome-wide screen identifies networks of positive and negative regulators of NOD2 induced IL-8 secretion. PPIs among hits from the siRNA screen (circles) from publically available databases, such as STRING and BIOGRID, are depicted by black lines. Arrows point from a kinase to its substrate. Underlined genes also affect NOD2 dependent NF-κB activity. Genes in boldface type were validated using independent siRNA pools. *, genes identified as regulators of NOD1-induced IL-8 mRNA (39). Superscript numbers indicate other figures to refer to for additional interacting proteins, not shown for simplicity. The COP1 complex, mentioned under “Results,” is highlighted in the dashed gray box.
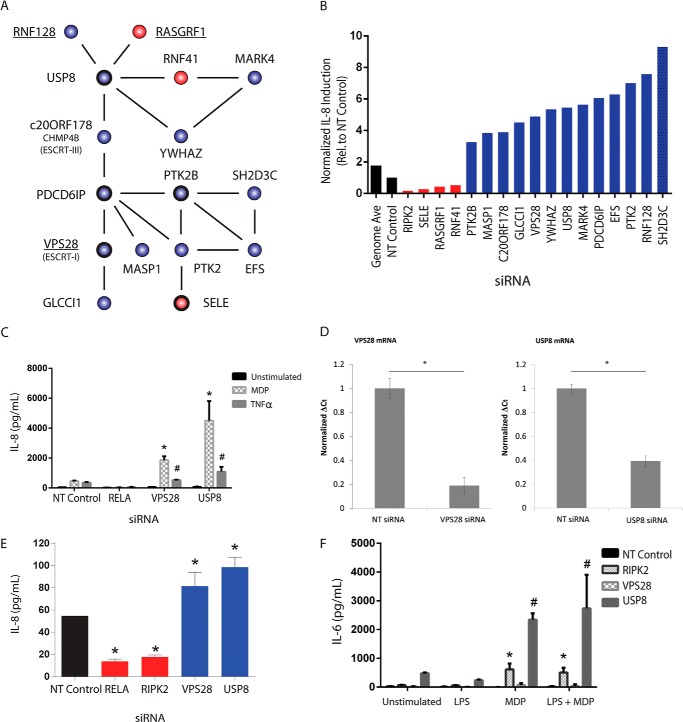
USP8 and VPS28 Negatively Regulate IL-8 Secretion
Several groups have reported an important role for protein ubiquitination in the control of NOD2 signaling. Therefore, we chose to examine a subnetwork of interacting IL-8 regulators centered around the ubiquitin-specific protease USP8 (Fig. 4A). Knockdown of USP8 and several of its interacting proteins significantly affected IL-8 secretion after stimulation with MDP (Fig. 4B). Among the IL-8 regulators present in the PPI network, both USP8 and VPS28 (vacuolar protein sorting 28) were validated as negative regulators of IL-8 secretion using independent siRNAs (Fig. 4, C and D). We went on to test the effect of silencing these two genes on IL-8 secretion in a cell-based bacterial infection assay using NOD2-expressing HEK293 cells exposed to S. aureus, a Gram-positive bacteria known to activate NOD2 signaling (53–55). Knockdown of either USP8 or VPS28 significantly enhanced IL-8 secretion in response to bacteria, consistent with a role for these two genes in the negative regulation of IL-8 secretion (Fig. 4E). Given that USP8 is strongly expressed in monocytes and macrophages (56), we went on to assess whether USP8 and VPS28 silencing affects cytokine secretion, using an immortalized murine macrophage assay. Because mice lack IL-8, we examined the production of the proinflammatory cytokine IL-6, which is secreted by LPS primed macrophages after MDP stimulation in a NOD2-dependent manner (53). Knockdown of either USP8 or VPS28 increased secretion of IL-6 relative to NT siRNA controls (Fig. 4F). Together, these data support a role for USP8 and VPS28 in the negative regulation of cytokine secretion.
FIGURE 4.
USP8 negatively regulates NOD2-induced IL-8 secretion. A, PPIs (lines) between identified IL-8 regulators (circles) revealed a subnetwork of highly connected proteins centered around USP8. Gene knockdown either decreased (red) or increased (blue) IL-8 secretion. Genes in boldface type were validated using independent siRNA pools. Underlined proteins also affected NOD2 dependent NF-κB activity. B, the normalized effect of silencing each gene in the subnetwork on IL-8 secretion in the primary screen is shown. C, gene silencing of USP8 and VPS28 in NOD2-expressing HEK293 cells using independent siRNA pools enhanced IL-8 secretion in response to stimulation with either the NOD2 agonist MDP or TNFα. D, mRNA levels of USP8 and VPS28 were assessed by quantitative real-time PCR to confirm knockdown. E, knockdown of USP8 or VPS28 enhances IL-8 secretion in response to bacterial exposure. NOD2-expressing HEK293 cells were transiently transfected with NT, USP8, or VPS28 siRNAs and 48 h later infected with S. aureus. Gentamycin was used to kill extracellular bacteria after 30 min, and IL-8 levels in the supernatant were measured by ELISA 17 h later. F, immortalized mouse macrophages were stimulated with MDP with or without LPS priming 48 h after transfection with the indicated siRNAs. Levels of the proinflammatory cytokine IL-6 were measured by ELISA 18 h after stimulation. C–F, knockdown experiments were performed in triplicate. Error bars, S.D. * and #, p values of <0.05 using Student's t tests against stimulation-specific NT siRNA controls.
