Background: The N-terminal domain of an insect β-glucan recognition protein (N-βGRP) stimulates innate immune responses.
Results: N-βGRP forms soluble and insoluble protein-carbohydrate complexes involving specific protein-protein interactions, which activates the prophenoloxidase pathway.
Conclusion: Assembly of βGRP oligomers stimulated by binding to microbial polysaccharide triggers phenoloxidase activation.
Significance: βGRP interactions stimulate an innate immune response by a novel mechanism.
Keywords: Analytical Ultracentrifugation, Innate Immunity, Insect Immunity, Mass Spectrometry (MS), Pattern Recognition Receptor (PRR), Protein Aggregation, Protein Assembly, Protein Cross-linking, Carbohydrate-binding Protein, Hemolymph
Abstract
Insect β-glucan recognition protein (βGRP), a pathogen recognition receptor for innate immune responses, detects β-1,3-glucan on fungal surfaces via its N-terminal carbohydrate-binding domain (N-βGRP) and triggers serine protease cascades for the activation of prophenoloxidase (pro-PO) or Toll pathways. Using biophysical and biochemical methods, we characterized the interaction of the N-terminal domain from Manduca sexta βGRP2 (N-βGRP2) with laminarin, a soluble form of β-1,3-glucan. We found that carbohydrate binding by N-βGRP2 induces the formation of two types of protein-carbohydrate complexes, depending on the molar ratio of carbohydrate to protein ([C]/[P]). Precipitation, analytical ultracentrifugation, and chemical cross-linking experiments have shown that an insoluble aggregate forms when the molar ratio of carbohydrate to protein is low ([C]/[P] ∼ 1). In contrast, a soluble complex, containing at least five N-βGRP2 molecules forms at a higher molar ratio of carbohydrate/protein ([C]/[P] >5). A hypothesis that this complex is assembled partly due to protein-protein interactions was supported by chemical cross-linking experiments combined with LC-MS/MS spectrometry analysis, which permitted identification of a specific intermolecular cross-link site between N-βGRP molecules in the soluble complex. The pro-PO activation in naive plasma was strongly stimulated by addition of the insoluble aggregates of N-βGRP2. The soluble complex with laminarin formed in the plasma also stimulated pro-PO activation, but at a lower level. Taken together, these results provide experimental evidence for novel mechanisms in which associations of βGRP with microbial polysaccharide promotes assembly of βGRP oligomers, which may form a platform needed to trigger the pro-PO pathway activation cascade.
Introduction
Insects rely on innate immune responses to fight against a broad spectrum of pathogens. These responses can be triggered by the recognition of pathogen-associated molecular patterns by pathogen recognition receptors. Recognition signals initiated by such interactions are amplified by extracellular serine protease cascades leading to at least two types of responses: prophenoloxidase (pro-PO)3 activation and subsequent melanization of pathogens, and activation of the Toll ligand, which leads to synthesis of antimicrobial peptides (1, 2).
Members of a family of pathogen recognition receptors in insect hemolymph known as β-1,3-glucan recognition proteins (βGRPs) or Gram-negative bacteria-binding proteins bind to β-1,3-glucan, a component of fungal cell walls (3–9). The βGRP/Gram-negative bacteria-binding proteins share a conserved domain architecture: an amino-terminal β-1,3-glucan binding domain (N-βGRP) and a carboxyl-terminal β-1,3-glucanase-like domain. N-βGRP can bind to curdlan (an insoluble linear β-1,3-glucan) (3), laminarin (a soluble β-1,6-branched β-1,3-glucan), and to the cell walls of yeast (8, 10), but interacts weakly with shorter β-1,3-glucan chains such as laminarihexaose (11, 12). N-βGRP promotes activation of the pro-PO pathway (10). The structure of insect N-βGRP and its carbohydrate-binding mechanism have been studied in recent years. Insect N-βGRPs have highly conserved amino acid sequences and all their tertiary structures adopt an immunoglobulin-like β-sandwich-fold, with two β-sheets forming “convex” and “concave” surfaces (11–14) (supplemental Fig. S1). These structural studies have included proposed modes for binding of β-1,3-glucan by N-βGRP. The crystal structure of N-βGRP complexed with laminarihexaose revealed that the carbohydrate-binding site is located on the convex β-sheet surface of the domain, with the bound laminarihexaose existing as a triple helical structure (14). However, much remains to be understood about the binding modes of a longer β-1,3-glucan that exhibited higher affinity to N-βGRP (11, 12) and the following molecular mechanisms underlying downstream signaling events in the pro-PO activation pathway, including activation of serine protease zymogens.
In a recent study, we demonstrated that the binding of laminarin to N-βGRP from Plodia interpunctella (Pi-N-βGRP) induces formation of a protein-carbohydrate macrocomplex containing multiple Pi-N-βGRP molecules and suggested that this complex is an initiation signal for activation of serine protease cascades that promote immune responses (12). Here, to better understand biological and biochemical aspects of such complex formation of βGRP, we studied the molecular interaction between a soluble β-1,3-glucan and N-βGRP from Manduca sexta, one of the most common model insects for invertebrate biochemistry, molecular biology, and innate immunity. In the past, we have identified two βGRPs from M. sexta (Ms-βGRP1 and -βGRP2) (4, 8). Another pathogen recognition receptor (microbial-binding protein), a new member of β-1,3-glucanase-related protein superfamily to which the βGRPs belong, has also been identified from M. sexta (15). Whereas Ms-βGRP1 is constitutively expressed in fat body and secreted into hemolymph (4), the transcription of Ms-βGRP2 is up-regulated after immune challenges with yeast or bacteria (8), suggesting that Ms-βGRP2 is involved in an acute immune response against microbial infection. Because of this biological significance, we here studied the N-terminal domain from Ms-βGRP2 (N-βGRP2) and demonstrated that N-βGRP2 forms two types of high molecular mass complexes with laminarin. A soluble complex of N-βGRP2 and laminarin formed when the molar ratio of the carbohydrate to protein was high ([C]/[P] >5). However, when this ratio was low ([C]/[P] ∼ 1), N-βGRP2 and laminarin associated to form insoluble and irreversible aggregates. We characterized the structural properties of the soluble complex using size exclusion chromatography and chemical cross-linking experiments combined with LC-MS/MS analysis. The effects of the soluble and insoluble macromolecular associations on activation of the pro-PO pathway were also studied to gain insight into the biological relevance of the distinct complexes formed by N-βGRP2 with longer β-1,3-glucan molecules.
EXPERIMENTAL PROCEDURES
Materials
Laminarin from Laminaria digitata and trypsin (proteomics grade) were purchased from Sigma. β-1,3 to β-1,6 cross-link number ratio of laminarin from L. digitata was 7 (16). Laminarihexaose was from Megazyme. Laminaritetraose was from Seikagaku Corporation. Wheat starch was a kind gift from Dr. Yong-Cheng Shi (Department of Grain Science and Industry, Kansas State University). Gel filtration standards were purchased from Bio-Rad. Cross-linking reagent DTSSP (3,3′-dithiobis(sulfosuccinimidyl propionate)) was purchased from Thermo Scientific.
Expression and Purification of N-βGRP2
A DNA sequence encoding the amino-terminal 109 residues of βGRP2 from M. sexta (N-βGRP2) (8) was inserted via NcoI/XhoI sites into plasmid pET28a, for expression of recombinant N-βGRP2 with a carboxyl-terminal His6 tag. Escherichia coli strain BL21(DE3) cells transformed with this plasmid vector was cultured in LB media or with M9 minimal media supplemented with 1 g/liter of 15NH4Cl and 2 g/liter of d-[13C]glucose (Cambridge Isotope Laboratories) for 15N and 13C labeling, respectively. N-βGRP2 was expressed by induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. After lysis of the bacteria, soluble N-βGRP2 was purified using Ni2+ affinity chromatography, followed by size exclusion chromatography on a Superdex 75 column (GE healthcare). Purity of N-βGRP2 was confirmed by SDS-PAGE, and the amino acid sequence was confirmed by mass spectrometry.