Multiple IL-8 Regulators Are Located in CD Risk Loci
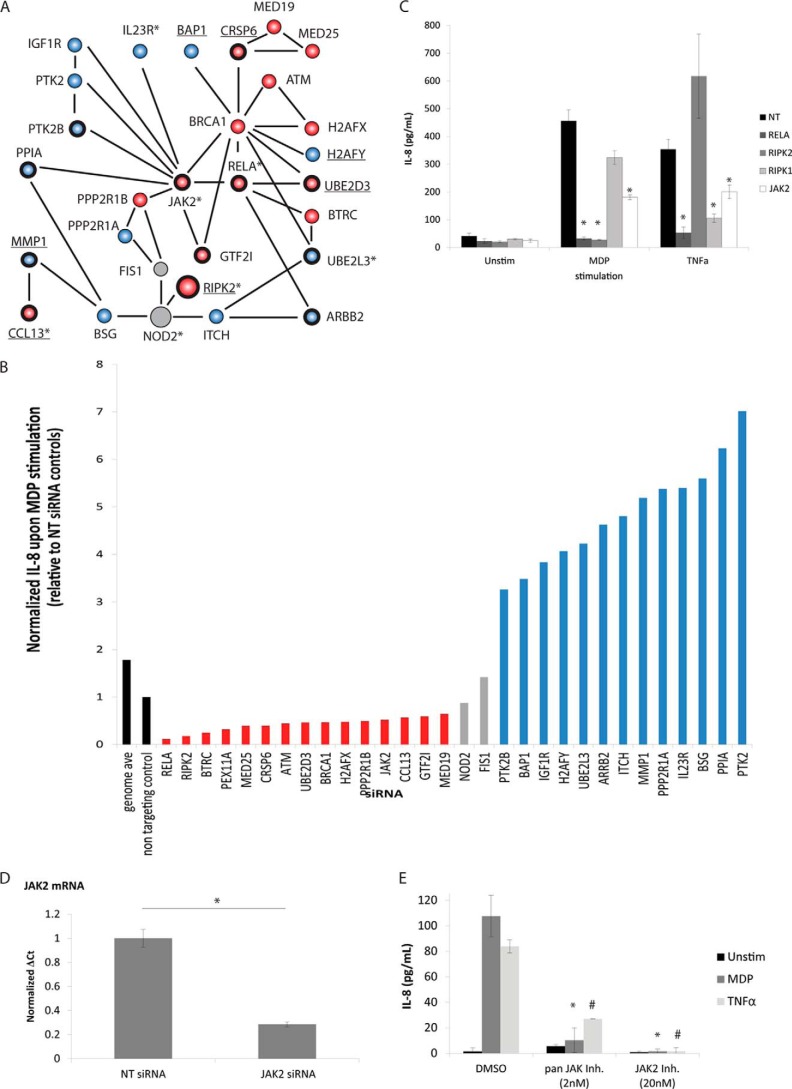
Given the strong association between NOD2 polymorphisms and CD risk, we investigated whether genes that affect NOD2-induced IL-8 secretion are associated with CD. To begin, we surveyed genes present in the 140 loci linked to CD by genome-wide association studies (GWAS) (57). 42 genes identified in our screen as IL-8 regulators are located within these CD loci, 17 of which were validated using independent siRNAs (Fig. 5 and supplemental Fig. S11). Interestingly, several of the IL-8 regulators present in CD risk loci can be linked to NOD2 via PPIs. Furthermore, many of the IL-8 regulators present in CD risk loci interact with other IL-8 regulators identified in the screen, building up biochemical and genetic evidence for a role for these genes in NOD2 signaling and CD pathogenesis. One particularly well connected subnetwork revolves around the tyrosine kinase JAK2 (Fig. 6, A–D). Interestingly, small molecule inhibitors of JAK2 further validate JAK2 as a positive regulator of NOD2-dependent MDP-induced IL-8 secretion (Fig. 6E). Furthermore, numerous genes implicated in either the NOD2 signaling pathway or CD by microarray (supplemental Table S7) or proteomic (supplemental Table S8) studies affect IL-8 secretion. Together, these analyses reveal that many putative NOD2/IL-8 pathway components can be linked to CD and suggest a possible role for multiple members of the NOD2/IL-8 pathway, and not just NOD2, in the pathogenesis of CD.
FIGURE 5.
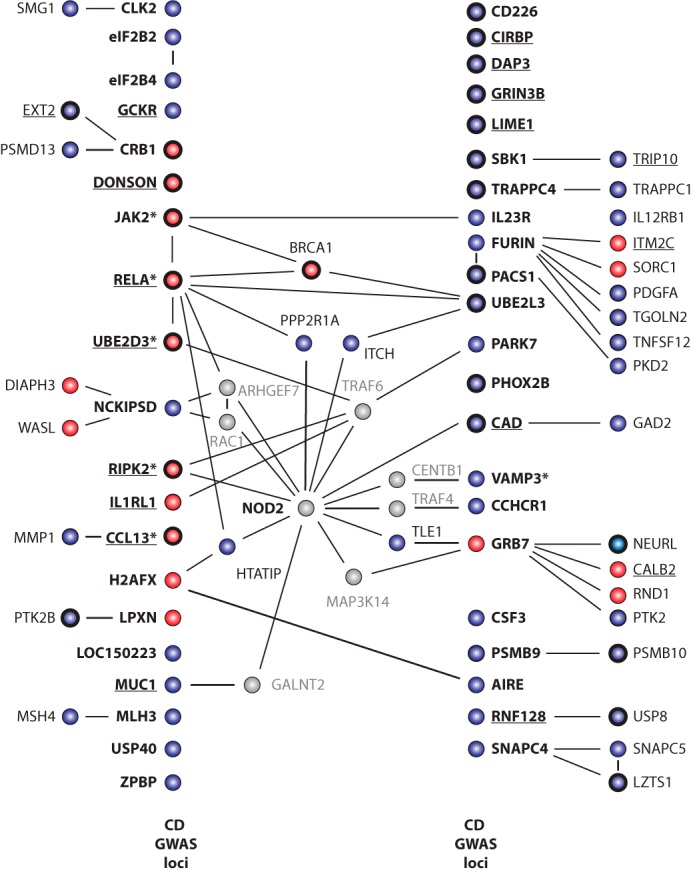
Many genes associated with Crohn disease risk affect NOD2 induced IL-8 secretion. Genes whose silencing either diminished (red) or enhanced (blue) NOD2-induced IL-8 secretion that are also present in loci associated with Crohn disease risk by GWAS are labeled in boldface type and organized into two columns, as indicated. Solid black lines, published PPIs, taken from the STRING and BIOGRID databases, between NOD2 regulators identified in the primary siRNA screen. In several cases, we highlight PPIs that link NOD2 with genes present in CD risk loci via a single step intermediate with a gene that was not a hit in our screen (gray). Boldface circles, genes supported by secondary validation experiments using independent siRNA pools. Knockdown of underlined genes affected both NF-κB induction and IL-8 production. *, “key genes” as nominated previously (57).
FIGURE 6.
A subnetwork of IL-8 regulators, many of which lie in loci associated with Crohn disease, are connected by PPIs. A, PPIs between IL-8 regulators identified in the siRNA screen were taken from publically available databases, such as STRING and BIOGRID, and are depicted by solid black. Each protein is depicted as a circle and is colored red or blue to indicate that it either positively or negatively regulates IL-8 secretion, respectively. Genes in boldface type were validated using independent siRNA, whereas underlined gene names indicate genes whose knockdown concordantly affected NF-κB activity. *, genes present in loci associated with CD risk by GWAS. B, silencing of the indicated genes increased (blue) or decreased (red) IL-8 levels in the primary screen after MDP stimulation relative to NT siRNA controls (black). C, JAK2 knockdown using independent siRNAs in a secondary validation screen decreases IL-8 induction in response to MDP and TNFα stimulation. Knockdown experiments were performed in triplicate. Error bars, S.D. *, p values <0.05 using Student's t tests against stimulation-specific NT siRNA controls. D, mRNA levels of JAK2 were assessed by quantitative real-time PCR to confirm knockdown. E, NOD2-expressing HEK293 cells were pretreated for 2 h with the indicated amount of either a pan-JAK inhibitor or the JAK2-specific inhibitor before stimulating with either MDP (20 ng/ml) or TNFα (10 ng/ml), and IL-8 levels were measured in the supernatant by ELISA 17 h later. Inhibitor experiments were performed in triplicate. Error bars, S.D. * and #, p values of <0.05 using Student's t tests against stimulation-specific vehicle (DMSO) controls.
DISCUSSION
IL-8 levels are typically very low in healthy tissues but can be rapidly and transiently induced in response to various stressors (58). Elaborate signaling networks have evolved to control IL-8 secretion and are critical in maintaining immune homeostasis necessary to balance host defense against excessive tissue damage associated with chronic inflammation. Here, we describe a genome-wide siRNA screen that identifies numerous genes that regulate NOD2-induced IL-8 secretion. Orthogonal data sets, such as PPI networks, offer additional biochemical evidence supporting roles for many of the identified IL-8 regulators and also provide mechanistic insight into the level at which these regulators may function. For instance, by comparing genes that influence NOD2-induced NF-κB activity with those that also regulate IL-8 secretion, we were able to distinguish concordant regulators from those that specifically affect IL-8 secretion. For example, silencing ZFP36L2 increased IL-8 secretion without affecting NF-κB induction. ZFP36L2 encodes a homolog of TTP (tristetraprolin), an RNA-binding protein known to bind to AU-rich elements in the 3′-UTR of IL-8 and negatively regulate its translation at a post-transcriptional level by destabilizing IL-8 mRNA (59). Notably, TTP knockdown did not influence IL-8 levels in our screen, presumably because it is not expressed in HEK cells (60). Similarly, silencing the RNA-binding protein SAMD4 enhanced IL-8 secretion, consistent with its published role in binding and destabilizing mRNA (61) and the presence of six consensus SAMD4 recognition elements in the 3′-UTR of IL-8 (62). Increasing our understanding of the various cellular mechanisms that influence IL-8 production and secretion is an important step forward in developing strategies to manipulate IL-8 levels for potential therapeutic applications.