NMR Spectroscopy and Structural Modeling
All NMR spectra were collected at 25 °C with a Varian VNMR 500 MHz equipped with 5-mm cryogenic triple resonance probes. For backbone resonance assignments, two-dimensional 1H-15N heteronuclear single quantum coherence spectrum (HSQC) and three-dimensional HNCA, HN(CO)CA, HNCACB, and HNCO were recorded on 1.5 mm 13C/15N-labeled N-βGRP2 prepared in 20 mm sodium phosphate buffer, pH 7.0, with 3 mm NaN3 in 90% H2O, 10% D2O. NMR spectra were processed using NMRPipe (17), and analyzed with Sparky (T. D. Goddard and D. G. Kneller, Sparky, University of California, San Francisco, CA) and CARA (18). The secondary structure of N-βGRP2 was predicted by the program TALOS+ (19) using the chemical shift of assigned 13Cα, 13Cβ, and 13C′ resonances. For titration experiments, a series of two-dimensional 1H-15N HSQC spectra of 0.3 mm 15N-labeled N-βGRP2 were collected in 50 mm sodium phosphate, pH 6.5, in the presence and absence of 200 mm NaCl. Laminarin or laminarihexaose prepared in the same buffer were titrated into the protein sample. The final molar ratio of carbohydrate to protein was 1 for laminarin and 10 for laminarihexaose.
Three-dimensional structural models of N-βGRP2 were generated using the homology modeling tool, I-TASSER (20, 21). The N-βGRP2 amino acid sequence including the C-terminal His tag was used as an input for processing by the I-TASSER algorithm.
Protein Precipitation Analysis
Various concentrations of N-βGRP2 (200, 95, 47, and 4 μm) and laminarin (∼1 mm) were incubated in microcentrifuge tubes for 2 h at room temperature. After centrifugation to pellet precipitates, the protein concentration in the supernatant was determined by absorbance measurement, using a molar extinction coefficient of 30,940 m−1 cm−1 at 280 nm for N-βGRP2.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted with an Optima XL-I ultracentrifuge (Beckman Coulter, Inc.) using an An-60 Ti rotor at 20 °C with 50 mm Tris-HCl, pH 7.3, containing 50 mm NaCl (22). Sedimentation was monitored by absorbance or interference optics using double-sector aluminum cells with a final loading of 400 μl/sector. Sedimentation was performed at 49,000 × g with scans made at 5-min intervals. Data were analyzed using DCDT+ software version 1.16. Sedimentation coefficients were calculated using g(s*) and dc/dt fitting functions in DCDT+ software. Buffer density and viscosity were calculated by SEDNTERP version 1.08.
Size Exclusion Chromatography
Size exclusion chromatography was performed on a Sephacryl S-100 column (GE Healthcare) equilibrated with 50 mm sodium phosphate, pH 6.5, with 200 mm NaCl to investigate formation of complexes between N-βGRP2 and laminarin. Gel filtration standards (Bio-Rad) containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.3 kDa) were used to determine the molecular mass of proteins. N-βGRP2 was incubated with a 20-fold molar excess of laminarin in the same buffer for 5 min and applied to the column and eluted at a flow rate of 1 ml/min.
Purification of a Soluble Complex Containing N-βGRP2 and Laminarin
A soluble complex of N-βGRP2 and laminarin was purified from the size exclusion chromatography described above. Centrifugal filter (Amicon Ultra-15) was used to concentrate the soluble complex and exchange a desired buffer as indicated in the following experiments.
Chemical Cross-linking and MS Analysis
To identify interacting regions in adjacent N-βGRP2 molecules within the N-βGRP2-laminarin soluble complex, the cross-linking reagent DTSSP was used to cross-link amino groups. The ϵ-amino groups from nine lysine residues as well as the N-terminal α-amino group of N-βGRP2 were considered as potential cross-linking sites for DTSSP. The purified soluble complex of N-βGRP2 and laminarin was prepared in 50 mm sodium phosphate buffer, pH 7.0, with 200 mm NaCl. DTSSP was added at a molar ratio of 100:1 (DTSSP to the N-βGRP2 soluble complex) with a final protein concentration of 5 μm. After 30 min of incubation at 25 °C, the cross-linking reaction was quenched by addition of Tris-HCl, pH 7.5, to a final concentration of 20 mm. Cross-linked samples were separated by SDS-PAGE under non-reducing conditions and visualized by silver staining using SilverQuest Silver Staining Kit (Invitrogen). Protein bands of interest were excised from the gel, and in-gel trypsin digestion was conducted following the manufacturer's protocol (Sigma) and the method described by Shevchenko et al. (23).
Digested peptide samples were analyzed using LC-MS/MS at the Nevada Proteomics Center (University of Nevada, Reno, NV). The peptides were separated and analyzed using a Michrom Paradigm Multi-dimensional Liquid Chromatography instrument (Michrom Bioresources) coupled with a Thermo LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptide samples dissolved in 100 μl of 0.1% formic acid were loaded onto a ZORBAX 300SB-C18 5μ (5 × 0.3 mm) trap column (Agilent Technologies), eluted from the trap, and then separated with a reverse phase Michrom Magic C18AQ column (3 μm, 200 Å, 0.2 × 150 mm) by a gradient elution using solvent A (0.1% formic acid) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 2 μl/min. The gradient was set from 5 to 40% solvent B for 90 min, increased to 80% solvent B in 10 s, and held at 80% solvent B for 1 min. MS spectra were recorded over the mass range of m/z 400–1600 with resolution of 60,000. The three most intense ions were isolated for fragmentation in the linear ion trap using collision-induced dissociation with a minimal signal of 500 and collision energy of 35.0 or using electron transferring dissociation with a minimal signal of 1000 and collision energy of 35.0. Dynamic exclusion was implemented with 2 repeat counts, repeat duration of 15 s, and exclusion duration of 90 s.
Intra- and intermolecular cross-linked peptides were identified by analyzing MS and MS/MS data with StavroX software (24). Potential cross-links were evaluated manually. The criteria for selection of a possible cross-linked peptide include a maximum mass deviation of 50 ppm between theoretical and experimental mass, the presence of sequential ions for a predicted cross-linked peptide. Cross-linked peptides identified in the N-βGRP2 monomer band and those with the measured distance between ϵ-amino groups of cross-linked lysine residues up to 12 Å apart in the structural model of the N-βGRP2 monomer are assumed to be intra-molecular cross-links. Cross-linked peptides involving the N-terminal α-amino group, however, are excluded from this criterion due to possible flexibility of the N-terminal region. Cross-linked peptides identified only in the N-βGRP2 dimer band and those with the measured distance between ϵ-amino groups of cross-linked lysine residues more than 12 Å apart in the structural model of N-βGRP2 monomer are defined as intermolecular cross-links. Oxidation of Met, deamidation of Gln, and Asn were added as potential modifications.
Pro-PO Activation Assay
Phenoloxidase activation assay was conducted using a method modified from Fabrick et al. (7) and Dai et al. (12). Hemolymph was collected from the day 2 fifth instar larvae of M. sexta. After hemocytes were removed by centrifugation at 8000 × g for 20 min at 4 °C, the plasma sample (10 μl) was incubated with 10 μg of N-βGRP2, the soluble N-βGRP2·laminarin complex, or insoluble N-βGRP2·laminarin aggregate in wells of a 96-well microplate. The volume of each sample well was brought to 100 μl with 50 mm sodium phosphate buffer, pH 6.5. After incubation for 1 h at room temperature, 100 μl of 5 mm dopamine hydrochloride was added. Phenoloxidase activity was determined by measuring the absorbance at 470 nm at 30-s intervals for 20 min. PO activity was presented as the slope of initial linear region (mOD/min). The assay was conducted with more than three technical replicates, and statistical analysis was performed using Prism 5 (GraphPad Software).
Immunoblot Analysis
Plasma samples supplemented with N-βGRP2 or the purified N-βGRP2·laminarin complex were separated by gel-filtration on Sephacryl S-100. Eluted fractions were analyzed by SDS-PAGE on 4–12% BisTris gels (Invitrogen) under reducing conditions. After transfer to nitrocellulose membrane, immunoblotting was conducted using rabbit anti-βGRP2 polyclonal antibodies (8) as the primary antibody (diluted 1:1000) and goat anti-rabbit IgG-alkaline phosphatase conjugate (Bio-Rad, diluted 1:3000) as the secondary antibody.