Other IL-8-specific regulatory mechanisms acting downstream of NF-κB-induced transcriptional activation are likely to influence processes such as IL-8 translation, storage, stability, and/or trafficking. Indeed, our screen revealed a number of genes implicated in the regulation of vesicle formation and trafficking as potential regulators of IL-8 secretion. In particular, we provide evidence that the ubiquitin-specific protease USP8 negatively regulates IL-8 secretion, consistent with studies demonstrating an important role for ubiquitin-mediated post-translational modification in the regulation of NOD2 signaling (63). Furthermore, our observation that USP8 negatively regulates IL-6 in BMDMs is intriguing because it suggests that some of the genes identified in our screen may play a more general role in regulating cytokine secretion beyond just IL-8.
Although the signaling pathways that lead to acute inflammation have become increasingly well characterized, less is known regarding the molecular mechanisms that mediate the resolution of inflammation. Negative regulators play a critical role in restoring homeostasis following pathogen clearance and in preventing chronic inflammation by maintaining the transient and self-limiting nature of the inflammatory response. Our screen has revealed many positive and negative regulators downstream of NF-κB activation, whose inhibition may provide new targets for the therapeutic modulation of the inflammatory response. Indeed, many of the validated hits are druggable enzymes (i.e. JAK2) for which small molecule inhibitors already exist.
Given that the NOD2 signaling pathway is just one of many pathways that culminate in the release of IL-8, we tested the effect of silencing a subset of hits from our primary screen on TNFα-induced IL-8 secretion, which occurs independently of NOD2. This counterscreen revealed that most of the hits identified were independent of the stimulus, supporting the idea that different receptors of the innate immune system use a relatively small number of specific components before converging on a more general and shared core pathway of cytokine release. This also suggests that, although we used MDP to induce the NOD2-dependent up-regulation of IL-8, many of the genes identified may be relevant in the regulation of other proinflammatory pathways.
siRNA screening is increasingly being used to identify genes that modulate a given biological process and help place previously unannotated genes into functional pathways (64). To date, only a limited number of RNAi screens have been used to study protein secretion. Two of these studies used Drosophila cells (65, 66), whereas the only reported screen using mammalian cells (67) examined the plasma membrane localization of a tagged viral protein. Our screen also complements several recently reported siRNA screens examining TLR-induced (68), LMP1-induced (41), or NOD2-induced (40) NF-κB activation or the induction of IL-8 mRNA upon activation of the related NOD1 receptor (39). Moreover, our screen extends these findings by being the first to systematically examine IL-8 induction in a genome-wide manner at the protein level. Furthermore, the previously published NOD1 screen used much higher amounts of NOD1 agonist delivered via a lipid-based transfection method, whereas our screen relied on endogenous routes of MDP entry and a more physiologically relevant amount of peptidoglycan agonist. As the number of published siRNA screens grows, key regulatory factors identified independently in multiple studies will emerge. Given the diversity of cell types that colonize the digestive tract, where NOD2 is known to have a physiologically relevant function, it will be important to examine the regulation of NOD2 in multiple cellular contexts because the cell type-specific gene expression patterns and regulatory mechanisms are likely to contribute to the heterogeneity of responses across cell types.
Although the etiology of CD remains elusive, it is thought to involve a complex interplay between genetic susceptibility, the immune response, the intestinal microbiome, and other environmental risk factors (69). To date, despite extraordinary progress in mapping genetic loci associated with CD risk (57), individual genetic markers are not able to accurately predict disease progression, severity, or therapeutic response with the one notable exception of NOD2 polymorphisms, which correlate with disease location and severity (70, 71). Given the strong genetic link between NOD2 polymorphisms and CD risk, it seems reasonable that mutations in other components of the NOD2 pathway may also affect CD risk. This idea is supported by the identification of a subset of CD patients with impaired MDP responses in the absence of mutations in NOD2 (72). Here, we identify numerous genes that affect MDP-induced IL-8 secretion present in loci associated with CD risk by GWAS, reinforcing a role for the NOD2/IL-8 signaling pathway, beyond just NOD2, in the pathogenesis of CD. Indeed, future studies may benefit from stratifying CD patients based on the status of various NOD2 pathway components rather than just NOD2 polymorphisms alone. Another potentially interesting application of our data set is in the prioritization of potentially causative genes present in loci identified by GWAS. Given that many CD risk loci identified by GWAS contain multiple genes and that SNP signals alone are rarely able to confidently predict the causative gene, the identification of genes in CD loci that functionally affect NOD2-induced IL-8 levels may help to prioritize candidate genes for future studies. For example, on-going efforts aimed at exploring the genetic architecture of CD via deep DNA sequencing approaches (38, 73) would probably benefit from having a more refined list of candidate genes, thereby reducing the search space for potential combinatorial interactions among putative causative variants that may be extremely rare or of modest effect size and therefore difficult to detect.