RESULTS
Structural Analysis of N-βGRP2
Recombinant N-βGRP2 expressed in E. coli was purified to homogeneity using Ni2+ affinity chromatography and size exclusion chromatography. The protein eluted as a monomer, based on the comparison of its elution volume with those of molecular weight standards (Fig. 1A). The two-dimensional 15N-1H HSQC spectrum for 15N-labeled N-βGRP2 showed excellent chemical shift dispersion (Fig. 1B), and the backbone chemical shifts of 1H, 15N, and 13C nuclei were assigned using a series of the multidimensional heteronuclear NMR methods; 94% of the backbone amide resonance of 109 non-proline residues could be assigned. The secondary structures of N-βGRP2 were predicted using the TALOS program (19) and the chemical shift assignments of 13Cα, 13Cβ, and 13C′ resonances (Fig. 1C), which agreed well with those of Pi-N-βGRP determined by solution NMR spectroscopy (12). A three-dimensional structural model of N-βGRP2 predicted using the I-TASSER server (C-score = 0.83 ± 0.08, expected root mean square deviation = 2.7 ± 2.0) (20, 21) aligned very well with N-βGRP from other insects, with root mean square deviations of the backbone Cα atoms between N-βGRP2 and Pi-N-βGRP (Protein Data Bank 2KHA (12)) being 0.38 Å (Fig. 1D).
FIGURE 1.
Purification, NMR backbone resonance assignments, and structural analysis of N-βGRP2. A, elution profile of N-βGRP2 from size exclusion chromatography. SDS-PAGE analysis of the sample before applying the size exclusion chromatography (lane 1) and the purified N-βGRP2 (lane 2) is shown on right. B, two-dimensional 1H-15N HSQC spectrum of 1.6 mm 13C/15N-labeled N-βGRP2 at 298 K on a Varian 500 MHz spectrometer equipped with a cryogenic triple resonance probe. Sequence specific assignments are indicated. C, secondary structure prediction for N-βGRP2 based on the TALOS+ program with obtained backbone chemical shift values. β-Strand probabilities are given by positive values and those for α-helix are by negative for clarity. Shown above the chart is the secondary structure topology obtained from the solution NMR structure of Pi-N-βGRP (PDB ID 2KHA (12)). D, comparison of structural model of N-βGRP2 generated using the I-TASSER server (green) with the NMR solution structure of Pi-N-βGRP (12) (blue).
Binding to Laminarin Induces the Association of N-βGRP2 Molecules
We carried out a series of NMR chemical shift titration experiments to study interactions between N-βGRP2 and laminarin. Laminarin is a water-soluble β-1,3-glucan with β-1,6-glucan branches. The β-1,3 to β-1,6 cross-linkage ratio varies depending on the biological source. Laminarin from L. digitata used in the present study has a cross-linkage ratio of 7 (16). Solubility of laminarin depends on the degree of β-1,6 branching, with more branched laminarin being more soluble. To calculate molarities of laminarin solutions, we adopted a value of 5.5 kDa for the molecular mass, as reported by the supplier (Sigma). Titration of laminarin into 15N-labeled N-βGRP2 solution did not result in chemical shift changes, but significant decreases of resonance peak intensities occurred (supplemental Fig. S2). On average, the resonance intensities decreased by more than 90% when the molar ratio of the carbohydrate to protein ([C]/[P]) was 0.5 in the absence of NaCl (supplemental Fig. S2C). There were no site-specific changes in resonance intensity, and almost all of the peaks, excluding the backbone NH group of C-terminal residues and side chain NH2 groups, disappeared from the spectrum when the laminarin was added at a 1:1 [C]/[P] ratio, as observed in our previous NMR study for Pi-N-βGRP (12). The presence of 0.2 m NaCl suppressed the decrease in resonance intensities to some extent (supplemental Fig. S2, B and C), but the addition of 0.3 mm laminarin resulted in peak disappearance similar to that observed in the absence of NaCl. Such peak disappearance can be caused by NMR line broadening due to large protein complex formations and/or conformational exchanges on millisecond time scale. However, we observed precipitation of N-βGRP2 during the titration experiments, which has not been reported for other NMR studies of insect βGRPs (12, 13) and this made the chemical shift titration approach inapplicable. Titration of N-βGRP2 with laminarihexaose, a shorter β-1,3-glucan, also resulted in no chemical shift perturbation, but resonance intensities were decreased when the molar ratio of oligosaccharide to protein was greater than 5 (data not shown). This result suggests weaker interaction of N-βGRP2 with laminarihexaose than with laminarin, consistent with other studies (11, 12).
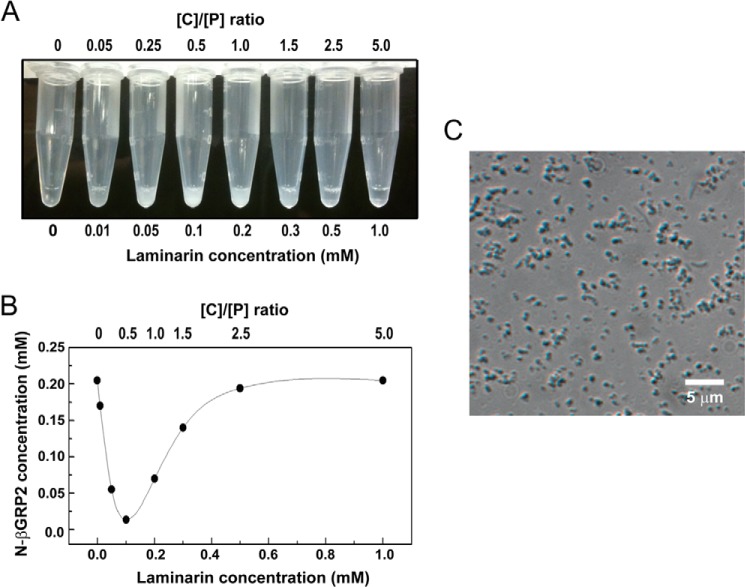
When the laminarin concentration was lower than or comparable with the protein concentration (0.2 mm), insoluble aggregation was visually confirmed (Fig. 2A). Maximum precipitation was observed when the molar ratio of the carbohydrate to protein was less than 1 (Fig. 2B). However, no precipitation was observed when N-βGRP2 was incubated with a molar excess of laminarin, at the higher [C]/[P] ratios (Fig. 2). Similar precipitation behavior of N-βGRP2 with laminarin was observed at different initial protein concentrations (4, 47, and 95 μm) (data not shown). The insoluble aggregates could not be re-solubilized by dilution or further addition of laminarin, laminarihexaose, or laminaritetraose, indicating that this aggregation occurs irreversibly. Microscopic observation showed that the insoluble aggregates of N-βGRP2 with laminarin formed evenly dispersed particles with a diameter of ∼0.5 μm (Fig. 2C).
FIGURE 2.
Complex phase behavior of N-βGRP2 with laminarin. A, protein precipitation was visually observed when laminarin concentration was low (10 μm to 0.3 mm) after incubating the N-βGRP2 at 0.2 mm with laminarin for 2 h at 25 °C. No precipitation was observed at higher concentrations of laminarin. B, after precipitates were pelleted by centrifugation, protein concentrations in the supernatant were determined using absorbance measurements. C, insoluble aggregates were observed by differential interference contrast microscopy.
We recently demonstrated using solution NMR and analytical ultracentrifugation methods that Pi-N-βGRP forms a soluble, high molecular mass complex upon binding to laminarin (12). To directly compare the effects of laminarin on N-βGRP2 and Pi-N-βGRP, sedimentation velocity analysis was performed (Fig. 3). N-βGRP2 (13.8 kDa) in the absence of laminarin had a sedimentation coefficient of 1.9 s. A mixture of 60 μm N-βGRP2 with 79 μm laminarin resulted in a broadening of the sedimentation peak, a decrease of signal height, and an increase of sedimentation coefficient to 5.5 s. Decrease of the signal height was due to precipitation or high order of aggregation. A further increase of laminarin concentration led to a slight increase of signal height and gave s values of 4.5 s when the laminarin concentration was 3.9 mm. Analysis of this sedimentation profile yielded an average molecular mass of 70 kDa. A very similar sedimentation profile was observed for Pi-N-βGRP when the laminarin concentration was much higher than the protein concentration.
FIGURE 3.
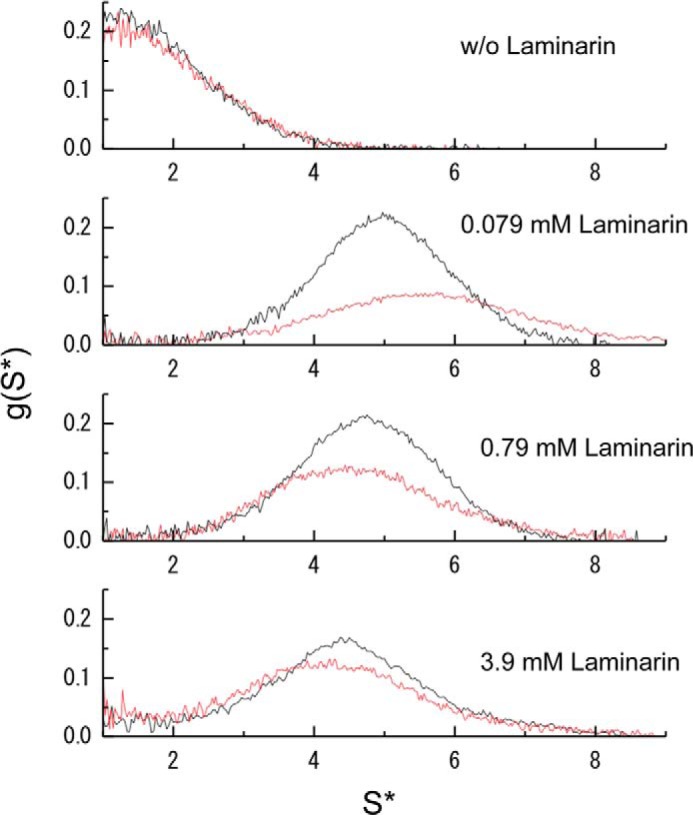
Sedimentation velocity profile for N-βGRP2 (red) and Pi-N-βGRP (black) in the presence of laminarin from L. digitata. Protein samples (60 μm) were subjected to ultracentrifugation at a speed of 49,000 rpm (Optima XL-I ultracentrifuge; Beckman). The data were analyzed as described under “Experimental Procedures.”
The results of these analytical ultracentrifuge and precipitation experiments are consistent with a model in which N-βGRP2 forms two types of macromolecular complexes with laminarin, depending on the molar ratio of the carbohydrate to protein. Aggregation or assembly to form insoluble particles of ∼0.5 μm diameter occurs at the lower molar ratio ([C]/[P] < 1), and a soluble complex (s = 4.5) forms at the higher molar ratio ([C]/[P] > 5).
Characterization of the Soluble Protein-Carbohydrate Complex
To structurally characterize the soluble N-βGRP2·laminarin complex, we first purified this complex using size exclusion chromatography (Fig. 4). After N-βGRP2 was incubated with a 20-fold molar excess of laminarin, the soluble protein-carbohydrate complex eluted as a single and relatively sharp peak at 57 ml, which corresponds to molecular mass of ∼75 kDa, based on a calibration curve obtained with molecular mass standard proteins. In contrast, N-βGRP2 in the absence of laminarin eluted at 77 ml, consistent with its existence as a monomer. This result suggests that association of the N-βGRP2 and laminarin leads to formation of a soluble macrocomplex with defined size and that the complex is stable due to strong protein-protein and protein-carbohydrate interactions.
FIGURE 4.
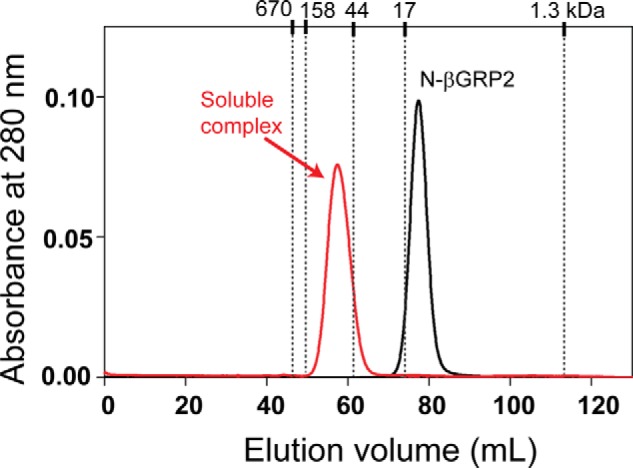
Elution profile of N-βGRP2·laminarin macrocomplex from Sephacryl S100 size exclusion chromatography in comparison with that of N-βGRP2 monomer. Elution volume of standards (thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.3 kDa)) is shown at the top of the chromatogram.
Chemical cross-linking of noncovalent protein complexes has been used to determine the stoichiometry of constituent monomers, and mass spectrometry analysis of the cross-linked species can identify interacting regions in the protein complexes (25–29). We have employed this combined approach to gain structural insights into molecular organization of the soluble N-βGRP2·laminarin macrocomplex. DTSSP is a cross-linking agent with a spacer arm of 12 Å that can be cleaved by reducing agents, and reacts with primary amine groups in proteins (25, 26). N-βGRP2 has nine ϵ-amino groups from lysine residues as well as the N-terminal α-amino group that could serve as targets for cross-linking by DTSSP if the residues are near 12 Å apart in the protein monomer or complex. Incubation of the soluble complex of N-βGRP2 and laminarin with 0.5 mm DTSSP yielded at least 4 higher molecular mass bands (Fig. 5, lane 5, b–e) in addition to the monomer species (13.8 kDa, a in Fig. 5). These bands correspond to molecular masses of 27, 44.6, 62.7, and 78.1 kDa, each of which approximately equals molecular masses expected for oligomers (from dimer to pentamer) of N-βGRP2. Most of these higher molecular mass species were reduced to monomer in the presence of 20 mm DTT (Fig. 5, lane 4). Although a band corresponding to the N-βGRP2 dimer was also observed when the N-βGRP2 monomer was treated with DTSSP in the absence of laminarin (Fig. 5, lanes 3), this band is of substantially weaker intensity than that for the soluble complex, probably suggesting a nonspecific inter-monomer cross-linking. This cross-linking experiment provides further evidence for the soluble complex of N-βGRP2 and laminarin, and demonstrates that the complex encompasses at least five N-βGRP2 molecules.
FIGURE 5.
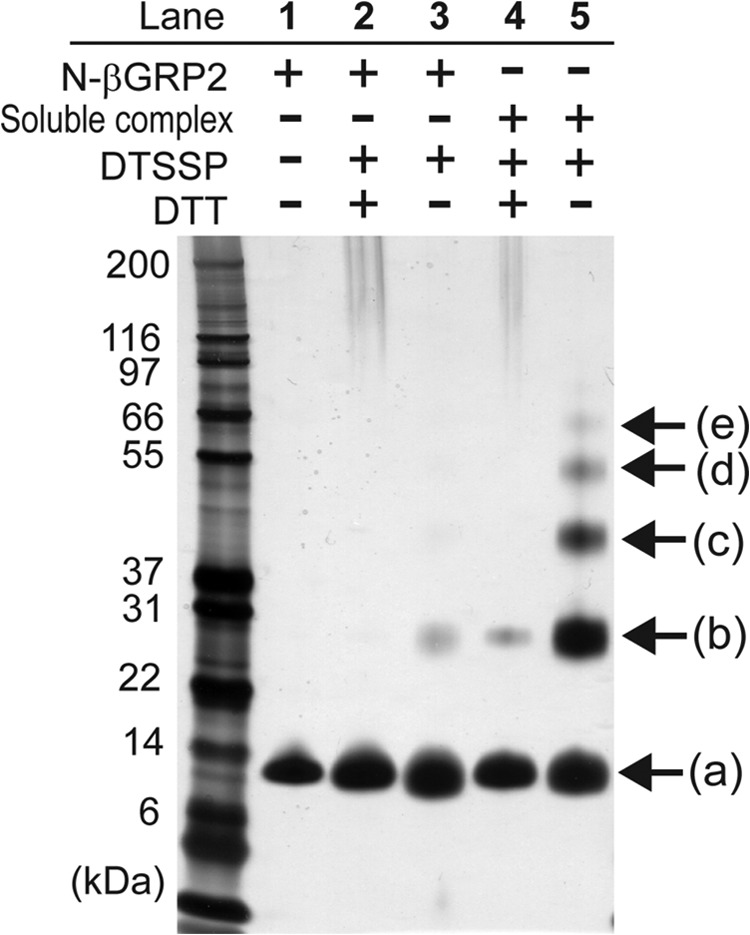
Chemical cross-linking of the soluble complex of N-βGRP2 and laminarin. The cross-linking reaction was performed by using 0.5 mm DTSSP for 30 min at 25 °C. The reaction products were analyzed by SDS-PAGE with silver staining. Lane 1, N-βGRP2; lane 2, N-βGRP2 + DTSSP with DTT; lane 3, N-βGRP2 + DTSSP under non-reducing conditions; lane 4, soluble complex + DTSSP with DTT; lane 5, soluble complex + DTSSP under non-reducing condition. N-βGRP2 monomer and higher molecular weight species formed are indicated by arrows, and those sizes were estimated to be 13.8 kDa for N-βGRP2 monomer a, 27 kDa for b, 44.6 kDa for c, 62.7 for d, 78.1 kDa for e, approximately corresponding to the sizes of N-βGRP2 oligomer (from dimer to pentamer).