In conclusion, the IL-8 regulators identified here provide an important resource for future hypothesis-based studies aimed at understanding the molecular mechanisms that regulate this important cytokine. Given that IL-8 secretion is deregulated in many inflammatory diseases, an increase in our understanding of how IL-8 secretion is regulated is critical to identify targets for potential immunomodulation-based therapeutics.
Supplementary Material
Acknowledgments
We are grateful to R. Jacob, M. Larsen, and S. Swaney (University of Michigan Center for Chemical Genomics) for assistance in carrying out the screen. We thank J. Whitfield for outstanding technical assistance, Y. He for providing the immortalized macrophage cell line, and C. Smibert (University of Toronto) for analysis of SAMD4 recognition elements.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK082437 (to C. M.) and 2R01DK61707 (to G. N.). This work was also supported by the Crohn's and Colitis Foundation of America and the Eli and Edythe L. Broad Foundation Medical Research Program for Inflammatory Bowel Disease.

This article contains supplemental Figs. S1–S11 and Tables S1–S8.
- CD
- Crohn disease
- MDP
- muramyl-dipeptide
- NT
- non-targeting
- PPI
- protein-protein interaction
- GWAS
- genome-wide association studies.
REFERENCES
- 1. Dinarello C. A. (2007) Historical insights into cytokines. Eur. J. Immunol. 37, S34–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen G., Shaw M. H., Kim Y. G., Nuñez G. (2009) NOD-like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol. 4, 365–398 [DOI] [PubMed] [Google Scholar]
- 3. Philpott D. J., Sorbara M. T., Robertson S. J., Croitoru K., Girardin S. E. (2014) NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol. 14, 9–23 [DOI] [PubMed] [Google Scholar]
- 4. Cho J. H. (2008) The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466 [DOI] [PubMed] [Google Scholar]
- 5. Van Limbergen J., Radford-Smith G., Satsangi J. (2014) Advances in IBD genetics. Nat. Rev. Gastroenterol Hepatol. 11, 372–385 [DOI] [PubMed] [Google Scholar]
- 6. Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 7. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 8. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 9. Abbott D. W., Yang Y., Hutti J. E., Madhavarapu S., Kelliher M. A., Cantley L. C. (2007) Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell Biol. 27, 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Y., Yin C., Pandey A., Abbott D., Sassetti C., Kelliher M. A. (2007) NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J. Biol. Chem. 282, 36223–36229 [DOI] [PubMed] [Google Scholar]
- 11. Hasegawa M., Fujimoto Y., Lucas P. C., Nakano H., Fukase K., Núñez G., Inohara N. (2008) A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 27, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshimura T., Matsushima K., Tanaka S., Robinson E. A., Appella E., Oppenheim J. J., Leonard E. J. (1987) Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc. Natl. Acad. Sci. U.S.A. 84, 9233–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waugh D. J., Wilson C. (2008) The interleukin-8 pathway in cancer. Clin. Cancer Res. 14, 6735–6741 [DOI] [PubMed] [Google Scholar]
- 14. Xie K. (2001) Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 12, 375–391 [DOI] [PubMed] [Google Scholar]
- 15. Brat D. J., Bellail A. C., Van Meir E. G. (2005) The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro. Oncol. 7, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beste M. T., Pfaffle-Doyle N., Prentice E. A., Morris S. N., Lauffenburger D. A., Isaacson K. B., Griffith L. G. (2014) Molecular network analysis of endometriosis reveals a role for c-Jun-regulated macrophage activation. Sci. Transl. Med. 10.1126/scitranslmed.3007988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanghetti E. A. (2013) The role of inflammation in the pathology of acne. J. Clin. Aesthet. Dermatol. 6, 27–35 [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Moran T., Swanson E., Julian C., Harris J., Bonen D. K., Hedl M., Nicolae D. L., Abraham C., Cho J. H. (2004) Regulation of IL-8 and IL-1β expression in Crohn's disease associated NOD2/CARD15 mutations. Hum. Mol. Genet. 13, 1715–1725 [DOI] [PubMed] [Google Scholar]
- 19. Lappalainen M., Paavola-Sakki P., Halme L., Turunen U., Färkkilä M., Repo H., Kontula K. (2008) Novel CARD15/NOD2 mutations in Finnish patients with Crohn's disease and their relation to phenotypic variation in vitro and in vivo. Inflamm. Bowel Dis. 14, 176–185 [DOI] [PubMed] [Google Scholar]
- 20. van Heel D. A., Hunt K. A., King K., Ghosh S., Gabe S. M., Mathew C. G., Forbes A., Playford R. J. (2006) Detection of muramyl dipeptide-sensing pathway defects in patients with Crohn's disease. Inflamm. Bowel Dis. 12, 598–605 [DOI] [PubMed] [Google Scholar]