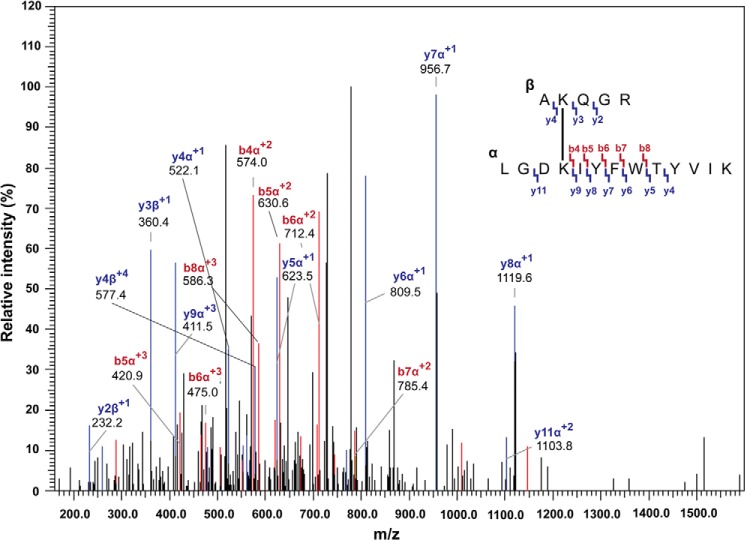
To identify the interacting region of N-βGRP2 molecules within the soluble complex, we digested the cross-linked dimer with trypsin and analyzed the peptide fragments with LC-MS/MS (Table 1). We determined that in the dimer, lysine 61 in the peptide (AK61QGR) is cross-linked to lysine 79 in the peptide (LGDK79IYFWTYVIK). The product ion spectrum of the identified intermolecular cross-link (Fig. 6) confirmed the sequence of the peptide and mapping of the cross-link sites between AK61QGR and LGDK79IYFWTYVIK. Lysine 61 is located on the concave surface of N-βGRP2, whereas lysine 79 is on the convex surface, and the measured distance between their ϵ-amino groups within the structural model of the N-βGRP2 monomer is 29.1 Å, significantly longer than the range of an intra-molecular cross-link with DTSSP. This dipeptide was not found in digested fragments of the cross-linked monomer. This result demonstrates that in the dimer, Lys61 in one N-βGRP2 is within ∼12 Å from Lys79 in an adjacent N-βGRP2 molecule.
TABLE 1.
DTSSP cross-linked peptides for the N-βGRP2·laminarin complex
Purified soluble complex of N-βGRP2 and laminarin were incubated at 25 °C with DTSSP at a molar ratio of the cross-linker to protein of 100:1, followed by SDS-PAGE with silver staining under non-reducing condition to separate cross-linked products (Fig. 6). Protein bands corresponding to the N-βGRP2 monomer and dimer were excised and in-gel digested with trypsin. Digested peptide fragments were then analyzed using LC-MS/MS, and intra- and intermolecular cross-linked peptides were identified with the software StavroX (24).
| Theoretical mass of cross-linked peptide | Observed mass of X-linked peptide | Error | Cross-linked residues | Dipeptides involved in cross-link | Cross-linking mode |
|---|---|---|---|---|---|
| Da | Da | ppm | |||
| 1682.766 | 1682.823 | 34.22 | α-NH2-Lys59 | MGER-(1–4) and DITKAKEGR-(56–64) | Intramolecular |
| 2378.189 | 2378.204 | −6.18 | Lys61-Lys79 | AKQGR-(60–64) and LGDKIYFWTYVIK-(76–88) | Intermolecular |
FIGURE 6.
Product ion spectrum obtained for the intermolecular cross-linked peptides AK61QGR and LGDK79IYFWTYVIK in the cross-linked dimer formed within the soluble complex of N-βGRP2-laminarin. Relatively abundant ions are labeled and colored red (b-type) and blue (y-type). The product ion map of the cross-linked peptides is also shown.
Effects of the N-βGRP2 Complexes with Laminarin on Pro-PO Activation
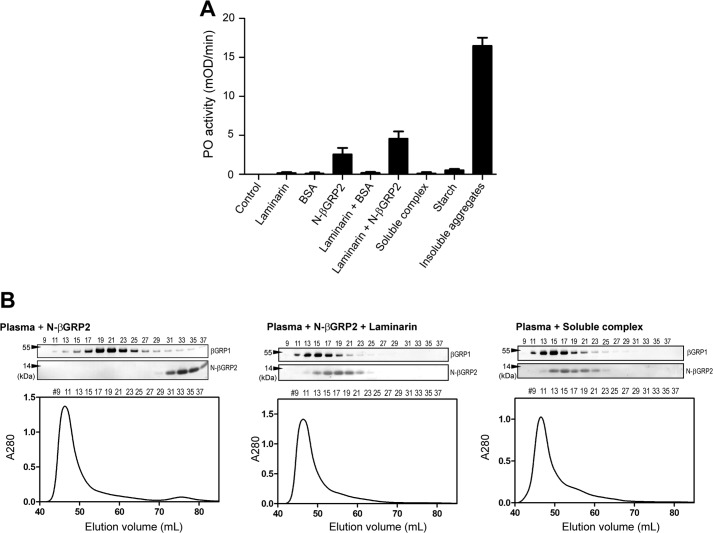
To gain insights into biological implications of the soluble and insoluble associations of N-βGRP2 with laminarin, we investigated their effects on the activation of pro-PO in plasma from M. sexta larvae. PO activity increased in plasma supplemented with purified N-βGRP2, and was further enhanced when both N-βGRP2 and laminarin were incubated with plasma sample (Fig. 7A), which is consistent with earlier studies (7, 8, 12). At this condition, the molar ratio of laminarin to N-βGRP2 is more than 20, which is high enough to form the soluble association we observed with purified components in the experiments described above. However, addition of the purified soluble complex to plasma did not stimulate significant PO activation. On the other hand, incubation of the insoluble complex with plasma led to a strong enhancement of PO activity, suggesting that the insoluble aggregates, formed when the molar ratio of protein to carbohydrate is high, is a potent activator of the pro-PO activation pathway. As the insoluble complex could not be resolubilized and was extensively washed with 50 mm sodium phosphate buffer before addition to plasma, it is not likely that the enhancement of PO activity was derived from the soluble complex and/or laminarin. Addition of starch (mixture of amylose and amylopectin) as a control material containing insoluble polysaccharide resulted in no pro-PO activation (Fig. 7A), indicating that the pro-PO activation by the insoluble complex of N-βGRP2 with laminarin is not simply due to the recognition of an insoluble phase in plasma.
FIGURE 7.
Activation of the prophenoloxidase pathway by N-βGRP2 and its multimeric complexes formed with laminarin. A, pro-PO activation assay. Ten microliters of plasma from day 2 5th instar larvae of M. sexta was incubated with 10 μg of N-βGRP2 with and without 100 μg of laminarin, soluble complex, or insoluble aggregate, in wells of a 96-well plate for 1 h at 25 °C. Phenoloxidase activity was determined using dopamine as a substrate by measuring the absorbance at 470 nm. The control contained no carbohydrate or protein, but 50 mm sodium phosphate buffer. BSA and starch were also used as negative controls. Values represent the mean slope (mOD/min) ± S.D. from triplicates. B, analysis of the complex formation in plasma by size exclusion chromatography combined with immunoblotting. The chromatography was performed for plasma with addition of N-βGRP2 (left), N-βGRP2 with laminarin (middle), and the purified soluble complex of N-βGRP2 and laminarin (right). Shown above each chromatograph is an immunoblot analysis of eluted fractions using anti-βGRP2 antibody.
We further tested whether the molecular assembly of N-βGRP2 with laminarin to form a larger complex occurs under the conditions of pro-PO activation. Plasma was supplemented with N-βGRP2 or the N-βGRP2·laminarin soluble complex, and after a 1-h incubation, the proteins in those plasma samples were separated by size exclusion chromatography. The resulting fractions were analyzed by SDS-PAGE and immunoblotting using the anti-βGRP2 antibody. N-βGRP2 eluted as a monomer in the absence of laminarin (Fig. 7B, left), but eluted much earlier when the protein was incubated with plasma in the presence of laminarin (Fig. 7B, middle), indicating that N-βGRP2 was associated to form a high molecular mass complex. This elution profile of N-βGRP2 was similar to that observed after the pre-formed soluble complex was incubated with the plasma (Fig. 7B, right). These results indicate that adding recombinant N-βGRP2 and laminarin to plasma results in formation of complexes as observed in the absence of plasma.
We also detected a 54-kDa plasma protein with the anti-βGRP2 antibodies (Fig. 7B). Our previous studies showed that βGRP2 is absent in the naive larvae and βGRP1 is constitutively present in hemolymph of the feeding stage larvae (4, 8). Due to their sequence similarity (57% identity), the antibody to βGRP1 cross-reacts with βGRP2 (8). Based on these results, the 54-kDa protein was deduced to be βGRP1, and this was further confirmed with immunoblotting analysis using anti-βGRP1 antibody (data not shown). The immunoblot analysis showed that βGRP1 eluted earlier in the presence of N-βGRP2 and laminarin and of the soluble complex (Fig. 7B, middle and right) when compared with it in the presence of N-βGRP2 (Fig. 7B, left), which indicates a higher molecular mass complex formation of natural βGRP1 in the presence of the soluble complex of N-βGRP2-laminarin.
DISCUSSION
Insoluble β-1,3-glucan, a major surface component of fungal cell walls, is detected by pattern recognition proteins to initiate innate immune responses to fungal infections in insects. In this study, we investigated the interactions between the amino-terminal carbohydrate-binding domain of the hemolymph plasma protein Ms-βGRP2 (N-βGRP2) and a soluble β-1,3-glucan, laminarin, by means of biophysical and biochemical methods. We identified two types of association between N-βGRP2 and laminarin. When the concentration of N-βGRP2 was greater than that of laminarin, insoluble complexes of N-βGRP2 and laminarin were formed, which appeared as fairly uniform particles when viewed by light microscopy. This preparation strongly stimulated the activation of pro-PO when added to plasma. However, when the concentration of laminarin was greater than that of N-βGRP2, we detected the formation of a soluble protein-carbohydrate complex (∼75 kDa), as previously observed for Pi-N-βGRP (12). This soluble complex was less effective in activation of pro-PO in plasma.
Macromolecular assembly of carbohydrate-binding proteins and their carbohydrate ligands can serve as effective and specific platforms to drive and control concomitant signaling events (30–33). Drosophila PGRP-LCx, another insect pathogen recognition receptor, forms a homodimer or heterodimer with PGRP-LCa, and their cytoplasmic domains recruit the adaptor molecule Imd, leading to downstream signaling in the immune deficiency pathway (32). Clustering of PGRP-LCx with DAP-type peptidoglycans on the bacterial surface was suggested based on the crystal structure of PGRP-LE complexed with a monomeric peptidoglycan (33). Many lectins interact with multivalent polysaccharides or glycoproteins to form highly organized, multidimensional cross-linked clusters (lattice-like structures), many of which form insoluble precipitates (34–36).
For insect βGRP, Kanagawa et al. (14) suggested from the crystal structure of N-βGRP complexed with triple helical laminarihexaoses that an unusually large carbohydrate binding surface on N-βGRP would enhance its affinity to carbohydrate. We propose that N-βGRP2 may also employ another strategy in which protein-protein interactions become established as a carbohydrate-protein complex is formed. This idea is supported by the formation of N-βGRP2 multimers only in the presence of laminarin. Such protein-protein interactions might increase overall avidity of carbohydrate binding, as observed for many lectins (31). Formation of oligomeric glucan-protein complexes has also been suggested for other β-glucan recognition proteins involved in immune responses both in vertebrates (37) and invertebrates (38).
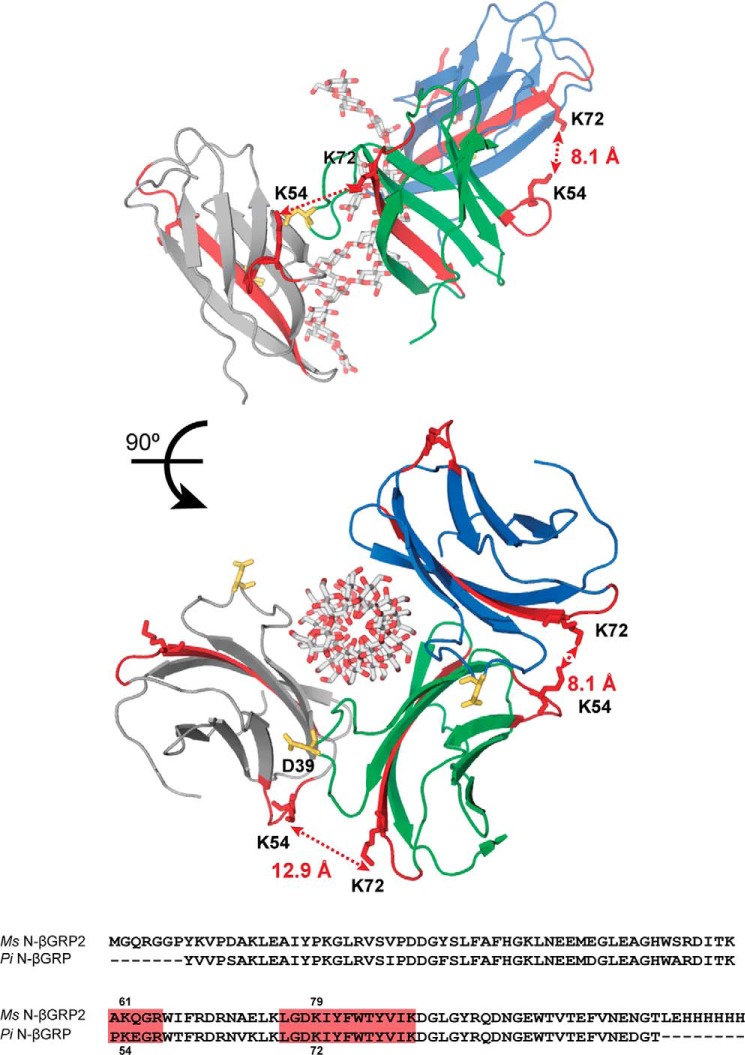
In the crystal structure of Plodia and Bombyx N-βGRPs complexed with laminarihexaose (14), protein molecules pack in a side-by-side fashion through electrostatic interactions in a loop that includes Asp39. We previously conducted site-directed mutagenesis of the corresponding aspartic acid residue (Asp45 in Ref. 12) and Glu46 in Ms-N-βGRP2) located in the contact “loop” region between the adjacent N-βGRP molecules in the crystals (Fig. 8), and demonstrated that the electrostatic interaction involving Asp45 contributes to self-association of the N-βGRP·laminarin complex (12). To characterize protein-protein interactions that accompany formation of the N-βGRP2·laminarin complex, we applied chemical cross-linking combined with mass spectrometry analysis and identified an inter-molecular cross-link formed between Lys61 and Lys79 in a cross-linked N-βGRP2 dimer formed in the presence of laminarin. Consistent with this finding, the distance between the ϵ-amino groups of lysine residues corresponding to Lys61 and Lys79 in adjacent proteins in the crystal structure of N-βGRP complexed with laminarihexaose is close enough to form a cross-linking bridge: 8.1 and 12.9 Å for Plodia N-βGRP (Fig. 8) and 13.9 and 17.6 Å for Bombyx N-βGRP. These results suggest that protein-protein interactions formed in the complex of N-βGRP2 and laminarin in our experiment may be similar to those observed in the crystals of N-βGRP formed with laminarihexaose.
FIGURE 8.