- 21. Kanazawa N., Okafuji I., Kambe N., Nishikomori R., Nakata-Hizume M., Nagai S., Fuji A., Yuasa T., Manki A., Sakurai Y., Nakajima M., Kobayashi H., Fujiwara I., Tsutsumi H., Utani A., Nishigori C., Heike T., Nakahata T., Miyachi Y. (2005) Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-κB activation: common genetic etiology with Blau syndrome. Blood 105, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 22. Miceli-Richard C., Lesage S., Rybojad M., Prieur A. M., Manouvrier-Hanu S., Häfner R., Chamaillard M., Zouali H., Thomas G., Hugot J. P. (2001) CARD15 mutations in Blau syndrome. Nat. Genet. 29, 19–20 [DOI] [PubMed] [Google Scholar]
- 23. Warner N., Burberry A., Franchi L., Kim Y. G., McDonald C., Sartor M. A., Núñez G. (2013) A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-κB signaling pathways. Sci. Signal 6, rs3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adami C., Brunda M. J., Palleroni A. V. (1993) In vivo immortalization of murine peritoneal macrophages: a new rapid and efficient method for obtaining macrophage cell lines. J. Leukoc. Biol. 53, 475–478 [DOI] [PubMed] [Google Scholar]
- 25. Yasumoto K., Okamoto S., Mukaida N., Murakami S., Mai M., Matsushima K. (1992) Tumor necrosis factor alpha and interferon γ synergistically induce interleukin 8 production in a human gastric cancer cell line through acting concurrently on AP-1 and NF-κB-like binding sites of the interleukin 8 gene. J. Biol. Chem. 267, 22506–22511 [PubMed] [Google Scholar]
- 26. Damgaard R. B., Nachbur U., Yabal M., Wong W. W., Fiil B. K., Kastirr M., Rieser E., Rickard J. A., Bankovacki A., Peschel C., Ruland J., Bekker-Jensen S., Mailand N., Kaufmann T., Strasser A., Walczak H., Silke J., Jost P. J., Gyrd-Hansen M. (2012) The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell 46, 746–758 [DOI] [PubMed] [Google Scholar]
- 27. Richmond A. L., Kabi A., Homer C. R., Marina-García N., Nickerson K. P., Nesvizhskii A. I., Sreekumar A., Chinnaiyan A. M., Nuñez G., McDonald C. (2012) The nucleotide synthesis enzyme CAD inhibits NOD2 antibacterial function in human intestinal epithelial cells. Gastroenterology 142, 1483–1492.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., Kai C., Yada T., Suzuki Y., Yamada T., Ozawa T., Kaneki K., Inoue T., Kobayashi M., Kodama T., Wada Y., Sekimizu K., Akimitsu N. (2014) Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406 [DOI] [PubMed] [Google Scholar]
- 29. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 30. Hisatsune J., Nakayama M., Isomoto H., Kurazono H., Mukaida N., Mukhopadhyay A. K., Azuma T., Yamaoka Y., Sap J., Yamasaki E., Yahiro K., Moss J., Hirayama T. (2008) Molecular characterization of Helicobacter pylori VacA induction of IL-8 in U937 cells reveals a prominent role for p38MAPK in activating transcription factor-2, cAMP response element-binding protein, and NF-κB activation. J. Immunol. 180, 5017–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eliopoulos A. G., Gallagher N. J., Blake S. M., Dawson C. W., Young L. S. (1999) Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274, 16085–16096 [DOI] [PubMed] [Google Scholar]
- 32. Manetsch M., Che W., Seidel P., Chen Y., Ammit A. J. (2012) MKP-1: a negative feedback effector that represses MAPK-mediated pro-inflammatory signaling pathways and cytokine secretion in human airway smooth muscle cells. Cell. Signal. 24, 907–913 [DOI] [PubMed] [Google Scholar]
- 33. Dauletbaev N., Eklove D., Mawji N., Iskandar M., Di Marco S., Gallouzi I. E., Lands L. C. (2011) Down-regulation of cytokine-induced interleukin-8 requires inhibition of p38 mitogen-activated protein kinase (MAPK) via MAPK phosphatase 1-dependent and -independent mechanisms. J. Biol. Chem. 286, 15998–16007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tao M., Scacheri P. C., Marinis J. M., Harhaj E. W., Matesic L. E., Abbott D. W. (2009) ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr. Biol. 19, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Till A., Rosenstiel P., Bräutigam K., Sina C., Jacobs G., Oberg H. H., Seegert D., Chakraborty T., Schreiber S. (2008) A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. J. Cell Sci. 121, 487–495 [DOI] [PubMed] [Google Scholar]
- 36. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chatr-Aryamontri A., Breitkreutz B. J., Heinicke S., Boucher L., Winter A., Stark C., Nixon J., Ramage L., Kolas N., O'Donnell L., Reguly T., Breitkreutz A., Sellam A., Chen D., Chang C., Rust J., Livstone M., Oughtred R., Dolinski K., Tyers M. (2013) The BioGRID interaction database: 2013 update. Nucleic Acids Res. 41, D816–D823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Momozawa Y., Mni M., Nakamura K., Coppieters W., Almer S., Amininejad L., Cleynen I., Colombel J. F., de Rijk P., Dewit O., Finkel Y., Gassull M. A., Goossens D., Laukens D., Lémann M., Libioulle C., O'Morain C., Reenaers C., Rutgeerts P., Tysk C., Zelenika D., Lathrop M., Del-Favero J., Hugot J. P., de Vos M., Franchimont D., Vermeire S., Louis E., Georges M. (2011) Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 43, 43–47 [DOI] [PubMed] [Google Scholar]
- 39. Yeretssian G., Correa R. G., Doiron K., Fitzgerald P., Dillon C. P., Green D. R., Reed J. C., Saleh M. (2011) Non-apoptotic role of BID in inflammation and innate immunity. Nature 474, 96–99 [DOI] [PubMed] [Google Scholar]
- 40. Lipinski S., Grabe N., Jacobs G., Billmann-Born S., Till A., Häsler R., Aden K., Paulsen M., Arlt A., Kraemer L., Hagemann N., Erdmann K. S., Schreiber S., Rosenstiel P. (2012) RNAi screening identifies mediators of NOD2 signaling: implications for spatial specificity of MDP recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 21426–21431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gewurz B. E., Towfic F., Mar J. C., Shinners N. P., Takasaki K., Zhao B., Cahir-McFarland E. D., Quackenbush J., Xavier R. J., Kieff E. (2012) Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 109, 2467–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hedvat C. V., Yao J., Sokolic R. A., Nimer S. D. (2004) Myeloid ELF1-like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J. Biol. Chem. 279, 6395–6400 [DOI] [PubMed] [Google Scholar]
- 43. Hitotsumatsu O., Ahmad R. C., Tavares R., Wang M., Philpott D., Turer E. E., Lee B. L., Shiffin N., Advincula R., Malynn B. A., Werts C., Ma A. (2008) The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kjellerup R. B., Kragballe K., Iversen L., Johansen C. (2008) Pro-inflammatory cytokine release in keratinocytes is mediated through the MAPK signal-integrating kinases. Exp. Dermatol. 17, 498–504 [DOI] [PubMed] [Google Scholar]
- 45. Rowlett R. M., Chrestensen C. A., Nyce M., Harp M. G., Pelo J. W., Cominelli F., Ernst P. B., Pizarro T. T., Sturgill T. W., Worthington M. T. (2008) MNK kinases regulate multiple TLR pathways and innate proinflammatory cytokines in macrophages. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G452–G459 [DOI] [PubMed] [Google Scholar]
- 46. Watari K., Nakaya M., Nishida M., Kim K. M., Kurose H. (2013) β-Arrestin2 in infiltrated macrophages inhibits excessive inflammation after myocardial infarction. PLoS One 8, e68351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharma D., Malik A., Lee E., Britton R. A., Parameswaran N. (2013) Gene dosage-dependent negative regulatory role of β-arrestin-2 in polymicrobial infection-induced inflammation. Infect. Immun. 81, 3035–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Popoff V., Adolf F., Brügger B., Wieland F. (2011) COPI budding within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3, a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen J., Wagner E. J. (2010) snRNA 3′ end formation: the dawn of the Integrator complex. Biochem. Soc. Trans. 38, 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lewis B. A., Reinberg D. (2003) The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116, 3667–3675 [DOI] [PubMed] [Google Scholar]
- 51. Wei M. L. (2006) Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 19, 19–42 [DOI] [PubMed] [Google Scholar]
- 52. Seward S. L., Jr., Gahl W. A. (2013) Hermansky-Pudlak syndrome: health care throughout life. Pediatrics 132, 153–160 [DOI] [PubMed] [Google Scholar]
- 53. Kim Y. G., Shaw M. H., Warner N., Park J. H., Chen F., Ogura Y., Núñez G. (2011) Cutting edge: Crohn's disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. J. Immunol. 187, 2849–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hruz P., Zinkernagel A. S., Jenikova G., Botwin G. J., Hugot J. P., Karin M., Nizet V., Eckmann L. (2009) NOD2 contributes to cutaneous defense against Staphylococcus aureus through α-toxin-dependent innate immune activation. Proc. Natl. Acad. Sci. U.S.A. 106, 12873–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deshmukh H. S., Hamburger J. B., Ahn S. H., McCafferty D. G., Yang S. R., Fowler V. G., Jr. (2009) Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect. Immun. 77, 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jostins L., Ripke S., Weersma R. K., Duerr R. H., McGovern D. P., Hui K. Y., Lee J. C., Schumm L. P., Sharma Y., Anderson C. A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S. L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J. P., Ahmad T., Amininejad L., Ananthakrishnan A. N., Andersen V., Andrews J. M., Baidoo L., Balschun T., Bampton P. A., Bitton A., Boucher G., Brand S., Büning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K. L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L. R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T. H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I. C., Lees C. W., Louis E., Mahy G., Mansfield J., Morgan A. R., Mowat C., Newman W., Palmieri O., Ponsioen C. Y., Potocnik U., Prescott N. J., Regueiro M., Rotter J. I., Russell R. K., Sanderson J. D., Sans M., Satsangi J., Schreiber S., Simms L. A., Sventoraityte J., Targan S. R., Taylor K. D., Tremelling M., Verspaget H. W., De Vos M., Wijmenga C., Wilson D. C., Winkelmann J., Xavier R. J., Zeissig S., Zhang B., Zhang C. K., Zhao H., Silverberg M. S., Annese V., Hakonarson H., Brant S. R., Radford-Smith G., Mathew C. G., Rioux J. D., Schadt E. E., Daly M. J., Franke A., Parkes M., Vermeire S., Barrett J. C., Cho J. H. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002) Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72, 847–855 [PubMed] [Google Scholar]