Ribbon model representations of three molecules of Pi-N-βGRPs (blue, green, and gray) with bound laminarihexaoses observed in the crystal structure (Preotein Data Bank 3AQZ) (14). Peptide fragments corresponding to cross-linked dipeptides identified in this study, AK61QGR and LGDK79IYFWTYVIK, are colored red. Distances between Lys54 and Lys72 of two neighboring N-βGRPs are indicated. Aspartic acid 39 (Asp45 in Ref. 12) in a long loop region is also shown in stick representation and colored yellow. Shown below is sequence alignment of Ms-N-βGRP2 and Pi-N-βGRP with cross-linked dipeptides shaded in red.
Laminarin is a β-1,3-linked glucose polymer with β-1,6 cross-link branches and contains 30 glucose residues on average, based on its average molecular mass (5.5 kDa). In contrast to insoluble linear polysaccharides such as curdlan and cellulose, laminarin is soluble in water, because its β-1,6 branches provide an extra degree of freedom by the rotation about the C-5 and C-6 bonds of the glucose ring (39). The crystal structure of N-βGRP complexed with laminarihexaoses arranged in a triple helical form has shown that N-βGRP interacts with β-1,3-glucans through six glucose residues, two from each laminarihexaose chain (14). Our chemical cross-linking experiment suggests that the mode of interaction between N-βGRP2 molecules bound to laminarin is consistent with that observed in the crystals of N-βGRP formed with laminarihexaose, which may indicate that binding of N-βGRP2 to laminarin chains promotes their association as a partial triple helical structure (although the β-1,6 branches would interfere with the helical structure formation to some extent). When the molar ratio of protein to carbohydrate is high (at the low [C]/[P] ratio), we predict that multiple N-βGRP2 molecules interacting with a helical form of laminarins assemble through protein-protein interaction to form a larger complex structure. We speculate that association of β-glucan chains to form triple helical structures, promoted by protein-protein interactions, leads to production of large complexes in which a significant loss of the motional freedom of glucan chains results in formation of the insoluble aggregates observed in this study. Hydrophobic association and hydrogen bond interactions between polysaccharide chains likely contribute to stabilize the association of polysaccharides in an insoluble form as occurs in curdlan and cellulose (40).
The insoluble complex of N-βGRP2 and laminarin may mimic assembly of βGRP2 upon binding to β-1,3-glucan on a fungal surface, and serves as a large and effective platform for downstream signaling events by recruiting serine proteases and prophenoloxidase, increasing their local concentrations. To our knowledge, this study represents the first report demonstrating that the insoluble complex of N-βGRP2 and β-1,3-glucan is a powerful activator for the pro-PO activation pathway in insect hemolymph (Fig. 7).
In the presence of excess laminarin (at a higher [C]/[P] ratio), on the other hand, the N-βGRP2 molecules bound to laminarins may be more dispersed, such that there are fewer protein-protein contacts, resulting in formation of smaller complexes, in which the laminarin chains retain sufficient flexibility to remain soluble. In our previous study (12), analytical ultracentrifuge results indicated that a Pi-N-βGRP·laminarin complex likely contained six protein and three laminarin molecules, consistent with our current observation of stable soluble complexes of ∼75 kDa, estimated by analytical ultracentrifuge and size exclusion chromatography. Such a soluble complex is possibly not large enough to serve as an effective platform for activation of downstream serine protease cascades.
Previous studies of a crayfish immune response demonstrated that activation of pro-PO in plasma by laminarin reached a maximum at relatively low laminarin concentration (∼1 μg/ml) and decreased at higher laminarin concentrations (41). In addition, when the concentration of laminarin was fixed, activation of crayfish pro-PO was greater at higher molar ratios of the glucan-binding protein to laminarin (42). We also previously observed that pro-PO activation by laminarin in M. sexta plasma can decrease as the laminarin concentration increases past an optimum level (43). In this study, the soluble complex of N-βGRP2 with laminarin formed in the plasma at a higher laminarin concentration (a higher [C]/[P] ratio) stimulated pro-PO activation, but at a significantly lower level compared with the insoluble complex (Fig. 7A). Thus, our result showing formation of the less active soluble complex may help to explain why high concentrations of laminarin lead to the inhibition or a lower level stimulation of pro-PO activation in crayfish (41, 42) and M. sexta (43).
It is likely that Toll activation can also be stimulated in a similar manner by formation of βGRP-glucan complexes resulting in activation of connected serine protease cascades for the Toll and pro-PO activation pathways (44–46). However, the molecular mechanism by which the downstream serine protease cascades are activated upon the β-1,3-glucan recognition by βGRP still remains to be established. This is partly because the molecular function of the C-terminal glucanase-like domain of βGRP and the unique modular domains of the initiating serine proteases (47, 48), and their interactions have not been characterized in detail. Intriguingly, within the higher order structure of N-βGRP2 observed in the crystal structure (Fig. 8), the carboxyl terminus of N-βGRP2 molecules positions away both from the carbohydrate-binding site and the protein-protein contact site. It is possible that the C-terminal glucanase-like domain can interact with the initiating proteases without interfering with protein complex organization mediated by N-βGRP. However, further biochemical and biophysical studies on full-length βGRP and the initiating serine proteases are necessary to better understand the molecular basis underlying this initial step for insect innate immune responses. The questions remaining to be answered include whether full-length βGRP associates to form protein complexes observed here, whether the C-terminal domain may mediate protein interactions with the initiating protease, and which modular domains in the initiating protease mediate the interaction with βGRP resulting in protease autoactivation.
Supplementary Material
Acknowledgments
We thank Maureen Gorman, David Quilici, and Rebekah Woolsey for helpful comments on the manuscript, and Lisa Brummett for technical assistance. The mass spectrometry experiment was performed by the Nevada Proteomics Center at the University of Nevada, which is supported by Grant 5P20 RR016464-11 from the National Center for Research Resources and Grant 8P20GM103440-11 from the NIGMS, National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grant GM41247. This is contribution number 14-185-J from the Kansas Agricultural Experiment Station.

This article contains supplemental Figs. S1–S2.
All chemical shifts for N-terminal domain of βGRP2 (N-βGRP2) were deposited in the BioMagResBank under accession number 19669.