- 59. Balakathiresan N. S., Bhattacharyya S., Gutti U., Long R. P., Jozwik C., Huang W., Srivastava M., Pollard H. B., Biswas R. (2009) Tristetraprolin regulates IL-8 mRNA stability in cystic fibrosis lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 296, L1012–L1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cao H. (2004) Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications. Biochemistry 43, 13724–13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smibert C. A., Lie Y. S., Shillinglaw W., Henzel W. J., Macdonald P. M. (1999) Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 5, 1535–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., Na H., Irimia M., Matzat L. H., Dale R. K., Smith S. A., Yarosh C. A., Kelly S. M., Nabet B., Mecenas D., Li W., Laishram R. S., Qiao M., Lipshitz H. D., Piano F., Corbett A. H., Carstens R. P., Frey B. J., Anderson R. A., Lynch K. W., Penalva L. O., Lei E. P., Fraser A. G., Blencowe B. J., Morris Q. D., Hughes T. R. (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tigno-Aranjuez J. T., Abbott D. W. (2012) Ubiquitination and phosphorylation in the regulation of NOD2 signaling and NOD2-mediated disease. Biochim. Biophys. Acta 1823, 2022–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Echeverri C. J., Perrimon N. (2006) High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 7, 373–384 [DOI] [PubMed] [Google Scholar]
- 65. Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R., Perrimon N., Malhotra V. (2006) Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 [DOI] [PubMed] [Google Scholar]
- 66. Wendler F., Gillingham A. K., Sinka R., Rosa-Ferreira C., Gordon D. E., Franch-Marro X., Peden A. A., Vincent J. P., Munro S. (2010) A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 29, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Simpson J. C., Joggerst B., Laketa V., Verissimo F., Cetin C., Erfle H., Bexiga M. G., Singan V. R., Hériché J. K., Neumann B., Mateos A., Blake J., Bechtel S., Benes V., Wiemann S., Ellenberg J., Pepperkok R. (2012) Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat. Cell Biol. 14, 764–774 [DOI] [PubMed] [Google Scholar]
- 68. Chiang C. Y., Engel A., Opaluch A. M., Ramos I., Maestre A. M., Secundino I., De Jesus P. D., Nguyen Q. T., Welch G., Bonamy G. M., Miraglia L. J., Orth A. P., Nizet V., Fernandez-Sesma A., Zhou Y., Barton G. M., Chanda S. K. (2012) Cofactors required for TLR7- and TLR9-dependent innate immune responses. Cell Host Microbe 11, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xavier R. J., Podolsky D. K. (2007) Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 [DOI] [PubMed] [Google Scholar]
- 70. Adler J., Rangwalla S. C., Dwamena B. A., Higgins P. D. (2011) The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am. J. Gastroenterol. 106, 699–712 [DOI] [PubMed] [Google Scholar]
- 71. Cleynen I., González J. R., Figueroa C., Franke A., McGovern D., Bortlík M., Crusius B. J., Vecchi M., Artieda M., Szczypiorska M., Bethge J., Arteta D., Ayala E., Danese S., van Hogezand R. A., Panés J., Peña S. A., Lukas M., Jewell D. P., Schreiber S., Vermeire S., Sans M. (2013) Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut 62, 1556–1565 [DOI] [PubMed] [Google Scholar]
- 72. Seidelin J. B., Broom O. J., Olsen J., Nielsen O. H. (2009) Evidence for impaired CARD15 signalling in Crohn's disease without disease linked variants. PLoS One 4, e7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rivas M. A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C. K., Boucher G., Ripke S., Ellinghaus D., Burtt N., Fennell T., Kirby A., Latiano A., Goyette P., Green T., Halfvarson J., Haritunians T., Korn J. M., Kuruvilla F., Lagacé C., Neale B., Lo K. S., Schumm P., Törkvist L., National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC), United Kingdom Inflammatory Bowel Disease Genetics Consortium, International Inflammatory Bowel Disease Genetics Consortium, Dubinsky M. C., Brant S. R., Silverberg M. S., Duerr R. H., Altshuler D., Gabriel S., Lettre G., Franke A., D'Amato M., McGovern D. P., Cho J. H., Rioux J. D., Xavier R. J., Daly M. J. (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.