- βGRP
- β-glucan recognition protein
- N-βGRP
- N-terminal carbohydrate binding domain of β-glucan recognition protein
- pro-PO
- prophenoloxidase
- PGRP
- peptidoglycan recognition protein
- DTSSP
- 3,3′-dithiobis(sulfosuccinimidyl propionate)
- HSQC
- heteronuclear single quantum coherence
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Lemaitre B., Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 2. Yu X.-Q., Zhu Y.-F., Ma C., Fabrick J. A., Kanost M. R. (2002) Pattern recognition proteins in Manduca sexta plasma. Insect Biochem. Mol. Biol. 32, 1287–1293 [DOI] [PubMed] [Google Scholar]
- 3. Ochiai M., Ashida M. (2000) A pattern-recognition protein for β-1,3-glucan. J. Biol. Chem. 275, 4995–5002 [DOI] [PubMed] [Google Scholar]
- 4. Ma C., Kanost M. R. (2000) A β1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 275, 7505–7514 [DOI] [PubMed] [Google Scholar]
- 5. Kim Y.-S., Ryu J.-H., Han S.-J., Choi K.-H., Nam K.-B., Jang I.-H., Lemaitre B., Brey P. T., Lee W.-J. (2000) Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 275, 32721–32727 [DOI] [PubMed] [Google Scholar]
- 6. Zhang R., Cho H. Y., Kim H. S., Ma Y. G., Osaki T., Kawabata S., Söderhäll K., Lee B. L. (2003) Characterization and properties of a 1,3-β-d-glucan pattern recognition protein of Tenebrio molitor larvae that is specifically degraded by serine protease during prophenoloxidase activation. J. Biol. Chem. 278, 42072–42079 [DOI] [PubMed] [Google Scholar]
- 7. Fabrick J. A., Baker J. E., Kanost M. R. (2003) cDNA cloning, purification, properties, and function of a β-1,3-glucan recognition protein from a pyralid moth, Plodia interpunctella. Insect Biochem. Mol. Biol. 33, 579–594 [DOI] [PubMed] [Google Scholar]
- 8. Jiang H., Ma C., Lu Z. Q., Kanost M. R. (2004) β-1,3-glucan recognition protein-2 (βGRP-2) from Manduca sexta: an acute-phase protein that binds β-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 34, 89–100 [DOI] [PubMed] [Google Scholar]
- 9. Gottar M., Gobert V., Matskevich A. A., Reichhart J. M., Wang C., Butt T. M., Belvin M., Hoffmann J. A., Ferrandon D. (2006) Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabrick J. A., Baker J. E., Kanost M. R. (2004) Innate immunity in a pyralid moth: functional evaluation of domains from a β-1,3-glucan recognition protein. J. Biol. Chem. 279, 26605–26611 [DOI] [PubMed] [Google Scholar]
- 11. Mishima Y., Quintin J., Aimanianda V., Kellenberger C., Coste F., Clavaud C., Hetru C., Hoffmann J. A., Latgé J. P., Ferrandon D., Roussel A. (2009) The N-terminal domain of Drosophila Gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors. J. Biol. Chem. 284, 28687–28697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dai H., Hiromasa Y., Takahashi D., VanderVelde D., Fabrick J. A., Kanost M. R., Krishnamoorthi R. (2013) An initial event in the insect innate immune response: structural and biological studies of interactions between β-1,3-glucan and the N-terminal domain of β-1,3-glucan recognition protein. Biochemistry 52, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahasi K., Ochiai M., Horiuchi M., Kumeta H., Ogura K., Ashida M., Inagaki F. (2009) Solution structure of the silkworm βGRP/GNBP3 N-terminal domain reveals the mechanism for β-1,3-glucan-specific recognition. Proc. Natl. Acad. Sci. U.S.A. 106, 11679–11684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanagawa M., Satoh T., Ikeda A., Adachi Y., Ohno N., Yamaguchi Y. (2011) Structural insights into recognition of triple-helical β-glucans by an insect fungal receptor. J. Biol. Chem. 286, 29158–29165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y., Sumathipala N., Rayaprolu S., Jiang H. (2011) Recognition of microbial molecular patterns and stimulation of prophenoloxidase activation by a β-1,3-glucanase-related protein in Manduca sexta larval plasma. Insect Biochem. Mol. Biol. 41, 322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hrmova M., Fincher G. B. (1993) Purification and properties of three (1–3)-β-d-gllucanase isoenzymes from young leaves of barley (Hordeum vulgare). Biochem. J. 289, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 18. Keller R. (2004) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment. Ph.D. thesis, Swiss Federal Institute of Technology, Zurich, Switzerland [Google Scholar]
- 19. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiromasa Y., Fujisawa T., Aso Y., Roche T. E. (2004) Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 279, 6921–6933 [DOI] [PubMed] [Google Scholar]
- 23. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 24. Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C. H., Krauth F., Fritzsche R., Kühn U., Sinz A. (2012) StavroX: a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23, 76–87 [DOI] [PubMed] [Google Scholar]
- 25. Bennett K. L., Kussmann M., Björk P., Godzwon M., Mikkelsen M., Sørensen P., Roepstorff P. (2000) Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping-a novel approach to assess intermolecular protein contacts. Protein Sci. 9, 1503–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petrotchenko E. V., Borchers C. H. (2010) Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 29, 862–876 [DOI] [PubMed] [Google Scholar]
- 27. Bhat S., Sorci-Thomas M. G., Calabresi L., Samuel M. P., Thomas M. J. (2010) Conformation of dimeric apolipoprotein A-I Milano on recombinant lipoprotein particles. Biochemistry 49, 5213–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. D'Ambrosio C., Talamo F., Vitale R. M., Amodeo P., Tell G., Ferrara L., Scaloni A. (2003) Probing the dimeric structure of porcine aminoacylase 1 by mass spectrometric and modeling procedures. Biochemistry 42, 4430–4443 [DOI] [PubMed] [Google Scholar]
- 29. Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) Intermolecular contact between globular N-terminal-fold and C-terminal domain of apoA-I stabilizes its lipid-bound conformation. J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 30. Sacchettini J. C., Baum L. G., Brewer C. F. (2001) Multivalent protein-carbohydrate interactions: a new paradigm for supermolecular assembly and signal transduction. Biochemistry 40, 3009–3015 [DOI] [PubMed] [Google Scholar]
- 31. Brewer C. F., Miceli M. C., Baum L. G. (2002) Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 12, 616–623 [DOI] [PubMed] [Google Scholar]
- 32. Choe K. M., Lee H., Anderson K. V. (2005) Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim J. H., Kim M. S., Kim H. E., Yano T., Oshima Y., Aggarwal K., Goldman W. E., Silverman N., Kurata S., Oh B. H. (2006) Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281, 8286–8295 [DOI] [PubMed] [Google Scholar]
- 34. Bhattacharyya L., Fant J., Lonn H., Brewer C. F. (1990) Binding and precipitating activities of Lotus tetragonolobus isolectins with l-fucosyl oligosaccharides. Formation of unique homogeneous cross-linked lattices observed by electron microscopy. Biochemistry 29, 7523–7530 [DOI] [PubMed] [Google Scholar]
- 35. Mandal D. K., Brewer C. F. (1992) Cross-linking activity of the 14-kilodalton β-galactoside-specific vertebrate lectin with asialofetuin: comparison with several galactose-specific plant lectins. Biochemistry 31, 8465–8472 [DOI] [PubMed] [Google Scholar]
- 36. Gupta D., Bhattacharyya L., Fant J., Macaluso F., Sabesan S., Brewer C. F. (1994) Observation of unique cross-linked lattices between multiantennary carbohydrates and soybean lectin: presence of pseudo-2-fold axes of symmetry in complex type carbohydrates. Biochemistry 33, 7495–7504 [DOI] [PubMed] [Google Scholar]
- 37. Hanashima S., Ikeda A., Tanaka H., Adachi Y., Ohno N., Takahashi T., Yamaguchi Y. (2014) NMR study of short β(1–3)-glucans provides insights into the structure and interaction with Dectin-1. Glycoconj. J. 31, 199–207 [DOI] [PubMed] [Google Scholar]
- 38. Ueda Y., Ohwada S., Abe Y., Shibata T., Iijima M., Yoshimitsu Y., Koshiba T., Nakata M., Ueda T., Kawabata S. (2009) Factor G utilizes a carbohydrate-binding cleft that is conserved between horseshoe crab and bacteria for the recognition of β-1,3-d-glucans. J. Immunol. 183, 3810–3818 [DOI] [PubMed] [Google Scholar]
- 39. Bacic A., Fincher G. B., Stone B. A. (2009) Chemistry, Biochemistry, and Biology of 1–3β-Glucans and Related Polysaccharides, Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 40. Bergenstråhle M., Wohlert J., Himmel M. E., Brady J. W. (2010) Simulation studies of the insolubility of cellulose. Carbohydr. Res. 345, 2060–2066 [DOI] [PubMed] [Google Scholar]
- 41. Söderhäll K., Unestam T. (1979) Activation of serum prophenoloxidase in arthropod immunity. The specificity of cell wall glucan activation and activation by purified fungal glycoproteins of crayfish phenoloxidase. Can. J. Microbiol. 25, 406–414 [DOI] [PubMed] [Google Scholar]
- 42. Duvic B., Söderhäll K. (1990) Purification and characterization of a β-1,3-glucan binding protein from plasma of the crayfish Pacifastacus leniusculus. J. Biol. Chem. 265, 9327–9332 [PubMed] [Google Scholar]
- 43. Tong Y., Kanost M. R. (2005) Manduca sexta Serpin-4 and Serpin-5 inhibits the prophenol oxidase activation pathway. J. Biol. Chem. 280, 14923–14931 [DOI] [PubMed] [Google Scholar]
- 44. An C., Ishibashi J., Ragan E. J., Jiang H., Kanost M. R. (2009) Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J. Biol. Chem. 284, 19716–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., Park B. J., Zhang R., Zhang J., Söderhäll K., Ha N. C., Lee B. L. (2008) Molecular control of phenoloxidase-induced melanin synthesis in an insect. J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 46. Cerenius L., Kawabata S., Lee B. L., Nonaka M., Söderhäll K. (2010) Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 35, 575–583 [DOI] [PubMed] [Google Scholar]
- 47. Ji C., Wang Y., Guo X., Hartson S., Jiang H. (2004) A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta. J. Biol. Chem. 279, 34101–34106 [DOI] [PubMed] [Google Scholar]
- 48. Buchon N., Poidevin M., Kwon H.-M., Guillou A., Sottas V., Lee B. L., Lemaitre B. (2009) A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.